ABSTRACT
In classical Hodgkin lymphoma (cHL), the highly abundant CD4+ T cells in the vicinity of tumor cells are considered essential for tumor cell survival, but are ill-defined. Although they are activated, they consistently lack expression of activation marker CD26. In this study, we compared sorted CD4+CD26- and CD4+CD26+ T cells from cHL lymph node cell suspensions by RNA sequencing and T cell receptor variable gene segment usage analysis. This revealed that although CD4+CD26- T cells are antigen experienced, they have not clonally expanded. This may well be explained by the expression of exhaustion associated transcription factors TOX and TOX2, immune checkpoints PDCD1 and CD200, and chemokine CXCL13, which were amongst the 100 significantly enriched genes in comparison with the CD4+CD26+ T cells. Findings were validated in single-cell RNA sequencing data from an independent cohort. Interestingly, immunohistochemistry revealed predominant and high frequency of staining for TOX and TOX2 in the T cells attached to the tumor cells. In conclusion, the dominant CD4+CD26- T cell population in cHL is antigen experienced, polyclonal, and exhausted. This population is likely a main contributor to the very high response rates to immune checkpoint inhibitors in cHL.
KEYWORDS: Hodgkin, CD26, polyclonal, exhaustion, TOX
Introduction
Classical Hodgkin lymphoma (cHL) is characterized by a low number of tumor cells, called Hodgkin-Reed Sternberg (HRS) cells, surrounded by a heterogeneous inflammatory infiltrate.1,2 HRS cells evade anti-tumor immune responses by shaping the tumor micro-environment (TME) and by inhibiting immune cells.3–5 The highly abundant CD4+ T cells also play a critical role in the pathogenesis of cHL. CD4+ T cells are actively recruited by the HRS cells and can form so-called rosettes in a subset of cases.2,6–9 In each rosette, the CD4+ T cells physically interact with an HRS cell, providing it with pro-survival signals and shielding it from cytotoxic CD8+ T cells and NK cells.4 Recently, we have shown that rosetting CD4+ T cells communicate with the HRS cell through formation of the immunological synapse, with a central role for HLA class II-T cell receptor (TCR) interactions.10 In addition, CD4+ T cells have been implicated as key players in the response to programmed cell death-1 (PD-1) immune checkpoint inhibition. Membranous HLA class II expression by HRS cells was predictive for complete remission and increased progression-free survival.11 Moreover, CD4+ TCR diversity significantly increased in the blood of patients who achieved complete remission after PD-1 inhibition.12 Thus, CD4+ T cells residing in proximity to HRS cells are emerging as crucial players in cHL pathogenesis and response to PD-1 blockade.
In recent years, studies on the characterization of the cHL TME have suggested that the CD4+ T cells are mainly CD4+ regulatory T cells (Tregs) and exhausted T-effector cells.3,13 Exhausted T cells are characterized by increased expression of inhibitory cell surface receptors, reduced secretion of cytokines, reduced cytotoxicity and reduced proliferative potential. Indeed, expression of immune checkpoint molecules PD-1, CTLA-4 and/or LAG-3 has been identified in variable proportions of cHL CD4+ T cells.13–15 However, detailed and unbiased characterization of CD4+ T cells in the cHL TME has remained challenging due to the difficulty of separating CD4+ T cells in proximity to HRS cells from more distant CD4+ T cells in lymph nodes.16 The CD4+ T cells close to the HRS cells are known to express several activation-associated cell surface markers, including CD38 and CD69, but not CD26, while CD4+ T cells more distant from HRS cells do express CD26.17–20 CD26, also known as dipeptidyl peptidase IV (DPP4), is a proteolytic enzyme that is upregulated after stimulation under normal physiological conditions and plays a role in co-stimulation.21,22 However, CD4+CD26- T cells in cHL remain CD26 negative after activation and can also not or only moderately induce expression of T cell activation-associated cytokines, suggesting a functionally unresponsive or anergic state.17,20 Moreover, the CD4+CD26- T cell population expresses more Treg and Th17 cell associated markers compared to CD4+ CD26+ T cells.20
In this study, we aimed to further characterize the CD4+CD26- T cells in the TME that are in the direct vicinity of the malignant HRS cells. We compared CD4+CD26- to CD4+CD26 + T cells sorted from cHL lymph node derived cell suspensions and characterized their gene expression profiles. We established differentially expressed gene signatures, most likely CD4+ T cell subset lineages and TCR variable (V) gene segment usage to assess clonality. In addition, we validated expression patterns of selected genes in previously published single-cell RNA-seq (scRNA-seq) data.13 Finally, we investigated whether genes that were specifically enriched in CD4+CD26- T cells were expressed in T cells rosetting the HRS cells.
Materials and methods
Patients, tissue, and cell suspensions
For bulk RNA-seq, cryopreserved cell suspensions derived from lymph nodes of 19 cHL patients were retrieved from the cell bank of the department of Pathology and Medical Biology, University Medical Center Groningen, Groningen, The Netherlands. Patient characteristics are summarized in Supplemental Table 1. Patients were selected based on membranous HLA class II positivity (>90%) on the HRS cells as determined by immunohistochemistry for HLA-DP, DQ, DR (clone: CR3/43; 1:200; Dako; Santa Clara, CA, USA) on corresponding formalin fixed paraffin embedded (FFPE) tissue sections. Material was used in accordance with the ethical principles of the Declaration of Helsinki. The medical ethical review board of the UMCG approved the protocol under #RR202100080.
Cell sorting
All 19 cell suspensions were stained for CD4 (clone: Edu-2) and CD26 (clone: 2A6) as described previously.20 CD4+CD26- and CD4+CD26+ T cells were sorted using a MoFlo sorter (BD Biosciences, CA, USA) with a 70 µm nozzle. Purity of sorted populations was checked and was at least 94% or higher for all 38 sorted populations.
RNA isolation and bulk RNA sequencing
Total RNA was extracted from the 38 populations using a miRNeasy Mini or Micro kit (Qiagen; Hilden, Germany) according to manufacturer’s instructions. The concentration and quality of total RNA was determined using a Fragment Analyzer and all samples had an RNA integrity number of 8.5 or above. Three of the 38 populations, all CD4+CD26+, were excluded from RNA-seq due to an insufficient amount of RNA (<10 ng total). RNA libraries were made after depletion of rRNA (rRNA depletion kit, NEB #E6310) using the NEBNext Ultra II Directional RNA library prep kit for Illumina at GenomeScan (Leiden, The Netherlands) according to manufacturer’s instructions (NEB #E7760S/L). Paired-end sequencing with a read length of 151 nucleotides was performed on an Illumina NovaSeq6000 sequencer (lllumina; CA, USA) aiming at ~25 million paired reads per sample. Image analysis, base calling, and quality check was performed with the Illumina data analysis pipeline RTA3.4.4 and Bcl2fastq v2.20.
RNA sequencing analysis
RNA-seq reads were mapped to the human reference genome GRCh37 using Hisat2.23 Reads were counted into ensembl v75 genes with Htseq-count. One patient was excluded from further analysis due to a low number of aligned unique reads in both sorted populations (Supplemental Table 2). Analysis and visualization of RNA-seq data were performed in the R statistical environment (version 4.0.2). Normalization and differential expression analysis (DEA) was performed by R/Bioconductor package DESeq2.24 Genes were considered differentially expressed when the adjusted p-value ≤0.05 and −1< log2foldchange>1. Genes of interest were selected based on the following criteria: (i) protein coding gene; (ii) described presence in T cells, according to RNA levels in the Schmiedel dataset (see: https://dice-database.org),25 and function, as found in PubMed searches; (iii) availability of antibodies; and (iv) moderate-to-high expression levels. Gene set variation analysis (GSVA) was utilized to measure enrichment for specific CD4+ T cell subsets within each sorted population with the R/Bioconductor package GSVA.26 CD4 subset-specific gene sets were retrieved from a previous study.25 DESeq2ʹs median of ratios, Regularized log transformation (rlog) and Variance Stabilizing Transformed (vst) values were used for data visualization, clustering and GSVA, respectively.
T cell receptor V segment usage
The TCR diversity codeset currently included in the nCounter CAR-T characterization panel (NanoString Technologies, Seattle, WA, USA) was used to determine V segment usage in CD4+CD26+ and CD4+CD26- T cell subsets of 11/19 cHL patients with sufficient RNA left after RNA-seq. The panel included probes for 45 TCR alpha V (TRAV) segments, 46 TCR beta V (TRBV) segments, some additional T-cell-specific genes including amongst others CD45RA and CD45RO specific probes and probes for 10 reference genes, 8 negative controls to control for background signal, and 6 controls to correct for hybridization efficiencies. The full list of 143 genes can be found in Supplemental Table 3. Probes were hybridized overnight with 80–100 ng RNA in a thermocycler at 67°C. The RNA probe complexes were loaded on an nCounter cartridge and analyzed on an nCounter SPRINT platform according to manufacturer’s instructions (NanoString Technologies). Reporter Code Count (RCC) files were normalized to correct for differences in hybridization efficiency between samples using the internal control probes with nSolver Analysis software (NanoString nCounter Technologies). An additional normalization step was performed to correct for differences in hybridization efficiency between individual TCR alpha V (TRAV) segment and TCR beta V (TRBV) segment probes using a synthetic reference sample provided by NanoString. As a final step, counts of individual TRAV/TRBV probes were expressed as a percentage relative to the sum of all TRAV/TRBV counts. To compare absolute CD45RA and CD45RO counts between samples, data were normalized for RNA input using the reference genes.
Single-cell RNA sequencing data
Previously obtained scRNA-seq data of 16 HLA class II positive and 5 HLA class II negative diagnostic cHL cases was re-analyzed to validate expression patterns in CD4+CD26- and CD4+CD26+ T cells.13 Patient characteristics are summarized in Supplemental Table 1. Cells from all 21 cases were combined in R (version 3.6.1), and filtering and normalization was performed as previously described.13 The CD4+ T cell subset was selected for analysis based on the expression of canonical marker genes (CD3+CD4+CD8-CD56-CD19-), and the co-expression pattern of genes identified in the bulk RNA-seq analysis was then examined in this population. To examine associations between expression of CD26 and the other marker genes, a 2 × 2 contingency table summarizing the number of cells with each co-expression pattern was calculated for each marker gene (e.g. for gene G, the number of CD26+G+, CD26+G-, CD26-G+, and CD26-G- cells). Odds ratios with 95% confidence intervals were then calculated from the contingency tables using the epitools package (version 0.5–10.1) to determine the likelihood of each marker gene being co-expressed with CD26. To determine linear correlation coefficients (rho) between markers of interest in the scRNA-seq data, pairwise Spearman correlations were calculated in R using log-normalized counts from the CD4+ T cells, comparing the expression of each pair of genes across all cells.
Immunohistochemistry
Frozen sections were stained with anti-CD26 (clone: 2A6) as previously described.20 FFPE tissue sections were stained for TOX, TOX2, NFIA, CXCL13, PD-1, CD200, and CTTN. CD30 and TARC were included as tumor cell markers and CD4 to identify CD4+ T cells. Immunohistochemistry (IHC) for CD30 (clone: BerH2; Ventana Medical Systems; AZ, USA), CD4 (clone: SP35; Ventana Medical Systems) and PD-1 (clone: NAT105; Ventana Medical Systems) was performed using an automated stainer following the manufacturer’s protocol (Ventana Benchmark Ultra; Ventana Medical Systems). IHC for TARC, TOX, TOX2, NFIA, CXCL13, CD200 and CTTN was performed according to standard lab procedures. Briefly, slides were deparaffinized in xylene, which was followed by a heat-induced antigen retrieval using a 10 mM citrate buffer at pH 6.0 (for NFIA), a 1 mM EDTA (ethylene diamine tetracetic acid) buffer at pH 8.0 (for TOX2 and CXCL13) or a 10 mM Tris (tris-hydroxymethyl-aminomethane)/1 mM EDTA buffer at pH 9.0 (for TARC, TOX, CD200 and CTTN). Primary antibodies and dilutions used included anti-TARC (polyclonal; 1:400; R&D Systems; MN, USA), TOX (clone: NAN448B; 1:400; Abcam; Cambridge, UK), TOX2 (polyclonal; 1:40; Atlas Antibodies; Stockholm, Sweden), NFIA (polyclonal; 1:400; Atlas Antibodies), CXCL13 (polyclonal; 1:80; R&D Systems), CD200 (clone: EPR22412-229; 1:500; Abcam) and CTTN (polyclonal; 1:50; Atlas Antibodies). The primary antibody against TOX2 was incubated overnight at 4°C, all the other antibodies were incubated for one hour at room temperature. Primary antibodies were detected using appropriate secondary and tertiary HRP conjugated antibodies and visualized with diaminobenzidine (DAB).
Staining was evaluated by two individual observers and discrepant cases were discussed until consensus was reached. Evaluation was focused on tumor cell containing areas as identified by CD30 and TARC positivity. CD4 staining was used to confirm presence of rosettes and to identify density of CD4+ T cells within the TME. Staining patterns of proteins selected from the RNA-seq analysis were evaluated and staining within rosettes was scored as 0 (0–25% of cells within rosette positive), 1 (25–50% of cells within rosette positive), 2 (50–75% of cells within rosette positive) or 3 (75–100% of cells within rosette positive). We assessed all rosettes within the tumor cell areas. For heterogeneous staining patterns, the average of the two most divergent scores was used. In addition, we identified whether markers were enriched within the tumor cell areas compared to the rest of the TME.
Code availability
Scripts used for data analysis are available upon request.
Data sharing statement
Bulk RNA-seq BAM files are deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB46009.
Results
Sorting and RNA-sequencing of CD4+ T cell subsets in cHL
CD4+ T cells of 19 HLA class II positive cHL cell suspensions were sorted based on membranous CD26 expression. The mean percentage of CD4+CD26- T cells was 70% (range 36–92%) (Supplemental Table 2). In 14/19 cases, the percentage of CD4+CD26- T cells was at least 2-fold higher compared to the percentage of CD4+CD26+ T cells, showing that CD4+CD26- T cells were usually dominant in the cHL TME. RNA was isolated from the sorted CD4+CD26- and CD4+CD26+ T cells, which was used for TCR V segment usage analysis and RNA-seq. The total number of aligned RNA-seq reads ranged from 49 to 96 × 106 (mean: 65 × 106) (Supplemental Table 2). The fraction of mapped reads marked as a duplicate ranged from 12.8– to 2.4% (mean: 25.7%). Both samples of one patient were excluded from further analysis due to a low number of aligned unique reads. The number of unique reads of the samples included in the subsequent analyses ranged from 28–71 × 106 (mean: 50 × 106). Subsequent analyses based on RNA-seq compared CD4+CD26- and CD4+CD26+ T cells using GSVA to identify similarities with specific CD4+ T cell subset lineages and DEA to identify distinct gene expression profiles. Findings were validated in an independent scRNA-seq cohort and on the protein level using immunohistochemistry.
CD4+CD26- T cells in cHL share characteristics with antigen experienced CD4+ T cell subsets
The GSVA scores for eight distinct CD4+ T cell subset gene signatures described by Schmiedel et al.25 were determined for each sorted T cell subset. CD4+CD26- T cells were mainly enriched for memory Treg and T follicular helper (Tfh) gene signatures in comparison with CD4+CD26+ T cells, and also showed some overlap with Th17 cells (Figure 1). CD4+CD26+ T cells were mainly enriched for naïve CD4+ T cells and Th1/17 cells, and to a lesser extent for naïve Treg and Th2 cells, indicating that the CD4+CD26- T cells had a memory signature, while CD4+CD26+ T cells had a naïve signature. To further support the GSVA results, we analyzed the expression levels of CD45RA and CD45RO using transcript specific probes included in the TCR diversity codeset. Consistent with the GSVA results, we observed a significant decrease of naïve T cell marker CD45RA and a significant increase of the memory T cell marker CD45RO in CD4+CD26- T cells compared to CD4+CD26+ T cells (Supplemental Figure S1), as was already shown in the literature.17,18 Altogether, this indicated that CD4+CD26- T cells are more antigen experienced than CD4+CD26+ T cells.
Figure 1.
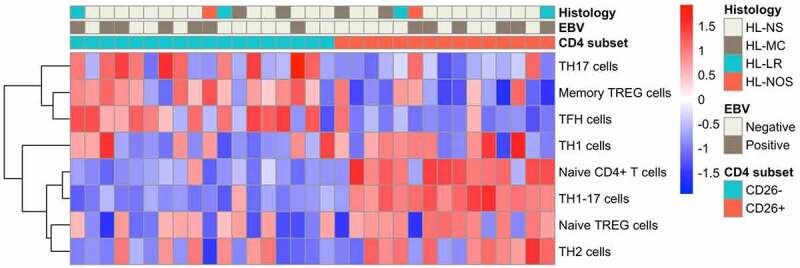
Supervised hierarchical clustering of GSVA-scores for CD4+ T cell subset gene signatures. Heatmap of the supervised hierarchical clustering of GSVA-scores for eight CD4+ T cell subset gene sets in 18 CD4+CD26- T cell populations and 15 CD4+CD26+ T cell populations. Supervised clustering is based on CD4 subset. NS = Nodular Sclerosis; MC = Mixed Cellularity; LR = Lymphocyte Rich; NOS = Not Otherwise Specified.
T cell receptor variable gene segment usage in CD4+CD26- T cells is polyclonal
To assess TCR diversity, we analyzed TCR alpha and TCR beta V segment usage. The TCR V segment usage of CD4+CD26- and CD4+CD26+ T cells both showed a polyclonal pattern with no apparent difference in TCR diversity for any of the TRAV and TRBV segments in CD4+CD26- and CD4+CD26+ T cells (Figure 2). So although CD4+CD26- T cells show characteristics of antigen experienced CD4+ T cells, no clonal TCR V segment usage pattern was observed.
Figure 2.
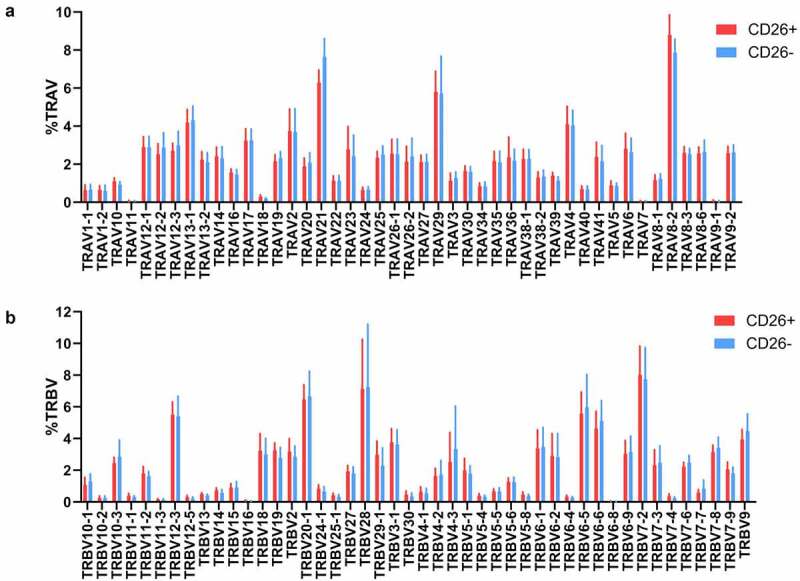
T cell receptor V segment usage in CD26- and CD26+CD4+ T cells. Relative abundance of (a) expressed T cell receptor alpha variable (TRAV) segments and (b) expressed T cell receptor beta variable (TRBV) segments in CD4+CD26+ and CD4+CD26- T cells. Visualized is mean with standard deviation of 11 paired CD4+ T cell populations.
The CD4+CD26- T cells have a distinct gene expression signature
Principal component analysis (PCA) considering the whole-gene expression profile revealed that samples separated based on CD26 expression status along PC1, the axis representing the highest variance in the data (Supplemental Figure S2). This already suggested that CD4+CD26- and CD4+CD26+ T cells have distinct gene signatures. Comparison of the RNA-seq data of CD4+CD26- T cells to those of the CD4+CD26+ T cells resulted in identification of 567 differentially expressed genes (DEGs) (adjusted p-value <0.05 and −1< log2foldchange>1) (Supplemental Table 4). Of these, 100 genes were significantly upregulated and 467 genes were significantly downregulated in CD4+CD26- T cells compared to CD4+CD26+ T cells. Unsupervised hierarchical clustering of the 567 DEGs revealed two clusters with a clear separation between CD4+CD26- and CD4+CD26+ T cells (Figure 3a). Within each T cell subset, no further clustering was observed for histological subtype or EBV status. The top DEG was DPP4 (CD26; log2foldchange = 5.83; padj = 5.45x10−45) consistent with our sorting strategy (Figure 3b). As we were interested in characterizing the CD4+CD26- T cells residing in proximity to the HRS cells we focused on the 100 genes significantly upregulated in this T cell population. Based on our selection criteria, we selected seven genes for subsequent analysis: TOX, TOX2, NFIA, PDCD1 (=PD-1), CD200, CXCL13 and CTTN (Supplemental Figure S3). Thus, CD4+CD26- T cells have increased expression of genes encoding for exhaustion associated transcription factors TOX and TOX2, transcription factor nuclear factor I A (NFIA), immune checkpoints PD-1 and CD200, chemokine CXCL13 and actin-binding filament protein cortactin (CTTN).
Figure 3.
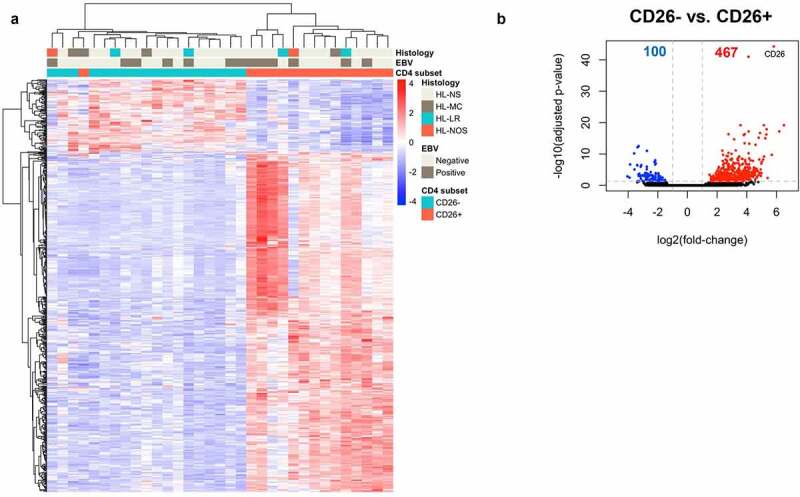
Genes differentially expressed between CD26- and CD26+CD4+ T cells. (a) Heatmap showing the unsupervised hierarchical clustering of 567 genes differentially expressed between CD4+CD26- and CD4+CD26+ T cells. Of the 567 genes, 100 were upregulated and 467 downregulated in CD4+CD26- T cells. Rows are genes, columns are samples. (b) Volcano plot for CD4+CD26- versus CD4+CD26+ . – log10(adjusted p-values) are plotted against log2(foldchange). Grey dashed lines indicate an adjusted p-value of <0.05 and a log2(foldchange) greater than 1 in both directions. NS = Nodular Sclerosis; MC = Mixed Cellularity; LR = Lymphocyte Rich; NOS = Not Otherwise Specified.
Co-expression patterns of genes of interest
To study co-expression of the genes of interest within individual CD4+ T cells, we used scRNA-seq data from 16 previously published HLA class II positive cHL cases.13 The seven selected genes were all significantly less frequently co-expressed with CD26 in CD4+ T cells, with odds ratios (OR) below 0.5 (Figure 4a). Moreover, the expression levels of the selected genes showed positive correlations to one another and negative correlations to CD26 expression across the CD4+ T cell population, albeit with low rho values that are likely due to the inherent ‘drop-out’ of expression values presented in scRNA-seq data (Supplemental Figure 4A). Both observations confirm the differential expression patterns as observed in our bulk RNA-seq analysis. Expression patterns in the scRNA-seq data showed thatTOX2 and CXCL13 were most frequently expressed in CD4+CD26- T cells (Figure 4b). In addition, CD4+CD26- T cells often co-expressed TOX2 with at least one of the other selected genes and in part of the CD4+ CD26- T cells we observed co-expression with three to four of the genes of interest. As expected, the majority of CD4+ CD26+ T cells did not express any of the selected genes (Figure 4b). Similar results were obtained in five HLA class II negative cHL cases (Supplemental Figure 4B,C).
Figure 4.
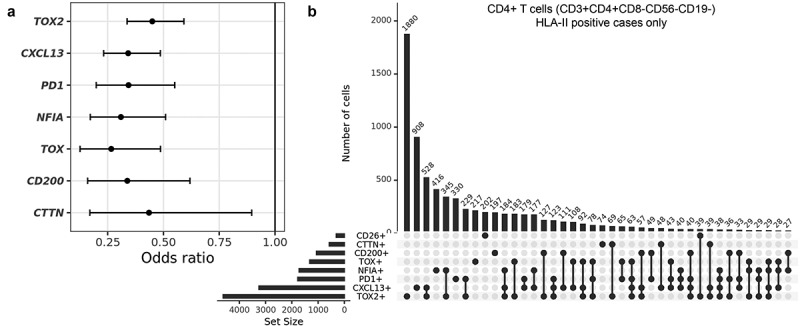
Characterization of co-expression patterns in CD4+ T cells using single-cell RNA-seq data from HLA class II positive cHL cases. (a) Odds ratio (OR) of expression of the selected genes in CD4+CD26+ versus CD4+CD26- T cells. Indicated are the OR with 95% confidence intervals. (b) UpSet plot showing co-expression patterns of CD26 with the seven selected genes in single CD4+ T cells. A total of 13014 cells was evaluated.
Rosetting T cells express TOX and TOX2
To investigate protein expression of the selected genes in the cHL microenvironment, immunohistochemistry was performed on tissue of the 19 cHL cases used for bulk RNA-seq (Figure 5a-f). The majority of cells surrounding the HRS cells did not express CD26, in line with previous studies.17,20 CD26 was expressed in a nodular or scattered pattern, with respectively only CD26+ cells outside the tumor cell areas or a low number of CD26+ cells scattered throughout the tissue with occasionally a positive cell close to a tumor cell. If expressed, TOX, TOX2, CD200, PD-1 and CXCL13 positive cells were enriched in the tumor cell areas in which the CD4+CD26- T cells reside. NFIA positive cells were in general scattered throughout the TME and CTTN did not show a T-cell-specific staining pattern. Staining patterns for all selected markers except CTTN were scored according to the percentage positive cells in the rosetting T cells (Supplemental Figure S5; Supplemental Table 2). TOX and TOX2 were the most sensitive markers for rosetting T cells with >50% of rosetting cells staining positive in 63% and 79% of the cHL cases, respectively (Figure 5g). TOX2 positive cells were in general more abundant in the cHL TME than TOX positive cells. CD200 stained positive in >50% of the rosetting cells in 44% of the cHL cases. In addition, HRS cells expressed CD200 in 78% of evaluated cases. PD-1 stained positive in >50% of rosetting cells in 35% of the cases, while in the majority of the remaining cases PD-1 staining was completely absent. Lower percentages of positive rosetting T cells were observed for NFIA and CXCL13. For NFIA only 21% of cases showed enrichment in rosettes. The number of CXCL13 positive cells was limited in the majority of cases, but they were usually in proximity of the tumor cells. In HLA class II negative cases staining patterns were similar to those observed in HLA class II positive cases. Again, TOX, TOX2 and CD200 were most frequently expressed in rosettes, although percentages of positive rosetting T cells was slightly lower in HLA class II negative cHL cases (Supplemental Table 5).
Figure 5.
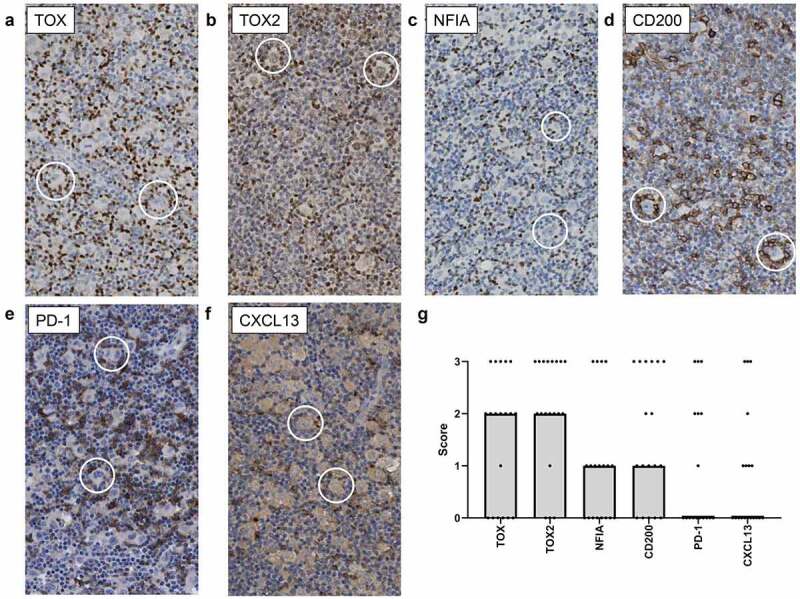
Representative images of immunohistochemical staining in cHL tissue to define rosette specific expression pattern. Representative images of (a) TOX, (b) TOX2, (c) NFIA, (d) CD200, (e) PD-1 and (f) CXCL13 staining. In each picture two rosettes (HRS cell with surrounding lymphocytes) are highlighted with a circle. (g) Visualization of the immunohistochemistry scores per marker. Points represent the scores of each of the 19 cHL cases used for sorting and subsequent bulk RNA-seq analysis. The bars represent the median score of all cases.
Discussion
Using bulk and single-cell RNA-seq approaches, we have characterized the CD4+CD26- T cells in the cHL TME. Our results show that these CD4+CD26- T cells are antigen experienced and polyclonal. Markers that were most prominently enriched in CD4+CD26- compared to CD4+CD26+ T cells and were expressed in the majority of rosetting CD4+ T cells were thymocyte selection-associated high-mobility group box (TOX) and TOX2, which are exhaustion associated transcription factors.
The finding that CD4+CD26- T cells in the TME of cHL displays an antigen experienced gene expression profile fits well with cHL associated T cell types that have been described in multiple previous reports.3,13,20,27 Our group has recently established that initial interaction between T cells and HRS cells is fast and involved a large proportion of T cells,10 which is consistent with antigen experienced T cells recognizing a broad spectrum of HRS cell derived antigens. These interactions occur by means of the immunological synapse, resulting in formation of T cell rosettes and low-level IL-2 production, suggesting that these T cells get activated, but only to some extent.10 This low-level activation state is consistent with expression of early activation markers CD38 and CD69, while CD25 and CD26 are missing.17,19,20 In the current study, we describe that these low-level activated T cells do not clonally expand, as seen by the polyclonal TCR V segment usage. This is in line with previous studies describing that rosetting T cells in HL are polyclonal.28,29
Our gene expression data suggest that these antigen experienced T cells consist of memory Treg cells and/or Tfh cells. However, further evidence for a contribution of Tfh cells in the cHL TME is lacking in literature and characteristic markers like BCL-6 and CXCR530,31 are not present in our differentially expressed gene set. In addition, although Tfh cell markers CD200 and PD-1 are expressed at higher levels, the intensity of staining for those markers is usually lower in CD4+CD26- T cells in the HL TME compared to CD4+ T cells in germinal centers within the same tissue. In contrast, memory Treg cells have commonly been described and are actively recruited by HRS cells that attract them by secreting high amounts of chemokines CCL17/TARC and CCL22/MDC.6–8 Thus, memory Treg constitute a major source of CD4+ T cells in the cHL TME, but it appears their gene expression is modulated to a profile that combines both memory Treg and Tfh features.
Besides the already known characteristic feature of being CD26-, we identified two additional proteins enriched in CD4+ T cells in the TME. These proteins, i.e. TOX and TOX2, were also frequently expressed by rosetting T cells in cHL. The TOX protein family consists of four members that all function as transcription factors: TOX (also known as TOX1), TOX2, TOX3 and TOX4.32 TOX and/or TOX2 are involved in many early lymphoid developmental processes, including positive selection in thymocytes, early development of CD4+ T and NK cells, development of innate lymphoid cells and lymph node organogenesis.33–35 More important in the setting of our study, they are essential for the development of Tfh cells and the induction of exhaustion in both CD4+ and CD8+ T cells.36–41 Corresponding with our DEG, TOX can induce the expression of the Tfh defining chemokine CXCL13, and immune checkpoint molecule PD-1, which is a well-established marker of exhaustion.42,43 Of note, in our immunohistochemistry analysis, T cells expressing TOX and/or TOX2 outnumbered cells expressing CXCL13 and PD-1, confirming a previous report that the exhaustion phenotype is a dominant feature in the cHL TME.3 In line with this induction of exhaustion by TOX and TOX2, is the inability of CD4+ CD26- T cells to upregulate production of several cytokines upon in vitro stimulation.20 It is well known that TOX and TOX2 are both induced by chronic antigen stimulation of the TCR.37,40 It would be interesting to study if this is also the case in HL, given the importance of HLA class II-TCR interactions in HL rosetting.10
Increased TOX levels in T cells are very prominent in our study in cHL and have also been described in solid malignancies and B cell non-Hodgkin lymphoma.43–46 Interestingly, several lines of evidence support a role of TOX and TOX2 in CD8+ T cells in sensitivity to immune checkpoint blockade. Knockdown of TOX in CD8+ T cells in a patient-derived xenograft mouse model of hepatocellular cancer decreased tumor growth, alleviated the CD8+ T cell exhaustion, increased CD8+ T cell infiltration and improved responses to PD-1 blockade therapy.43 In addition, CD8+ chimeric antigen receptor (CAR) T cells with a combined deficiency in TOX and TOX2 were more active and promoted profound tumor regression and prolonged survival in a melanoma mouse model.39 If these results in CD8+ T cells can be extrapolated to CD4+ T cells in cHL several important implications arise. First, cHL has a very high response rate to PD-1 blockade with objective response rates as high as 87% in the relapsed and refractory setting with also excellent efficacy in first-line treatment combined with concomitant or sequential de-intensified chemotherapy regimens in ongoing trials.47–50 Reversal of exhaustion in CD4+ T cells is likely responsible for these very high response rates because of their high abundance, their importance to survival of HRS cells and their predictive role in response to PD-1 blockade.4,10–12 Second, the extent of exhaustion in CD4+ T cells is expected to be predictive for immune checkpoint blockade outcome and it would be interesting to study TOX and TOX2 expression in this setting. Finally, targeting TOX and/or TOX2 with small molecule inhibitors,51 might revert T cell exhaustion, thereby being an attractive way to further improve immunotherapy results in cHL.
In conclusion, we have extensively characterized CD4+CD26- T cells in the TME of cHL. These T cells are antigen experienced, polyclonal and probably originate from memory Treg cells. They are enriched for exhaustion associated transcription factors TOX and TOX2, which also induce expression of immune checkpoints. Targeting of TOX and TOX2 might reverse T cell exhaustion and thereby provides an interesting opportunity for immunotherapy.
Supplementary Material
Acknowledgments
J.V. was funded by the Graduate School of Medical Sciences, University of Groningen, University Medical Center Groningen, The Netherlands. C.S. was funded by a Large Scale Applied Research Project from Genome Canada (Grant No. 13124), Genome BC (Grant No. 271LYM) and the Canadian Cancer Society Research Institute (Grant No. 705288).
Disclosure statement
A.D. receives research funding from Millenium/Takeda. C.S. is an advisory board member for Curis Inc., AbbVie, Seattle Genetics, and Roche, reports receiving commercial research grants from Bristol-Myers Squibb and Trillium Therapeutics, and remuneration from Bayer and Juno Therapeutics. No potential conflicts of interest were disclosed by the other authors.
Author contributions
J.V., L.V., A.v.d.B and A.D. designed the study; J.R.P., M.M.T. and K.K. performed bulk RNA-seq analysis; L.C., T.A. and C.S. performed single-cell RNA-seq analysis; J.V., M.M. and L.V. acquired data (e.g. sorting, RNA isolation, nanostring, IHC); A.D. and L.K. analyzed nanostring data; J.V., J.R.P., L.V., A.v.d.B. and A.D. wrote the manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Nagasaki J, Togashi Y, Sugawara T, Itami M, Yamauchi N, Yuda J, Sugano M, Ohara Y, Minami Y, Nakamae H, et al. The critical role of CD4+ T cells in PD-1 blockade against MHC-II-expressing tumors such as classic Hodgkin lymphoma. Blood Adv. 2020;4(17):4069–10. doi: 10.1182/bloodadvances.2020002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poppema S, Bhan AK, Reinherz EL, Posner MR, Schlossman SF.. In situ immunologic characterization of cellular constituents in lymph nodes and spleens involved by Hodgkin’s disease. Blood. 1982;59(2):226–232. doi: 10.1182/blood.V59.2.226.226. [DOI] [PubMed] [Google Scholar]
- 3.Cader FZ, Schackmann RCJ, Hu X, Wienand K, Redd R, Chapuy B, Ouyang J, Paul N, Gjini E, Lipschitz M, et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatory T-cell-rich and exhausted T-effector microenvironment. Blood. 2018;132(8):825–836. doi: 10.1182/blood-2018-04-843714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Sattarzadeh A, Diepstra A, Visser L, van Den Berg A. van den Berg A. The microenvironment in classical Hodgkin lymphoma: an actively shaped and essential tumor component. Semin Cancer Biol. 2014;24:15–22. doi: 10.1016/j.semcancer.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Hollander P, Rostgaard K, Smedby KE, Molin D, Loskog A, de Nully Brown P, Enblad G, Amini R-M, Hjalgrim H, Glimelius I, et al. An anergic immune signature in the tumor microenvironment of classical Hodgkin lymphoma is associated with inferior outcome. Eur J Haematol. 2018;100(1):88–97. doi: 10.1111/ejh.12987. [DOI] [PubMed] [Google Scholar]
- 6.Niens M, Visser L, Nolte IM, van der Steege G, Diepstra A, Cordano P, Jarrett RF, Te Meerman GJ, Poppema S, van Den Berg A, et al. Serum chemokine levels in Hodgkin lymphoma patients: highly increased levels of CCL17 and CCL22. Br J Haematol. 2008;140(5):527–536. doi: 10.1111/j.1365-2141.2007.06964.x. [DOI] [PubMed] [Google Scholar]
- 7.Ishida T, Ishii T, Inagaki A, Yano H, Komatsu H, Iida S, Inagaki H, Ueda R. Specific recruitment of CC chemokine receptor 4-positive regulatory T cells in Hodgkin lymphoma fosters immune privilege. Cancer Res. 2006;66(11):5716–5722. doi: 10.1158/0008-5472.CAN-06-0261. [DOI] [PubMed] [Google Scholar]
- 8.Iellem A, Mariani M, Lang R, Recalde H, Panina-Bordignon P, Sinigaglia F, D’Ambrosio D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2001;194(6):847–853. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart AE, Williams AR, Habeshaw JA. Rosetting and other reactions of the Reed-Sternberg cell. J Pathol. 1977;122(2):81–90. doi: 10.1002/path.1711220205. [DOI] [PubMed] [Google Scholar]
- 10.Veldman J, Visser L, Huberts-Kregel M, Muller N, Hepkema B, van Den Berg A, Diepstra A. Rosetting T cells in Hodgkin lymphoma are activated by immunological synapse components HLA class II and CD58. Blood. 2020;136(21):2437–2441. doi: 10.1182/blood.2020005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, Sasse S, Younes A, Fanale M, Santoro A, et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J Clin Oncol. 2018;36(10):942–950. doi: 10.1200/JCO.2017.77.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cader FZ, Hu X, Goh WL, Wienand K, Ouyang J, Mandato E, Redd R, Lawton LN, Chen P-H, Weirather JL, et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat Med. 2020;26(9):1468–1479. doi: 10.1038/s41591-020-1006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoki T, Chong LC, Takata K, Milne K, Hav M, Colombo A, Chavez EA, Nissen M, Wang X, Miyata-Takata T, et al. Single-Cell Transcriptome Analysis Reveals Disease-Defining T-cell Subsets in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Cancer Discov. 2020;10(3):406–421. doi: 10.1158/2159-8290.CD-19-0680. [DOI] [PubMed] [Google Scholar]
- 14.Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, Hu X, Redd R, Freeman GJ, Neuberg D, et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood. 2017;130(22):2420–2430. doi: 10.1182/blood-2017-03-770719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel SS, Weirather JL, Lipschitz M, Lako A, Chen PH, Griffin GK, Armand P, Shipp MA, Rodig SJ. The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative. Blood. 2019;134(23):2059–2069. doi: 10.1182/blood.2019002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromm JR, Kussick SJ, Wood BL. Identification and purification of classical Hodgkin cells from lymph nodes by flow cytometry and flow cytometric cell sorting. Am J Clin Pathol. 2006;126(5):764–780. doi: 10.1309/7371XK6F6P7474XX. [DOI] [PubMed] [Google Scholar]
- 17.Poppema S. Immunology of Hodgkin’s disease. Baillieres Clin Haematol. 1996;9(3):447–457. doi: 10.1016/S0950-3536(96)80020-5. [DOI] [PubMed] [Google Scholar]
- 18.Poppema S. The nature of the lymphocytes surrounding Reed-Sternberg cells in nodular lymphocyte predominance and in other types of Hodgkin’s disease. Am J Pathol. 1989;135:351–357. [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka T, Camerini D, Seed B, Torimoto Y, Dang NH, Kameoka J, Dahlberg HN, Schlossman SF, Morimoto C. Cloning and functional expression of the T cell activation antigen CD26. J Immunol. 1992;149:481–486. [PubMed] [Google Scholar]
- 20.Ma Y, Visser L, Blokzijl T, Harms G, Atayar C, Poppema S, van Den Berg A. The CD4+CD26- T-cell population in classical Hodgkin’s lymphoma displays a distinctive regulatory T-cell profile. Lab Invest. 2008;88(5):482–490. doi: 10.1038/labinvest.2008.24. [DOI] [PubMed] [Google Scholar]
- 21.Fleischer B. CD26: a surface protease involved in T-cell activation. Immunol Today. 1994;15(4):180–184. doi: 10.1016/0167-5699(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 22.Klemann C, Wagner L, Stephan M, Von Horsten S. Cut to the chase: a review of CD26/dipeptidyl peptidase-4ʹs (DPP4) entanglement in the immune system. Clin Exp Immunol. 2016;185(1):1–21. doi: 10.1111/cei.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550,014–0550-8. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175(6):1701. 1715.e16. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14(1):7,2105–14-7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrarini I, Rigo A, Zamo A, Vinante F. Classical Hodgkin lymphoma cells may promote an IL-17-enriched microenvironment. Leuk Lymphoma. 2019;60(14):3395–3405. doi: 10.1080/10428194.2019.1636983. [DOI] [PubMed] [Google Scholar]
- 28.Trümper L, Jung W, Daus H, Mechtersheimer G, Von Bonin F, Pfreundschuh M. Assessment of clonality of rosetting T lymphocytes in Hodgkin’s disease by single-cell polymerase chain reaction: detection of clonality in a polyclonal background in a case of lymphocyte predominance Hodgkin’s disease. Ann Hematol. 2001;80(11):653–661. doi: 10.1007/s002770100370. [DOI] [PubMed] [Google Scholar]
- 29.Roers A, Montesinos-Rongen M, Hansmann ML, Rajewsky K, Küppers R. Amplification of TCRbeta gene rearrangements from micromanipulated single cells: t cells rosetting around Hodgkin and Reed-Sternberg cells in Hodgkin’s disease are polyclonal. Eur J Immunol. 1998;28(8):2424–2431. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Roider T, Seufert J, Uvarovskii A, Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm J-P, Herbst SA, Bruch P-M, et al. Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels. Nat Cell Biol. 2020;22(7):896–906. doi: 10.1038/s41556-020-0532-x. [DOI] [PubMed] [Google Scholar]
- 31.Ji LS, Sun XH, Zhang X, Zhou ZH, Yu Z, Zhu XJ, Huang LY, Fang M, Gao YT, Li M, et al. Mechanism of Follicular Helper T Cell Differentiation Regulated by Transcription Factors. J Immunol Res. 2020;2020:1826587. doi: 10.1155/2020/1826587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang C, Huang S, Zhao Y, Chen S, Li Y. TOX as a potential target for immunotherapy in lymphocytic malignancies. Biomark Res. 2021;9(1). 20,021-00275-y. doi: 10.1186/s40364-021-00275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205(1):245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, Oostendorp RAJ, Leung W. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood. 2014;124(26):3905–3913. doi: 10.1182/blood-2014-06-582965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Li Z. TOX gene: a novel target for human cancer gene therapy. Am J Cancer Res. 2015;5:3516–3524. [PMC free article] [PubMed] [Google Scholar]
- 36.Balanca CC, Salvioni A, Scarlata CM, Michelas M, Martinez-Gomez C, Gomez-Roca C, Sarradin V, Tosolini M, Valle C, Pont F, et al. PD-1 blockade restores helper activity of tumor-infiltrating, exhausted PD-1hiCD39+ CD4 T cells. JCI Insight. 2021;6(2). doi: 10.1172/jci.insight.142513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, Von Hoesslin M, Cullen JG, Fan Y, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571(7764):265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 38.Scott AC, Dundar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571(7764):270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo H, Chen J, Gonzalez-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8 + T cell exhaustion. Proc Natl Acad Sci U S A. 2019;116(25):12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat Immunol. 2019;20(7):890–901. doi: 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, Zhao X, Wang X, Feng H, Gou M, Jin W, Wang X, Liu X, Dong C. The Transcription Factor Tox2 Drives T Follicular Helper Cell Development via Regulating Chromatin Accessibility. Immunity. 2019. 839.e5;51(5):826. doi: 10.1016/j.immuni.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Yoshitomi H, Kobayashi S, Miyagawa-Hayashino A, Okahata A, Doi K, Nishitani K, Murata K, Ito H, Tsuruyama T, Haga H, et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat Commun. 2018;9(1):3762. 018-06187-0. doi: 10.1038/s41467-018-06187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, He Q, Shen H, Xia A, Tian W, Yu W, Sun B. TOX promotes the exhaustion of antitumor CD8(+) T cells by preventing PD1 degradation in hepatocellular carcinoma. J Hepatol. 2019;71(4):731–741. doi: 10.1016/j.jhep.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Kim K, Park S, Park SY, Kim G, Park SM, Cho JW, Kim DH, Park YM, Koh JW, Kim HR, et al. Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer. Genome Med. 2020;12(1) 22,020-00722-9 doi: 10.1186/s13073-020-00722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, Liang C, Zhao Y, Deng T, Tan J, Lu Y, Liu S, Li Y, Chen S. Increased TOX expression concurrent with PD-1, Tim-3, and CD244 in T cells from patients with non-Hodgkin lymphoma. Asia Pac J Clin Oncol. 2021. doi: 10.1111/ajco.13545. [DOI] [PubMed] [Google Scholar]
- 46.Maestre L, Garcia-Garcia JF, Jimenez S, Reyes-Garcia AI, Garcia-Gonzalez A, Montes-Moreno S, Arribas AJ, Gonzalez-Garcia P, Caleiras E, Banham AH, et al. High-mobility group box (TOX) antibody a useful tool for the identification of B and T cell subpopulations. PLoS One. 2020;15(2):e0229743. doi: 10.1371/journal.pone.0229743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merryman RW, Armand P, Wright KT, Rodig SJ. Checkpoint blockade in Hodgkin and non-Hodgkin lymphoma. Blood Adv. 2017;1(26):2643–2654. doi: 10.1182/bloodadvances.2017012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brockelmann PJ, Goergen H, Keller U, Meer J, Ordemann R, Halbsguth TV, Sasse S, Sökler M, Kerkhoff A, Mathas S, et al. Efficacy of Nivolumab and AVD in Early-Stage Unfavorable Classic Hodgkin Lymphoma: the Randomized Phase 2 German Hodgkin Study Group NIVAHL Trial. JAMA Oncol. 2020;6(6):872–880. doi: 10.1001/jamaoncol.2020.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramchandren R, Domingo-Domenech E, Rueda A, Trneny M, Feldman TA, Lee HJ, Provencio M, Sillaber C, Cohen JB, Savage KJ, et al. Nivolumab for Newly Diagnosed Advanced-Stage Classic Hodgkin Lymphoma: safety and Efficacy in the Phase II CheckMate 205 Study. J Clin Oncol. 2019;37(23):1997–2007. doi: 10.1200/JCO.19.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voltin CA, Mettler J, van Heek L, Goergen H, Müller H, Baues C, Keller U, Meer J, Trautmann-Grill K, Kerkhoff A, et al. Early Response to First-Line Anti-PD-1 Treatment in Hodgkin Lymphoma: a PET-Based Analysis from the Prospective, Randomized Phase II NIVAHL Trial. Clin Cancer Res. 2021;27(2):402–407. doi: 10.1158/1078-0432.CCR-20-3303. [DOI] [PubMed] [Google Scholar]
- 51.Agrawal V, Su M, Huang Y, Hsing M, Cherkasov A, Computer-Aided ZY. Discovery of Small Molecule Inhibitors of Thymocyte Selection-Associated High Mobility Group Box Protein (TOX) as Potential Therapeutics for Cutaneous T-Cell Lymphomas. Molecules. 2019;24(19):3459. doi: 10.3390/molecules24193459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


