Abstract
How the liver’s extracellular matrix (ECM) protein composition and stiffness cooperatively regulate primary human hepatocyte (PHH) phenotype is unelucidated. Here, we utilize protein microarrays and high content imaging with single-cell resolution to assess PHH attachment/functions on 10 major liver ECM proteins in single and two-way combinations robotically spotted onto polyacrylamide gels of 1 kPa or 25 kPa stiffness. Albumin, cytochrome-P450 3A4 (CYP3A4), and hepatocyte nuclear factor alpha (HNF4α) positively correlate with each other and cell density on both stiffnesses. The 25 kPa stiffness supports higher average albumin and HNF4α expression after 14 days, while ECM protein composition significantly modulates PHH functions across both stiffnesses. Unlike previous rodent data, PHH functions are highest only when collagen-IV or fibronectin are mixed with specific proteins, whereas non-collagenous proteins without mixed collagens downregulate functions. Combination of collagen-IV and hyaluronic acid retains high CYP3A4 on 1 kPa, whereas collagens-IV and -V better retain HNF4α on 25 kPa over 14 days. Adapting ECM conditions to 96-well plates containing conjugated hydrogels reveals novel regulation of other functions (urea, CYP1A2/2A6/2C9) and drug-mediated CYP induction by the ECM protein composition/stiffness. This high-throughput pipeline can be adapted to elucidate ECM’s role in liver diseases and facilitate optimization of engineered tissues.
Keywords: collagen, drug screening, drug-drug interaction, cytochrome P450, induction
Graphical Abstract
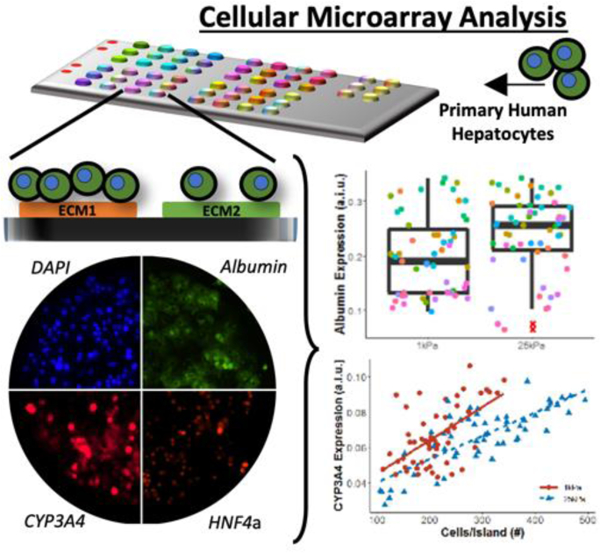
High-throughput cell microarrays and high content imaging with single-cell resolution elucidates synergistic and novel effects of extracellular matrix (ECM) protein composition and stiffness on diverse functions of primary human hepatocytes (PHH), which is useful for drug screening, disease modeling, and regenerative medicine. Select ECM conditions adapted to multiwell plates improve PHH functions and drug responsiveness for 2 weeks in culture.
1. Introduction
The liver’s extracellular matrix (ECM) is composed of diverse proteins such as collagens, glycoproteins, and proteoglycans[1], and has a Young’s modulus of ≤1 kPa.[2, 3] Upon sustained injury to the liver due to xenobiotics, hepatitis B/C viruses, alcohol, and over-nutrition, the ECM’s composition/stiffness can change dramatically due to the secretion of excessive collagen-I by activated (myofibroblastic) hepatic stellate cells.[4] However, how the protein composition and stiffness of the ECM regulate the functions of human liver cells in physiology and disease has not been fully elucidated, and doing so in vivo in rodents is challenging due to the presence of several confounding variables (e.g., fluid flow, non-parenchymal interactions, and soluble factors) and species-specific differences in liver functions.[5] Therefore, to mitigate the limitations with animal studies, cultures of primary human hepatocytes (PHHs) are used routinely for drug screening, disease modeling, and mechanistic inquiries.[6] However, PHHs display a precipitous decline in phenotypic functions when cultured on collagen-I adsorbed onto tissue culture polystyrene (TCPS) and glass.[7] Sandwiching hepatocytes within two layers of gelled collagen-I can induce the reformation of bile canaliculi but other functions still show rapid decline[8], potentially due to an excessive amount of collagen-I that is more akin to liver fibrosis. Similarly, culturing hepatocytes with tumor-derived murine Matrigel™ can induce some functions for ~1 week [7], though comparisons to human liver ECM are challenging. In contrast, culturing PHHs on decellularized human liver ECM can also transiently improve phenotypic functions;[9] however, such ECM is typically variable in quality due to the unpredictable conditions of the transplant-rejected human livers and the harsh cellular dissociation methods.[10]
In contrast to complex ECM gels such as those discussed above, recombinant and purified ECM proteins can be useful to elucidate how such proteins affect the hepatic phenotype individually and in specific combinations, often in unexpected ways. Previously, rat hepatocytes cultured on different combinations of recombinant ECM proteins for 7 days displayed varying albumin staining on ECM combinations, and such differences were often due to the unexpected interactions between different ECM proteins.[11] In addition to the protein composition of the ECM, the hepatocyte phenotype in vitro is highly sensitive to the underlying substrate’s stiffness. Specifically, primary mouse hepatocytes[3], primary rat hepatocytes[12, 13], mouse embryonic stem cell (ESC)-derived-hepatocytes[14], PHHs[15], and human ESC-derived hepatocytes[16] all displayed higher levels of phenotypic functions when cultured on softer ECM gels versus stiffer substrates. However, how the protein composition and stiffness of ECM synergize to regulate PHH functions over prolonged culture remains largely unelucidated.
Since conventional plate-based approaches are too costly for interrogating the large combinational search space of liver ECM proteins and stiffnesses, we previously utilized a high-throughput ECM microarray that facilitates the independent modulation of cell-cell interactions, ECM composition, substrate stiffness, and soluble factors.[17–19] A dehydrated polyacrylamide (PA) hydrogel is used for the contact deposition of proteins with a robotic spotter. Upon hydration, the proteins are retained in the spots, and the hydrophilic PA resists protein adsorption and prevents cell adhesion in all areas except on the domains containing the ECM proteins. We subsequently developed a high content imaging pipeline to enable single-cell measurements from the microarray.[18] Here, we sought to use this microarray platform and high content imaging pipeline to elucidate the combinatorial effects of 10 major ECM proteins present in the liver and liver-like substrate stiffnesses on the attachment and phenotypic protein staining patterns (albumin, CYP3A4, HNF4α) of PHHs over 2 weeks in culture, while using collagen-I as the control ECM utilized commonly for PHH culture.[7] We then validated select ECM regulators of PHH staining patterns obtained from the microarrays on hydrogels conjugated to the bottom of 96-well plates, which also allowed us to evaluate secreted hepatic markers, metabolism of different cytochrome P450 (CYP) enzyme substrates, and drug-mediated CYP induction towards determining the utility of PHHs cultured onto biophysically tuned ECM substrates for use in the drug development pipeline.
2. Results
2.1. PHH attachment and retention over 2 weeks on ECM microarrays
Circular islands of ECM proteins in triplicates were spotted via a robotic spotter onto glass slides with conjugated PA of 1 kPa and 25 kPa stiffnesses; a 450 µm diameter for the spotted islands was chosen since it was found previously to induce robust formation of homotypic contacts between PHHs.[7] Islands/spots contained 10 human ECM proteins present in the liver, including collagen I (C1), collagen III (C3), collagen IV (C4), collagen V (C5), decorin (DC), fibronectin (FN), hyaluronic acid (HA), laminin (LN), lumican (LU), and tenascin C (TC) - Figure 1A. This list includes structural proteins (C1, C3, C4, C5), basement-like membrane proteins (C4, LN), glycoproteins (FN, TC), proteoglycans (HA, DC), and proteins upregulated in disease (LU), and represents the most abundant proteins by percentage from the 150+ proteins identified in native liver tissue.[20–24] We restricted our investigations to single and two-way combinations of the above proteins at single concentrations to keep the scope manageable and because we were able to use these conditions previously to reveal unique ECM regulators of liver progenitor differentiation.[19] Finally, we conducted all microarray studies in 3 PHH donors across three time-points in culture (day 1, 7, and 14). Therefore, overall, we tested 990 total conditions (55 total ECM protein combinations x 2 stiffnesses x 3 PHH donors x 3 time-points) prior to making key conclusions about the role of the ECM in regulating PHH functions in vitro.
Figure 1. High-throughput extracellular matrix (ECM) microarrays for probing primary human hepatocyte (PHH) attachment and phenotype.
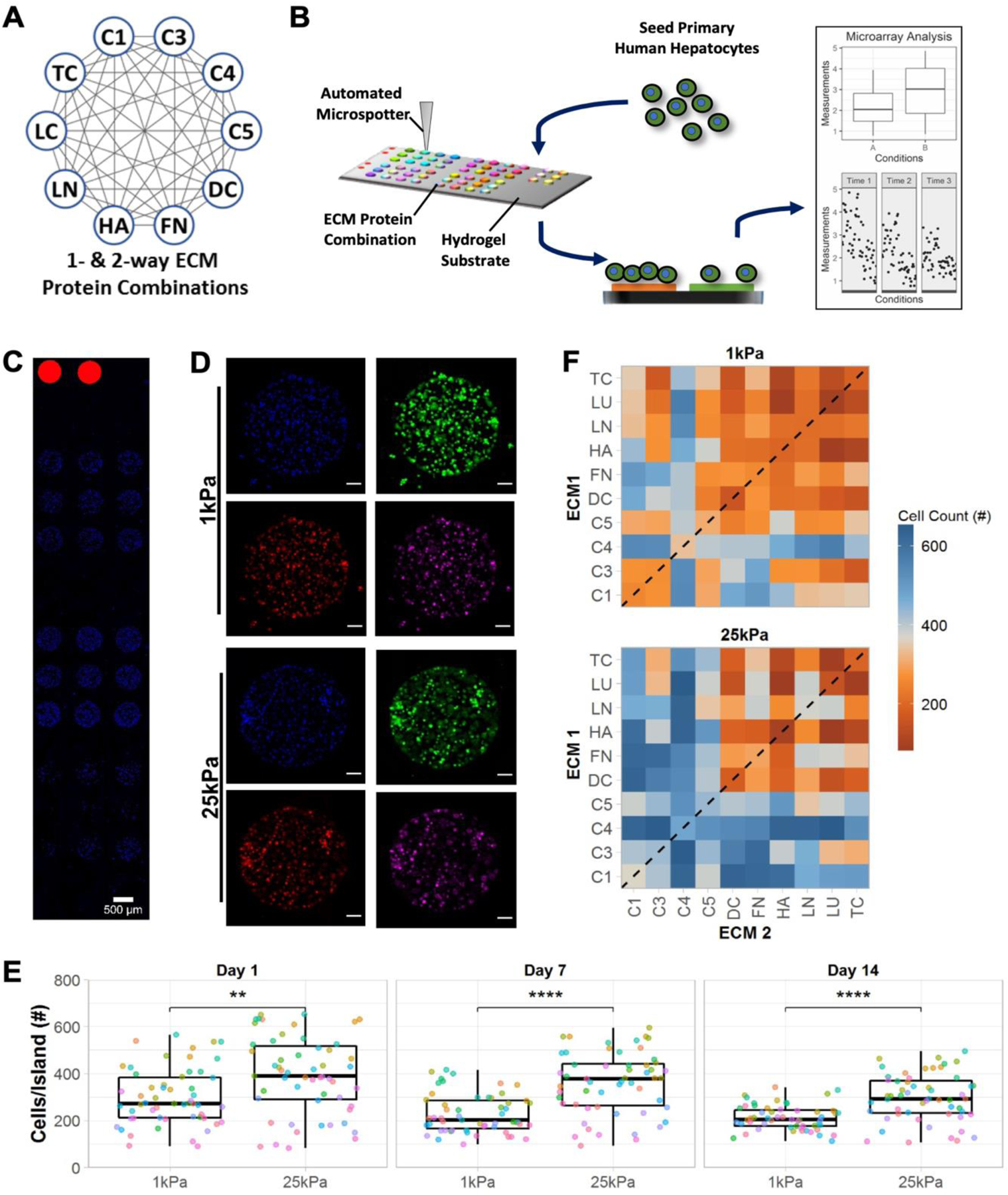
A) Single- and two-way combinations of 10 liver-inspired recombinant ECM proteins. Each circle represents a single protein (collagens 1 [C1], 3 [C3], 4 [C4], 5 [C5] decorin [DC], fibronectin [FN], hyaluronic acid [HA], laminin [LN], lumican [LC], and tenascin [TC]) and each grey line represents a two-way protein combination; a total of 55 compositions of ECM proteins were utilized in this study. B) Schematic of the experimental workflow, including: fabrication of microarrays using an automated microspotter to transfer pre-made ECM protein solutions to a polyacrylamide (PA) hydrogel of a tunable stiffness conjugated to a glass slide; seeding of PHHs that attach only to the ECM protein ‘islands’ and not to the PA gel; differential attachment of PHHs and expression of PHH markers on ECM islands as assessed via high-content imaging; plotting and statistical analysis of single cell imaging data across the various ECM conditions. C) Representative section of an ECM microarray demonstrating the differential attachment of PHHs to ECM islands of 450 µm diameter; nuclei are counterstained with DAPI and rhodamine-labeled dextran spots on microarrays are utilized for alignment. Scale bar = 500 µm. D) Representative islands of 4-color-stained PHHs on the microarrays of two different PA stiffnesses, 1 kPa and 25 kPa (Young’s moduli). Scale bars = 50 µm. E) Box plots showing the median number of cells per ECM island (center bar) along with the interquartile range on both microarray stiffnesses over 14 days of culture (each dot represents the mean cell number measured across islands of a specific ECM composition); mean distributions were compared across the stiffnesses using the Wilcoxon Test. **p<0.01 and ****p<0.0001. F) Heatmap of mean cell number across islands of single- (dashed line) and two-way combinations of the ECM proteins on the two stiffnesses after 14 days of culture. Similar heatmaps after 1 and 7 days of culture are shown in Figure S1A, Supporting Information. For panels E and F, data displayed represents summarized single-cell measurements across three PHH donors per condition (n = 22–24 ECM islands across representative microarrays).
PHHs were seeded on arrays, allowed to attach overnight, unattached cells were washed, and then cultures were fixed on days 1, 7, and 14 post-seeding to assess phenotypic markers via high-content imaging (Figure 1B). DAPI (nuclear counterstain) was used to obtain PHH counts on each ECM island; representative DAPI-stained PHHs on ECM islands are shown in Figure 1C. PHH phenotype on ECM islands was assessed at single-cell resolution via immunostaining for albumin, HNF4α, and CYP3A4 (Figure 1D).
PHH attachment and retention on ECM islands was higher on 25 kPa stiffness versus 1 kPa stiffness on days 1, 7, and 14, albeit average PHH numbers/island declined over time on both stiffnesses (Figure 1E); nonetheless, 25 kPa substrates retained on average ~246 PHHs/island versus 194 PHHs/island on 1 kPa substrates. In addition to substrate stiffness, PHH attachment was also significantly influenced by ECM island protein composition (Figure 1F). Compositions containing collagens (e.g., C4 and C1) led to better PHH attachment overall than compositions containing proteoglycans (e.g., LU and DC) (Figure S1, Supporting Information).
2.2. PHH phenotypic marker expression on ECM microarrays
PHHs on ECM islands spotted onto 1 kPa and 25 kPa microarrays stained positive for intracellular albumin (Figure 2A), a surrogate marker that is routinely used to assess hepatocyte health and protein synthesis/secretion capability.[6] After 1 day of PHH seeding, mean albumin from single-cell fluorescent intensity measurements was found to be higher on 1 kPa microarrays versus 25 kPa ones (Figure S2A, Supporting Information); however, by day 14, these trends were reversed (Figure 2B), thereby showing that 25 kPa microarrays were better able to sustain PHH albumin expression than 1 kPa ones over prolonged culture. Heatmaps of albumin expression were created to elucidate the effects of ECM protein compositions for a given stiffness on PHH albumin expression (Figure 2C and Figure S2B, Supporting Information). C4 in single- or two-way combinations was identified as a positive regulator of albumin expression across both stiffnesses. More broadly, the addition of a collagenous protein to an ECM mixture was found to be important for high albumin expression. For non-collagenous proteins, effects were more dependent on both composition and stiffness (e.g., LN,LU), except for FN,TC, which led to relatively high albumin expression across both stiffnesses. Lastly, a positive correlation between albumin expression and cell density (cells/island) was observed (Figure 2D), which is consistent with previous literature showing that increased PHH homotypic interactions, with the concomitant increase in junction formation, leads to higher albumin production.[7]
Figure 2. Albumin expression in primary human hepatocytes (PHHs) on extracellular matrix (ECM) microarrays.
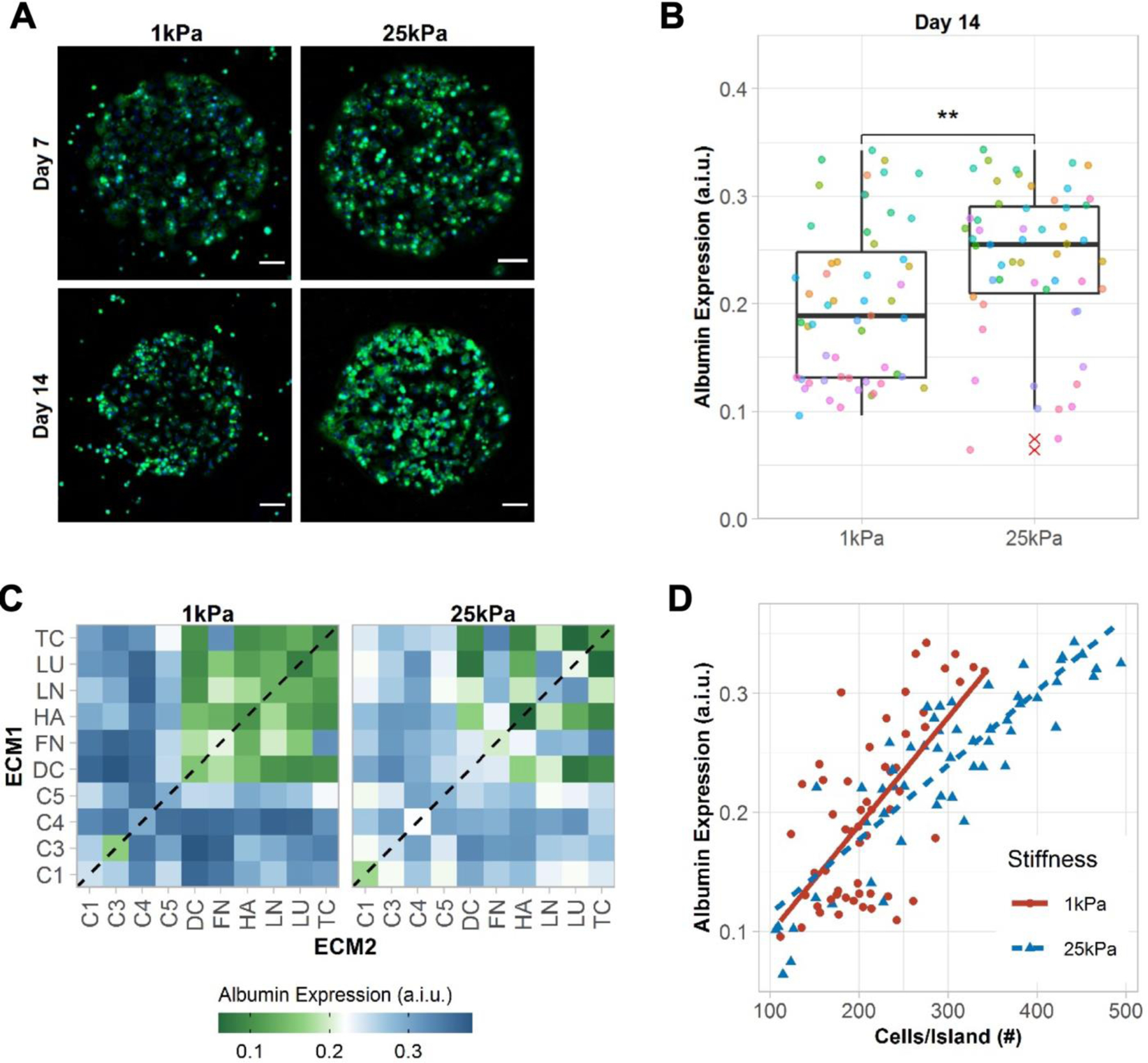
A) Representative images of albumin expression in PHHs after 7 and 14 days of culture on the microarrays of two different stiffnesses, 1 kPa and 25 kPa. Scale bars = 50 µm. B) Box plots show the median albumin expression (center bar) and interquartile range (IQR) for ECM conditions separated for each microarray stiffness at 14 days of culture (each dot represents the mean albumin expression of an individual ECM composition; red asterisks represent outliers beyond 1.5*IQR). Similar box plots for 1 and 7 days of culture are shown in Figure S2A, Supporting Information. A.i.u. = arbitrary intensity units. Comparison of means was performed using the Wilcoxon Test. **p<0.01. C) Heatmap of mean albumin expression on single- (dashed line) and two-way combinations of the ECM proteins after 14 days of culture. Similar heatmaps for 1 and 7 days of culture are shown in Figure S2B, Supporting Information. D) Correlation of mean cells per ECM island and mean albumin expression. Positive correlations were observed using spearman’s rank correlation for 1 kPa (rs=0.58, p=6.1e-06) and 25 kPa (rs=0.86, p=2.2e-16). For panels B-D, data displayed represents summarized single-cell measurements across three PHH donors per condition (n = 22–24 ECM islands across representative microarrays).
PHHs were positive for intracellular CYP3A4 on both 1 kPa and 25 kPa stiffnesses (Figure 3A); CYP3A4 is the most abundant CYP enzyme in the liver and metabolizes about half of the drugs metabolized by the CYP family of enzymes.[25] CYP3A4 expression in PHHs was significantly higher on 1 kPa microarrays after 1 day of seeding than on the 25 kPa microarrays; however, differences became insignificant over 7 and 14 days of culture (Figure S3A, Supporting Information). As with albumin, CYP3A4 expression increased proportionally with both increases in cell density per ECM island and albumin expression irrespective of microarray stiffness (Figure 3B), which suggests that more differentiated PHHs with better cell-cell contacts simultaneously upregulate both albumin and CYP3A4. When comparing stiffnesses, while controlling for cell density per island or for albumin expression, CYP3A4 expression was higher on 1 kPa microarrays than on the 25 kPa microarrays (Figure 3B).
Figure 3. CYP3A4 expression in primary human hepatocytes (PHHs) on extracellular (ECM) microarrays.
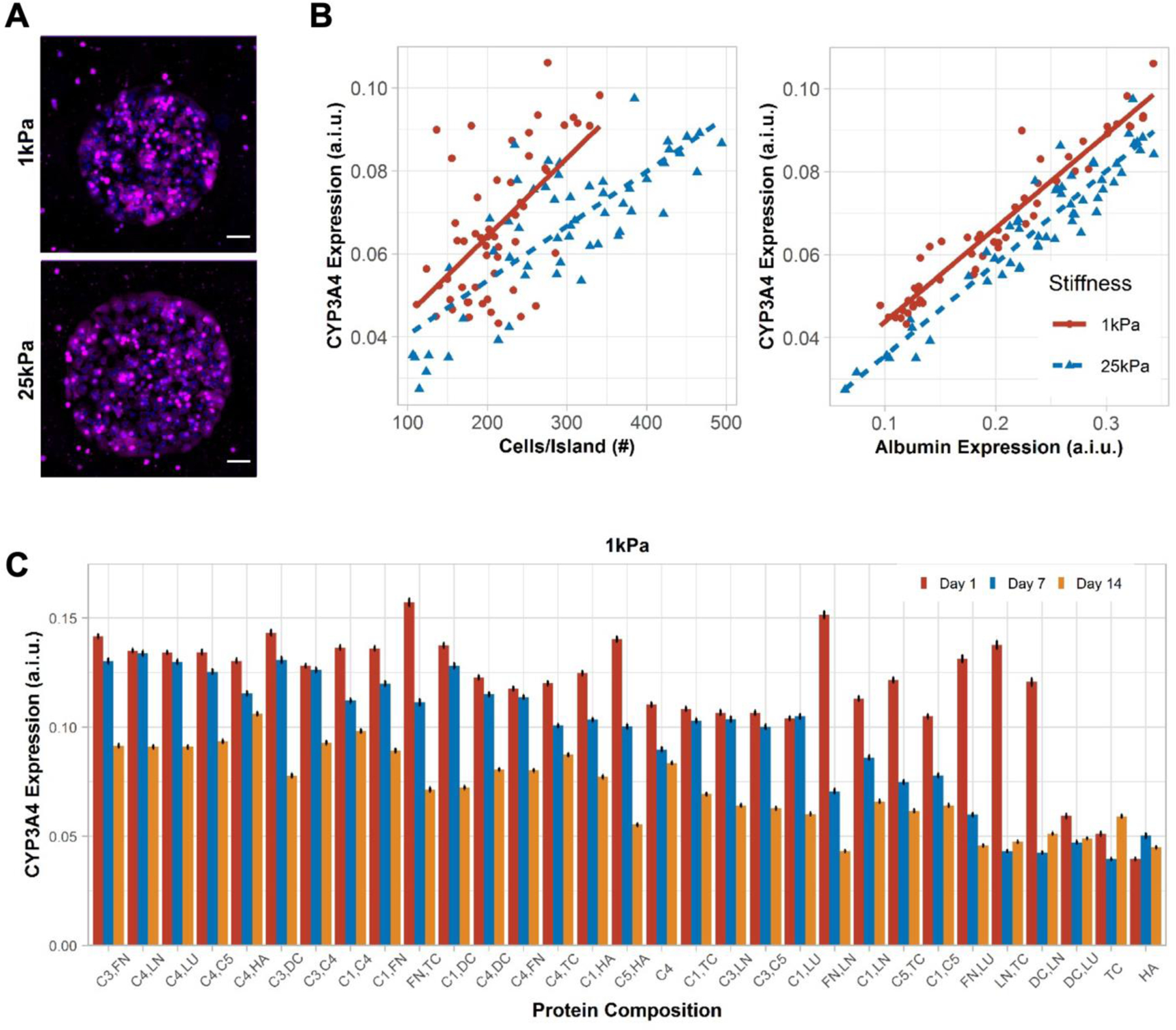
A) Representative images of CYP3A4 expression in PHHs after 14 days of culture on the microarrays of two different stiffnesses, 1 kPa and 25 kPa. Scale bars = 50 µm. B) Positive correlation was observed between mean cells per ECM island and mean CYP3A4 expression on 1 kPa (rs=0.51, p=9.4e-05) and 25 kPa (rs=0.72, p<2.2e-16) stiffnesses. Similarly, mean albumin expression positively correlates with mean CYP3A4 expression on 1kPa (rs=0.97, p<2.2e-16) and 25 kPa (rs=0.91, p<2.2e-16) stiffnesses. A.i.u. = arbitrary intensity units. C) Bar plot shows the mean CYP3A4 expression in PHHs on those ECM compositions that retained greater than 200 cells per island after 14 days of culture on 1 kPa stiffness. Similar bar plot on microarray of 25 kPa stiffness is shown in Figure S3B, Supporting Information. Error bars represent standard error of mean. For panels B-C, data displayed represents summarized single-cell measurements across three PHH donors per condition (n = 22–24 ECM islands across representative microarrays).
CYP3A4 expression also varied as a function of ECM protein composition on both 1 kPa (Figure 3C) and 25 kPa microarrays (Figure S3B, Supporting Information). While some ECM protein compositions, especially those containing C1, C3, and C4 (e.g., C3,FN; C4,FN), maintained CYP3A4 expression at relatively high and steady levels for 7 days on 1 kPa microarrays, by 14 days of culture, CYP3A4 expression on such ECM compositions had declined by ~25–70% of day 7 levels. Nonetheless, the C4,HA combination on 1 kPa microarrays led to the steadiest and one of the highest CYP3A4 expression over 14 days. In contrast, CYP3A4 expression was ~2–3-fold lower on other ECM proteins (e.g., FN,DC; LN,LU; C5,HA) on 1 kPa microarrays. CYP3A4 expression on 25 kPa microarrays mirrored the trends above on 1 kPa microarrays, albeit the overall range of the CYP3A4 expression gradient on different ECM compositions was blunted on the 25 kPa microarrays.
PHHs were positive for nuclear HNF4α on both 1 kPa and 25 kPa stiffnesses (Figure 4A); HNF4α is a transcription factor in the liver and a master regulator of diverse liver functions.[26] HNF4α expression was similar on both stiffnesses, except for a transient difference at the 7 day time-point; furthermore, a slight decrease in HNF4α expression was observed on both stiffnesses (Figure S4A, Supporting Information). Additionally, HNF4α expression increased proportionally with increases in a) cell density per ECM island, b) albumin expression, and c) CYP3A4 expression (Figure 4B). While some ECM protein compositions, especially those containing C1, C3, and C4 (e.g., C4,LN; C3,FN; C3,C4), led to high HNF4α expression in PHHs on day 1 and retention of expression after 14 days to within 60–70% of day 1 levels on 1 kPa microarrays, other ECM proteins led to a more precipitous decline over time (Figure 4C). On 25 kPa microarrays, ECM compositions containing C1, C3, and C4 (e.g., C4,LN; C3,FN; C3,C4) also caused higher HNF4α expression as compared to other proteins; however, while HNF4α expression levels on 25 kPa microarrays were typically slightly lower on day 1 than on 1kPa microarrays, certain ECM compositions (e.g., C4,C5; C3,LU) on 25 kPa microarrays maintained HNF4α levels within ~20% over 14 days (Figure 4D). Heat maps of HNF4α expression over time on both stiffnesses are shown in Figure S4B, Supporting Information.
Figure 4. HNF4α expression in primary human hepatocytes (PHHs) on extracellular (ECM) microarrays.
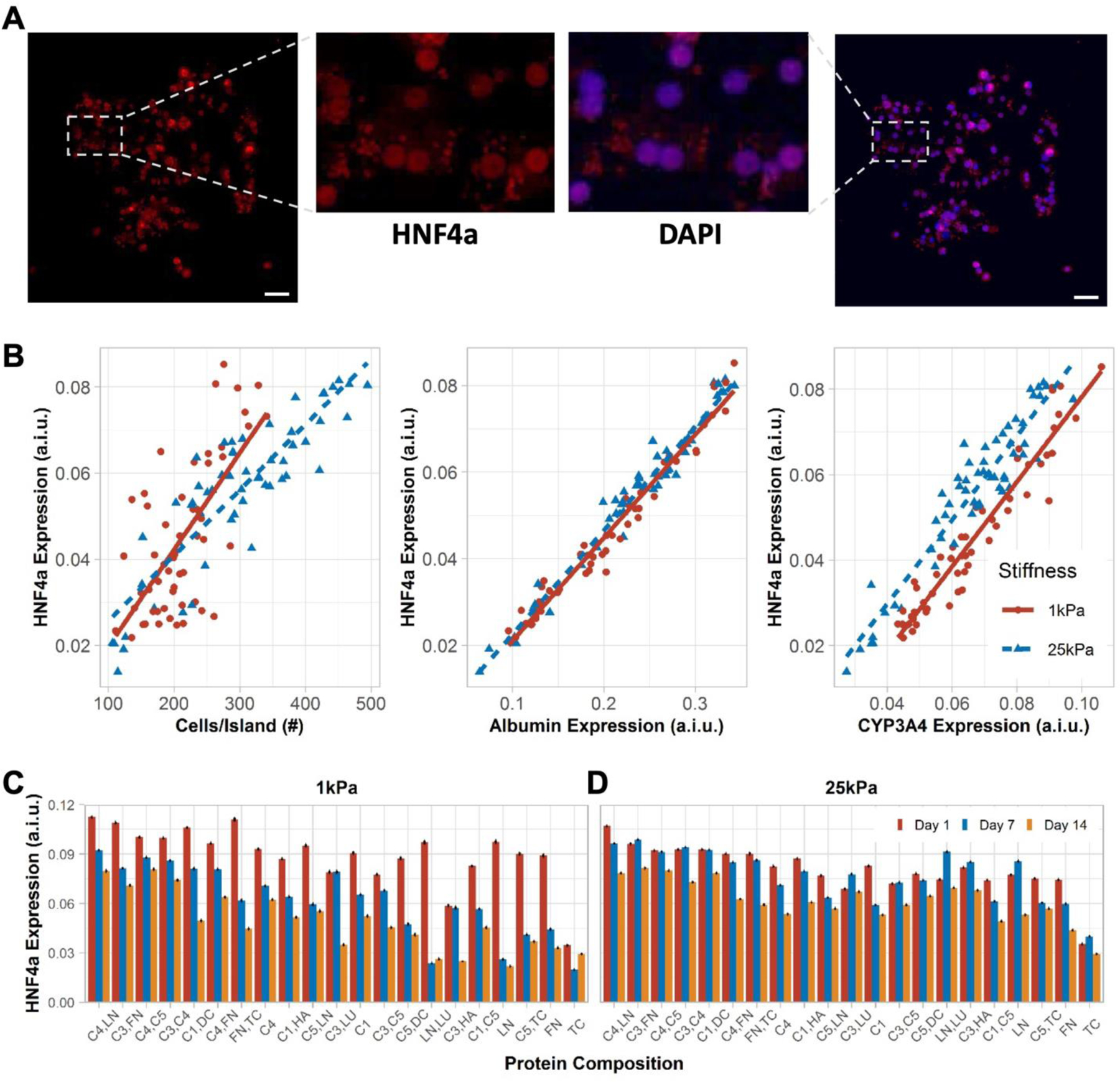
A) A representative island of PHHs shows HNF4α (transcription factor) expression overlaying with the nuclear stain. Scale bars = 50 µm. B) Positive correlation was observed between mean cells per ECM island and mean HNF4α expression on 1 kPa (rs=0.59, p=3.2e-06) and 25 kPa (rs=0.85, p<2.2e-16) stiffnesses. Similarly, mean albumin expression positively correlates with mean HNF4α expression on 1 kPa (rs=0.98, p<2.2e-16) and 25 kPa (rs=0.97, p<2.2e-16) stiffnesses. Lastly, mean CYP3A4 expression positively correlates with mean HNF4α expression on 1 kPa (rs=0.96, p<2.2e-16) and 25 kPa (rs=0.9, p<2.2e-16) stiffnesses. A.i.u. = arbitrary intensity units. C) Bar plots show the mean HNF4α expression in PHHs on representative ECM compositions after 14 days of culture on 1 kPa and D) 25 kPa stiffnesses. Error bars represent standard error of mean. For panels B-D, data displayed represents summarized single-cell measurements across three PHH donors per condition (n = 22–24 ECM islands across representative microarrays).
2.3. Rank ordering ECM effects on PHH phenotypic marker expression on microarrays
Next, we performed linear regression analysis on cell density per island and albumin expression after 7 days of culture as a function of the ECM composition on the two different stiffnesses, while using C1-alone as the control ECM since it is widely used for PHH culture.[6] This analysis showed that when C4 was mixed with other ECM proteins (LU, C3, LN, C5, DC, FN, C1, HA), cell attachment (Figure 5A) and albumin expression (Figure 5C) on 1 kPa microarrays were enhanced versus C1-alone. In contrast, non-collagenous proteins on their own or in combinations without collagens (e.g., HA, DC, LN, LU, TC) led to lower cell attachment and/or lower albumin expression on 1 kPa microarrays than C1-alone. FN alone did not cause a statistically significant downregulation in cell attachment/retention or albumin expression on 1 kPa microarrays but when mixed with LU or HA, led to downregulation of albumin on 1 kPa microarrays. For C3, when it was mixed with FN or DC, cell attachment/retention and albumin expression were enhanced relative to C1-alone on 1 kPa microarrays, whereas when C3 was used on its own or mixed with C1, HA, LU, or TC, attachment/retention and albumin expression were statistically similar to C1-alone. Finally, for 25 kPa microarrays, most ECM protein compositions caused similar or higher cell attachment/retention (Figure 5B) and albumin expression (Figure 5D) after 7 days of culture relative to C1-alone, with some key exceptions (LU,TC; HA,TC; HA,LU; HA; DC,LU; DC,TC; TC; DC,HA; and LU) that caused a downregulation in cell attachment/retention and/or albumin expression.
Figure 5. Rank ordering the effects of extracellular matrix (ECM) protein composition and stiffness on primary human hepatocyte (PHH) attachment and albumin expression.
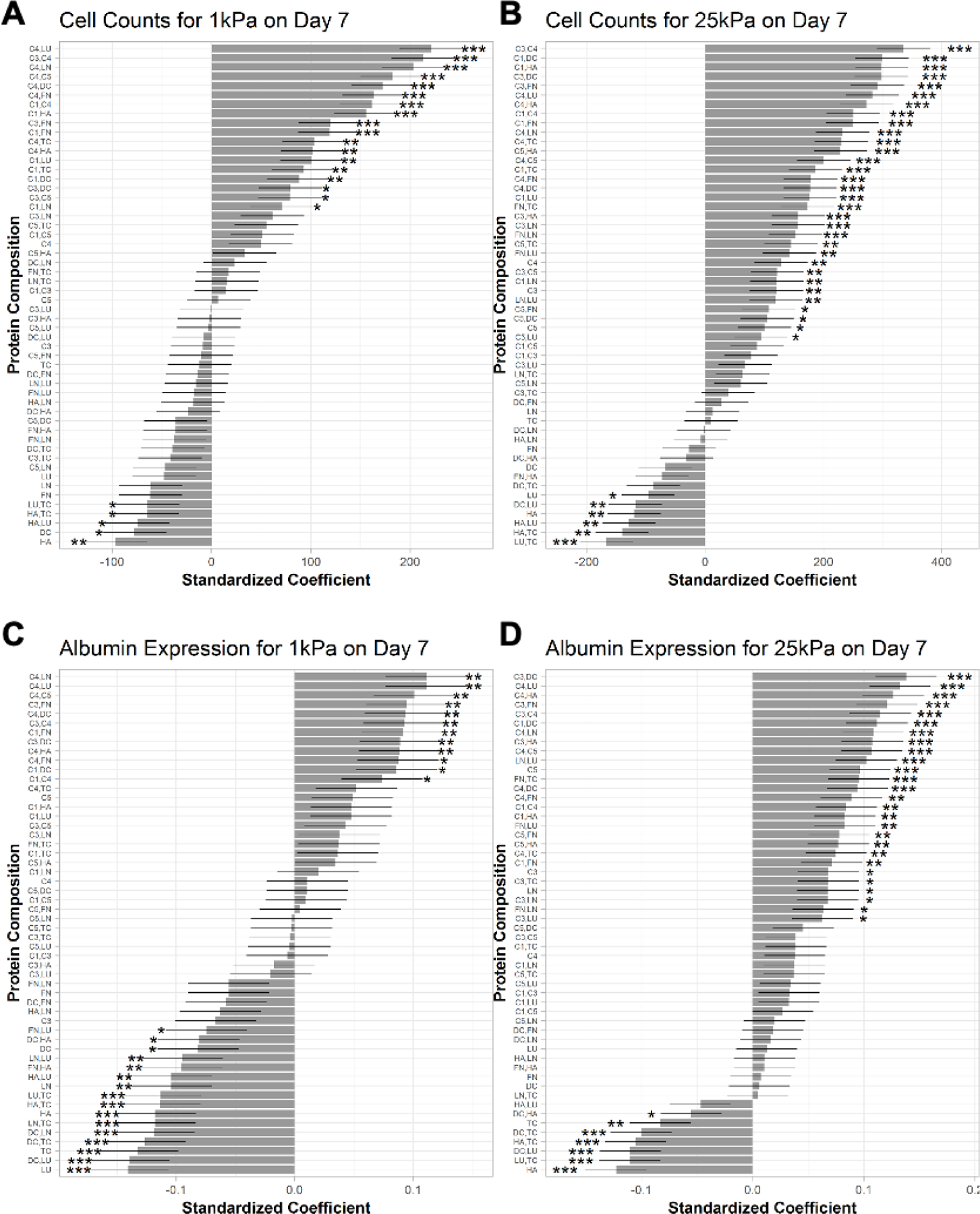
A) Multiple linear regression model for the effects of the ECM protein combinations on mean cells per ECM island after 7 days of culture on microarrays of 1 kPa and B) 25 kPa stiffnesses. C) Effects of the ECM protein combinations on mean albumin expression after 7 days of culture on 1 kPa and D) 25 kPa stiffnesses. Conditions are ranked in descending order based on the corresponding standardized coefficient. Similar rank orderings for CYP3A4 and HNF4α are shown in Figure S5, Supporting Information. Error bars represent standard error. *p ≤ 0.01, **p ≤ 0.001, and ***p ≤ 0.0001. For all panels, data displayed represents summarized single-cell measurements across three PHH donors per condition (n = 22–24 ECM islands across representative microarrays).
When similar regression analysis as above was performed on HNF4α expression in PHHs on microarrays after 7 days of culture, C4 mixed with LN or LU led to the highest expression followed by C4 mixed with C5, C3, HA, C1, FN, or DC relative to C1-alone on 1 kPa microarrays (Figure S5A, Supporting Information); similarly, FN mixed with C1 or C3 also led to higher HNF4α expression than C1-alone. In contrast, all the non-collagenous proteins (LU, DC, TC, LN, FN, HA) on their own or mixed with each other, as well as some collagenous proteins and combinations (C3; C5,DC; C3,HA; and C5,TC), led to downregulation of HNF4α relative to C1-alone on 1 kPa microarrays; all the other ECM combinations led to statistically similar HNF4α expression as C1-alone. Finally, for 25 kPa microarrays, most of the ECM protein compositions caused similar or higher HNF4α expression after 7 days of culture (C4,LU; C3,DC; C4,HA, and C3,FN had highest HNF4α expression) relative to C1-alone, with some key exceptions (LU,TC; DC,LU; HA,TC; DC,TC; HA,LU; TC; HA; DC,HA; and FN,HA) that caused a downregulation in HNF4α expression (Figure S5B, Supporting Information).
When similar regression analysis as above was performed on CYP3A4 expression in PHHs on microarrays after 7 days of culture, C4 mixed with LN or LU led to the highest expression followed by C4 mixed with C5, C3, HA, DC, C1, or FN relative to C1-alone on 1 kPa microarrays (Figure S5C, Supporting Information). Similarly, FN mixed with C3 or C1, C1 or C3 mixed with DC, and C1 mixed with HA also led to higher CYP3A4 expression than C1-alone. In contrast, non-collagenous proteins, LU, TC, LN and some of their combinations (LN,TC; LU,TC) led to the downregulation of CYP3A4 relative to C1-alone on 1 kPa microarrays; DC mixed with LU or LN or TC, and HA mixed with LU or TC also led to the downregulation of CYP3A4. Finally, for the 25 kPa microarrays, most of the ECM protein compositions caused similar or higher expression of CYP3A4 (Figure S5D, Supporting Information) after 7 days of culture (C3,DC; C4,LU; C3,FN; and C4,HA had highest CYP3A4 expression) relative to C1-alone, with some key exceptions (HA; DC,LU; HA,TC; LU,TC; and DC,TC) that caused the downregulation of CYP3A4.
2.4. PHH phenotype and drug responses in hydrogel-conjugated 96-well plates
From the microarray data, we first selected those ECM conditions that led to >200 cells retained per island over 14 days of culture. Next, linear regression analysis above was used to select those ECM compositions that led to the highest PHH functions (e.g., C4,LN; C3,FN; and C1,DC). Lastly, we selected those ECM compositions that led to similar or lower PHH functions than C1-alone on microarrays, as well as LN,LU, which displayed stiffness dependence in its effects on PHH functions. The ECM compositions selected using the criteria above were then adsorbed to commercially available Matrigen™ plates at a constant total coating density for all combinations. PHH attachment and spreading were more dependent on ECM composition as opposed to stiffness (Figure 6A). For instance, C4,LN retained confluent PHH monolayers over 14 days of culture, whereas the addition of C3 in the place of LN resulted in more aggregate formation on the 1 kPa stiffness; similarly, C3,FN maintained the monolayers over 7 days of culture as opposed to proteoglycan combinations such as LN,LU.
Figure 6. Phenotype of primary human hepatocytes (PHHs) in hydrogel-conjugated 96-well plates coated with select combinations of extracellular matrix (ECM) proteins.
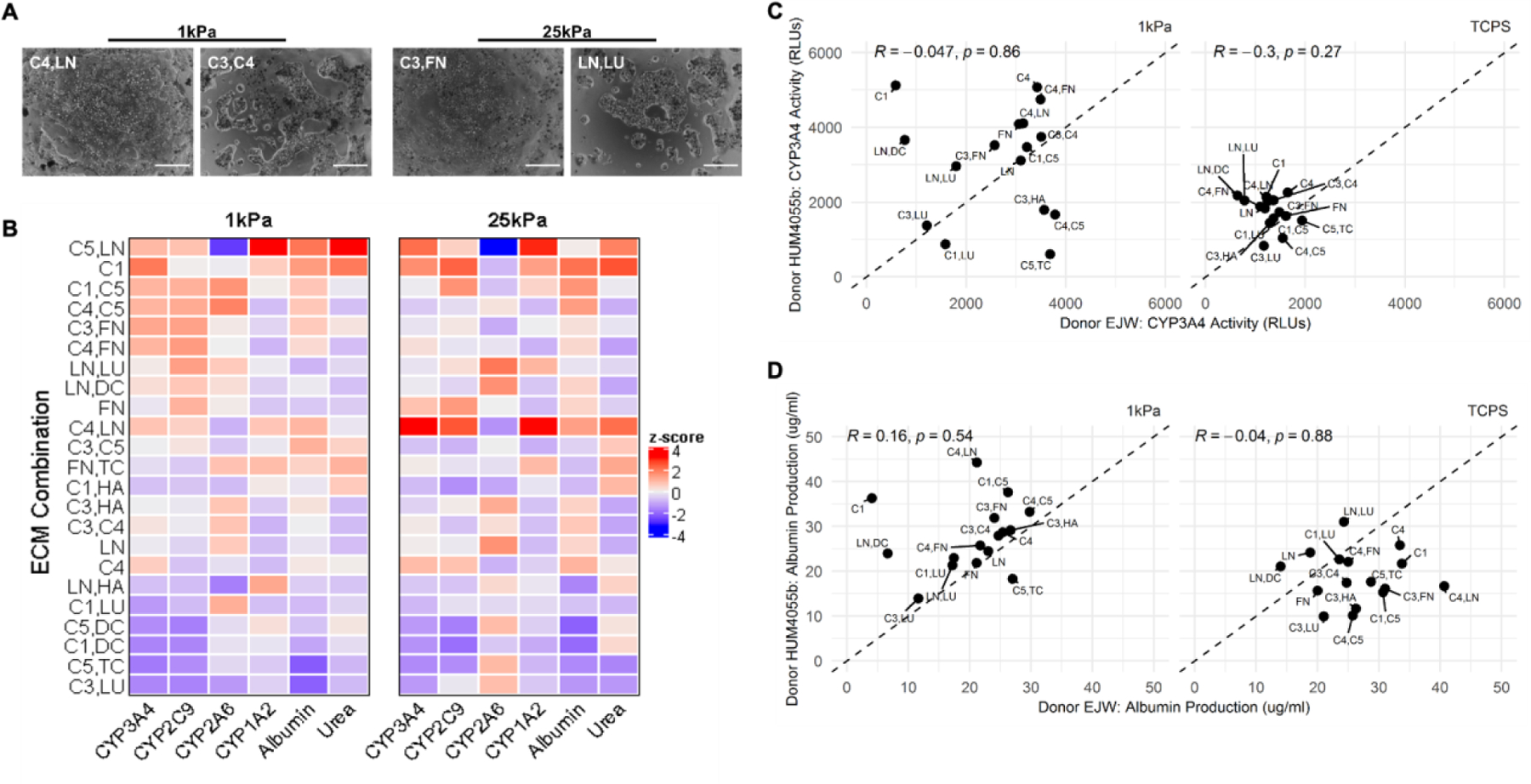
A) Select phase contrast images of PHHs on two stiffnesses (1 kPa and 25 kPa) coated with different combinations of ECM proteins. Scale bars = 50 µm. B) Cluster analysis for all measured functions of PHHs adhered to the selected combinations of ECM proteins on the two stiffnesses after 14 days of culture for a representative PHH donor. C) Scatterplot analysis of PHH donor-to-donor variability on different combinations of ECM proteins after 7 days of culture on 1 kPa (left) and tissue culture polystyrene (TCPS, no conjugated hydrogel) control wells (right) for CYP3A4 enzyme activities and D) albumin secretion.
We measured the secretions of albumin and urea and the activities of 4 CYPs using prototypical substrates over 14 days of culture in PHHs cultured on the selected ECM conditions within the 96-well plates. Consistent with the microarray results, ECM compositions containing collagens led to some of the highest albumin secretion on both stiffnesses (Figure 6B). Specifically, on the 1 kPa stiffness, albumin secretion was highest on C5, LN; C1; C4,LN; and C3,C5, while on the 25 kPa stiffness, albumin secretion was highest on C1; C1,C5; C4,C5; C4,LN; and C4. Across both the stiffnesses, albumin secretion was lowest on LN,HA; C1,LU; C5,DC; C1,DC; C5,TC; and C3,LU. Urea secretion (a marker of ammonia detoxification) was highest on C5,LN; C1; C3,FN; C3,C5; C1,HA; C4; C5,DC and FN,TC on the 1 kPa stiffness. Similar trends were observed on 25 kPa stiffness except C4,LN induced higher urea secretion on 25 kPa stiffness versus the 1 kPa stiffness. Across both stiffnesses, urea secretion was lowest on C3,HA; C3,C4; LN; C5,TC; and C3,LU.
CYP3A4 activities were highest on C1,C5; C4,C5; C3,FN; C4,FN; C4,LN; C5,LN; C1; and C4 on the 1 kPa stiffness. On the 25 kPa stiffness, the highest CYP3A4 activities were observed on C4,LN; C5,LN; C1; FN; and C4. Across both stiffnesses, CYP3A4 was lowest on LN,HA; C1,LU; C5,DC; C1,DC; C5,TC; and C3,LU. CYP2C9 activities were highest on C1,C5; C4,C5; C3,FN; C4,FN; C5,LN; LN,LU; and FN on the 1 kPa stiffness. On the 25 kPa stiffness, the highest CYP2C9 activities were observed on C1; C4,LN; FN; and C1,C5. Across both stiffnesses, CYP2C9 was lowest on LN,HA; C5,DC; C1,DC; and C5,TC.
CYP2A6 activities were highest on C1,C5 and C4,C5 on the 1 kPa stiffness, and highest on LN,LU; LN,DC; C3,HA; and LN on the 25kPa stiffness. Across both stiffnesses, CYP2A6 was lowest on C5,LN; C4,LN; and C1,HA. Lastly, CYP1A2 activities were highest on C5,LN and LN,HA on the 1 kPa stiffness, and highest on C4,LN; C5,LN; and C1 on the 25 kPa stiffness. Across both stiffnesses, CYP1A2 was lowest on C4,C5; C4,FN; C3,HA; C3,C4; and LN.
Overall, on the 1 kPa stiffness, all measured PHH functions were relatively well retained for 14 days and had typically higher than average functional levels on C1; C1,C5; C3,FN; C4,C5; and C4,FN, whereas the lowest functional levels were observed on C1,DC; C5,TC; and C3,LU. On the 25 kPa stiffness, all measured PHH functions, except for CYP2A6, were relatively well retained for 14 days and had typically higher than average functional levels on C1 and C4,LN, whereas the lowest function levels were observed on C1,DC; C5,TC; and C3,LU, except for CYP2A6, which was better retained on these ECM compositions.
We next compared PHH functions on selected ECM compositions on the 1 kPa stiffness to the same ECM compositions adsorbed to TCPS. Overall, CYP3A4 activity was downregulated on average on TCPS as compared to the 1 kPa stiffness, and PHHs were more sensitive to ECM composition on the 1 kPa stiffness (Figure 6C). When comparing functions of two PHH donors on the 1kPa stiffness, several ECM compositions (e.g., C3,LU; LN; C1,C5; C3,C4) yielded similar functional levels in both PHH donors, whereas some ECM compositions, such as C1; LN,DC; and C5,TC led to disparate responses across the two PHH donors (Figure 6C). For albumin secretion, donor dependency was observed on 1 kPa stiffness and TCPS; specifically, one of the two donors displayed reduced albumin secretion on TCPS as compared to the 1 kPa stiffness, whereas the other donor had similar overall albumin secretion levels across both stiffnesses (Figure 6D). On the 1 kPa stiffness, both donors secreted similar albumin levels on the majority of ECM compositions, with some key exceptions, such as C1; LN,DC; and C4,LN (Figure 6D).
Next, we measured the time-course of PHH functions on selected ECM compositions on the 1 kPa stiffness that led to high and low functions as determined above. Albumin secretion was highest (~3-fold relative to the lowest performing condition) on C3,FN and C4,FN, followed by C1,HA, and lowest on C5,TC (Figure 7A). Urea secretion was the highest (~1.5-fold) on C1,HA, and C3,FN, followed by C4,FN, and C5,TC (Figure 7B). CYP3A4 activities on all of the tested ECM compositions declined over time, but were better retained over time (~30–50% activity of day 1 activity retained on day 14) on C4,FN, and C3,FN, while being least stable on C5,TC (Figure 7C). CYP2C9 activities on different ECM compositions showed similar trends as the CYP3A4 activities, albeit declined more precipitously over time (Figure 7D). CYP2A6 activities were higher on C3,FN over 7 days of culture but then were similar across all ECM compositions by 14 days of culture; furthermore, CYP2A6 activities were stable on all ECM compositions for 7 days of culture and then declined by 25–50% by 14 days of culture (Figure 7E). Lastly, CYP1A2 activities were similar and stable for 14 days of culture across the ECM compositions tested (Figure 7F).
Figure 7. Time-course of the phenotype of primary human hepatocytes (PHHs) in hydrogel-conjugated 96-well plates coated with select combinations of extracellular matrix (ECM) proteins.
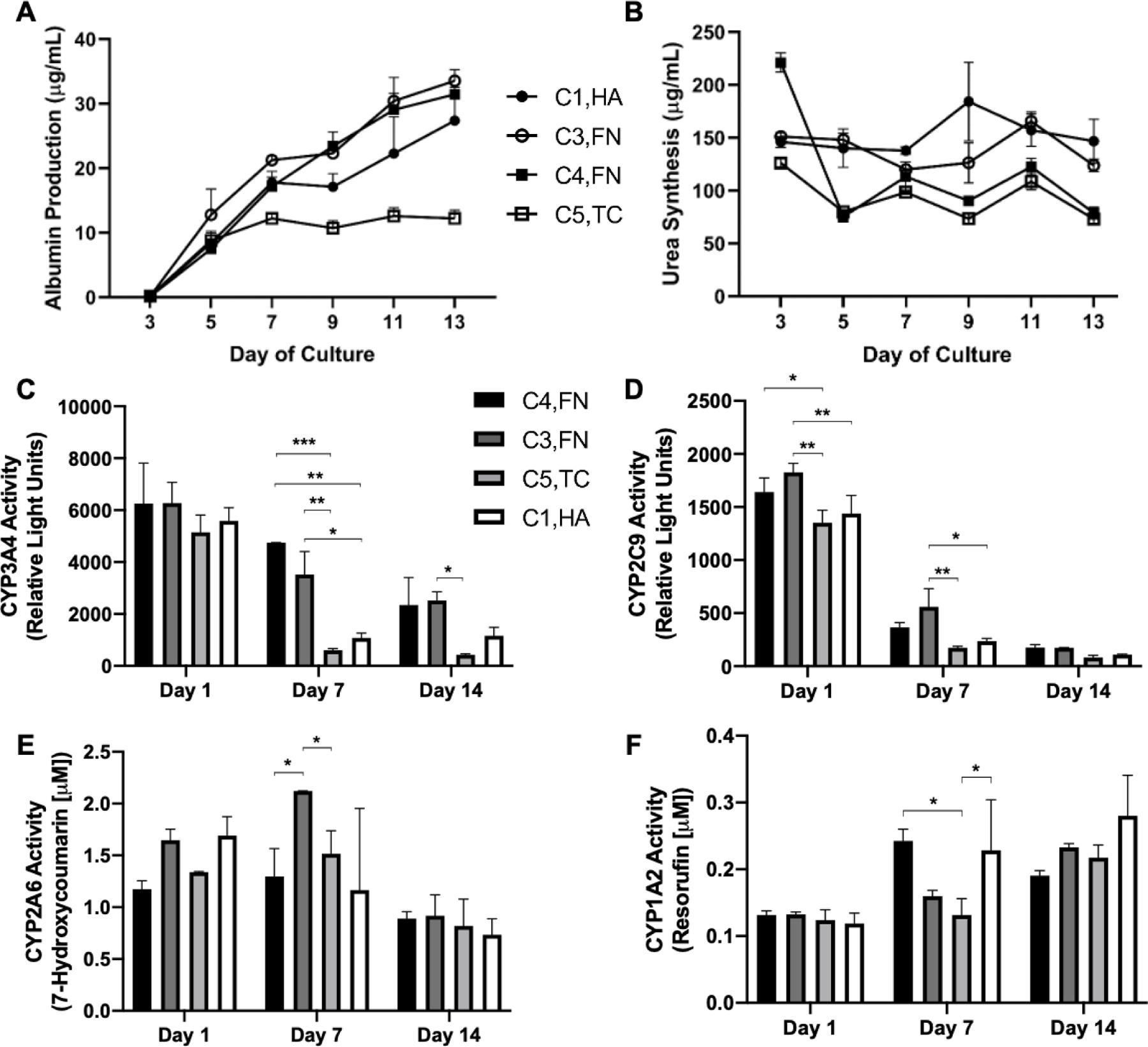
A) Albumin secretion, B) urea synthesis, C) CYP3A4 enzyme activity, D) CYP2C9 activity, E) CYP2A6 activity, F) and CYP1A2 activity in PHHs attached to 1 kPa wells coated with selected combinations of ECM proteins. Data from a representative PHH donor is shown. Error bars represent standard deviation. *p ≤ 0.01, **p ≤ 0.001, and ***p ≤ 0.0001.
Lastly, two ECM combinations that led to high PHH functions on both 1 kPa and 25 kPa stiffnesses were selected for drug-mediated CYP induction studies, which are used in pharmaceutical practice to assess the potential for drug-drug interactions.[27] PHHs on the above stiffnesses and TCPS were cultured for 7 days, incubated for 2 days with prototypical drugs, and then probed for the activities of key CYP enzymes. For phenobarbital-mediated CYP3A4 induction via the constitutive androstane receptor (CAR), PHHs displayed higher CYP3A4 activity on the 1 kPa stiffness relative to the 25 kPa stiffness, irrespective of ECM composition (Figure 8A). However, while PHHs on the 1 kPa stiffness showed higher phenobarbital-induced CYP3A4 activity relative to TCPS for C4,LN, these trends were reversed on C5,LN. Additionally, PHHs on TCPS displayed higher phenobarbital-induced CYP3A4 activities on both tested ECM compositions. Rifampin, an activator of the pregnane X receptor (PXR), induced CYP2C9 activity in PHHs cultured on all stiffnesses, but rifampin-induced CYP2C9 activities were maximal at the 1 kPa and 25 kPa stiffnesses versus TCPS (Figure 8B). Some stiffness dependency was observed for the C5,LN composition such that PHHs on the 1 kPa stiffness displayed higher rifampin-induced CYP2C9 activity as compared to the 25 kPa stiffness. Finally, CYP1A2 activity was induced by omperazole, an activator of the aryl hydrocarbon receptor (AHR), at similar levels across all stiffnesses and ECM compositions tested (Figure 8C).
Figure 8. Drug-mediated induction of cytochrome-P450 enzyme activities of primary human hepatocytes (PHHs) cultured in hydrogel-conjugated or tissue culture polystyrene (TCPS) 96-well plates coated with select combinations of extracellular matrix (ECM) proteins.
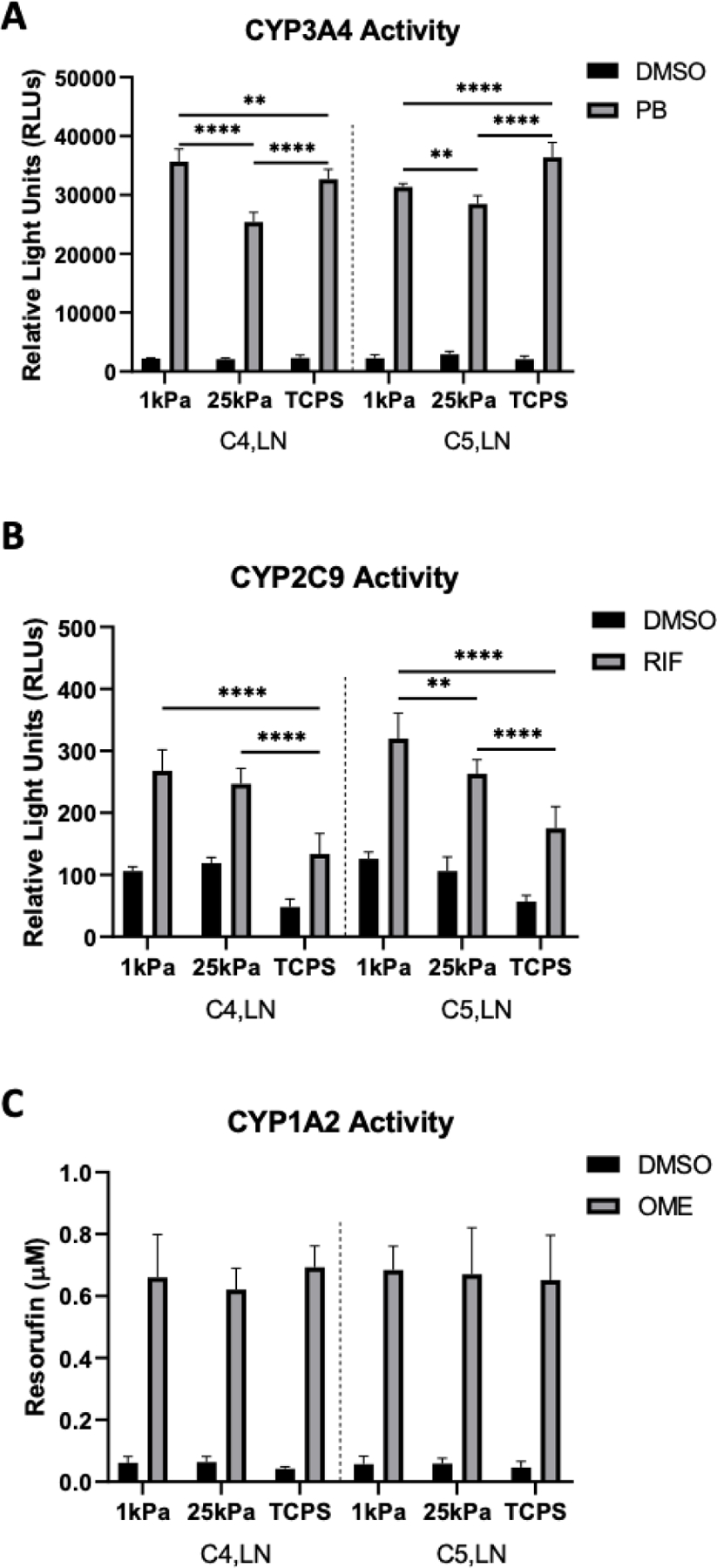
A) CYP3A4 enzyme activity after the incubation of PHHs for 48 hours with 1 mM phenobarbital (PB) or dimethyl sulfoxide (DMSO) vehicle control. B) CYP2C9 activity after the incubation of PHHs for 48 hours with 25 µM rifampin (RIF) or DMSO vehicle control. C) CYP1A2 induction after the incubation of PHHs for 48 hours with 50 µM omeprazole (OME) or DMSO vehicle control. Data from a representative PHH donor is shown. Error bars represent standard deviation. Two-way ANOVA with Tukey’s Test was used for multiple comparisons. *p<0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
3. Discussion
ECM is known to be a key regulator of hepatic functions in physiology and disease.[1] While there have been previous efforts in recapitulating the protein composition and stiffness of ECM independently for short-term hepatocyte culture, how these components interact to regulate the functions of PHHs over prolonged culture has not been investigated. While such an investigation can be carried out using conventional multiwell plates, the process is labor-intensive and costly. In contrast, high-throughput ECM microarrays allow for the independent modulation of cell-cell interactions, ECM composition/stiffness, and soluble factor incubations. Here, we utilized this platform along with a custom high-content imaging pipeline to uncover for the first time how 10 major ECM proteins present in the liver, individually and in two-way combinations, regulate the phenotype of PHHs on liver-like stiffnesses. We then adapted our findings to a hydrogel-conjugated multi-well format that led to higher PHH functions and drug-mediated CYP induction for 2 weeks as compared to adsorbed collagen-I on TCPS.
We robotically spotted proteins onto our microarray into circular islands of 450 µm in diameter, which has been found previously to induce the robust formation of homotypic contacts between PHHs.[7] For the stiffness of the culture substrate, we chose 1 kPa (Young’s modulus) to represent the stiffness similar to native liver tissue and 25 kPa to determine the effects on cell attachment and functions relative to 1 kPa, but still orders of magnitude lower than the stiffness of plastic or glass (~GPa). PA was chosen as the polymer to modulate substrate stiffness since it resists cell attachment, and therefore, homotypic interactions can be defined for all the ECM protein islands on the entire microarray. For proteins, we chose 10 human ECM proteins present in the liver, which included structural proteins (C1, C3, C4, C5), basement-like membrane proteins (C4, LN), glycoproteins (FN, TC), proteoglycans (HA, DC), and proteins upregulated in disease (LU).[22, 23] While the liver has a larger repertoire of proteins (150+), the above proteins represent the bulk of proteins by percentage.[24] We restricted our investigations to single and two-way combinations of the above proteins at single concentrations (55 total ECM combinations x 2 stiffnesses) to keep the scope manageable and because such a strategy has still provided key insights on liver progenitor differentiation in our previous work.[19] Lastly, all of our microarray studies were executed in 3 PHH (unpooled) donors across three time points in culture (day 1, 7, and 14).
For PHH phenotype, we evaluated nuclei via DAPI to obtain cell counts over time; albumin as a marker of liver’s protein synthesis capability and widely utilized for appraising PHH functions across multiple platforms[28]; CYP3A4 since it metabolizes ~50% of drugs on the market [25]; and HNF4α since it is the master transcription factor that regulates diverse hepatic functions[26] and has been shown previously to be sensitive to substrate stiffness in primary mouse hepatocyte cultures.[3] For the multi-well plate studies, we further appraised urea levels in supernatants as well as the activities of additional CYP enzymes (CYP1A2, 2A6, 2C9) to elucidate how the protein composition of the ECM and its stiffness affect major functions of PHHs over 2 weeks in culture.
We found that the attachment/retention of PHHs on the ECM islands was on average higher on the 25 kPa stiffness as compared to the 1 kPa stiffness over 14 days of culture; this result is consistent with the literature in that cells attach better to stiffer surfaces due to stronger traction forces[29], though for hepatocytes, too much spreading on stiffer surfaces (e.g., glass or polystyrene) has been correlated to de-differentiation.[30, 31] However, PHH attachment was significantly influenced by the protein composition of the ECM islands, in that the compositions containing 3 of the 4 collagens (C4>C1>C3) led to better attachment of PHHs than the compositions containing only non-collagenous proteins (FN>LN>DC>HA>TC>LU), though some non-collagenous protein combinations, especially those containing LN, FN, and LU, still retained at least 200 cells per island on the 25 kPa stiffness even after 14 days of culture. Furthermore, cell density positively correlated with PHH functions on both stiffnesses, which is consistent with previous work showing that the homotypic interactions between PHHs are critical for the maintenance of their phenotype.[7] Lastly, the three measured PHH functions (albumin, CYP3A4, and HNF4α) correlated positively and strongly with each other, which suggests a common underlying regulatory mechanism.
Despite lower average cell attachment, all three PHH functions were on average (across 3 PHH donors) and on a per cell basis higher on the 1 kPa stiffness than the 25 kPa stiffness after 1 day of culture, which is consistent with previous literature in short-term cultures of rodent hepatocytes.[3] However, unlike previous studies that did not utilize PHHs nor prolonged hepatic culture, here we found that by 14 days of culture, substrates with 25 kPa stiffness led to higher albumin and HNF4α expression on average and on a per cell basis than the 1 kPa substrates. In contrast, average CYP3A4 expression was statistically similar across both stiffnesses even after 14 days of culture, which suggests that the functional differences across the two stiffnesses were not entirely due to the differences in cell retention/attachment after 14 days of culture.
The protein composition of the ECM islands significantly regulated the functions of PHHs across both stiffnesses. Linear regression analysis after 7 days of culture elucidated that when C4 was mixed with specific ECM proteins (LU, C3, LN, C5, DC, FN, C1, HA), cell attachment and PHH functions were at their highest levels relative to the standard utilized in pharmaceutical practice, C1-alone. However, C4 on its own did not enhance the functions of PHHs compared to C1-alone, which contrasts with previous results in primary rat hepatocytes[11], in which C4 alone had the highest effect on albumin expression after 7 days of culture as compared to the other 31 combinations of ECM proteins. Non-collagenous proteins (LU, DC, TC, LN, FN, HA) on their own or mixed, as well as when mixed with some collagenous proteins (e.g., C5,DC; C3,HA; C5,TC), led to lower cell attachment and/or lower functions of PHHs than C1-alone. Interestingly, FN did not significantly modulate the functions of PHHs when presented on its own, which contrasts with previous results with primary rat hepatocytes[11], in which FN alone was a positive regulator of hepatic albumin expression. However, when combined with HA, FN downregulated the functions of PHHs, whereas when combined with C1 or C3, FN enhanced the functions of PHHs relative to C1-alone. Lastly, while C3 individually was found to be a negative regulator of rat hepatocyte functions previously[11], here we found that C3 enhanced PHH functions, but only on 25k Pa substrates, which were not previously tested with rodent hepatocytes.
Temporally, the C4,HA combination on the 1 kPa microarrays led to the steadiest and one of the highest CYP3A4 expression over 14 days of culture. CYP3A4 expression on the 25 kPa microarrays mirrored the trends on the 1 kPa microarrays, though the overall range of the CYP3A4 expression gradient on the different ECM compositions was blunted on the 25 kPa microarrays relative to the 1 kPa microarrays. While specific ECM compositions (C4,LN; C3,FN; C3,C4) led to the retention of HNF4α expression in the nucleus after 14 days of cultures to within 60–70% of day 1 levels on both stiffnesses, only specific ECM compositions (e.g., C4,C5; C3,LU) on the 25 kPa microarrays maintained HNF4α levels within ~20% of day 1 levels even after 14 days of culture. These results contrast with previous results in which HNF4α and other functions were typically found to be higher in hepatocytes from different species and origins when the cells were cultured on softer ECM gels versus stiffer substrates. [3, 12, 13, 15, 16]. Therefore, de novo investigation of the effects of varying protein composition and stiffness of ECM on specific cell types over prolonged culture is necessary to reveal unexpected synergies as we have done for PHHs here.
From the microarray results, we selected the top, middle, and low performing ECM conditions and adapted them to commercially available 96-well plates with conjugated hydrogels of 1 kPa and 25 kPa stiffnesses. Such multiwell plates allowed us to maintain select ECM compositions entirely in separate wells as opposed to microarrays in which the PHHs adhered to islands of different ECM protein compositions secrete molecules into the same common medium, thereby leading to confounding paracrine crosstalk. Despite this limitation of the microarrays, here we found that the trends observed from the microarrays were consistent with those observed on the multiwell plates. Specifically, ECM compositions containing collagens led to some of the highest PHH functions on both stiffnesses, while functions were lowest when TC, LU, or DC were mixed with C1, C3, or C5. However, we observed similarities and differences across the effects of ECM compositions on diverse functions of PHHs. For instance, after 14 days of culture, urea secretion as well as CYP3A4/2C9/1A2 activities were expressed at higher-than-average levels on C5,LN across both stiffnesses, but albumin levels were on par with average secretion on C5,LN on the 25 kPa stiffness; furthermore, CYP2A6 showed the lowest activities on C5,LN on both stiffnesses. In contrast, while CYP2A6 was downregulated on C4,LN on the 25 kPa stiffness, other functions were upregulated, whereas the opposite trends were observed on C3,LU. While the mechanism underlying these functional differences are not known, this example illustrates the importance of measuring different PHH functions on combinatorial microenvironments due to unexpected synergies. Future work could be aimed at investigating how the cooperation of the protein composition and stiffness of the ECM regulates hepatic molecular pathways and functions, such as those found in the different functional liver zones that have varying ECM compositions.[32, 33]
When comparing the responses of different PHH donors on the softer substrates, several ECM compositions were identified that led to similar and high PHH functional output for 14 days of culture across both donors (LN; C4; C1,C5; C3,C4; C3,FN; C4,FN). In contrast, C1-alone was effective for inducing high functions in only 1 PHH donor, which may partly explain why PHH donors/lots display variable functions (besides any genetic polymorphisms) in conventional culture platforms that rely only on adsorbed or gelled C1. With the use of liver-like stiffness and specific ECM compositions, the functional differences across different PHH donors due to genetic and/or epigenetic differences could be better studied, all the while minimizing donor-to-donor variations introduced solely by the non-physiologic culture method.
While PHHs were previously shown to display a rapid decline in phenotypic functions[7], here we found that the PHHs maintained some functions at relatively stable levels for at least 2 weeks on select ECM protein compositions on softer hydrogels within 96-well plates. Specifically, urea synthesis, CYP1A2, and CYP2A6 were remarkably stable on the ECM compositions tested. Albumin secretion increased over 2 weeks on some of the ECM compositions and was highest on C3,FN and C4,FN. CYP3A4, however, declined over 2 weeks on all of the tested ECM compositions but was nonetheless better retained on C3,FN and C4,FN. CYP2C9 was the least stable of all CYPs tested, though activities were still modulated by the protein composition of the ECM. Our observations with different CYP isoforms indicate the role ECM plays in their regulation.
PHHs adhered to select ECM proteins on the 1 kPa and 25 kPa stiffnesses within hydrogel-conjugated 96-well plates maintained their in vivo-like responsiveness to prototypical drugs for CYP induction. Drug-induced CYP2C9 activities were higher on the 1 kPa stiffness than 25 kPa stiffness and TCPS irrespective of the ECM composition, though trends were dependent on the protein composition of the ECM for CYP3A4 induction. CYP1A2 was inducible on all the tested stiffnesses and ECM compositions to statistically similar levels. Even with their declining functions, PHH monocultures with or without an ECM gel overlay are utilized during drug development for a first-tier screen to evaluate those drugs that strongly induce CYP enzymes and/or cause severe toxicity to PHHs.[34–36] Therefore, our simple-to-implement approach with select combinations of ECM proteins and stiffness coupled with the commercially available hydrogel-conjugated plates could help improve the first-tier assays with better retention of PHH functions and drug-mediated CYP inducibility across multiple PHH donors than is possible with C1-coated TCPS.
Even though our studies did not include the entire complexity of the human liver ECM, some interesting correlations to in vivo findings could be made. For instance, C4 was consistently identified as a protein that, when mixed with other proteins, but not alone, led to the highest PHH functions across both stiffnesses, which may be due to its critical role in the native liver.[1] In contrast, primary rat hepatocytes displayed the highest functions on C4 alone while C4,FN downregulated functions of primary rat hepatocytes[11] whereas the same combination enhanced the functions of PHHs here, which may be due to species-specific differences in how combinatorial ECM regulates hepatic functions. C1 on its own led to low PHH functions, especially on the 25 kPa stiffness, which is consistent with excessive production of C1 by activated (myofibroblastic) hepatic stellate cells in liver fibrosis/cirrhosis that leads to PHH dysfunction.[37, 38] Similarly, we observed lower CYP3A4 activities on the 25 kPa stiffness on several, but not all, ECM compositions, which may be due to dysregulation of CYP3A4 in the stiffening liver as in fibrosis/cirrhosis[38, 39] but only in the presence of specific ECM protein compositions. Lastly, we found that LU and TC on their own led to low PHH functions irrespective of stiffness, which is consistent with marked upregulation of these proteins in liver fibrosis.[23, 40] TC, for example, has been implicated in activating hepatic stellate cells; our results here show that it may dysregulate PHH functions directly, though when its concentration is lowered and it is mixed with C4, TC’s deleterious effects on PHHs are mitigated to some extent, which is a novel finding here.
While our studies here are the first to elucidate the synergistic role of ECM protein composition and stiffness on diverse functions of PHHs over prolonged culture, with several differences identified from previous literature that evaluated the factors above in isolation over short-term hepatic culture, the liver microenvironment is more complex with the presence of heterotypic interactions between PHHs and liver non-parenchymal cells (NPC), as well as gradients of O2 and soluble factors induced across the liver sinusoid due to the flowing blood from the portal triad to the central vein. These additional cues likely synergize with the ECM to further modulate PHH functions in physiology and disease. Indeed, co-culture with both liver- and non-liver-derived NPCs has been shown to induce major functions in PHHs[41] and could be needed along with ECM composition and stiffness to stabilize CYP2C9 and CYP3A4 that could not be completely stabilized with only a biophysically tuned substrate here. Towards elucidating the synergies between the above-mentioned cues, our microarray platform is modular in that it is amenable to a) the seeding of different cell types, including co-cultures on the islands, b) incubation with different soluble factors to investigate interactions with ECM on cellular phenotype and c) inclusion within modular bioreactors[42] to induce fluid flow and thus gradients of O2, hormones, and other soluble factors. We anticipate that, in the absence of detailed mechanistic insights into all the various molecular regulators of multiple human liver cell types, high-throughput investigations of the role of distinct and combinatorial microenvironmental cues will be critical for optimizing human liver platforms for drug screening and ultimately regenerative medicine, as well as to elucidate the role of ECM components and their interactions in liver physiology and disease states.
4. Conclusion
We utilized a high-throughput ECM microarray platform to elucidate for the first time the synergistic role of ECM protein composition and stiffness on the phenotype of PHHs over prolonged culture. Adaption of select ECM compositions and physiological stiffness to 96-well plates produced a simple-to-implement strategy to better maintain several PHH functions than possible with C1-alone adsorbed to TCPS, which is still routinely utilized in pharmaceutical practice for the first-tier screening of compound libraries for CYP induction and/or drug toxicity. More broadly, our high-throughput approach could help elucidate the role of ECM in liver diseases (e.g., alcoholic and non-alcoholic fatty liver diseases, hepatitis B/C viral infections, and hepatocellular carcinoma) as well as facilitate the optimization of engineered liver tissues for cell-based therapy.
5. Experimental Section
5.1. Preparation of PA hydrogels
PA hydrogels were prepared as previously described.[43] Briefly, pre-cleaned glass slides were salinized with 2% (v/v) 3-(trimethoxysilyl)proply methacrylate in ethanol for 30 minutes and dried on a hot plate at 110°C for 5–15 minutes. Pre-polymer solution was prepared with appropriate acrylamide/bisacrilamide percentages (w/v) for the desired Young’s moduli (as described below) alongside a photoinitiator solution of 20% (w/v) Irgacure 2959 (BASF, Luwigshafen, Germany) in methanol. To reduce polymer network differences that could affect the interaction and retention of ECM proteins, acrylamide/bisacrilamide percentages (w/v) of 4/0.40 and 8/0.55 were used for 1 kPa and 25 kPa substrates, respectively, to achieve similar porosity.[19, 44] About 100 µL of 9:1 pre-polymer:photoinitiator solution was pipetted onto each glass slide and a 22 x 60 mm coverslip was placed on the top to prevent inhibition of the polymerization reaction by O2. An ultraviolet (UV) cross-linker was used to expose the slides to 365 nm UV A for 10 minutes (4 W m−2). Hydrogels were then immersed in deionized H2O (dH2O), glass coverslips were removed, and hydrogels on the glass slides were dehydrated at 50°C for 15–30 minutes and stored at room temperature (RT) until microspotting.
5.2. Microarray fabrication
Microarrays were prepared at a stiffness of 1 kPa or 25 kPa, as measured via nanoindentation (Optics11 Life Piuma Nanoindenter). Furthermore, an OmniGrid Micro automated microspotter (Digilab, MA) created array grids of 10 ECM proteins in one- and two-way combinations, for a total of 55 combinations (Figure 1A). The ECM proteins included human collagen I (C1; Millipore, Burlington, MA), collagen III (C3; Millipore), collagen IV (C4; Abcam, Cambridge, MA), collagen V (C5; Abcam), decorin (DC; R&D Systems, Minneapolis, MN), fibronectin (FN, Millipore-Sigma, Burlington, MA), hyaluronic acid (HA; Lifecore Biomedical, Chaska, MN), laminin (LN, Millipore), lumican (LU, Acro Biosystems, Newark, DE), and tenascin C (TC; R&D Systems); these ECM proteins were selected based on our previous work with liver progenitors[19] and also since these are major ECM proteins present in the liver.[24] Each microspot or ‘island’ was spotted with a pin diameter of 450 µm and 1 mm center-to-center spacing. Single proteins were spotted at 250 ng µL−1 while 125 ng µL−1 per protein was utilized for two-way combinations. The retention of ECM proteins on PA microarrays was previously characterized using fluorescently labeled C1.[11, 45] Lastly, rhodamine-labeled dextran spots were spotted for microarray alignment.
5.3. PHH culture
Prior to cell seeding, the microarrays were placed in a 4-chamber rectangular culture dish and incubated with 4 mL of 1% (v/v) penicillin/streptomycin (Corning Life Sciences, Tewksbury, MA) in 1X phosphate buffered saline (PBS; Corning) under UV sterilization for 30 minutes. Cryopreserved PHHs (lots included HUM4055, 54 year old Caucasian female, Lonza, Walkersville, MD; HUM4192, 16 year old Asian female, Lonza; and EJW, 29 year old Caucasian female, BioIVT, Baltimore, MD) were thawed and seeded at a density of 1 million cells per microarray in a serum-free cell culture medium consisting of 1X Dulbecco’s Modified Eagle’s Medium (DMEM, Corning) supplemented with 15 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid] buffer (Corning), 1% penicillin/streptomycin, 1% ITS+ (insulin, transferrin, selenous acid, linoleic acid, bovine serum albumin; Corning), 7 ng mL−1 glucagon (Sigma-Aldrich, St. Louis, MO) and 0.1 µM dexamethasone (Sigma-Aldrich). Cells were allowed to attach to microarray for 12–18 hours to enable the highly adhesive ECM islands to become confluent with cells, after which the microarrays were rinsed 3x with 1X DMEM containing 1% penicillin/streptomycin to remove the unattached cells. Culture medium exchanges (same serum-free medium formulation as above) were executed every 2 days.
5.4. Microarray immunostaining
PHHs on the microarrays were fixed in 4% (w/v) paraformaldehyde in 1X PBS for 10 minutes at RT. Subsequently, samples were blocked and permeabilized using 5% (v/v) donkey serum with 0.3% (v/v) Triton X-100 (Amresco, Solon, OH) in 1X PBS for 1 hour at RT. Samples were further incubated for 1 hour at RT with a combination of the following primary antibodies diluted in dilution buffer consisting of a 1% (v/v) bovine serum albumin and 0.3% (v/v) Triton X-100 in 1X PBS: goat anti-albumin (1:200 from stock, Abcam, Cambridge, MA), rabbit anti-HNF4α (1:200 from stock, ThermoFisher, Waltham, MA), and mouse anti-CYP3A4 (1:200 from stock; GeneTex, Irvine, CA). Samples were washed three times with 1X PBS and a final wash in dH2O in preparation for incubation with secondary antibodies for 1 hour at RT. Secondary antibodies included Alexa Fluor™ (AF) 455 (4 µg mL−1, ThermoFisher), AF567 (4 µg mL−1, ThermoFisher), and AF647 (4 µg mL−1, ThermoFisher). Samples were mounted in Fluoromount-G with DAPI (Southern Biotech, Birmingham, AL) and sealed with a coverslip.
5.5. Imaging and image analysis
Microarrays were imaged using an automated IX83 microscope (Olympus America, Center Valley, PA) with a high sensitivity 4.2MP sCMOS camera (ORCA-Flash4.0 LT+, Hamamatsu, Skokie, IL). Images were first converted to 8-bit Tiff files in Fiji [46] and then rotated to align the rhodamine-labeled dextran microspots for subsequent cropping. Microarrays were then divided into individual island images using a custom MATLAB script and analyzed using CellProfiler software[47] for mean fluorescent intensity at a single-cell resolution reported as arbitrary intensity units (a.i.u). Output measurements for each channel were compiled into R statistical software for data visualization and statistical analysis. Mean fluorescent intensities for all cells subjected to an individual ECM protein composition/stiffness condition at a specific time-point (e.g., C1 on 1 kPa substrates at day 1 of culture) were summarized across the 3 PHH donors. For each donor, every sacrificial time point was conducted using 2–3 microarrays per stiffness and triplicate islands per ECM condition (~22–24 individual ECM islands total) across independent experiments and reported as a mean ± standard error of the mean (SEM) in the corresponding figures.
5.6. PA-conjugated multi-well plate studies
Hydrogel bound 96-Softwell™ plates were acquired with Easy Coat™ composition from Matrigen (Brea, CA) at 1 kPa and 25 kPa stiffnesses. Young’s moduli were measured using a spherical tip (1 mm diameter) indentation and generation of a force-displacement curve by the manufacturer. The ECM compositions were pre-formulated in a deep-well plate to a concentration of 25 µg mL−1 total concentration (e.g., 12.5 µg mL−1 per protein in two-way combinations). The solution containing the ECM proteins was coated to Softwell and tissue culture polystyrene (TCPS) control plates at 50 µL per well for 2 hours at 37°C and then washed twice with dH2O prior to seeding of the cells. The PHH suspension was prepared using the same methods as for microarray experiments above and seeded at a density of 50,000 cells per well with overnight attachment (12–18 hours). Non-adherent cells were washed 2x with media and culture medium was changed and collected every 2 days for ~2 weeks in culture.
5.7. Biochemical assays
Albumin was measured using a sandwich-based enzyme linked immunosorbent assay (ELISA, Bethyl Laboratories, Montgomery, TX) with horseradish peroxidase detection and 3,3’5,5’-tetramethylbenzidine substrate (TMB, Rockland Immunochemicals, Boyertown, PA). Urea concentration was measured from collected supernatants using diacetyl monoxime with acid and heat (Stanbio Labs, Boerne, TX).[48] Absorbance of samples was read on the Synergy H1 multimode plate reader (Biotech, Winooski, VT).
CYP3A4 and CYP2C9 enzyme activities were measured after the incubation of cultures for 1 hour with luciferin-IPA (Promega Life Sciences, Madison, WI) or for 3 hours with luciferin-H (Promega), respectively, followed by the processing of collected supernatants per the manufacturer’s recommendations; luminescence was quantified with the Synergy H1 multimode reader. CYP1A2 and CYP2A6 enzymatic activities were measured by incubating cultures for 1 hour with 50 µM coumarin (Sigma-Aldrich) or for 3 hours with 5 µM 7-ethoxyresorufin (Sigma-Aldrich), respectively. The CYP2A6-generated metabolite, 7-hydroxycoumarin (7-HC), and CYP1A2-generated metabolite, resorufin, in culture supernatants were quantified using fluorescence measurements (excitation/emission 355/460 nm for 7-HC and 550/585 nm for resorufin) on the Synergy H1 multimode reader.
5.8. Drug-mediated CYP induction studies
Rifampin and omeprazole (Sigma-Aldrich) were dissolved in 100% (v/v) dimethyl sulfoxide (DMSO, Corning) while phenobarbital (Sigma-Aldrich) was dissolved in 1X PBS. Cultures were incubated for 2 days with inducer drugs (rifampin at 25 µM, omeprazole at 50 µM, and phenobarbital at 1 mM) or solvent only (0.1% v/v DMSO and 0.2% 1X PBS), followed by the assessment of CYP activities as described above.
5.9. Data analysis
Cellular microarray data was collected and analyzed from 2–3 independent experiments across 3 different PHH donors (donor demographics provided in ‘PHH culture’ section). Data processing was performed using R. Correlation between microarray features was performed using Spearman’s rank correlation test after assessing normality via Shapiro-Wilk tests and q-q plots. Multiple linear regression modeling was performed in R using the lm function and collagen-I (C1) condition was taken to be the reference group. For all hypothesis testing, p<0.05 was used for determining significance. Statistical significance for drug-mediated CYP induction assays was determined using one-way analysis of variance followed by a Tukey’s multiple comparison test for more than two groups. Error bars represent standard error of the mean (SEM) for microarray results and standard deviation (SD) for the multiwell plate studies.
Supplementary Material
Acknowledgements
Funding was provided by the US National Institutes of Health, grant 1R01DK125471-01A1 to S.R.K and G.U.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Chase P. Monckton, Department of Biomedical Engineering, University of Illinois at Chicago, 851 South Morgan Street, Chicago, Illinois, 60607, USA
Aidan Brougham-Cook, Department of Bioengineering, University of Illinois at Urbana-Champaign, 2112 Everitt Laboratory, 1406 West Green Street, Urbana, Illinois, 61801, USA.
Kerim B. Kaylan, Department of Bioengineering, University of Illinois at Urbana-Champaign, 2112 Everitt Laboratory, 1406 West Green Street, Urbana, Illinois, 61801, USA
Gregory H. Underhill, Department of Bioengineering, University of Illinois at Urbana-Champaign, 2112 Everitt Laboratory, 1406 West Green Street, Urbana, Illinois, 61801, USA
Salman R. Khetani, Department of Biomedical Engineering, University of Illinois at Chicago, 851 South Morgan Street, Chicago, Illinois, 60607, USA
Data Availability Statement
Data and image processing and modeling scripts available on request from the authors.
References
- [1].Bedossa P, Paradis V, The Journal of pathology 2003, 200, 504. [DOI] [PubMed] [Google Scholar]
- [2].Mueller S, Sandrin L, Hepat Med 2010, 2, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Desai SS, Tung JC, Zhou VX, Grenert JP, Malato Y, Rezvani M, Espanol-Suner R, Willenbring H, Weaver VM, Chang TT, Hepatology 2016, 64, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arriazu E, Ruiz de Galarreta M, Cubero FJ, Varela-Rey M, Perez de Obanos MP, Leung TM, Lopategi A, Benedicto A, Abraham-Enachescu I, Nieto N, Antioxid Redox Signal 2014, 21, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shih H, Pickwell GV, Guenette DK, Bilir B, Quattrochi LC, Human and Experimental Toxicology 1999, 18, 95. [DOI] [PubMed] [Google Scholar]
- [6].Underhill GH, Khetani SR, Cell Mol Gastroenterol Hepatol 2018, 5, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khetani SR, Bhatia SN, Nature biotechnology 2008, 26, 120. [DOI] [PubMed] [Google Scholar]
- [8].Sivaraman A, Leach JK, Townsend S, Iida T, Hogan BJ, Stolz DB, Fry R, Samson LD, Tannenbaum SR, Griffith LG, Current Drug Metabolism 2005, 6, 569. [DOI] [PubMed] [Google Scholar]
- [9].Sellaro TL, Ranade A, Faulk DM, McCabe GP, Dorko K, Badylak SF, Strom SC, Tissue Engineering, Part A 2010, 16, 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mattei G, Magliaro C, Pirone A, Ahluwalia A, Artif Organs 2017, 41, E347. [DOI] [PubMed] [Google Scholar]
- [11].Flaim CJ, Chien S, Bhatia SN, Nature methods 2005, 2, 119. [DOI] [PubMed] [Google Scholar]
- [12].You J, Park S-A, Shin D-S, Patel D, Raghunathan VK, Kim M, Murphy CJ, Tae G, Revzin A, Tissue engineering. Part A 2013, 19, 2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Natarajan V, Berglund EJ, Chen DX, Kidambi S, RSC Adv 2015, 5, 80956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li L, Sharma N, Chippada U, Jiang X, Schloss R, Yarmush ML, Langrana NA, Annals of Biomedical Engineering 2008, 36, 865. [DOI] [PubMed] [Google Scholar]
- [15].Deegan DB, Zimmerman C, Skardal A, Atala A, Shupe TD, J Mech Behav Biomed Mater 2015, 55, 87. [DOI] [PubMed] [Google Scholar]
- [16].Mittal N, Tasnim F, Yue C, Qu Y, Phan D, Choudhury Y, Tan M-H, Yu H, ACS Biomaterials Science & Engineering 2016, 2, 1649. [DOI] [PubMed] [Google Scholar]
- [17].Malta DF, Reticker-Flynn NE, da Silva CL, Cabral JM, Fleming HE, Zaret KS, Bhatia SN, Underhill GH, Acta Biomater 2016, 34, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kaylan KB, Ermilova V, Yada RC, Underhill GH, Sci Rep 2016, 6, 23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kourouklis AP, Kaylan KB, Underhill GH, Biomaterials 2016, 99, 82. [DOI] [PubMed] [Google Scholar]
- [20].Baiocchini A, Montaldo C, Conigliaro A, Grimaldi A, Correani V, Mura F, Ciccosanti F, Rotiroti N, Brenna A, Montalbano M, D’Offizi G, Capobianchi MR, Alessandro R, Piacentini M, Schinina ME, Maras B, Del Nonno F, Tripodi M, Mancone C, PLoS One 2016, 11, e0151736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arteel GE, Naba A, JHEP Rep 2020, 2, 100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bedossa P, Paradis V, J Pathol 2003, 200, 504. [DOI] [PubMed] [Google Scholar]
- [23].Krishnan A, Li X, Kao WY, Viker K, Butters K, Masuoka H, Knudsen B, Gores G, Charlton M, Lab Invest 2012, 92, 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO, BMC Cancer 2014, 14, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zanger UM, Schwab M, Pharmacol Ther 2013, 138, 103. [DOI] [PubMed] [Google Scholar]
- [26].Hwang-Verslues WW, Sladek FM, Curr Opin Pharmacol 2010, 10, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Underhill GH, Khetani SR, Drug Metab Dispos 2018, 46, 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR, Hepatology 2015, 61, 1370. [DOI] [PubMed] [Google Scholar]
- [29].Semler EJ, Ranucci CS, Moghe PV, Advances in biochemical engineering/biotechnology 2006, 102, 1. [DOI] [PubMed] [Google Scholar]
- [30].Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE, Science (New York, NY) 1994, 264, 696. [DOI] [PubMed] [Google Scholar]
- [31].Sun P, Zhang G, Su X, Jin C, Yu B, Yu X, Lv Z, Ma H, Zhang M, Wei W, Li W, Cell Rep 2019, 29, 3212. [DOI] [PubMed] [Google Scholar]
- [32].Kietzmann T, Redox Biol 2017, 11, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Reid LM, Fiorino AS, Sigal SH, Brill S, Holst PA, Hepatology 1992, 15, 1198. [DOI] [PubMed] [Google Scholar]
- [34].Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho C-S, Choi Y-J, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CEP, Gómez-Lechón MJ, Groothuis GMM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Häussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhütter H-G, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EHK, Stieger B, Stöber R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG, Archives of toxicology 2013, 87, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Badolo L, Jensen B, Sall C, Norinder U, Kallunki P, Montanari D, Xenobiotica 2015, 45, 177. [DOI] [PubMed] [Google Scholar]
- [36].Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de Graaf D, Toxicological Sciences 2008, 105, 97. [DOI] [PubMed] [Google Scholar]
- [37].Perepelyuk M, Terajima M, Wang AY, Georges PC, Janmey PA, Yamauchi M, Wells RG, AJP: Gastrointestinal and Liver Physiology 2013, 304, G605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Davidson MD, Kukla DA, Khetani SR, Integrative Biology 2017, 9, 662. [DOI] [PubMed] [Google Scholar]
- [39].Woolsey SJ, Mansell SE, Kim RB, Tirona RG, Beaton MD, Drug Metab Dispos 2015, 43, 1484. [DOI] [PubMed] [Google Scholar]
- [40].Yamada S, Ichida T, Matsuda Y, Miyazaki Y, Hatano T, Hata K, Asakura H, Hirota N, Geerts A, Wisse E, Liver 1992, 12, 10. [DOI] [PubMed] [Google Scholar]
- [41].Bhatia SN, Balis UJ, Yarmush ML, Toner M, FASEB Journal 1999, 13, 1883. [DOI] [PubMed] [Google Scholar]
- [42].Allen JW, Bhatia SN, Biotechnology and bioengineering 2003, 82, 253. [DOI] [PubMed] [Google Scholar]
- [43].Kaylan KB, Kourouklis AP, Underhill GH, J Vis Exp 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ, Nat Mater 2014, 13, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reticker-Flynn NE, Malta DF, Winslow MM, Lamar JM, Xu MJ, Underhill GH, Hynes RO, Jacks TE, Bhatia SN, Nature Communications 2012, 3, 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Nat Methods 2012, 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM, Genome biology 2006, 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Davidson MD, Khetani SR, Toxicol Sci 2020, 174, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and image processing and modeling scripts available on request from the authors.


