Abstract
The pirate butterfly Catacroptera cloanthe (Stoll, 1781) (Nymphalidae: Kallimini) is a monotypic genus of butterfly that occupies grassland and savanna habitats in Sub-Saharan Africa, and exhibits seasonal variation. Genome skimming by Illumina sequencing allowed the assembly of a 78.8% AT-rich complete circular mitogenome of 15,204 bp from C. cloanthe. The mitogenome has a typical butterfly gene order consisting of 13 protein-coding genes, two rRNAs, 22 tRNAs, and a control region. Catacroptera cloanthe COX1 begins with an atypical CGA start codon, while COX2, ND3, ND4, and ND5 end with incomplete T or TA stop codons, completed by the addition of the poly-A tail during mRNA processing. Bayesian phylogenetic reconstruction placed Catacroptera cloanthe as sister to Mallika jacksoni in the monophyletic tribe Kallimini, which was consistent with previous phylogenetic hypotheses.
Keywords: Illumina sequencing, mitogenomes, Lepidoptera, Nymphalidae, Kallimini
The pirate butterfly Catacroptera cloanthe (Stoll, 1781) exhibits seasonal variation and occupies grassland and savanna habitats in Sub-Saharan Africa (Henning et al. 2009; Williams 2020; Woodhall 2020). It was originally placed in the genus Papilio but later reclassified as the monotypic genus (a genus containing only one species), Catacroptera (Karsch 1894). The larval host plants, geographic distribution, and seasonal variation are well documented in this species (Williams 2020; Woodhall 2020). Based on molecular phylogenetic analysis, C. cloanthe has been placed as the sister taxon to another monotypic butterfly genus, the Jackson’s leaf butterfly, Mallika jacksoni (Wahlberg et al. 2005, 2009). Here, I report the complete mitochondrial genome sequence of C. cloanthe from specimen Cclo2016.1, collected in Sibwesa, Mpanda, Tanzania (GPS 6.59103S, 30.65727E) in February 2016. It was pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (http://www.wallisroughley.ca/, Jason Gibbs, Jason.Gibbs@umanitoba.ca) with voucher number WRME0507737.
A DNEasy Blood and Tissue Kit (Qiagen, Düsseldorf, Germany) were used to extract DNA from a single specimen leg (McCullagh and Marcus 2015). DNA was sonicated and a fragment library was prepared as described previously (Peters and Marcus 2017) using a NEBNext Ultra II DNA Library Prep Kit. Then Illumina NovaSeq6000 (San Diego, CA) sequencing was performed (Marcus 2018). The mitogenome of Catacroptera cloanthe (GenBank accession no. MW722786) was assembled using Geneious version 10.2.6 by mapping the SRA library (15,236,066 paired 150 bp reads; GenBank SRA PRJNA706380) to a Mallika jacksoni reference mitogenome (Lepidoptera: Nymphalidae, MT704828) (Alexiuk et al. 2020a), using the medium sensitivity settings for five iterations. Mitogenome annotation was performed by aligning the Catacroptera genome with the previously annotated and published mitogenome from Mallika jacksoni (Alexiuk et al. 2020a) in Geneious version 10.2.6, and protein-coding genes were identified based on sequence homology.
The assembled circular mitogenome of C. cloanthe was 15,204 bp and composed of 8011 paired reads with the nucleotide composition: 39.3% A, 13.2% C, 7.9% G, and 39.5% T. The gene structure and order in C. cloanthe are typical of the arrangement found in most butterfly mitogenomes (Park et al. 2016). The C. cloanthe protein-coding gene start codons include: ATG (ATP6, COX2, COX3, CYTB, ND1, ND4, and ND4L), ATT (ND2 and ND6), ATC (ATP8 and ND3), CGA, an atypical COXI start codon found in many other insects (Liao et al. 2010), and ATA (ND5), a start codon infrequently used in insect mitochondria but is frequently used in other animal groups (Okimoto et al. 1990; Han et al. 2016; Alexiuk et al. 2020b). The mitogenome contains three protein-coding genes (COX2, ND3, and ND5) with single-nucleotide (T) stop codons, and one protein-coding gene (ND4) with a two-nucleotide (TA) stop codon completed by post-transcriptional addition of 3′ A residues. All structures of the tRNAs were verified using ARWEN version 1.2 (Laslett and Canback 2008) and have typical cloverleaf secondary structures with the exception for trnS (AGN) whose dihydrouridine arm is replaced by a loop, whereas the control region and mitochondrial rRNAs are typical for Lepidoptera (McCullagh and Marcus 2015).
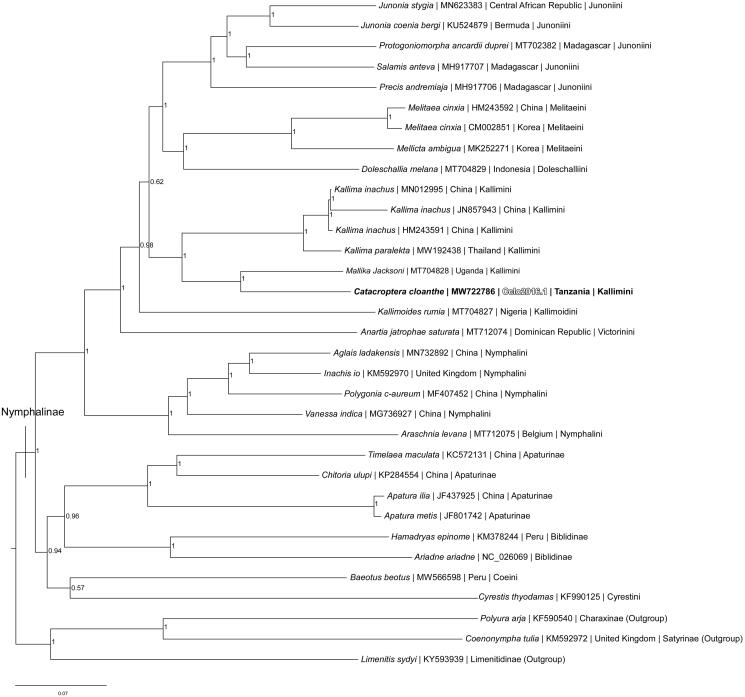
Phylogenetic reconstruction was completed using the complete mitogenome of C. cloanthe, 29 mitogenomes from the subfamily Nymphalinae, and three outgroup species from other subfamilies (Charaxinae, Limenitidinae, and Satyrinae) within the family Nymphalidae (Wahlberg et al. 2005, 2009; Hamilton et al. 2020; Lalonde and Marcus 2020; Payment et al. 2020; Alexiuk et al. 2020a; Lalonde 2021). Mitogenome sequences were aligned in CLUSTALX version 2.1 (Thompson et al. 1997; Larkin et al. 2007) and analyzed using Bayesian Inference with the GTR + I + G model selected using jModeltest version 2.1.1 (Darriba et al. 2012) in MrBayes version 3.2.7 (Ronquist and Huelsenbeck 2003; Ronquist et al. 2012) (Figure 1). Phylogenetic analysis places C. cloanthe as the sister taxon to Mallika jacksoni within the monophyletic tribe Kallimini, which is consistent with recent phylogenetic analyses (Wahlberg et al. 2005, 2009).
Figure 1.
Bayesian Inference phylogeny (GTR + I + G model, average deviation of split frequencies = 0.000639) of the Catacroptera cloanthe mitogenome, 29 additional mitogenomes from 11 tribes within subfamily Nymphalinae, and three outgroup species from other subfamilies within family Nymphalidae (Charaxinae (Polyura arja), Satyrinae (Coenonympha tulia), and Limenitidinae (Limenitis sydyi)) produced by 10 million MCMC iterations in MrBayes with sampling every 1000 generations. The Bayesian posterior probability values determined by MrBayes are given at each node.
Acknowledgments
I would like to thank Jeffrey Marcus for his constructive criticism of this manuscript and Genome Quebec for assistance with library preparation and sequencing.
Funding Statement
This work received support from the Natural Sciences and Engineering Research Council of Canada (NSERC) under [Grant RGPIN-2016-06012] and the University of Manitoba under the University Research Grants Program.
Ethics statement
Ethics approval was not required for this study.
Author Contributions
M.L. contributed to the conception and design of research, the analysis of the and interpretation of the data, drafting, and revising of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession nos. MW722786 and MW722785. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA706380, SRX10228918, and SAMN18128423, respectively.
References
- Alexiuk MR, Marcus JM, Lalonde MML.. 2020a. The complete mitochondrial genome of the Jackson’s leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5:3316–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiuk MR, Marcus JM, Lalonde MML.. 2020b. The complete mitochondrial genome and phylogenetic analysis of the European map butterfly Araschnia levana (Insecta: Lepidoptera: Nymphalidae). Mitochondr DNA Part B. 5:3264–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton RV, Marcus JM, Lalonde MML.. 2020. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae. Mitochondr DNA Part B. 5(3):3306–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Wang G, Xue T, Chen Y, Li J.. 2016. The F-type complete mitochondrial genome of Chinese freshwater mussels Cuneopsis pisciculus. Mitochondrial DNA Part A. 27(5):3376–3377. [DOI] [PubMed] [Google Scholar]
- Henning GA, Terblanche RF, Ball JB.. 2009. South African red data book: butterflies. Pretoria, South Africa: South African National Biodiversity Institute. [Google Scholar]
- Karsch F. 1894. Ueber einige neue oder ungenügend bekannte, durch Herrn G. Zenker auf der deutschen Forschungsstation Yaúnde im Hinterlande von Kamerun gesammelte Nymphaliden. Mitt Mus Natkd Berl Dtsch Entomol. 39(1):1–10. [Google Scholar]
- Lalonde MML. 2021. Phylogenetic analysis of the complete mitochondrial genome of the graphic beauty butterfly Baeotus beotus (Doubleday 1849) (Lepidoptera: Nymphalidae: Nymphalinae: Coeini). Mitochondr DNA Part B. 6(4):1516–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde MML, Marcus JM.. 2020. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniomorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondr DNA Part B. 5:3261–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. [DOI] [PubMed] [Google Scholar]
- Liao F, Wang L, Wu S, Li YP, Zhao L, Huang GM, Niu CJ, Liu YQ, Li MG.. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh BS, Marcus JM.. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae. J Asia-Pacific Entomol. 18(4):749–755. [Google Scholar]
- Okimoto R, Macfarlane JL, Wolstenholme DR.. 1990. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 18(20):6113–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I.. 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826. [DOI] [PubMed] [Google Scholar]
- Payment JE, Marcus JM, Lalonde MML.. 2020. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondr DNA Part B. 5:3415–3417. [Google Scholar]
- Peters MJ, Marcus JM.. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae. Syst Entomol. 42(1):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG.. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N, Brower AVZ, Nylin S.. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc Lond. 86(2):227–251. [Google Scholar]
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, Brower AVZ.. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 276(1677):4295–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC. 2020. Afrotropical butterflies. Book afrotropical butterflies. Vol. 2021. Metamorphosis. http://www.lepsocafrica.org/?p=publications&s=atb.
- Woodhall S. 2020. Butterflies of South Africa a field guide. 2 ed. London: Penguin Random House (Pty) Ltd. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession nos. MW722786 and MW722785. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA706380, SRX10228918, and SAMN18128423, respectively.