Abstract
The discovery that gut-microbiota plays a profound role in human health has opened a new avenue of basic and clinical research. Application of ecological approaches where the bacterial 16S rRNA gene is queried has provided a number of candidate bacteria associated with coronary artery disease and hypertension. We examine the associations between gut microbiota and a variety of cardiovascular disease (CVD) including atherosclerosis, coronary artery disease, and blood pressure. These approaches are associative in nature and there is now increasing interest in identifying the mechanisms underlying these associations. We discuss three potential mechanisms including: gut permeability and endotoxemia, increased immune system activation, and microbial derived metabolites. In addition to discussing these potential mechanisms we highlight current studies manipulating the gut microbiota or microbial metabolites to move beyond sequence-based association studies. The goal of these mechanistic studies is to determine the mode of action by which the gut microbiota may affect disease susceptibility and severity. Importantly, the gut microbiota appears to have a significant effect on host metabolism and CVD by producing metabolites entering the host circulatory system such as short-chain fatty acids and trimethylamine N-Oxide. Therefore, the intersection of metabolomics and microbiota research may yield novel targets to reduce disease susceptibility. Finally, we discuss approaches to demonstrate causality such as specific diet changes, inhibition of microbial pathways, and fecal microbiota transplant.
Keywords: Microbiota, Cardiovascular diseases, Atherosclerosis, Hypertension
1. Introduction
We are beginning to appreciate the role of commensal microbiota in cardiovascular disease (CVD) risk1,2 and these microbiota are located in a variety of niches within the body including the skin, oral, and gut, and are composed of bacteria, viruses, and fungi. There has been a particular focus on the gut as it is heavily colonized with microbes. It is estimated that more than 70% of all the microbes in the human body are present in the colon alone3 and trillions of commensal microbes including bacteria, bacteriophage, fungi, archaea, and unicellular eukaryotes live in the large intestine that forms a highly diverse dynamic interdependent complex ecological community known as the intestinal microbiota.4 The healthy gut microbiota depends on the host for their energy need and provides a range of health benefits by providing nutrients, improved barrier function, shaping host immune system, and preventing diseases including CVD.5
The gut microbiota is not the only commensal community studied in relation to CVD risk. There is also evidence that the oral microbiota, which contains more than 10 000 bacterial species from 22 phyla6 is also associated with CVD. Data from large epidemiological studies associated poor oral hygiene with increased risk of CVD,7 and several oral bacteria including Porphyromonas gingivalis have been detected in atherosclerotic plaques.8,9 What is not clear is if the association between oral microbiota and CVD indicates a specific mechanism or metabolic profile leading to increased disease or is a marker of other important factors to consider such as access to healthcare. Additional work remains to determine the mechanism(s) underlying the oral microbe–CVD associations and thus the majority of the review focuses on the gut microbiota and CVD.
The combined complexity of the microbiota is only beginning to be appreciated. Maternal transmission,10 diet,11 host genetics,12 aging,13 and sex14 all impact the composition of the microbiota. Recent large-scale sequencing efforts have identified tremendous interindividual variation in the microbiota which totals in excess of 45 million genes in the oral and gut niches. Moreover, nearly 50% of all genes were ‘unique to a single metagenomic sample’15 and only ∼550 000, less than 2%, overlapped between the oral and gut niches. This heterogeneity combined with the possibility of copy number variants within individual bacterial species highlight just how much of the microbiota remains to be characterized.
In this review, we focus on the association between microbiota and CVD, specifically coronary artery disease (CAD) and hypertension. We examine the steps to establishing causality and then discuss the underlying mechanisms by which gut microbiota may affect CVD. Finally, we examine potential therapeutics for CVD using gut microbiota and microbial metabolites.
2. Microorganisms and CVD risk
For over 50 years, the potential role of infectious microorganisms, including bacteria and viruses, as potential risk factors for CVD has been appreciated through a number of epidemiological studies.16–18 Infection and the subsequent inflammatory processes are thought to induce the onset, progression, and rupture of atherosclerotic plaques.19 The mechanisms by which infection aggravates atherosclerosis is direct cell invasion that accelerates plaque growth through local effects, or indirect systemic production of inflammatory cytokines that promote development of atherosclerosis.20 Representative infectious agents associated with atherosclerosis include Chlamydia pneumonia, Helicobacter pylori, P. gingivalis, and Influenza A virus.21
Although there are a number of pathogenic bacteria associated with CVD, C. pneumonia, a Gram-negative and intracellular bacteria, is an exemplar. C. pneumonia was the first proposed bacteria of CVD etiology responsible for the induction of inflammation in the vascular wall of CVD patients.22 In addition, antibiotics targeting C. pneumonia are thought to have anti-inflammatory effects, and may contribute to atherosclerotic plaque stability.21 For example, C. pneumoniae infection in rabbits accelerated the thickness of the tunica intimal wall and atherosclerosis, and treatment with antibiotics reduced the extent of atherosclerosis.23 This effect is not universal as studies in mice have not shown a similar reduction in lesion size by antibiotics after infection with C. pneumoniae.24 Nevertheless, there are examples of pathogenic bacteria potentially having direct effects on CVD. We now discuss how commensal bacteria of the microbiota may also affect CVD risk.
3. Microbiota and CVD associations
CVD includes a number of pathologies and diseases such as heart failure, stroke, peripheral artery disease, aortic valve disease, atherosclerosis, and hypertension but we limit our discussion to CAD and hypertension. In a number of case–control studies, distinct shifts of microbial composition have been identified in fecal samples from patients with CVD (Table 1). It is not entirely clear whether these microbial communities are causal to CVD or if they are simply affected by other environmental factors contributing to the development of CVD. In the following section, we provide a summary of a number of recently reported associations between gut microbiota and CAD or hypertension.
Table 1.
Reported alterations in the gut microbiota in various CVD cohorts
| Condition | Ethnicity | Technique | Associated microbiota changes | References |
|---|---|---|---|---|
| Myocardial infarction | European (Swedish) | Metagenomic sequencing | Increased: Collinsella | 36 |
| Decreased: Eubacterium, Roseburia | ||||
| CAD/T2D | European (Spanish) | 16S rRNA V2-V3 region | Increased: Enterobacteriaceae, Streptococcus, and Desulfovibrio | 30 |
| Decreased: Faecalibacterium prausnitzii and Bacteroides fragilis | ||||
| CVD | European (American) | 16S rRNA V4 region | Increased: Prevotella and Tyzzerella | 27 |
| Decreased: Alloprevotella, Catenibacterium | ||||
| CVD | Asian (Indian) | 16S rRNA & Metagenomic sequencing | Increased: Proteobacteria, Actinobacteria, Propionibacterium phages, Pseudomonas phages, Rhizobium phages, Lymphocystis virus, and Torque Teno viruses | 28 |
| CAD | Asian (Japanese) | 16S rRNA V3-V4 region | Increased: Firmicutes/bateriodetes ratio and Lactobacillales | 26 |
| Decreased: Bacteroides and Prevotella | ||||
| CAD | Asian (Chinese) | Metagenomic sequencing | Increased: Streptococcus sp. M334 and M143, and Clostridium sp. HGF2 | 33 |
| CVD | Asian (Chinese) | Metagenomic sequencing | Increased: Escherichia coli, Klebsiella spp, Enterobacter aerogenes, Streptococcus sp, Lactobacillus salivarius, Solobacterium moorei, Atropobium parvulum, Ruminococcus gnavus, and Eggerthella lenta | 31 |
| Decreased: Roseburia intestinalis, Faecalibacterium cf. prausnitzii, Bacteriodes spp, Prevotella copri, and Alistipes shahii | ||||
| Hypertension | Asian (Chinese) | Metagenomic sequencing | Increased: Prevotella, Klebsiella, Porphyromonas, and Actinomyces | 42 |
| Decreased: Faecalibacterium, Oscillibacter, Roseburia, Bifidobacterium, Coprococcus, and Butyrivibrio | ||||
| Hypertension | Asian (Chinese) | Metagenomic sequencing | Increased: Klebsiella, Clostridium, Streptococcus, Parabacteroides, Eggerthella, and Salmonella | 32 |
| Decreased: Faecalibacterium, Roseburia, and Synergistetes | ||||
| Hypertension | European (Finns) | Metagenomic sequencing | Increased: Blautia, Cellulomonas, Collinsella, and Desulfovibrio, Dielma, Eisenbergiella, Holdemania, Megasphaera, Phascolarctobacterium, Ruthenibacterium, Sutterella, and Turicibacter | 45 |
| Decreased: Lactobacillusa, and Citrobacter, Coprobacillus | ||||
| Hypertension | European/African (American) | 16S rRNA V3-V4 region | Increased: Anaerovorax, Clostridium IV, Oscillibacter, and Sporobacter | 43 |
| Decreased: Akkermansia, Ruminococcus, Anaerovorax, Sporobacter, and Asaccharobacter |
3.1 Gut microbiota and CAD
Dysbiosis refers to an imbalance in the microbial community in the human body. Alterations in the composition of the gut microbiota associated with the risk of CVD have been the focus of the majority of microbiota studies. Indeed, identifying patterns of specific composition of microbiota associated with CVD susceptibility contribute greatly to understanding the pathogenesis of diseases. The composition of the microbiota associated with aortic lesion size is distinct from the oral and gut microbiota.25 Frequently, studies utilize a case–control design to identify the microbial differences among subjects with CVD compared to controls. Foremost, there have been several consistent results reported from a number of cohorts (Table 1).26–28 For example, Strepcococcus spp., exhibiting pathogenic expansion in intestinal dysbiosis,29 showed increased abundance in stool samples from various CVD patients.30–33 In addition, the abundance of Faecalibacterium spp. and Roseburia spp., which are known to have anti-inflammatory effects and reduce atherosclerotic events in mice and humans,34,35 were relatively depleted in CAD patients.30,31 Finally, metagenome studies have shown depletion of butyrate-producing bacteria including Faecalibacterium prausnitzii and Roseburia intestinalis, and a high abundance of Enterobacteriaceae and Streptococcus in atherosclerotic patients.31 Deeper sequencing approaches have also identified alterations in the metagenomic profile of the gut microbiome of CVD patients. These data demonstrate enrichment of genes encoding peptidoglycan biosynthesis, while healthy cohorts are enriched in phytoene dehydrogenase genes.36 Thus, not only are the composition of the microbiota altered in CVD but the underlying functional capacity of the microbiota may be altered.
3.2 Gut microbiota and hypertension
Similar to CAD there is increasing interest in how gut microbiota may affect blood pressure and development of hypertension. Studies of angiotensin II infused mice, spontaneous hypertensive rats, and hypertensive patients have identified a distinct microbial composition of gut microbiota compared to controls including decreased microbial abundance, diversity, and low intestinal epithelial integrity.37–39 Broad-based antibiotic treatment in mice reduced intestinal dysbiosis and attenuated hypertension in angiotensin II-infused mice.40 Furthermore, angiotensin II-induced hypertension is alleviated in germ-free mice compared to conventionally raised mice, suggesting that the gut microbiota is important for the development of hypertension.41 Studies in humans have clearly demonstrated that there is an inverse association between measures of α-diversity and hypertension.63 In addition to reduced diversity, there is an increased abundance of opportunistic pathogenic taxa such as Klebsiella, and Streptococcus in subjects with hypertension,32,42,43 while several taxa are associated with high blood pressure (see Table 1) and may affect intestinal cell inflammation.44 In a longitudinal cohort, CARDIA, containing both Caucasians and African American subjects, identified a slightly different profile of bacteria positively associated with hypertension: Anaerovorax, Clostridium IV, Oscillibacter, and Sporobacter.43 Conversely, a number of studies have identified Bacteroides thetaiotaomicron as associated with reduced blood pressure in both hypertensive and healthy subjects.43–46 Although these differences between hypertensive subjects and healthy cohorts do not indicate causality, they do suggest that the composition and diversity of the gut microbiota are associated with clinical features of hypertension. Importantly, having the microbiota assessed in longitudinal cohorts such as CARDIA allows for epidemiologists to assess the risk of developing hypertension (or CAD) based on the current microbiota composition and thereby allowing for the assessment of the clinical utility of these data for predicting disease.
4. Limitations of association-based microbiota studies and CVD risk
A majority of microbiome-based studies utilize analysis of the bacterial 16S rRNA gene. For details on the methods and analytical techniques needed for 16S analysis, we direct the reader to several excellent reviews.47–50 16S rRNA studies have provided tremendous insight into the specific bacteria associated with CVD and undeniability have transformed our understanding of disease risk but there are limitations. Commonly used 16S rRNA gene sequencing techniques often do not reach species or sub-species level resolution, and the analysis often excludes less abundant microbial taxa. Thus, studies focused primarily on abundance neglect the functional pathways of microbes that contribute to disease risk, although the disease can be driven by microbes that can represent a small fraction of the microbial community. Additionally, many well-described cohorts designed to longitudinally assess CVD risk have not routinely collected fecal samples. Thus, many of the current reports are cross-sectional in nature and therefore precludes assessment of the relative risk of developing CVD on current composition of the microbiota which is critical to developing therapeutic strategies.
We now appreciate some of the biases and intrinsic issues associated with 16S analysis. These include a number of items and several have recently been discussed in the context of hypertension.51 First, the quality of DNA obtained from samples can be varied by differences in sample collection method (sample type, collection time, or processing method), storage and processing techniques (DNA extraction, library construction, sequencing depth, or quality filters). Second, from a technical point of view, amplification bias, improper or no internal sequencing control, contamination, or non-standard databases for mapping can lead to alteration in microbial composition independent of actual microbial changes. Finally, the results of these analyzes are usually expressed as a proportion rather than an absolute number of the microbes per gram of sample, and the proportion of specific microbial taxa in a sample may not be related to the risk of disease. Numerous large-scale efforts are underway to standardize microbiome methods and protocols in larger cohorts in an attempt to address some of these issues.52–54 More recently, there is interest in utilizing whole-genome sequencing of microbiota samples termed ‘metagenomics’, to provide much better taxonomic resolution down to species or strain level. As metagenomics results in sequences of genes contained in the sample, it provides an opportunity for functional profiling of the metabolic pathways present in community55 while also providing the taxonomic details available in 16S studies. While metagenomics provides a complete view of the microbial communities, it is expensive and the analytical approaches are computationally complex and time-consuming.
5. Towards function and mechanistic understanding of the microbiota in CVD
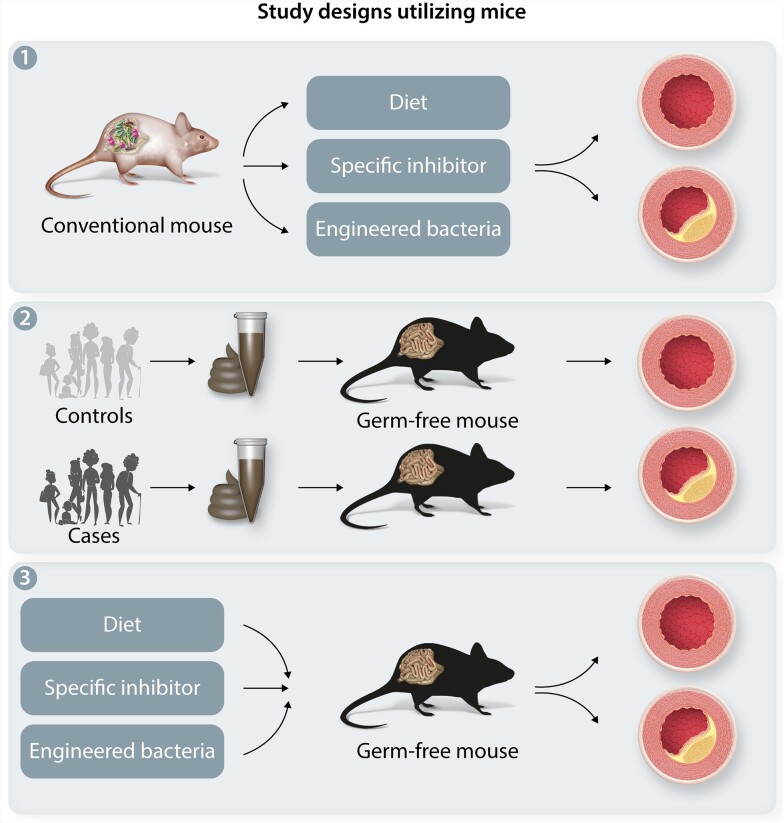
Although the microbiota is complex, there are a number of approaches to establishing causality and infer mechanism(s) of microbiota and specific bacteria for CVD (Figure 1). In particular, studies utilizing microbial transplantation in model organism can demonstrate the direct role of the gut microbial taxa in the risk of CVD. Specifically, the microbiota can be ablated from model organisms such as mice using antibiotic cocktails or germ-free animals. This provides a naïve state by which individual or complex communities can be added and then examined for hypertension or CVD.
Figure 1.
Towards Function and Mechanistic Understanding of the Microbiota in CVD. Examples of ways to move from association to causation. (A) Examples of microbiome approaches include where conventional mice are provided an agent to perturb the microbiota (either expand specific taxa or inhibit a pathway). (B) Examples of forward microbiome approaches include a comparison of the gut microbiome between controls and CVD subjects from human cohorts. These fecal microbiota samples are then transplanted into germ-free or antibiotic-treated animals to determine the gut microbiota community effect on BP. (C) Diet, specific bacterial enzyme inhibitor, or engineered microbiota can be introduced to animal model to establish a causal relation.
For example, studies utilizing germ-free animals have been critical to further our understanding of the functional role of the microbiota in disease susceptibility.56 Germ-free mice can also receive fecal microbiota transfers from samples collected in case/control studies. For example, blood pressure of germ-free mice increased when they received a transplant of stool from hypertensive patients as compared to mice receiving samples from healthy donors. Blood pressure elevations after FMT have also been observed in conventional mouse models.57 Germ-free mice administered stool samples from hypertensive patients or hypertensive rats have an elevation of blood pressure.42,58 These studies provide evidence that gut microbiota can affect the development of hypertension. Studies have been performed using two strains of mice that differ in atherosclerosis susceptibility and report that a portion of atherosclerosis can be attributed to the microbiota.59 It is important to note that the complexity of the microbiota can produce unexpected results such as increased blood pressure in Dahl salt-sensitive rats receiving fecal transplants from salt-resistant normotensive rats.60 The latter indicates that the relationship between microbiota and host biology is more complex than simply classifying microbiota as ‘good’ or ‘bad’.
In addition to studies examining the transplantation of fecal samples, there have been attempts at understanding the role of specific bacteria in CVD. For example, colonization of germ-free ApoE-deficient (ApoE−/−) mice with Roseburia intestinalis isolated from humans reduced the levels of inflammatory markers and atherosclerosis providing evidence of the causative role of Roseburia intestinalis in CVD.35 As the literature expands with similar studies, novel pathways and therapeutic targets may be identified. In the following section, we provide an overview of studies utilizing these functional approaches to provide potential mechanisms for how the microbiota affects CVD risk. In particular, we focus on host:microbe effects on the immune system, gut permeability and microbial derived metabolites. It is important to note that by its nature the gut microbiota is a diverse community and thus it is likely that there are multiple mechanisms for its effects on CVD.
5.1 Effect of gut microbiota on cardiovascular health via modulation of immune function
Gut microbiota is a strong modulator of host immunity and host immune response plays a key role in a wide range of pathology including CVD. For example, atherosclerosis which underlies many forms of CAD is considered a chronic inflammatory disease with the involvement of both innate and adaptive immunity.61 Several gut bacteria have been reported to influence distinct immune cells and there is an indication that T-cells are important in these processes.62–65 How these systemic alterations in adaptive immunity impact specific microbiota and CAD or hypertension remains to be fully elucidated.
In mice, there is growing evidence suggesting that the gut microbiota can affect atherosclerosis and inflammatory pathways. For example, microbiota transfer from mice prone to inflammatory dysregulation (Caspase 1−/−) into atherosclerosis prone low-density lipoprotein receptor deficient (LDLR−/−) mice does in fact increase atherosclerotic plaque formation compared to those who received microbiota from LDLR−/− mice.66 One possible mechanism of this effect is that gut microbiota elevated systemic inflammatory cytokines interleukin (IL)-1β, IL-2, and interferon (IFN)-γ. On the other hand, transplants with Bacteroides vulgatus and Bacteroides dorei attenuated atherosclerotic lesions and decreased plasma tumor necrosis factor-α (TNFα) level.67 Similarly, Lactobacillus plantarum ATCC 14917 supplementation inhibited atherosclerotic lesion formation by decreasing serum oxidized LDL (OxLDL), TNFα, and IL-1β production in the aorta.68 Higher abundance of the Roseburia and Blautia among others was associated with a decreased atherosclerotic lesion in mice and reduced plasma total cholesterol, TNFα, and IL-1β concentration.69
Similarly, hypertension is associated with inflammation and a number of alterations in immune system (reviewed in Norlander et al.70). In hypertensive humans, decreased relative abundances of butyrate-producing Roseburia and Faecalibacterium was observed along with increased TNFα:IFN-γ ratio, TNFα and IL-6 production in the isolated peripheral blood mononuclear cells compared to normotensive people. Additionally, high-salt diet-induced hypertension is associated with depleted gut Lactobacillus abundance, increased IFN-γ+ CD4 T cells and serum IFN-γ level, and decreased TGF-β1+ CD4 T-cells and serum TGF-β1 concentration.71 These findings do suggest that therapies that modulate the gut microbiota may simultaneously affect inflammatory processes and subsequently CVD.
5.2 Gut permeability and intestinal barrier dysfunction
The cells of the intestine serve as a critical barrier to the bacteria in the gut. This function is maintained by tight junctions between epithelial cells, mucus production, and mucosal immunity. When the intestinal barrier is compromised, lipopolysaccharides (LPS) derived from Gram-negative bacteria can enter the host circulation resulting in endotoxemia. Endotoxemia constitutes a strong risk factor of early atherogenesis72 and circulating LPS can bind to the host's Toll-like receptor (TLR) resulting in an inflammatory response in the host.73 Patients with CVD have higher levels of endotoxin in the blood compared to normal individuals.74,75 Translocation of LPS from the intestine is supported by a higher concentration of endotoxin in the hepatic vein compared to blood drawn directly from the ventricle.76 There is evidence that specific Bacteria may alter gut permeability and endotoxemia. Administration of live A. muciniphila reduced intestinal permeability, circulating endotoxin and aortic atherosclerosis in ApoE−/− mice.77 A recent proof-of-concept study in humans found that administration of pasteurized A. muciniphila for 3 months reduced circulating LPS in obese patients.78
5.3 Microbial metabolites and CVD
Seminal studies by Hazen and colleagues79 have demonstrated that gut microbiota may in fact generate metabolites that affect overall health or CVD pathogenesis. We are beginning to appreciate that many gut-derived metabolites can act on organs such as the liver via portal circulation and be metabolized by host enzymes. Microbial metabolites also help us understand the underlying mechanism by which gut bacterial taxa may influence host biology and thus health and disease (Table 2; Figure 2). We will briefly discuss the association between trimethylamine N-Oxide (TMAO) and CVD, and focus on other microbial metabolites such as short-chain fatty acids (SCFAs), tryptophan metabolites, and bile acid metabolites in this review.
Table 2.
Mechanism of microbiota-related metabolites on CVD
| Metabolites | Foods rich in metabolites | Bacteria producing metabolites | Mechanism | Study group | References |
|---|---|---|---|---|---|
| TMAO | Red meat, eggs, fish, poultry | Anaerococcus, Clostridium, Desulfovibrio, Edwardsiella, Proteus, Providencia, and others161 | Cholesterol accumulation | ApoE−/− mice | 162 |
| Foam cell formation | ApoE−/− mice | 162 | |||
| Platelet hyper-reactivity | Human and germ-free mice | 79 | |||
| Vascular endothelial dysfunction | Human and germ-free mice | 79 | |||
| Fibrosis and remodeling | Human and germ-free mice | 79 | |||
| Activation of PERK-FoxO1 signaling | ob/ob mice, In vitro studies (primary rat hepatocytes and HEK239T cells) |
163 | |||
| SCFAs |
Fermented foods (cheese, butter, yoghurt) |
Anaerostipes, Blautia, Coprococcus, Eubacterium, Faecalibacterium, Marvinbryantia, Megasphaera, Roseburia, Ruminococcus, and others164,165 | Lowering blood pressure by binding SCFA binding G protein-coupled receptor (GPR41, GPR43, GPR109A) | GPR41−/− mice, GPR43/GPR109A−/− mice, and germ-free mice | 97 , 99 |
| Tryptophan metabolites | |||||
| Indole, ILA, IPA, IAA | Red meat, eggs, fish, poultry | Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, Peptostreptococcus, Ruminococcus, Ruminiclostridium, and others105,106 | Act on AHR found in intestinal immune cells and thereby alter innate and adaptive immune responses | GF and conventional mouse | 110 |
| Indoxyl sulfate | Induces expression of proinflamatory cytokine IL-6 | Hypertensive rats | 113 | ||
| Induces expression of monocyte chemo-attractant protein-1 | Cell culture | 166 | |||
| Caused endothelial cell senescence, | Cell culture | 167 | |||
| Proliferation and migration of vascular smooth muscle cells | Cell culture | 168 | |||
TMAO, Trimethylamine N-oxide; SCFA, short-chain fatty acid; IPA, indolepropionic acid; ILA, indolelactic acid; IAA, indoleacetic acid.
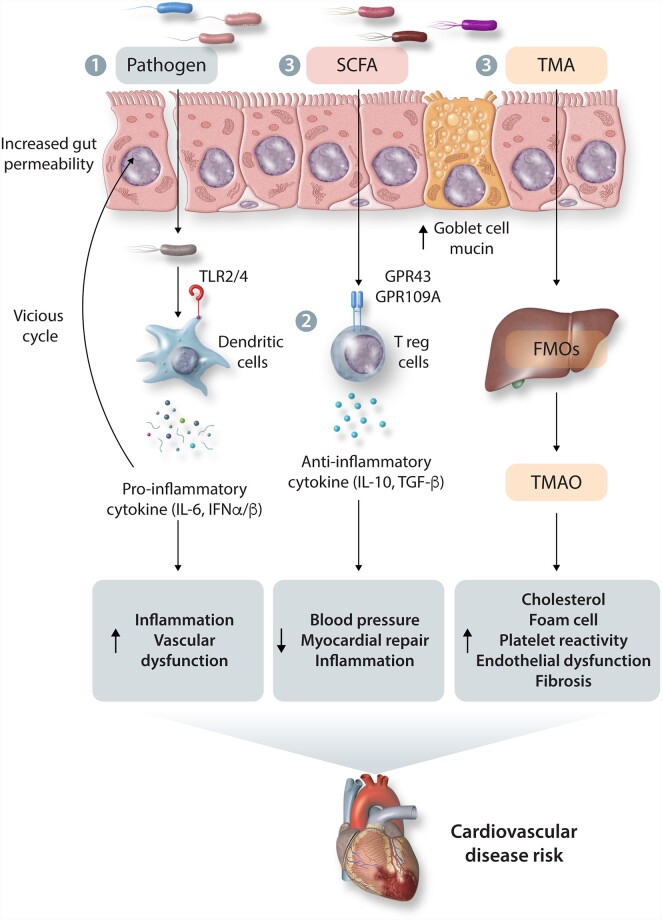
Figure 2.
Overview of potential mechanisms of microbiota–host interactions and CVD. 1- Pathogenic bacteria or bacterial components (LPS) can enter the host circulation resulting in endotoxemia and an inflammatory response. 2- local and systemic inflammatory processes can affect CVD development 3- Metabolites such as SCFAs and TMA can either bind host receptors or be further metabolized to pro- or anti- CVD molecules. SCFAs, short-chain fatty acids; GPR, G protein-coupled receptor; TLR, toll-like receptor; TMA, trimethylamine; TMAO, trimethylamine N-oxide; FMOs, flavin-containing monooxygenases; IL, interleukin; IFN, interferon; TGF-B, transforming growth factor beta.
5.3.1 Trimethylamine-oxide
Many studies have been conducted to investigate metabolites associated with the pathogenesis of CVD, and one of the most studied and reviewed metabolites to date is TMAO. We briefly report the association between TMAO and CVD since there are several well written published reviews which provide detailed reviews of TMAO.80 Dietary choline and L-carnitine are known to be converted to trimethylamine (TMA) by microbial enzymes (TMA lyase) contained in the genomes of specific gut microbiota. TMA is then absorbed in the intestine, delivered to the liver via portal vein, and then converted to TMAO by hepatic flavin monooxygenase 3 (FMO3).81 In subsequent studies combining data from more than 4000 subjects who underwent coronary angiography, TMAO elevation is associated with death, MI, and stroke over a 3-year period.82,83 These prognostic effects also have been evaluated in patients with a history of diabetes,84 chronic kidney disease, heart failure, MI, and peripheral arterial disease.80
Although the association between TMAO levels and various CAD events has been reproduced in many studies, there are studies where TMAO is not associated with CAD. For example, a recent study showed that TMAO levels were not associated with atherosclerosis in the Framingham Heart Study Offspring cohort (1215 individuals) or supporting animal studies,85 or CARDIA.86 One small clinical trial examined FMT of low TMAO producing vegan donors into subjects at risk of CVD but failed to observe a significant reduction in TMAO levels.87 Understanding what role, if any differences in the microbiota or diet, play in these disparate results remains to be determined.
Evidence supporting the role of TMAO in the development of hypertension is not yet clear. Preclinical studies have shown that experimental hypertensive rats have higher intestinal permeability and portal blood TMA level in the colon tissue,88 and TMAO treatment increased plasma aquaporin-2 concentration which elicits greater water reabsorption, and eventually leads to hypertension.89 Apart from these animal studies, evidence has been established in humans through a systematic review that high TMAO plasma levels are associated with high blood pressure risk in 8 studies with 11 750 individuals and 6176 hypertensive cases.90 However, the studies included in this systematic review recruited most of the participants were from the United States. Further large-scale prospective cohorts are expected to characterize the association, especially the causality in the general population.
5.3.2 Short-chain fatty acids
The main products of microbial enzyme reaction of dietary fiber are SCFAs, such as acetate, butyrate, and propionate. The main butyrate producers in the human colon are Firmicutes (phylum), with Lachnospiraceae (family), and the Ruminococcaceae (family) the two most abundant groups.91,92 An additional pathway in lactic acid-utilizing bacteria exists where lactate and acetate are converted to butyrate.91 A number of other phyla produce butyrate and the efficiency of this production may reflect expression of specific genes such as butyryl-CoA transferase, butyryl-CoA dehydrogenase, and butyrate kinase.93 Two species are well characterized with regard to SCFA production: Bifidobacterium species produce acetate and lactate94 and Akkermansia muciniphila produces acetate and propionate.92,95
It is known that the most direct route through which SFCA modulates the risk of CVD is the regulation of blood pressure. Initial clinical intervention study showed that fiber intake reduced blood pressure and that SCFAs are involved in blood pressure control.96 SCFA metabolites lower blood pressure by modulating the SCFA binding G protein-coupled receptor (GPR) 41 or olfactory receptor 78 in vitro and in vivo.44,97,98 Supplementing the diet with SCFAs protects against the development of hypertension and involves the SCFA receptor GPR43/GPR109A, which also regulates the abundance of Treg cells in mice.99–101 Thus, SCFAs may regulate cardiovascular homeostasis by activating receptors in the cells of the cardiovascular system. Supporting these mechanistic studies are data from a controlled trial that showed that butyrate (600 mg/day) significantly reduced diastolic blood pressure measured after a 10-min rest period in 15 patients with type 2 diabetes.102 Although SCFAs are among the most frequently published intestinal microbial-derived metabolites, more clinical trials and mechanistic studies are needed to validate the effects of SCFAs on CVD and its risk factors.
5.3.3 Other microbial metabolites
Identification of microbial derived metabolites that affect host physiology is an active area of investigation. There is particular interest in small molecules derived from dietary tryptophan.103,104 In the gut, tryptophan is catabolized by a variety of pathways associated with several classes of gut bacteria including Lactobacillus, Bacteroides, Bifidobacterium, and Clostridium to produce a number of metabolites such as tryptamine, indole, indolelactic acid (ILA), indolepropionic acid (IPA), indoleacetic acid (IAA), indolealdehyde (IAld), and metabolite 3-methylindole (skatole).105–108 These tryptophan metabolites have diverse biological effects including enhancing the intestinal epithelial barrier, stimulating gastrointestinal motility, helping the secretion of gut hormones, exerting anti-inflammatory and anti-oxidative effects in the systemic circulation, and modulating gut microbial composition.109 Some of these effects may be through the aryl hydrocarbon receptor (AHR) and thereby alter innate and adaptive immune responses in a ligand-specific fashion110 which could modulate CVD risk. Not all tryptophan metabolites are associated with beneficial effects on gut health or CVD risk. In particular, indoxyl sulfate is associated with aortic calcification,111 increased carotid intima-media thickness,112 increases expression of cytokines in vascular cells.113 Further understanding if compositional differences in the microbiota or if specific bacteria modulate the metabolism of tryptophan or other nutrients may further shed light on the underlying mechanisms by which the microbiota affect CVD risk.
6. Therapeutic potential for modulating microbiota on CVD risk
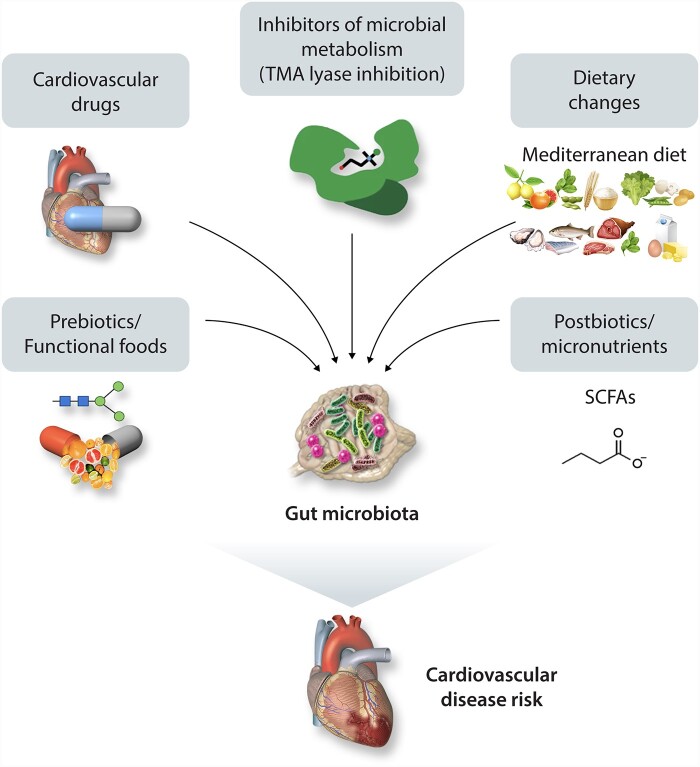
The association between altered composition of intestinal microbiota in CVD patients, production of microbial metabolites, and CVD risk mentioned above suggest that gut microbiota may be a significant modulator of CVD, and their relationship has become a potential target for new therapeutics (Figure 3). We review a number of potential strategies to module the microbiota and CVD risk including diet, inhibition of microbial pathways, and fecal microbiota transplants.
Figure 3.
Therapeutic interventions for improving CVD. Current strategies for improving cardiovascular disease by manipulating intestinal microbiota, including bacterial TMA lyase enzyme inhibitors, fecal microbial transplantation, prebiotics, probiotics, and dietary interventions.
6.1 Diet and prebiotics in the CVD–microbiota axis
Diet has been considered to be the most direct driving factor for gut microbiota composition.114–117 In many cases, diet is a factor modulating CAD, hypertension118 and gut microbiota. Epidemiological, clinical, and experimental studies have demonstrated that diet and nutrition play a central role in the prevention of CVD.119 Based on these data, the American Heart Association has made formal dietary recommendations (https://www.heart.org/en/healthy-living/healthy-eating/eat-smart/nutrition-basics/aha-diet-and-lifestyle-recommendations, last accessed on 02/06/21). Some of these specific recommendations are now known to affect the microbiota.
An example of these are recommendations to reduce salt intake as certain humans and model organisms are sensitive to a high-salt diet.60,120 We now appreciate that high salt intake influences gut microbial diversity in rodents and humans.121–123 High salt intake altered microbiota diversity and increased the abundance of Erwinia genus and Corynebacteriaceae family in mice122 and depleted the abundance of Lactobacillus murinus in human. Mechanistically, a high salt diet promotes local and systemic tissue inflammation via increases of pro-inflammatory cytokines and increased gut permeability in both human and animal studies.121,124,125 Some of the effects of dietary salt may be attributed to specific bacteria such as Bacteroides fragilis, which through a variety of intermediate metabolic effects ultimately activates the mineralocorticoid receptor and increases blood pressure.126
In addition to specific dietary components that affect CVD risk and the microbiota, there is evidence that dietary patterns also play a role. For example, one of the most studied diets in CVD research is the Mediterranean diet (MeD). Epidemiological studies found that MeD has a cardio-protective effect compared to western diets.127,128 Several studies129,130 have demonstrated that MeD elicits beneficial microbiota profiles and microbial metabolite production. For example, one study129 found that higher consumption of MeD is associated with increased levels of Prevotella, fiber degrading Firmicutes, fecal SCFAs, and lower urinary TMAO compared to a western diet. Similarly, another study130 reported that closer adherence to the Mediterranean dietary pattern and greater consumption of plant-based nutrients such as vegetable proteins and polysaccharides were associated with a lower ratio of Firmicutes: Bacteroidetes and Streptococcus; higher Catenibacterium, Bifidobacterium, and fecal SCFAs. One recent study131 found that MeD with probiotics containing Bifidobacterium longum and Lactobacillus rhamnosus increased gut microbial diversity and decreased Bacteroidetes-to-Firmicutes ratio along with other health benefits including improved BMI, fasting glucose, and homeostasis, which was not fully observed in MeD alone indicating that at least a part of the health benefit of MeD depends on the gut microbial composition.
Dietary fiber is an important macronutrient in the context of gut microbiota and CVD. Large population-based observational studies132 found that a diet containing high dietary fiber was associated with a reduced CVD risk, which was largely mediated via the reduction of LDL-cholesterol. Clinical trials found that ingestion of soluble fiber (2–10 g/day) was associated with a significant 7% LDL-cholesterol reduction133 in a dose-dependent manner.134 Several gut bacteria utilize dietary fiber and produce SCFAs which prevent CVD as discussed earlier in this manuscript. In addition to SCFAs production, dietary soluble fibers bind to the bile acids in the gut and block reabsorption of the bile acids, which leads to increased bile acid synthesis in the liver, and ultimately increased LDL clearance from blood.135,136
Growing evidence showing that prebiotics supplementation reduces CVD, specifically blood pressure and atherosclerosis, by manipulating the gut microbiota, which supports prebiotic interventions to prevent or treat CVD. Prebiotics are the non-digestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or the activity of one or a limited number of bacterial species already resident in the colon.137 Inulin, a linear β-2,1 fructosyl-fructose polydisperse carbohydrate material, feeding decreased atherosclerosis in the aortic root of mice and normalized the altered microbial abundance of Bifidobacterium, Lactobacillus, Akkermansia, Allobaculum, and Coprococcus.138 These results are supported by another study where inulin supplementation increased Akkermansia and Bifidobacterium abundance, decreased bacterial taxa involved in secondary bile acid metabolism, and reversed endothelial dysfunction.139 β-glucan is a glucose polysaccharide that can sequester cholesterol, scavenges reactive oxygen species, and produces SCFAs when digested by gut microbiota.140 In addition to serving as an energy source for gut bacteria, β-glucans are immunostimulatory through activation of β-glucan receptors, such as dectin-1 or CR3 on the intestinal macrophages.141,142 Oat β-glucan supplementation increased high-density lipoprotein (HDL)-cholesterol and decreased plasma triglyceride (TG) and atherosclerosis alone with enrichment of the genus Akkermansia in the gut.143
6.2 Targeting the microbiota to affect CVD using small molecules
The approach of manipulating intestinal microbial communities and their metabolic pathways has not yet reached clinical practice, but some studies show promising results. Targeting microbial enzymes (TMA lyases) that convert nutrients such as choline or carnitine into TMA regulates TMAO levels and experimental studies have found several chemical compounds that can trigger the modulatory effects on CVD. One example is 3,3-dimethyl-1-butanol (DMB), which is a TMA lyase inhibitor found in natural products such as olive oil, which can decrease plasma TMAO levels without perturbing microbial cell viability in in-vivo mouse models.144,145 Mice receiving DMB have been shown to have reduced atherosclerotic lesion, attenuated foam cell formation, and alleviated progression of CVD.145 Chemically synthesized compounds have been used to inhibit TMA lyase in mice and these molecules also showed selective targeting and sustained inhibition of TMA-lyase in the host.146
In addition to TMAO, there are a number of other microbial pathways whose modulation or inhibition may affect CVD risk. One possible pathway to target is the microbial cholesterol dehydrogenase enzyme (ismA gene), which converts cholesterol to the sterol coprostanol.147 Coprostanol is not absorbed as efficiently as cholesterol in the gastrointestinal tract and thus this pathway contributes to lowering blood cholesterol levels.148 Individuals carrying coprostanol-forming microorganisms have significantly lower cholesterol levels in stools and lower plasma total cholesterol, with effects comparable to those attributed to variations in human genes involved in lipid homeostasis. Therefore, altering abundance of the bacteria-containing ismA or increasing its expression could reduce CVD risk by lowering intestinal and serum cholesterol levels.
An alternative approach is to use small molecules, such as cyclic d, l-α-peptides, which modulate growth of specific bacteria. Initial experiments have been promising as these compounds remodel the microbiota of high-fat diet-fed Ldlr−/− mice to resemble a low-fat diet gut microbiota and inhibited atherosclerosis development.149 The effects of this modulation are broad and include decreased plasma cholesterol, suppressed pro-inflammatory cytokines such as IL-6, TNFα, and IL-1B, and altered levels of SCFAs and bile acids in the feces and plasma. Identification of similar compounds that affect host phenotype by targeting microbial processes is an exciting and active area of research.
6.3 Fecal microbiota transplantation
While most often used for mechanistic studies, fecal microbiota transplantation (FMT) has also been used as a therapy for patients with Clostridium difficile infection150 and ulcerative colitis.151 This process includes the collection of stools collected from healthy donors or the recipients themselves (self FMT) prior to administration into the intestine of patients suffering from disease or related dysbiosis. These studies are complex and to date have not been extensively studied with regards to CVD endpoints. While the clinical utility of such approach is under debate for CVD,152 the therapeutic effect of FMT has been reported effective against insulin resistance and small intestinal permeability.153,154
7. Concluding remarks and future perspectives
Many studies have confirmed the link between gut microbiota and CVD and we are beginning to understand the underlying mechanisms of these associations. One exciting aspect of this work is related to metabolomics. In some sense, the gut microbiota is an intermediate trait between diet (environmental factor) and CVD risk (clinical trait) that produces metabolites, some of which play an important role in the pathogenesis of CVD. Recent studies have confirmed that specific microbial taxa are associated with CVD, and that various gut-derived metabolites, including TMAO or SCFAs, can promote or attenuate CVD. However, much still remains to be investigated. For example, only bacterial communities are being studied extensively and other members in gut microbiota such as virus, fungal, or archaea are not widely studied and thus their roles in human disease remain underappreciated. Therefore, combining these members in the analysis of the microbiota–CVD association might open us a new door for potential therapeutics.
Perhaps the most important questions remain—how will these exciting results be applied clinically (if at all)? Studies utilizing the LifeLines-DEEP population cohort have identified that candidate SNPs explain 3–7% of the variation in HDL and triglycerides while adding 16S microbial diversity explains another 4–6% of the variation in HDL and triglycerides.155 Interestingly, when genetic and microbiome data are combined they explain significantly more variation in HDL and triglycerides. Thus, there is an indication that obtaining microbiota data may further characterize patients as we move towards a model of precision medicine. For example, application of machine learning to datasets containing gut microbiota, genetics, and diet together156 predicted better postprandial plasma TG, glucose, and insulin responses. In addition to direct causal pathways, it is likely that specific bacteria or microbiota components will be casually implicated or simply act as moderating variables for both genetic studies157,158 and drug therapies.159,160 Thus, the microbiota remains a critical part of the movement towards personalized medicine. Ultimately utilizing microbiota data that has been vetted to be clinically relevant may in fact refine our risk predictions and therapeutic interventions.
Authors’ contributions
B.J.B. supervised all portion of the review process, interpreted the results, and mentored manuscript writing. M.N.H. and M.K. conducted the literature search, extracting the information and drafting the manuscript. M.N.H. and M.K. also addressed co-authors comments and concerns. B.J.B., M.N.H., and M.K. critically revised the manuscript. B.J.B. had primary responsibility for final content. All authors read and approved the final manuscript.
Acknowledgements
This research was supported in part by the National Institutes of Health grant 5R01HL128572 (BJB), United States Department of Agriculture (USDA) project 2032-51530-025-00D (BJB). The USDA is an equal opportunity employer. Figures were created using Biorender.com.
Conflict of interest: none declared.
References
- 1. Velmurugan G, Ramprasath T, Gilles M, Swaminathan K, Ramasamy S.. Gut microbiota, endocrine-disrupting chemicals, and the diabetes epidemic. Trends Endocrinol Metab 2017;28:612–625. [DOI] [PubMed] [Google Scholar]
- 2. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J, MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ley RE, Peterson DA, Gordon JI.. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006;124:837–848. [DOI] [PubMed] [Google Scholar]
- 4. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R.. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sekirov I, Russell SL, Antunes LC, Finlay BB.. Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. [DOI] [PubMed] [Google Scholar]
- 6. Keijser B, Zaura E, Huse S, Van der Vossen J, Schuren F, Montijn R, Ten Cate J, Crielaard W.. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 2008;87:1016–1020. [DOI] [PubMed] [Google Scholar]
- 7. de Oliveira C, Watt R, Hamer M.. Toothbrushing, inflammation, and risk of cardiovascular disease: results from Scottish Health Survey. Bmj 2010; 340:c2451–c2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi C, Viereck J, Hua N, Phinikaridou A, Madrigal AG, Gibson IIF, Hamilton JA, Genco CA.. Porphyromonas gingivalis accelerates inflammatory atherosclerosis in the innominate artery of ApoE deficient mice. Atherosclerosis 2011;215:52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang T, Kurita-Ochiai T, Hashizume T, Du Y, Oguchi S, Yamamoto M.. Aggregatibacter actinomycetemcomitans accelerates atherosclerosis with an increase in atherogenic factors in spontaneously hyperlipidemic mice. FEMS Immunol Med Microbiol 2010;59:143–151. [DOI] [PubMed] [Google Scholar]
- 10. Van Daele E, Knol J, Belzer C.. Microbial transmission from mother to child: improving infant intestinal microbiota development by identifying the obstacles. Crit Rev Microbiol 2019;45:613–648. [DOI] [PubMed] [Google Scholar]
- 11. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD.. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE.. Human genetics shape the gut microbiome. Cell 2014;159:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, Tsuchiya S, Dohi O, Yoshida N, Kamada K, Ishikawa T, Handa O, Konishi H, Okuda K, Tsujimoto Y, Ohnogi H, Itoh Y.. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol 2019;54:53–63. [DOI] [PubMed] [Google Scholar]
- 14. Sinha T, Vich Vila A, Garmaeva S, Jankipersadsing SA, Imhann F, Collij V, Bonder MJ, Jiang X, Gurry T, Alm EJ, D’Amato M, Weersma RK, Scherjon S, Wijmenga C, Fu J, Kurilshikov A, Zhernakova A.. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes 2019;10:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tierney BT, Yang Z, Luber JM, Beaudin M, Wibowo MC, Baek C, Mehlenbacher E, Patel CJ, Kostic AD.. The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe 2019;26:283–295 e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pankey GA. Effect of viruses on the cardiovascular system. Am J Med Sci 1965;250:103–114. [DOI] [PubMed] [Google Scholar]
- 17. Cluff LE, Reynolds RC, Page DL, Breckenridge JL.. Staphylococcal bacteremia and altered host resistance. Ann Intern Med 1968;69:859–873. [DOI] [PubMed] [Google Scholar]
- 18. Saikku P, Leinonen M, Mattila K, Ekman MR, Nieminen MS, Mäkelä PH, Huttunen JK, Valtonen V.. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 1988;332:983–986. [DOI] [PubMed] [Google Scholar]
- 19. Libby P, Ridker PM, Hansson GK, Leducq Transatlantic Network on A. Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol 2009;54:2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pant S, Deshmukh A, Gurumurthy GS, Pothineni NV, Watts TE, Romeo F, Mehta JL.. Inflammation and atherosclerosis–revisited. J Cardiovasc Pharmacol Ther 2014;19:170–178. [DOI] [PubMed] [Google Scholar]
- 21. Pothineni NVK, Subramany S, Kuriakose K, Shirazi LF, Romeo F, Shah PK, Mehta JL.. Infections, atherosclerosis, and coronary heart disease. Eur Heart J 2017;38:3195–3201. [DOI] [PubMed] [Google Scholar]
- 22. Kuo CC, Grayston JT, Campbell LA, Goo YA, Wissler RW, Benditt EP.. Chlamydia pneumoniae (TWAR) in coronary arteries of young adults (15-34 years old). Proc Natl Acad Sci U S A 1995;92:6911–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muhlestein JB, Anderson JL, Hammond EH, Zhao L, Trehan S, Schwobe EP, Carlquist JF.. Infection with Chlamydia pneumoniae accelerates … lerosis and treatment with azithromycin prevents it in a rabbit model. Circulation 1998;97:633–636. [DOI] [PubMed] [Google Scholar]
- 24. Blessing E, Campbell LA, Rosenfeld ME, Chesebro B, Kuo CC.. A 6 week course of azithromycin treatment has no beneficial effect on atherosclerotic lesion development in apolipoprotein E-deficient mice chronically infected with Chlamydia pneumoniae. J Antimicrob Chemother 2005;55:1037–1040. [DOI] [PubMed] [Google Scholar]
- 25. Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Backhed F.. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A 2011;108: 4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So A, Kasahara K, Yodoi K, Matsumoto T, Mizoguchi T, Ogawa W, Hirata K.. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atheroscler Thromb 2016;23:908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, Correa A, He J.. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa Heart Study Participants. Circ Res 2016;119:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J.. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One 2014;9:e105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taur Y, Pamer EG.. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis 2013;26:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanchez-Alcoholado L, Castellano-Castillo D, Jordan-Martinez L, Moreno-Indias I, Cardila-Cruz P, Elena D, Munoz-Garcia AJ, Queipo-Ortuno MI, Jimenez-Navarro M.. Role of gut microbiota on cardio-metabolic parameters and immunity in coronary artery disease patients with and without type-2 diabetes mellitus. Front Microbiol 2017;8:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K.. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 2017;8:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, Han X, Huang Y, Zhao L, Li P, Fang Z, Zhou J, Guan X, Ding Y, Wang S, Khan M, Xin Y, Li S, Ma Y.. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol 2017;7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Q, Liu Z, Zhong S, Li R, Xia H, Jie Z, Wen B, Chen X, Yan W, Fan Y, Guo Z, Meng N, Chen J, Yu X, Zhang Z, Kristiansen K, Wang J, Xu X, He K, Li G.. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep 2016;6:22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P.. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, Mehrabian M, Denu JM, BäCkhed F, Lusis AJ, Rey FE.. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 2018;3:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Backhed F, Nielsen J.. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012;3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M.. Gut dysbiosis is linked to hypertension. Hypertension 2015;65:1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sata Y, Marques FZ, Kaye DM.. The emerging role of gut dysbiosis in cardio-metabolic risk factors for heart failure. Curr Hypertens Rep 2020;22:38. [DOI] [PubMed] [Google Scholar]
- 39. Adnan S, Nelson JW, Ajami NJ, Venna VR, Petrosino JF, Bryan RM Jr., Durgan DJ.. Alterations in the gut microbiota can elicit hypertension in rats. Physiol Genomics 2017;49:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pevsner-Fischer M, Blacher E, Tatirovsky E, Ben-Dov IZ, Elinav E.. The gut microbiome and hypertension. Curr Opin Nephrol Hypertens 2017;26:1–8. [DOI] [PubMed] [Google Scholar]
- 41. Karbach SH, Schonfelder T, Brandao I, Wilms E, Hormann N, Jackel S, Schuler R, Finger S, Knorr M, Lagrange J, Brandt M, Waisman A, Kossmann S, Schafer K, Munzel T, Reinhardt C, Wenzel P.. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J Am Heart Assoc 2016;5:e003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, Zhang W, Weldon R, Auguste K, Yang L, Liu X, Chen L, Yang X, Zhu B, Cai J.. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR Jr., Shikany JM, Lloyd-Jones DM, Launer LJ, Fodor AA, Meyer KA.. Gut microbiota composition and blood pressure. Hypertension 2019;73:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K, Mohammed M, Handberg EM, Richards EM, Pepine CJ, Raizada MK.. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci (Lond) 2018;132:701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palmu J, Salosensaari A, Havulinna AS, Cheng S, Inouye M, Jain M, Salido RA, Sanders K, Brennan C, Humphrey GC, Sanders JG, Vartiainen E, Laatikainen T, Jousilahti P, Salomaa V, Knight R, Lahti L, Niiranen TJ.. Association between the gut microbiota and blood pressure in a population cohort of 6953 individuals. J Am Heart Assoc 2020;9:e016641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verhaar BJH, Collard D, Prodan A, Levels JHM, Zwinderman AH, Backhed F, Vogt L, Peters MJL, Muller M, Nieuwdorp M, van den Born BJH.. Associations between gut microbiota, faecal short-chain fatty acids, and blood pressure across ethnic groups: the HELIUS study. Eur Heart J 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boers SA, Jansen R, Hays JP.. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur J Clin Microbiol Infect Dis 2019;38:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gołębiewski M, Tretyn A.. Generating amplicon reads for microbial community assessment with next-generation sequencing. J Appl Microbiol 2020;128:330–354. [DOI] [PubMed] [Google Scholar]
- 49. Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K.. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017;5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mandal RS, Saha S, Das S.. Metagenomic surveys of gut microbiota. Genomics Proteomics Bioinformatics 2015;13:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marques FZ, Jama HA, Tsyganov K, Gill PA, Rhys-Jones D, Muralitharan RR, Muir J, Holmes A, Mackay CR.. Guidelines for transparency on gut microbiome studies in essential and experimental hypertension. Hypertension 2019;74:1279–1293. [DOI] [PubMed] [Google Scholar]
- 52. Amos GCA, Logan A, Anwar S, Fritzsche M, Mate R, Bleazard T, Rijpkema S.. Developing standards for the microbiome field. Microbiome 2020;8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, McDonnell SA, Nolan JA, Sutton TDS, Dalmasso M, McCann A, Ross RP, Hill C.. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 2018;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brumfield KD, Huq A, Colwell RR, Olds JL, Leddy MB.. Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One 2020;15:e0228899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thomas T, Gilbert J, Meyer F.. Metagenomics-a guide from sampling to data analysis. Microb Informatics Exp 2012;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kennedy EA, King KY, Baldridge MT.. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol 2018;9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim TT, Parajuli N, Sung MM, Bairwa SC, Levasseur J, Soltys CM, Wishart DS, Madsen K, Schertzer JD, Dyck JRB.. Fecal transplant from resveratrol-fed donors improves glycaemia and cardiovascular features of the metabolic syndrome in mice. Am J Physiol Endocrinol Metab 2018;315:E511–E519. [DOI] [PubMed] [Google Scholar]
- 58. Toral M, Robles-Vera I, de la Visitacion N, Romero M, Yang T, Sanchez M, Gomez-Guzman M, Jimenez R, Raizada MK, Duarte J.. Critical role of the interaction gut microbiota - sympathetic nervous system in the regulation of blood pressure. Front Physiol 2019;10:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL.. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 2015;290:5647–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B.. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics 2015;47:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ross R. Atherosclerosis—an inflammatory disease. N Engl J Med 1999;340:115–126. [DOI] [PubMed] [Google Scholar]
- 62. Round JL, Mazmanian SK.. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci 2010;107:12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K.. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011;331:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H.. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- 65. Gil-Cruz C, Perez-Shibayama C, De Martin A, Ronchi F, van der Borght K, Niederer R, Onder L, Lutge M, Novkovic M, Nindl V, Ramos G, Arnoldini M, Slack EMC, Boivin-Jahns V, Jahns R, Wyss M, Mooser C, Lambrecht BN, Maeder MT, Rickli H, Flatz L, Eriksson U, Geuking MB, McCoy KD, Ludewig B.. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 2019;366:881–886. [DOI] [PubMed] [Google Scholar]
- 66. Brandsma E, Kloosterhuis NJ, Koster M, Dekker DC, Gijbels MJJ, van der Velden S, Rios-Morales M, van Faassen MJR, Loreti MG, de Bruin A, Fu J, Kuipers F, Bakker BM, Westerterp M, de Winther MPJ, Hofker MH, van de Sluis B, Koonen DPY.. A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circ Res 2019;124:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, Hoshi N, Hatano N, Ozawa G, Sasaki N, Mizoguchi T, Amin HZ, Hirota Y, Ogawa W, Yamada T, Hirata KI.. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 2018;138:2486–2498. [DOI] [PubMed] [Google Scholar]
- 68. Hassan A, Din AU, Zhu Y, Zhang K, Li T, Wang Y, Xu S, Lei H, Yu X, Wang G.. Anti-atherosclerotic effects of Lactobacillus plantarum ATCC 14917 in ApoE(-/-) mice through modulation of proinflammatory cytokines and oxidative stress. Appl Microbiol Biotechnol 2020;104:6337–6350. [DOI] [PubMed] [Google Scholar]
- 69. Wu M, Yang S, Wang S, Cao Y, Zhao R, Li X, Xing Y, Liu L.. Effect of berberine on atherosclerosis and gut microbiota modulation and their correlation in high-fat diet-fed ApoE-/- mice. Front Pharmacol 2020;11:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Norlander AE, Madhur MS, Harrison DG.. The immunology of hypertension. J Exp Med 2018;215:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu J, Li T, Wu H, Shi H, Bai J, Zhao W, Jiang D, Jiang X.. Lactobacillus rhamnosus GG strain mitigated the development of obstructive sleep apnea-induced hypertension in a high salt diet via regulating TMAO level and CD4(+) T cell induced-type I inflammation. Biomed Pharmacother 2019;112:108580. [DOI] [PubMed] [Google Scholar]
- 72. Wiedermann CJ, Kiechl S, Dunzendorfer S, Schratzberger P, Egger G, Oberhollenzer F, Willeit J.. Association of endotoxemia with carotid atherosclerosis and cardiovascular disease. J Am Coll Cardiol 1999;34:1975–1981. [DOI] [PubMed] [Google Scholar]
- 73. Hug H, Mohajeri MH, La Fata G.. Toll-like receptors - regulators of the immune response in the human gut. Nutrients 2018; 10:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ibrahim M, Behairy M, El-Ashry M, Mostafa AE.. Cardiovascular risk of circulating endotoxin level in prevalent hemodialysis patients. Egypt Heart J 2018;70:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hsu CC, Wei TS, Huang CC, Chen YM.. Endotoxemia is associated with acute coronary syndrome in patients with end stage kidney disease. BMC Nephrol 2017;18:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Peschel T, Schönauer M, Thiele H, Anker SD, Schuler G, Niebauer J.. Invasive assessment of bacterial endotoxin and inflammatory cytokines in patients with acute heart failure. Eur J Heart Fail 2003; 5:609–614. [DOI] [PubMed] [Google Scholar]
- 77. Li J, Lin S, Vanhoutte PM, Woo CW, Xu A.. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe-/- mice. Circulation 2016;133:2434–2446. [DOI] [PubMed] [Google Scholar]
- 78. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen JP, de Vos WM, Cani PD.. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 2019;25:1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL.. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 2016;165:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Witkowski M, Weeks TL, Hazen SL.. Gut microbiota and cardiovascular disease. Circ Res 2020;127:553–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ.. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 2013;17:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL.. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang Z, Tang WH, Buffa JA, Fu X, Britt EB, Koeth RA, Levison BS, Fan Y, Wu Y, Hazen SL.. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur Heart J 2014;35:904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang WH, Wang Z, Li XS, Fan Y, Li DS, Wu Y, Hazen SL.. Increased trimethylamine N-oxide portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin Chem 2017;63:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koay YC, Chen YC, Wali JA, Luk AWS, Li M, Doma H, Reimark R, Zaldivia MTK, Habtom HT, Frank AE, Fusco-Allison G, Yang J, Holmes A, Simpson SJ, Peter K, O'Sullivan JF, Plasma levels of TMAO can be increased with ‘healthy’ and ‘unhealthy’ diets and do not correlate with the extent of atherosclerosis but with plaque instability. Cardiovasc Res 2020;117:435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Meyer KA, Benton TZ, Bennett BJ, Jacobs DR Jr., Lloyd-Jones DM, Gross MD, Carr JJ, Gordon-Larsen P, Zeisel SH.. Microbiota-dependent metabolite trimethylamine N-oxide and coronary artery calcium in the coronary artery risk development in young adults study (CARDIA). J Am Heart Assoc 2016;5:e003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smits LP, Kootte RS, Levin E, Prodan A, Fuentes S, Zoetendal EG, Wang Z, Levison BS, Cleophas MCP, Kemper EM, Dallinga‐Thie GM, Groen AK, Joosten LAB, Netea MG, Stroes ESG, de Vos WM, Hazen SL, Nieuwdorp M.. Effect of vegan fecal microbiota transplantation on carnitine‐and choline‐derived trimethylamine‐N‐oxide production and vascular inflammation in patients with metabolic syndrome. J Am Heart Assoc 2018;7:e008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jaworska K, Huc T, Samborowska E, Dobrowolski L, Bielinska K, Gawlak M, Ufnal M.. Hypertension in rats is associated with an increased permeability of the colon to TMA, a gut bacteria metabolite. PLoS One 2017;12:e0189310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu M, Han Q, Yang J.. Trimethylamine-N-oxide (TMAO) increased aquaporin-2 expression in spontaneously hypertensive rats. Clin Exp Hypertens 2019;41:312–322. [DOI] [PubMed] [Google Scholar]
- 90. Ge X, Zheng L, Zhuang R, Yu P, Xu Z, Liu G, Xi X, Zhou X, Fan H.. The gut microbial metabolite trimethylamine N-oxide and hypertension risk: a systematic review and dose-response meta-analysis. Adv Nutr 2020;11:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Louis P, Flint HJ.. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009;294:1–8. [DOI] [PubMed] [Google Scholar]
- 92. Louis P, Flint HJ.. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017;19:29–41. [DOI] [PubMed] [Google Scholar]
- 93. Vital M, Howe AC, Tiedje JM.. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L.. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Derrien M, Vaughan EE, Plugge CM, de Vos WM.. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–1476. [DOI] [PubMed] [Google Scholar]
- 96. Streppel MT, Arends LR, van 't Veer P, Grobbee DE, Geleijnse JM.. Effect of dietary fiber intake on blood pressure - a meta-analysis of randomized, controlled clinical trials. Arch Intern Med 2005;165:150–156. [DOI] [PubMed] [Google Scholar]
- 97. Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE, Pluznick JL.. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics 2016;48:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ.. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A 2013;110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kaye DM, Shihata WA, Jama HA, Tsyganov K, Ziemann M, Kiriazis H, Horlock D, Vijay A, Giam B, Vinh A, Johnson C, Fiedler A, Donner D, Snelson M, Coughlan MT, Phillips S, Du XJ, El-Osta A, Drummond G, Lambert GW, Spector TD, Valdes AM, Mackay CR, Marques FZ.. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation 2020;141:1393–1403. [DOI] [PubMed] [Google Scholar]
- 100. Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M, Tan JK, Kuruppu S, Rajapakse NW, El-Osta A, Mackay CR, Kaye DM.. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 2017;135:964–977. [DOI] [PubMed] [Google Scholar]
- 101. Bartolomaeus H, Balogh A, Yakoub M, Homann S, Marko L, Hoges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, Haase N, Kraker K, Hering L, Maase M, Kusche-Vihrog K, Grandoch M, Fielitz J, Kempa S, Gollasch M, Zhumadilov Z, Kozhakhmetov S, Kushugulova A, Eckardt KU, Dechend R, Rump LC, Forslund SK, Muller DN, Stegbauer J, Wilck N.. Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation 2019;139:1407–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Roshanravan N, Mahdavi R, Alizadeh E, Jafarabadi MA, Hedayati M, Ghavami A, Alipour S, Alamdari NM, Barati M, Ostadrahimi A.. Effect of butyrate and inulin supplementation on glycemic status, lipid profile and glucagon-like peptide 1 level in patients with type 2 diabetes: a randomized double-blind, placebo-controlled trial. Horm Metab Res 2017;49:886–891. [DOI] [PubMed] [Google Scholar]
- 103. Yu E, Ruiz-Canela M, Guasch-Ferré M, Zheng Y, Toledo E, Clish CB, Salas-Salvadó J, Liang L, Wang DD, Corella D.. Increases in plasma tryptophan are inversely associated with incident cardiovascular disease in the Prevencion con Dieta Mediterranea (PREDIMED) Study. J Nutr 2017;147:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hayashi T, Yamashita T, Watanabe H, Kami K, Yoshida N, Tabata T, Emoto T, Sasaki N, Mizoguchi T, Irino Y, Toh R, Shinohara M, Okada Y, Ogawa W, Yamada T, Hirata KI.. Gut microbiome and plasma microbiome-related metabolites in patients with decompensated and compensated heart failure. Circ J 2018;83:182–192. [DOI] [PubMed] [Google Scholar]
- 105. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL.. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017;551:648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ.. Major phenylpropanoid‐derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 2013;57:523–535. [DOI] [PubMed] [Google Scholar]
- 107. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L.. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013;39:372–385. [DOI] [PubMed] [Google Scholar]
- 108. Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh C-S, Colonna M.. Lactobacillus reuteri induces gut intraepithelial CD4+ CD8αα+ T cells. Science 2017;357:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Roager HM, Licht TR.. Microbial tryptophan catabolites in health and disease. Nat Commun 2018;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K.. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep 2018;23:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA, Group EUTW, on behalf of the European Uremic Toxin Work Group (EUTox). Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol 2009;4:1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sato B, Yoshikawa D, Ishii H, Kikuchi R, Arima T, Takeshita K, Inoue Y, Suzuki S, Tanaka M, Kumagai S, Matsumoto M, Hayashi M, Ando H, Amano T, Matsubara T, Niwa T, Murohara T.. Indoxyl sulfate, a uremic toxin, and carotid intima-media thickness in patients with coronary artery disease. Int J Cardiol 2013;163:214–216. [DOI] [PubMed] [Google Scholar]
- 113. Adelibieke Y, Yisireyili M, Ng H-Y, Saito S, Nishijima F, Niwa T.. Indoxyl sulfate induces IL-6 expression in vascular endothelial and smooth muscle cells through OAT3-mediated uptake and activation of AhR/NF-κB pathway. Nephron Exp Nephrol 2014;128:1–8. [DOI] [PubMed] [Google Scholar]
- 114. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ.. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE.. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 2007;73:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Louis P, Scott KP, Duncan SH, Flint HJ.. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 2007;102:1197–1208. [DOI] [PubMed] [Google Scholar]
- 117. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, Zhao L.. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 2010;4:232–241. [DOI] [PubMed] [Google Scholar]
- 118. Jama HA, Beale A, Shihata WA, Marques FZ.. The effect of diet on hypertensive pathology: is there a link via gut microbiota-driven immunometabolism? Cardiovasc Res 2019;115:1435–1447. [DOI] [PubMed] [Google Scholar]
- 119. Torres N, Guevara-Cruz M, Velázquez-Villegas LA, Tovar AR.. Nutrition and Atherosclerosis. Arch Med Res 2015;46:408–426. [DOI] [PubMed] [Google Scholar]
- 120. Ha SK. Dietary salt intake and hypertension. Electrolyte Blood Press 2014;12:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Frätzer C, Krannich A, Gollasch M, Grohme DA, Côrte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Müller DN.. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017;551:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bier A, Braun T, Khasbab R, Di Segni A, Grossman E, Haberman Y, Leibowitz AA.. High salt diet modulates the gut microbiota and short chain fatty acids production in a salt-sensitive hypertension rat model. Nutrients 2018;10:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang C, Huang Z, Yu K, Ding R, Ye K, Dai C, Xu X, Zhou G, Li C.. High-salt diet has a certain impact on protein digestion and gut microbiota: a sequencing and proteome combined study. Front Microbiol 2017;8:1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA.. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013;496:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yi B, Titze J, Rykova M, Feuerecker M, Vassilieva G, Nichiporuk I, Schelling G, Morukov B, Chouker A.. Effects of dietary salt levels on monocytic cells and immune responses in healthy human subjects: a longitudinal study. Transl Res 2015;166:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yan X, Jin J, Su X, Yin X, Gao J, Wang X, Zhang S, Bu P, Wang M, Zhang Y, Wang Z, Zhang Q.. Intestinal flora modulates blood pressure by regulating the synthesis of intestinal-derived corticosterone in high salt-induced hypertension. Circ Res 2020;126:839–853. [DOI] [PubMed] [Google Scholar]
- 127. Estruch R, Ros E, Salas-Salvadó J, Covas M-I, Corella D, Arós F, Gómez-Gracia E, Ruiz-Gutiérrez V, Fiol M, Lapetra J, Lamuela-Raventos RM, Serra-Majem L, Pintó X, Basora J, Muñoz MA, Sorlí JV, Martínez JA, Martínez-González MA.. Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med 2013;368:1279–1290. [DOI] [PubMed] [Google Scholar]
- 128. De Lorgeril M, Renaud S, Salen P, Monjaud I, Mamelle N, Martin J, Guidollet J, Touboul P, Delaye J.. Mediterranean alpha-linolenic acid-rich diet in secondary prevention of coronary heart disease. Lancet 1994;343:1454–1459. [DOI] [PubMed] [Google Scholar]
- 129. De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D.. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016;65:1812–1821. [DOI] [PubMed] [Google Scholar]
- 130. Garcia-Mantrana I, Selma-Royo M, Alcantara C, Collado MC.. Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front Microbiol 2018;9:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pellegrini M, Ippolito M, Monge T, Violi R, Cappello P, Ferrocino I, Cocolin LS, De Francesco A, Bo S, Finocchiaro C.. Gut microbiota composition after diet and probiotics in overweight breast cancer survivors: a randomized open-label pilot intervention trial. Nutrition 2020;74:110749. [DOI] [PubMed] [Google Scholar]
- 132. Bazzano LA, He J, Ogden LG, Loria CM, Whelton PK.. Dietary fiber intake and reduced risk of coronary heart disease in US men and women: the National Health and Nutrition Examination Survey I Epidemiologic Follow-up Study. Arch Intern Med 2003;163:1897–1904. [DOI] [PubMed] [Google Scholar]
- 133. Brown L, Rosner B, Willett WW, Sacks FM.. Cholesterol-lowering effects of dietary fiber: a meta-analysis. Am J Clin Nutr 1999;69:30–42. [DOI] [PubMed] [Google Scholar]
- 134. Anderson JW, Allgood LD, Lawrence A, Altringer LA, Jerdack GR, Hengehold DA, Morel JG.. Cholesterol-lowering effects of psyllium intake adjunctive to diet therapy in men and women with hypercholesterolemia: meta-analysis of 8 controlled trials. Am J Clin Nutr 2000;71:472–479. [DOI] [PubMed] [Google Scholar]
- 135. Kritchevsky D, Story JA.. Binding of bile salts in vitro by nonnutritive fiber. J Nutr 1974;104:458–462. [DOI] [PubMed] [Google Scholar]
- 136. Andersson M, Ellegard L, Andersson H.. Oat bran stimulates bile acid synthesis within 8 h as measured by 7alpha-hydroxy-4-cholesten-3-one. Am J Clin Nutr 2002;76:1111–1116. [DOI] [PubMed] [Google Scholar]
- 137. Gibson GR, Roberfroid MB.. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 1995;125:1401–1412. [DOI] [PubMed] [Google Scholar]
- 138. Hoffman JB, Petriello MC, Morris AJ, Mottaleb MA, Sui Y, Zhou C, Deng P, Wang C, Hennig B.. Prebiotic inulin consumption reduces dioxin-like PCB 126-mediated hepatotoxicity and gut dysbiosis in hyperlipidemic Ldlr deficient mice. Environ Pollut 2020;261:114183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Catry E, Bindels LB, Tailleux A, Lestavel S, Neyrinck AM, Goossens JF, Lobysheva I, Plovier H, Essaghir A, Demoulin JB, Bouzin C, Pachikian BD, Cani PD, Staels B, Dessy C, Delzenne NM.. Targeting the gut microbiota with inulin-type fructans: preclinical demonstration of a novel approach in the management of endothelial dysfunction. Gut 2018;67:271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Nakashima A, Yamada K, Iwata O, Sugimoto R, Atsuji K, Ogawa T, Ishibashi-Ohgo N, Suzuki K.. Beta-glucan in foods and its physiological functions. J Nutr Sci Vitaminol (Tokyo) 2018;64:8–17. [DOI] [PubMed] [Google Scholar]
- 141. Brown GD, Gordon S.. A new receptor for β-glucans. Nature 2001;413:36–37. [DOI] [PubMed] [Google Scholar]
- 142. Chan GC-F, Chan WK, Sze DM-Y.. Sze DM-Y. The effects of β-glucan on human immune and cancer cells. J Hematol Oncol 2009;2:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ryan PM, London LE, Bjorndahl TC, Mandal R, Murphy K, Fitzgerald GF, Shanahan F, Ross RP, Wishart DS, Caplice NM, Stanton C.. Microbiome and metabolome modifying effects of several cardiovascular disease interventions in apo-E(-/-) mice. Microbiome 2017;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chen ML, Yi L, Zhang Y, Zhou X, Ran L, Yang J, Zhu JD, Zhang QY, Mi MT.. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. mBio 2016;7:e02210–02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, DiDonato AJ, Fu X, Hazen JE, Krajcik D, DiDonato JA, Lusis AJ, Hazen SL.. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 2015;163:1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, Barrington WT, Russell MW, Reed JM, Duzan A, Lang JM, Fu X, Li L, Myers AJ, Rachakonda S, DiDonato JA, Brown JM, Gogonea V, Lusis AJ, Garcia-Garcia JC, Hazen SL.. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med 2018;24:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kriaa A, Bourgin M, Mkaouar H, Jablaoui A, Akermi N, Soussou S, Maguin E, Rhimi M.. Microbial reduction of cholesterol to coprostanol: an old concept and new insights. Catalysts 2019;9:167. [Google Scholar]
- 148. Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP, Xavier RJ.. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe 2020;28:245–257 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, Moore NE, Aleman Muench GR, Wu J, Chen W, Pinto AFM, Maryanoff BE, Saghatelian A, Soroosh P, Torkamani A, Leman LJ, Ghadiri MR.. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol 2020;38:1288–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA, Russell G, Surawicz C. Fecal Microbiota Transplantation W. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 2011;9:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, Lee CH.. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015;149:102–109 e106. [DOI] [PubMed] [Google Scholar]
- 152. Woodworth MH, Carpentieri C, Sitchenko KL, Kraft CS.. Challenges in fecal donor selection and screening for fecal microbiota transplantation: a review. Gut Microbes 2017;8:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, Herrema H, Ackermans M, Serlie MJM, de Brauw M, Levels JHM, Sales A, Gerdes VE, Ståhlman M, Schimmel AWM, Dallinga-Thie G, Bergman JJ, Holleman F, Hoekstra JBL, Groen A, Bäckhed F, Nieuwdorp M.. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 2020;69:502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Craven L, Rahman A, Nair Parvathy S, Beaton M, Silverman J, Qumosani K, Hramiak I, Hegele R, Joy T, Meddings J, Urquhart B, Harvie R, McKenzie C, Summers K, Reid G, Burton JP, Silverman M.. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability. Am J Gastroenterol 2020;115:1055–1065 [DOI] [PubMed] [Google Scholar]
- 155. Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, Franke L, Poon TW, Xavier RJ, Gevers D, Hofker MH, Wijmenga C, Zhernakova A.. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res 2015;117:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf J, Capdevila J, Hadjigeorgiou G, Davies R, Al Khatib H, Bonnett C, Ganesh S, Bakker E, Hart D, Mangino M, Merino J, Linenberg I, Wyatt P, Ordovas JM, Gardner CD, Delahanty LM, Chan AT, Segata N, Franks PW, Spector TD.. Human postprandial responses to food and potential for precision nutrition. Nat Med 2020;26:964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Yang Q, Lin SL, Kwok MK, Leung GM, Schooling CM.. The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a Mendelian randomization study. Am J Epidemiol 2018;187:1916–1922. [DOI] [PubMed] [Google Scholar]
- 158. Hughes DA, Bacigalupe R, Wang J, Ruhlemann MC, Tito RY, Falony G, Joossens M, Vieira-Silva S, Henckaerts L, Rymenans L, Verspecht C, Ring S, Franke A, Wade KH, Timpson NJ, Raes J.. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat Microbiol 2020;5:1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Vieira-Silva S, Falony G, Belda E, Nielsen T, Aron-Wisnewsky J, Chakaroun R, Forslund SK, Assmann K, Valles-Colomer M, Nguyen TTD, Proost S, Prifti E, Tremaroli V, Pons N, Le Chatelier E, Andreelli F, Bastard JP, Coelho LP, Galleron N, Hansen TH, Hulot JS, Lewinter C, Pedersen HK, Quinquis B, Rouault C, Roume H, Salem JE, Sondertoft NB, Touch S, MetaCardis C, Dumas ME, Ehrlich SD, Galan P, Gotze JP, Hansen T, Holst JJ, Kober L, Letunic I, Nielsen J, Oppert JM, Stumvoll M, Vestergaard H, Zucker JD, Bork P, Pedersen O, Backhed F, Clement K, Raes J, MetaCardis Consortium. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 2020;581:310–315. [DOI] [PubMed] [Google Scholar]
- 160. Tuteja S, Ferguson JF.. Gut microbiome and response to cardiovascular drugs. Circ Genom Precis Med 2019;12:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Qi J, You T, Li J, Pan T, Xiang L, Han Y, Zhu L.. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J Cell Mol Med 2018;22:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL.. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013;19:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chen S, Henderson A, Petriello MC, Romano KA, Gearing M, Miao J, Schell M, Sandoval-Espinola WJ, Tao J, Sha B, Graham M, Crooke R, Kleinridders A, Balskus EP, Rey FE, Morris AJ, Biddinger SB.. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell Metab 2019;30:1141–1151 e1145. [DOI] [PubMed] [Google Scholar]
- 164. Morrison DJ, Preston T.. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016;7:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L.. The role of short-chain fatty acids in health and disease. Adv Immunol 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- 166. Watanabe I, Tatebe J, Namba S, Koizumi M, Yamazaki J, Morita T.. Activation of aryl hydrocarbon receptor mediates indoxyl sulfate-induced monocyte chemoattractant protein-1 expression in human umbilical vein endothelial cells. Circ J 2013;77:224–230. [DOI] [PubMed] [Google Scholar]
- 167. Koizumi M, Tatebe J, Watanabe I, Yamazaki J, Ikeda T, Morita T.. Aryl hydrocarbon receptor mediates indoxyl sulfate-induced cellular senescence in human umbilical vein endothelial cells. J Atheroscler Thromb 2014; 21:904–916. [DOI] [PubMed] [Google Scholar]
- 168. Shimizu H, Hirose Y, Nishijima F, Tsubakihara Y, Miyazaki H.. ROS and PDFG-β receptors are critically involved in indoxyl sulfate actions that promote vascular smooth muscle cell proliferation and migration. Am J Physiol Cell Physiol 2009;297:C389–C396. [DOI] [PubMed] [Google Scholar]