Abstract
Background
Cryptosporidiosis outbreaks in South America are poorly documented. In March 2018, 51 cases of cryptosporidiosis were reported in Maripasoula, a village located in a remote forest area along the border between Surinam and French Guiana.
Method
To identify the origin of the epidemic, we performed epidemiological, microbiological, and environmental investigations. Only the cases involving diarrhoea and Cryptosporidium-positive stool were considered as bona fide, while cases involving diarrhoea and close contact with a confirmed case were classified as “possible”.
Results
We identified 16 confirmed cases and 35 possible ones. Confirmed cases comprised nine children (median age of 18 months, range: 6–21), one immunocompromised adult and six soldiers. One child required a hospitalisation for rehydration. All 16 Cryptosporidium stools were PCR positive, and sequencing of the gp60 gene confirmed only one Cryptosporidium hominis subtype IbA10G2. Tap water consumption was the only common risk factor identified. Contamination of the water network with Cryptosporidium parvum subtype IIdA19G2 was found.
Conclusion
Water quality is a major public health issue in Amazonian French Guiana, especially for population at risk (children, people with comorbidity, travelers). For them, alternative water supply or treatment should be implemented.
Introduction
The apicomplexan protozoan Cryptosporidium is distributed worldwide and is one of the most common waterborne pathogens [1]. It is the second leading cause of diarrheal death for children under the age of five, causing 12% of the total diarrhoea mortality burden [2]. Its transmission is fecal-oral and can occur through the ingestion of oocysts from untreated water (drinking or recreational) or food and by contact with infected persons or animals [3], with an incubation period of 5 to 7 days [4]. Ingestion of faecally-contaminated water has been responsible for large outbreaks [4,5]. Symptoms are non-specific and include diarrhoea, nausea, vomiting, lack of appetite and cramps, which complicates diagnosis of immunocompetent patients [6]. Symptoms may be protracted and severe in immunocompromised patients and in children–for whom they are frequently associated with severe watery diarrhoea and dehydration [7].
Cryptosporidium hominis and C. parvum are the primary causative agents of human cryptosporidiosis, but their prevalence varies in different regions of the world [3]. Transmission had already been reported along the Maroni river, a river flowing in the Amazonian forest between French Guiana and Surinam, although the mode of contamination was not well understood [8]. Maripasoula is an insulated border town between French Guiana and Surinam, an hour by plane or three days by canoe from the nearest road. A significant number of Maripasoula’s inhabitants live in precarious conditions. Epidemiological surveillance in Maripasoula consists in the monitoring of inhabitants requiring care for diarrhoea in the Delocalised Centre for Prevention and Care (CDPS). In March 2018, the physician in charge for infectious diseases at the CDPS alerted the Regional Health Authority in Cayenne about three successive immunocompetent children having cryptosporidiosis.
On April 24, 2018, epidemiological surveillance by the French army detected an increase in the incidence of cryptosporidiosis. This outbreak allowed further understanding of cryptosporidiosis transmission along the Maroni river, by identifying the mode of contamination and further developing appropriate countermeasures.
Materials and methods
Ethics statement
The study was approved by the ethics committee of the Andrée Rosemon Hospital, Cayenne (French Guiana). Patients’ medical charts were reviewed to collect demographic, exposure, clinical, biological, microbiological, and outcome data via anonymised and standardised forms. The collected database of medical charts was declared to the French Data Protection Authority (CNIL), no.1939018.
Case definition
A “confirmed case” was defined as a patient living or stationed in Maripasoula between January 1 and May 31, 2018, with gastrointestinal illness (diarrhoea or nausea or vomiting or abdominal pain), and a Cryptosporidium-positive stool test. Other patients with diarrhoea and in close contact with a confirmed case (same household and time) were defined as “possible cases” without further stool examination (Fig 1). Finally, “control cases” were subjects with similar environmental exposure (Maroni banks) but who did not present any symptom.
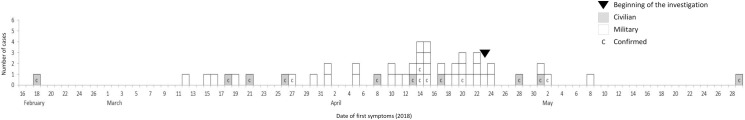
Fig 1. Epidemiological curve showing the distribution of confirmed (C) and possible cases over time (January 1—May 31, 2018).
Population study
To determine the most likely common source of contamination, we collected information from confirmed cases’ medical records regarding demographic characteristics, clinical manifestations, and laboratory results (Table 1). In addition, a questionnaire was used to collect information regarding food exposures, water consumption, contact with animals and relationship/contact between patients. Places of residence were also collected and placed on a map to identify areas of interest (Fig 2). Military and police personnel were fully available, and a retrospective cohort study was possible among confirmed and control cases. Poisson regression enabled us to identify the most probable source of contamination.
Table 1. Confirmed cases: Demographic characteristics, clinical manifestations and laboratory results.
MPA: Maripasoula.
| Civilians | Soldiers | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | |
| Date of first symptoms (2018) | 18 February | 18 March | 21 March | 26 March | 8 April | 13 April | 17 April | 28 April | 1 May | 29 May | 27 March | 14 April | 14 April | 15 April | 20 April | 02 May |
| length of illness | 1 day | 13 days | 26 days | 2 days | 11 days | 22 days | 3 days | Unknown | 8 days | 68 days | 17 days | Unknown | 19 days | 3 days | 5 days | 4 days |
| Age | 12 months | 6 months | 21 months | 18 months | 19 months | 19 months | 11 months | 12 months | 60 years | 20 months | 34 years | 48 years | 22 years | 30 years | 23 years | 21 years |
| Sex | Male | Female | Female | Female | Male | Male | Male | Male | Male | Male | Male | Male | Male | Male | Male | Male |
| Urban area | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA | MPA |
| District | Abdallah | Montagne | Montagne | Abdallah | Djakata | Djakata | Montagne | Abdallah | Abdallah | Sophie | Abdallah | Abdallah | Sophie | Abdallah | Sophie | Sophie |
| Comorbidity | No | No | No | No | No | No | No | Yes | Yes | No | No | No | No | No | No | No |
| Immunosuppressive factors | No | No | No | No | No | No | No | Yes (1) | Yes (2) | No | No | No | No | No | No | No |
| Symptoms | ||||||||||||||||
| Fever (3) | Yes | Yes | Yes | No | Yes | No | No | No | No | No | Yes | No | No | No | No | No |
| Diarrhoea | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Vomiting | Yes | No | No | No | Yes | No | No | No | Yes | Yes | No | No | Yes | Yes | No | No |
| Hospitalization | No | No | No | No | No | No | No | Yes | No | Yes | No | No | No | No | No | No |
| Nitazoxanide TUA | No | No | No | No | No | No | No | Yes | No | No | No | No | No | No | No | No |
| Coinfection | No | Yes | Yes | No | No | No | No | No | No | No | No | No | No | No | No | No |
| Sample date | - | 21 March | 22 March | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Species | - | Salmonella spp. | Norovirus | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Consumption of regular tap water | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Contact with animals | No | No | No | No | No | No | No | No | No | No | No | No | No | No | No | No |
MPA: Maripasoula
(1) Malnutrition
(2) Kidney transplant recipient
(3) Temperature > 38°C
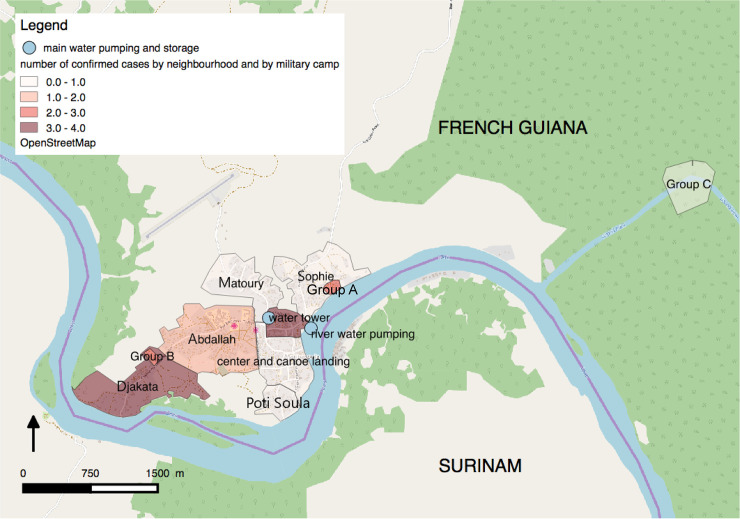
Fig 2. Map of Maripasoula.
Spatial distribution of cases and the water catchment area. The map was generated in QGIS software (QGIS Development Team (2020). QGIS Geographic information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org). The data on the map comes from the CDPS database and the military health service.
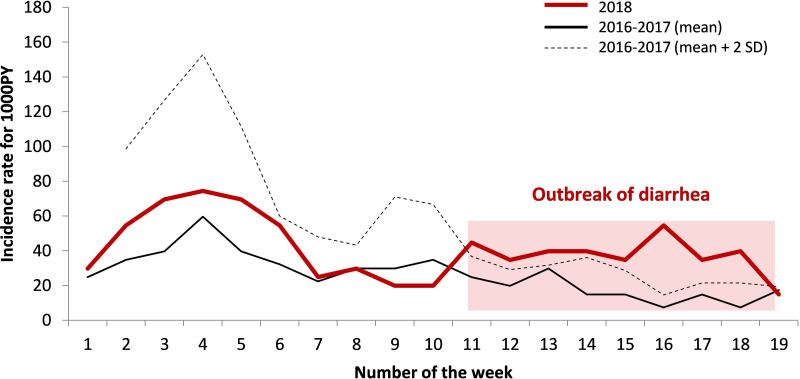
Data on diarrhoeal cases from the Maripasoula CDPS monitoring system were analysed during the outbreak and compared with the two previous years (Fig 3). We used the data from the French national institute for statistics and economic studies (Institut national de la statistique et des études économiques–INSEE) [9] concerning the official population of the municipality of Maripasoula which was accurate as of January 1, 2016 (12,798) to calculate the attack rate.
Fig 3. Incidence of diarrhoeal cases in the Maripasoula health centre in 2018 compared to the two previous years (for 1,000 persons-year).
The number of weeks correspond to the year 2018 i.e. from January 1 to May 13, 2018.
Fecal sample collections and parasite detection
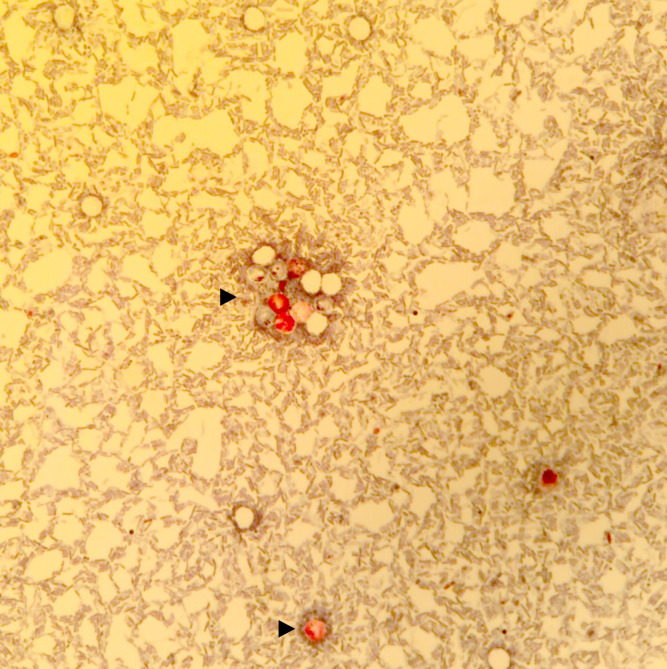
Between, January 1 and May 31, 2018, 103 faecal samples from Maripasoula were examined at the Parasitology Laboratory of Andrée Rosemon Hospital, Cayenne. A Bailanger’s concentration method and Kato-Katz technique were carried out for intestinal parasites identification. For the identification of Cryptosporidium oocysts, a modified Ziehl–Neelsen stain (Fig 4) was performed on the concentration pellet of the enrichment performed with the Paraprep L SAF kit (Diamodal, Vienna, Austria) [10,11]. To account for the possibility of other infectious aetiologies, stool specimens were also tested using bacterial culture and rapid diagnosis test screening for adenovirus, rotavirus (Adeno-Rota color, Servibio, Les Ulis, France) and norovirus (Norovirus GenI and GenII, Servibio, Les Ulis, France).
Fig 4. Stool sample after modified Ziehl-Neelsen stain.
Cryptosporidium spp. oocysts are stained fuchsia pink (500x magnification). Isolated or grouped oocysts are indicated by black arrows.
All stool samples were sent to the French national reference center in France. DNA extraction was directly performed from received stools without any previous concentration. Two molecular methods were performed after DNA extraction according to manufacturer’s instructions using a NucliSENS easyMAG device (bioMérieux, Marcy l’Etoile, France) [12]. An in-house real-time polymerase chain reaction (RT-PCR) assay was set up to enable the detection and identification of the most common Cryptosporidium species/genotypes based on differences in the melting temperatures of the PCR-probe complexes [13]. The technique was validated using DNA from characterised Cryptosporidium genus and positive controls for C. parvum and C. hominis [14]. All isolates were further subtyped using a gp60-based tool. GP60 subtypes were identified according to the protocol described by Sulaiman et al. [15]. Briefly, a nested PCR was performed using primers: AL3531 (5’-ATAGTCTCCGCTGTATTC-3’)/AL3533 (5’-GAGATATATCTTGGTGCG-3’) and secondly AL3532 (5’-TCCGCTGTATTCTCAGCC-3’)/LX0029 (5’-CGAACCACATTACAAATGAAGT-3’). Thermocycling conditions were: 94°C for 3 min, followed by 40 cycles of 94°C for 45 s, 54°C for 45s and 72°C for 60 s and a final step at 72°C for 7 min. Sequencing was performed using an AB3500 automated sequencer (Applied Biosystems, Illkirch, France) [16]. The nucleotide sequences obtained categorised C. parvum and C. hominis to many families of subtypes, by aligning the gp60 sequences obtained and reference sequences retrieved from GenBank.
Water collection and parasite detection
The drinking water in Maripasoula was pumped from two main sources: the Maroni river (≈70%) and the ground (≈30%). These sources were then mixed at the water tower and distributed throughout the town. Before storage and distribution, the water from the Maroni was sand filtered, flocculated, and chlorinated (>1.0 mg/L). The water from the ground was not treated and mixed with Maroni treated water. The wells were temporarily shut down between December 2017 and March 2018, but ground water was nevertheless sampled. Two samples were taken from the water tower on May 29, 2018: untreated water coming from the Maroni river (100 L) and post-treatment water (100 L). A peripheral tap water sample was taken from a water fountain (100 L) in the police settlement. For each sample, water was filtered through a standardised 1-micron filter Envirocheck sampling capsule (PALL, Saint-Germain-en-Laye, France). All samples were sent to the national reference center in France. Each received Envirocheck sampling capsule was eluted twice using 120 mL of an elution solution composed of 750 μl of Tween 80 (previously 10-fold diluted in PBS) (Sigma-aldrich, Vienne, Austria) and 750 μl of antifoam B emulsion (previously 10-fold diluted in PBS) (Sigma-aldrich, Vienne, Austria) in PBS 1X (Sigma-aldrich, Vienne, Austria) qsp 500mL.
Eluates were centrifugated at 2500 rpm for 30 minutes at +4°C. The oocysts potentially encountered in pellets of centrifugation were isolated with immunocapture using magnetic beads covered with specific antibodies from the walls of Cryptosporidium spp. oocysts (Isolate Cryptosporidium, TCS Bioscience Ltd, Buckingham, United Kingdom). They were then eluted (100 μl final) following the manufacturer’s recommendations. Ten microliters of the suspension were used for microscopic observation of the oocysts with immunofluorescence methods using a mixture of anti-Cryptosporidium monoclonal antibodies labeled with fluorescein isothiocyanate (FITC) (Crypto-Cell IF; Cellabs, Australia), following manufacturer’s recommendations.
DNA extraction from environmental samples was conducted on the 90μL remaining suspensions using QIAmpPowerFecal DNA Kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations. An RT-PCR amplifying 166 bp from the LIB13 locus was carried out to detect and differentiate the species present according to method described by Hadfield et al. [17]. Sequencing was performed as previously described for stool investigations [16].
Results
Outbreak and patients’ description
Between January 1 and May 31, 2018, we diagnosed 16 confirmed cases: nine in immunocompetent children, six among military and police personnel, and one was a 60-years-old immunocompromised man (a kidney transplant patient) (Fig 1 and Table 1). In addition, there were 35 possible cases among military and police personnel.
The global attack rate was 0.12% (16/12,798). It was 0.94% (9/962) in 0-5-aged-children, and 26% (41/157) among military and police personnel.
The median age was 18 months for children [range 6–21 months], and 28 years for military and police personnel [range 20–50 years]. One child had comorbidity and was hospitalized for severe malnutrition, and two needed hospitalization to be rehydrated, one of whom was treated with Nitazoxanide. No adult patient requires hospital care. Symptoms were diarrhoea (9/9), fever (4/9) and vomiting (3/9) among children, and diarrhoea (6/7), vomiting (5/7), abdominal pain (4/7), headache (2/7) and fever (1/7) among adults; median duration of symptoms were 11 and five days respectively. All patients eventually recovered.
Cryptosporidium hominis subtype IbA10G2 was found in all stool samples (Genbank accession number of the representative sequence OK032157). Two children had co-infections with Salmonella spp. and norovirus, respectively.
Confirmed civilian cases came from different districts of the city and did not share obvious similarities besides tap water consumption (Fig 2).
Retrospective cohort study
Military and police personnel were divided into three groups. Group A and B stayed in two different settlements in Maripasoula, without any contact, and with different food supply except the consumption of tap water. Group C was 30 minutes upstream from Maripasoula, their food was the same as Group B, but water was supplied from bottled-water. The retrospective cohort study showed that tap-water consumption was at risk for cryptosporidiosis (RR = 9.9, 95% CI = [2.5–65.7], Table 2). None of them had any contact with animals or with recreational water in the Maroni during the incubation period.
Table 2. Poisson regression about cryptosporidiosis factors of exposure, results from the retrospective cohort study among military and police personnel in Maripasoula (n = 157), French Guiana, 2018.
| Factor of exposure | Cases | Total number exposed | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|---|
| RR | 95%CI | p | RR | 95%CI | p | |||
| Tap water (DA = 157) | ||||||||
| no | 5 | 72 | ref | |||||
| yes | 41 | 85 | 1.8 | 1.5–2.3 | <10 −5 | 9.9 | 2.5–65.7 | 0.004 |
| Bottled water (DA = 132) | ||||||||
| no | 12 | 28 | ref | |||||
| yes | 24 | 104 | 0.7 | 0.5–1.0 | 0.04 | 0.6 | 0.3–1.2 | 0.12 |
| Local food (DA = 155) | ||||||||
| no | 29 | 129 | ref | |||||
| yes | 15 | 26 | 1.8 | 1.2–2.9 | <10−3 | 1.3 | 0.6–3.0 | 0.43 |
| Raw vegetables, fruits (DA = 153) | ||||||||
| no | 15 | 95 | ref | |||||
| yes | 27 | 58 | 1.6 | 1.2–2.0 | <10−3 | 1.2 | 0.6–3.0 | 0.62 |
| Contact with animals (DA = 157) | ||||||||
| no yes |
0 0 |
0 0 |
- | - | - | - | - | - |
| Recreational water (DA = 157) | ||||||||
| no yes |
0 0 |
0 0 |
- | - | - | - | - | - |
DA = data available; ref: reference
Investigation of the drinking water system
According to our investigation, Maroni’s surface water was at considerable risk of pollution: water was pumped from the middle of downtown Maripasoula, the water catchment point was located at few meters away from the riverbanks, and the security perimeter around the water catchment point was not respected, highly exposed to contamination from dirty and runoff waters of the town. A review of the water treatment system (sand filtration and chlorination) determined that it was ineffective in case of substantial contamination.
No oocyst was detected by microscopy. Two samples of the three were found positive by molecular methods for C. parvum: the post-treatment water (Ct: 38.03) and the drinking water (Ct: 39.01) from Maripasoula’s water distribution system. Ground water was found to be negative. Only one sample could be analysed (the post-treatment water, subtyping using gp60 gene), and revealed the presence of Cryptosporidium parvum subtype IIdA19G2 (Genbank accession number of the representative sequence OK032156).
Outbreak controls measures
In June 2018, the company responsible for the water supply in Maripasoula improved the water treatment system, specifically regarding the maintenance and repair of pumps, pre-treatment pH-adjustment to optimise the coagulation process and further improve water clarification, sand filters and tank cleaning (resulting in a temporary increase in water chlorination). Displacement of the water catchment point was planned to move further upstream on the Maroni river. Military camps were supplied with bottled water from May 24 to the end of June 2018, then UV lights system were installed. Only a new case of cryptosporidiosis was reported in September 2018. Stool sample genotyping revealed C. hominis subtype IbA10G2, the same genotype found during the epidemic. The sample was taken from a 9-month-old male who had presented diarrhoea for 24 hours. His only risk factor was the consumption of soups prepared with tap water, although the hypothesis of contact with an asymptomatic carrier could not be ruled out as a source of exposure.
Discussion
This outbreak of cryptosporidiosis in French Guiana was an opportunity to better understand cryptosporidiosis transmission in the Amazonian forest ecosystem. In the present study, the identification of C. hominis subtype IbA10G2 in all the stool samples suggested a common source of contamination. This subtype is the most commonly identified subtype in waterborne cryptosporidiosis outbreaks worldwide [18–20], and it has be shown to be more virulent than other subtypes resulting in intensive transmission [20]. Data in South America remains scarce. Environmental studies carried out in Colombia and Brazil have reported the presence of a great spectrum of Cryptosporidium spp. in raw and treated water as well as in animal reservoirs, including C. parvum [21–23]. Other molecular studies have reported genetic diversity of Cryptosporidium in human stool samples from Argentina, Brazil and Peru, each time C. hominis subtype IbA10G2 was identified; including two outbreaks in Peru and Columbia [20,24–27]. Therefore, it is the predominant subtype in South America. In our study, available evidence indicated a waterborne source of contamination through the public water supply network. Tap water was the unique common link between civilians, military personnel, and police personnel. Results of the retrospective cohort study were concordant with this statement. Human transmission was not suspected as none of the groups had contacts together. Contact with animals was not reported by any patients; which was in keeping with the subtype involved which excluded zoonotic transmission [28]. These findings were consistent with the deficiencies found by the investigation into the drinking water system. There must have been an initial contamination of the resource (which was exposed to the effluents), followed by a failure in the treatment system. Indeed, the environmental investigation found contamination of the water network with C. parvum subtype IIdA19G2. Isolation of a different species from that found in the stool of the confirmed cases can be explain by the sampling delay, two months after the outbreak, knowing that the concentration of cryptosporidia is highly variable in the environment. Isolating these protozoa from the water was laborious, there was no standardised method for catching and detecting parasites at the environmental level [11,29]. Molecular and genetic analyses have now been developed [30], but these techniques are dependent on the quality and number of the samples received. Unfortunately, only four samples could be processed because of logistical issues: we used all the filters available, and we had to send samples from the depths of the Amazonian forest to metropolitan France within 72 hours at 4°C, which is costly. Moreover, the detection threshold of the PCR we used was 100 times better for C. parvum than for C. hominis [31] which can explain why only C. parvum was found in the water samples. Due to all these constraints, the contamination of the water network as the source of the outbreak remained the main hypothesis.
This outbreak highlighted the need to enhance drinking water treatment. Slifko et al. reported that Cryptosporidium was resistant to disinfection with chlorine at concentrations typically applied in drinking water treatment plants (2 to 6 mg/L) [32]. However, alternative methods of disinfection are available such as pulsed UV light [33], or slow sand filtration [34], which is a pragmatic but effective method. The security of the resource is also primordial and should be moved upstream Maripasoula. However, in order to prevent these infections in low-resource populations, one of the inexpensive public advices for residents and visitors to these areas is to boil the water for one to three minutes to eliminate Cryptosporidium oocysts [35].
In this outbreak, the children affected by the epidemic were under two years old [36,37]. Cryptosporidium is among the leading causes of moderate-to-severe diarrhoea in children under two years-old, and may be under recorded when asymptomatic or associated with other pathogens [38]. Severe cases generally involve immunocompromised patients, infants or patients with comorbidities [38,39]. On the other hand, the military and police personnel were "naive" adults arriving in a new environment, and it could have been their first contact with the pathogen [40]. They were therefore more vulnerable, explaining the high attack rate observed among them.
Conclusion
We report the second epidemic of cryptosporidiosis reported in French Guiana since 2015 [8], with some severe cases among children, and a high attack rate among temporary residents (here the military and police personnel). Water quality is a real public health problem in Amazonian forest, especially for population at risk (children, people with comorbidity, travelers). For them, alternative water supply or improved treatment should be implemented.
Key learning points
This outbreak in the Amazonian forest highlights a waterborne transmission of Cryptosporidium hominis which is consistent with previously published data.
Water quality and water treatment is a real public health issue in Amazonian forest, especially for populations at risk and “naïve” adults.
For these particular populations it is necessary to set up means of prevention for cryptosporidiosis, such as bottled or boiled water consumption.
Top five papers
Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004–2010. Water Res. 2011;45: 6603–6614.
Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015;15: 85–94.
Chappell CL, Okhuysen PC, Sterling CR, Wang C, Jakubowski W, Dupont HL. Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg. 1999;60: 157–164.
Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, et al. Genetic Recombination and Cryptosporidium hominis Virulent Subtype IbA10G2. Emerg Infect Dis. 2013;19: 1573–1582.
Chyzheuskaya A, Cormican M, Srivinas R, O’Donovan D, Prendergast M, O’Donoghue C, et al. Economic Assessment of Waterborne Outbreak of Cryptosporidiosis. Emerg Infect Dis. 2017;23: 1650–1656.
Acknowledgments
We are sincerely grateful to Dr. Philippe Tabard, Dr Alice Sanna, Damien Brelivet (Agence Régionale de la Santé de la Guyane, Cayenne, French Guiana) and Luisiane Carvalho (CIRE), for their help with managing the outbreak management. Our thanks also go to Dr. Elise Martin and Dr. Fanny Henaff (Paediatric department,CHAR, Cayenne, French Guiana) who managed hospitalised cases; Dr. Paul Brousse and Basma Guarmit (CDPS, French Guiana) for their help with the investigation and data collection.
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004–2010. Water Res. 2011;45: 6603–6614. doi: 10.1016/j.watres.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 2.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388: 1459–1544. doi: 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15: 85–94. doi: 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horne S, Sibal B, Sibal N, Green HK. Cryptosporidium outbreaks: identification, diagnosis, and management. Br J Gen Pract. 2017;67: 425–426. doi: 10.3399/bjgp17X692501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mac Kenzie WR, Hoxie NJ, Proctor ME, Gradus MS, Blair KA, Peterson DE, et al. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331: 161–167. doi: 10.1056/NEJM199407213310304 [DOI] [PubMed] [Google Scholar]
- 6.Chalmers RM, Davies AP. Minireview: clinical cryptosporidiosis. Exp Parasitol. 2010;124: 138–146. doi: 10.1016/j.exppara.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 7.Mmbaga BT, Houpt ER. Cryptosporidium and Giardia Infections in Children: A Review. Pediatr Clin North Am. 2017;64: 837–850. doi: 10.1016/j.pcl.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 8.Mosnier E, Martin N, Razakandrainibe R, Dalle F, Roux G, Buteux A, et al. Cryptosporidiosis Outbreak in Immunocompetent Children from a Remote Area of French Guiana. Am J Trop Med Hyg. 2018;98: 1727–1732. doi: 10.4269/ajtmh.17-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilan démographique 2015—Insee Première—1581. [cited 23 Apr 2019]. Available: https://www.insee.fr/fr/statistiques/1908103
- 10.Casemore DP, Armstrong M, Jackson B, Nichols G, Thom BT. Screening for Cryptosporidium in stools. Lancet. 1984;1: 734–735. doi: 10.1016/S0140-6736(84)92245-1 [DOI] [PubMed] [Google Scholar]
- 11.Khurana S, Chaudhary P. Laboratory diagnosis of cryptosporidiosis. Trop Parasitol. 2018;8: 2–7. doi: 10.4103/tp.TP_34_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mary C, Chapey E, Dutoit E, Guyot K, Hasseine L, Jeddi F, et al. Multicentric evaluation of a new real-time PCR assay for quantification of Cryptosporidium spp. and identification of Cryptosporidium parvum and Cryptosporidium hominis. J Clin Microbiol. 2013;51: 2556–2563. doi: 10.1128/JCM.03458-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valeix N, Costa D, Basmaciyan L, Valot S, Vincent A, Razakandrainibe R, et al. Multicenter Comparative Study of Six Cryptosporidium parvum DNA Extraction Protocols Including Mechanical Pretreatment from Stool Samples. Microorganisms. 2020;8. doi: 10.3390/microorganisms8091450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalle F, Roz P, Dautin G, Di-Palma M, Kohli E, Sire-Bidault C, et al. Molecular characterization of isolates of waterborne Cryptosporidium spp. collected during an outbreak of gastroenteritis in South Burgundy, France. J Clin Microbiol. 2003;41: 2690–2693. doi: 10.1128/JCM.41.6.2690-2693.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43: 2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa D, Razakandrainibe R, Valot S, Vannier M, Sautour M, Basmaciyan L, et al. Epidemiology of Cryptosporidiosis in France from 2017 to 2019. Microorganisms. 2020;8. doi: 10.3390/microorganisms8091358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadfield SJ, Robinson G, Elwin K, Chalmers RM. Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of real-time PCR. J Clin Microbiol. 2011;49: 918–924. doi: 10.1128/JCM.01733-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalmers RM, Hadfield SJ, Jackson CJ, Elwin K, Xiao L, Hunter P. Geographic Linkage and Variation in Cryptosporidium hominis. Emerg Infect Dis. 2008;14: 496–498. doi: 10.3201/eid1403.071320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widerström M, Schönning C, Lilja M, Lebbad M, Ljung T, Allestam G, et al. Large Outbreak of Cryptosporidium hominis Infection Transmitted through the Public Water Supply, Sweden. Emerg Infect Dis. 2014;20: 581–589. doi: 10.3201/eid2004.121415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, et al. Genetic Recombination and Cryptosporidium hominis Virulent Subtype IbA10G2. Emerg Infect Dis. 2013;19: 1573–1582. doi: 10.3201/eid1910.121361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez C, López MC, Galeano LA, Qvarnstrom Y, Houghton K, Ramírez JD. Molecular detection and genotyping of pathogenic protozoan parasites in raw and treated water samples from southwest Colombia. Parasit Vectors. 2018;11: 563. doi: 10.1186/s13071-018-3147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Triviño-Valencia J, Lora F, Zuluaga JD, Gomez-Marin JE. Detection by PCR of pathogenic protozoa in raw and drinkable water samples in Colombia. Parasitol Res. 2016;115: 1789–1797. doi: 10.1007/s00436-016-4917-5 [DOI] [PubMed] [Google Scholar]
- 23.Inácio SV, Widmer G, de Brito RLL, Zucatto AS, de Aquino MCC, Oliveira BCM, et al. First description of Cryptosporidium hominis GP60 genotype IkA20G1 and Cryptosporidium parvum GP60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet Parasitol. 2017;233: 48–51. doi: 10.1016/j.vetpar.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 24.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, et al. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 2008;14: 1567–1574. doi: 10.3201/eid1410.071273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolando RFR, Silva S da, Peralta RHS, Silva AJ da, Cunha F de S, Bello AR, et al. Detection and differentiation of Cryptosporidium by real-time polymerase chain reaction in stool samples from patients in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2012;107: 476–479. doi: 10.1590/s0074-02762012000400006 [DOI] [PubMed] [Google Scholar]
- 26.Peralta RHS, Velásquez JN, Cunha F de S, Pantano ML, Sodré FC, Silva S da, et al. Genetic diversity of Cryptosporidium identified in clinical samples from cities in Brazil and Argentina. Mem Inst Oswaldo Cruz. 2016;111: 30–36. doi: 10.1590/0074-02760150303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvan-Diaz AL, Bedoya-Urrego K, Medina-Lozano A, Uran-Velasquez J, Alzate JF, Garcia-Montoya G. Common occurrence of Cryptosporidium hominis in children attending day-care centers in Medellin, Colombia. Parasitol Res. 2020;119: 2935–2942. doi: 10.1007/s00436-020-06782-5 [DOI] [PubMed] [Google Scholar]
- 28.Feng Y, Ryan UM, Xiao L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018. doi: 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 29.Hong S, Kim K, Yoon S, Park W-Y, Sim S, Yu J-R. Detection of Cryptosporidium parvum in environmental soil and vegetables. J Korean Med Sci. 2014;29: 1367–1371. doi: 10.3346/jkms.2014.29.10.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slifko TR, Smith HV, Rose JB. Emerging parasite zoonoses associated with water and food. Int J Parasitol. 2000;30: 1379–1393. doi: 10.1016/s0020-7519(00)00128-4 [DOI] [PubMed] [Google Scholar]
- 31.Costa D, Soulieux L, Razakandrainibe R, Basmaciyan L, Gargala G, Valot S, et al. Comparative Performance of Eight PCR Methods to Detect Cryptosporidium Species. Pathogens. 2021;10: 647. doi: 10.3390/pathogens10060647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rochelle PA, Marshall MM, Mead JR, Johnson AM, Korich DG, Rosen JS, et al. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl Environ Microbiol. 2002;68: 3809–3817. doi: 10.1128/AEM.68.8.3809-3817.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garvey M, Farrell H, Cormican M, Rowan N. Investigations of the relationship between use of in vitro cell culture-quantitative PCR and a mouse-based bioassay for evaluating critical factors affecting the disinfection performance of pulsed UV light for treating Cryptosporidium parvum oocysts in saline. J Microbiol Methods. 2010;80: 267–273. doi: 10.1016/j.mimet.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 34.Hijnen WAM, Dullemont YJ, Schijven JF, Hanzens-Brouwer AJ, Rosielle M, Medema G. Removal and fate of Cryptosporidium parvum, Clostridium perfringens and small-sized centric diatoms (Stephanodiscus hantzschii) in slow sand filters. Water Res. 2007;41: 2151–2162. doi: 10.1016/j.watres.2007.01.056 [DOI] [PubMed] [Google Scholar]
- 35.Chyzheuskaya A, Cormican M, Srivinas R, O’Donovan D, Prendergast M, O’Donoghue C, et al. Economic Assessment of Waterborne Outbreak of Cryptosporidiosis. Emerg Infect Dis. 2017;23: 1650–1656. doi: 10.3201/eid2310.152037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kłudkowska M, Pielok Ł, Frąckowiak K, Paul M. Intestinal coccidian parasites as an underestimated cause of travellers’ diarrhoea in Polish immunocompetent patients. Acta Parasitol. 2017;62: 630–638. doi: 10.1515/ap-2017-0077 [DOI] [PubMed] [Google Scholar]
- 37.Certad G, Viscogliosi E, Chabé M, Cacciò SM. Pathogenic Mechanisms of Cryptosporidium and Giardia. Trends Parasitol. 2017;33: 561–576. doi: 10.1016/j.pt.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 38.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382: 209–222. doi: 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 39.Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health. 2018;6: e758–e768. doi: 10.1016/S2214-109X(18)30283-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chappell CL, Okhuysen PC, Sterling CR, Wang C, Jakubowski W, Dupont HL. Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg. 1999;60: 157–164. doi: 10.4269/ajtmh.1999.60.157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.