Abstract
CDC weak oxidizer group 2 (WO-2) consists of nine phenotypically similar human clinical isolates received by the Centers for Disease Control and Prevention between 1989 and 1998. Four of the isolates were from blood, three were from sputum, and one each was from bronchial fluid and maxillary sinus. All are aerobic nonfermentative, motile gram-negative rods with one to eight polar flagella per cell. All grew at 25 and 35°C and were positive for catalase, urease (usually delayed 3 to 7 days), citrate, alkalinization of litmus milk, oxidization of glycerol (weakly), and growth on MacConkey agar and in nutrient broth without NaCl. All except one strain were oxidase positive with the Kovács method, and all except one isolate weakly oxidized d-glucose. All were negative for oxidation of d-xylose, d-mannitol, lactose, sucrose, maltose, and 20 other carbohydrates, esculin hydrolysis, indole production, arginine dihydrolase, and lysine and ornithine decarboxylase. Only two of nine isolates reduced nitrate. Broth microdilution susceptibilities were determined for all strains against 13 antimicrobial agents. Most of the strains were resistant to ampicillin, extended-spectrum cephalosporins, and aminoglycosides, including gentamicin, tobramycin, and amikacin, but they varied in their susceptibility to fluoroquinolones. High-performance liquid chromatographic and mass spectrometric analyses of the WO-2 group identified ubiquinone-8 as the major quinone component. The percent G+C of the WO-2 strains ranged from 65.2 to 70.7% (thermal denaturation method). All shared a common cellular fatty acid (CFA) profile, which was characterized by relatively large amounts (7 to 22%) of 16:1ω7c, 16:0, 17:0cyc, 18:1ω7c, and 19:0cyc11-12; small amounts (1 to 3%) of 12:0 and 14:0; and eight hydroxy acids, 2-OH-12:0 (4%), 2-OH-14:0 (trace), 3-OH-14:0 (12%), 2-OH-16:1 (1%), 2-OH-16:0 (3%), 3-OH-16:0 (4%), 2-OH-18:1 (2%), and 2-OH-19:0cyc (3%). This profile is similar to the CFA profile of Pandoraea, a recently described genus associated with respiratory infections in cystic fibrosis patients (T. Coenye et al., Int. J. Syst. Evol. Microbiol., 50:887–899, 2000). Sequencing of the 16S rRNA gene (1,300 bp) for all nine strains indicated a high level (≥98.8%) of homogeneity with Pandoraea spp. type strains. DNA-DNA hybridization analysis (hydroxyapatite method; 70°C) confirmed the identity of WO-2 with the genus Pandoraea and assigned three strains to Pandoraea apista and three to Pandoraea pnomenusa, and identified three additional new genomospecies containing one strain each (ATCC BAA-108, ATCC BAA-109, ATCC BAA-110). This study also shows that Pandoraea isolates may be encountered in blood cultures from patients without cystic fibrosis.
The Special Bacteriology Reference Laboratory of the Centers for Disease Control and Prevention (CDC) receives for identification and classification bacterial isolates from reference laboratories throughout the United States. Since 1989, we have received nine phenotypically similar unidentified clinical isolates from seven different U.S. state health departments. The phenotypic profile of these strains most closely resembles an unclassified isolate described by Trotter et al. (Trotter's bacillus) (17), Acidovorax temperans, and Burkholderia cepacia genomovar III bv. c reported by Vandamme et al. (18). A morphologic and biochemical characterization of these isolates was presented in 1996 at the 8th International Congress of Bacteriology and Applied Microbiology Division, International Union of Microbiological Societies, and the provisional designation CDC weak oxidizer group 2 (WO-2) was assigned to the group (R. S. Weyant, D. G. Hollis, M. I. Daneshvar, J. G. Jordan, and C. W. Moss, 8th Int. Congr. Bacteriol. Appl. Microbiol. Div., Int. Union Microbiol. Soc., p. 29, 1996).
Although a relatively small number of isolates have been received, the most common source of isolation for WO-2 strains was blood, suggesting the potential to cause invasive disease. Three of the strains were isolated from patients with underlying conditions, including chronic obstructive pulmonary disease and cystic fibrosis, suggesting that these strains may act as opportunistic pathogens.
In 2000, Coenye et al. described a new genus, Pandoraea, of gram-negative nonfermenters isolated primarily from sputa of cystic fibrosis patients and from soil. This new genus contains five named species (P. apista, P. pulmonicola, P. pnomenusa, P. sputorum, and P. norimbergensis) and one unnamed species (3). Presented herein is a polyphasic analysis of the WO-2 group, including morphologic, biochemical, cellular fatty acid (CFA) composition, isoprenoid quinone content, DNA-DNA hybridization, 16S rRNA gene sequencing, percent guanine-plus-cytosine (G+C) content, and in vitro antimicrobial susceptibility determinations. These results demonstrate that WO-2 should be assigned to the genus Pandoraea, with strains representing P. apista, P. pnomenusa, and three additional new genomospecies.
MATERIALS AND METHODS
Bacterial strains.
The strains studied, along with their sources and geographic origins, are presented in Table 1. Trotter's bacillus was submitted in 1988 to CDC for identification, and the type strain and reference strains of Pandoraea species and A. temperans were generously provided by E. Falsen, Culture Collection, University of Göteborg, Göteborg, Sweden. The reference strain of B. cepacia genomovar III bv. c, LGM 12614 (NCTC 13010), was obtained from the Bacteria Collection, University of Ghent, Ghent, Belgium. Unless otherwise indicated, strains were cultured on heart infusion agar supplemented with 5% rabbit blood (RBA) (BBL Microbiology Systems, Cockeysville, Md.) and incubated at 35°C in a candle jar. All strains were stored as suspensions in defibrinated rabbit blood in liquid nitrogen.
TABLE 1.
Sources and demographic and clinical information for WO-2 strains and other taxa in this study
| Strain | Date received | Sender location | Patient age, sex | Source/clinical diagnosis |
|---|---|---|---|---|
| G3307 | 1989 | California | 66 yr, female | Blood/COPDa |
| G3308 | 1989 | California | 75 yr, female | Bronchial wash/NGb |
| G5056 | 1990 | Texas | 46 yr, male | Blood/septicemia |
| G5084 | 1990 | Georgia | Unknown age, female | Maxillary sinus/NG |
| G7835 | 1992 | Hawaii | 76 yr, male | Blood/bacteremiac |
| G8107 | 1993 | Louisiana | 49 yr, male | Blood/NG |
| G9278 | 1994 | Ohio | 35 yr, male | Sputum/cystic fibrosis |
| G9805 | 1996 | Utah | 71 yr, female | Sputum/pneumonia |
| H652 | 1998 | Ohio | Unknown age, female | Sputum/cystic fibrosis |
| G1565 (Trotter's bacillus) | 1988 | Oklahoma | 5 yr, male | Lung mass/CGDd |
| G8098e (CCUG 11779T, A. temperans) | 1993 | Göteborg, Sweden | 68 yr, male | Urine/NG |
| G8099 (CCUG 22215, A. temperans) | 1993 | Gavle, Sweden | NG | Wound secretion/NG |
| H857 (LGMf 12614, NCTC 13010, B. cepacia genomovar III biovar c) | 1998 | Ghent, Belgium | NG | NG/cystic fibrosis |
| H1766 (CCUG 39680T, Pandoraea species) | 2000 | NG | NG | Garden soil |
| H1767 (CCUG 38412T, P. apista) | 2000 | Denmark | NG | Sputum/cystic fibrosis |
| H1768 (CCUG 39188T, P. norimbergensis) | 2000 | Nürnberg, Germany | NG | Oxic water layer above a sulfide-containing sediment |
| H1769 (CCUG 38742T, P. pnomenusa) | 2000 | United Kingdom | NG | Sputum/cystic fibrosis |
| H1770 (CCUG 38759T, P. pulmonicola) | 2000 | Canada | NG | Sputum/cystic fibrosis |
| H1771 (CCUG 39682T, P. sputorum) | 2000 | USA | NG | NG/cystic fibrosis |
COPD, chronic obstructive pulmonary disease.
NG, not given.
Post-mitral valve replacement.
CGD, chronic granulomatous disease.
Culture Collection, University of Göteborg, Göteborg, Sweden; T, type strain.
Bacteria Collection, Ghent University, Ghent, Belgium.
Phenotypic tests.
Biochemical testing was done using the methods of the CDC Special Bacteriology Reference Laboratory (22). With the exception of the optimum growth temperature tests, all biochemical tests were performed at 35°C in an aerobic incubator. The oxidase, catalase, and growth temperature tests were read after 1 day of incubation. All other tests were read after 1, 2, and 7 days of incubation. A final reading of the gelatin tests was done after 14 days of incubation. Five isolates were tested for growth on Trypticase soy agar with 5% sheep blood (TSAS; BBL). All strains were tested for growth on B. cepacia-selective medium (Mast Diagnostics, Mast Group Ltd., Merseyside, United Kingdom).
CFA analysis.
Cells were saponified, and the liberated fatty acids were methylated and analyzed by capillary gas-liquid chromatography (GLC) (22). CFA profiles were identified using a commercially available system (MIDI, Newark, Del.). The amide-linked hydroxy acids that were not totally released by this saponification procedure were completely released by a subsequent acid hydrolysis of the methanolic aqueous layer after the methylation step (22). The identification of fatty acids and determination of double-bond positions in monounsaturated acids were accomplished by GLC and GLC-mass spectrometry (GLC-MS). The confirmation of hydroxy acids was accomplished by both acetylation and GLC-MS analysis, as described previously (22).
Isoprenoid quinone analysis.
Isoprenoid quinones were extracted from 100 mg of lyophilized cells and were analyzed by reverse-phase high-performance liquid chromatography (HPLC) and MS (9, 10). The retention times were obtained by HPLC. Peaks were collected and analyzed directly by electron-impact and chemical-ionization MS.
DNA relatedness and percent G+C determination.
All strains were cultured on 20 to 30 RBA plates and incubated for 24 h at 35°C. Cells were harvested and lysed, and the DNA was isolated and purified according to the method of Brenner et al. (2). DNA from strains G3307, G5056, G5084, and G9805 was labeled with [32P]dCTP using a commercial nick translation kit (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) and tested for reassociation to unlabeled DNA from the same strain (homologous reaction) as well as to other WO-2 strains and to the type and reference strains (heterologous reactions). The G+C content of B. cepacia has been reported to be 67%; therefore, 70°C was chosen for the optimal reassociation temperature. Relative binding ratios ([percent heterologous DNA bound to hydroxyapatite/percent homologous DNA bound to hydroxyapatite] × 100) and percent divergence (the percentage of unpaired bases in related DNA sequences) were calculated as described by Brenner et al. (2). Divergence was calculated to the nearest 0.5%, with each decrease of 1°C in thermal stability of a heterologous DNA duplex due to approximately 1% unpaired bases within related DNA (1). All reactions were done in duplicate at the optimal temperature of 70°C. The percent G+C was determined for strains G3307, G5056, G5084, G9805, and H652 by the thermal denaturation method of Mandel et al. (8).
16S rRNA gene sequencing.
Purified genomic DNA was diluted to 1 μg/ml in sterile water. Ten microliters of the diluted DNA was used in a 100-μl PCR mixture containing a 200 μM concentration of deoxynucleoside triphosphate, 1 mM MgCl2, 1× PCR buffer II (Perkin-Elmer, Foster City, Calif.), 0.1 μM FD1 primer, 0.1 μM RD1 primer, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). The primers FD1 and RD1 were originally described by Weisburg et al. as suitable for amplifying the 16S rRNA gene of many eubacteria (21). We omitted the linker sequences from the primers, as previously described (4). The parameters for the amplification were 94°C for 5 min, 35 cycles of 94°C for 15 s, 50°C for 15 s, 72°C for 1.5 min, and finally a 72°C extension for 5 min before cooling to 4°C. The results of the PCR were checked by running 10 μl of each reaction mixture on a 1% agarose gel. The amplicon was then purified and concentrated by using a QiaQuick PCR Purification kit from Qiagen (Valencia, Calif.). Approximately 60 ng of PCR product was used for each sequencing reaction. The sequencing reaction consisted of 16S DNA, 8 μl of ABI BigDye terminator from a cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.), 3.2 pmol of primer, and sterile water to a volume of 20 μl. The primer set used for sequencing was derived from those designed by Stackebrandt and Charfreitag (16). The manufacturer's instructions for the cycle sequencing kit were followed for the thermal cycler conditions. The extension products from each sequencing reaction were purified through a Centrisep column (Princeton Separations, Adelphia, N.J.) and dried in a vacuum centrifuge for 20 min. The sequencing reaction mixtures were resolved on a 4.25% acrylamide–6 M urea gel electrophoresed on an ABI 377 automated sequencer (PE Applied Biosystems). The sequence data were edited and compiled using the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.). The 16S sequence was aligned using the Wisconsin Sequence Analysis Package program PILEUP with 16S sequences of proteobacteria retrieved from GenBank, and the multiple sequence alignment was edited by hand to remove regions that were not represented by all members. The edited alignment was used in TREECON (version 1.3b) (19) to derive a phylogenetic dendrogram using the nucleotide substitution model of Jukes and Cantor (7) and the neighbor-joining method of Saitou and Nei (15).
In vitro antimicrobial susceptibility tests.
Antimicrobial susceptibility profiles were determined by the broth microdilution method described by the National Committee for Clinical Laboratory Standards (NCCLS) (11, 12), except that the results were read after a 24-h incubation (5). The strains were streaked onto TSAS and incubated for 18 to 24 h at 35°C in ambient air. Growth was taken from the plate with a sterile cotton-tipped swab and suspended in a tube of cation-adjusted Mueller-Hinton broth (Difco, Detroit, Mich.) to a density equivalent to 1.0 McFarland standard. Eight milliliters of the broth suspension was added to 32 ml of sterile distilled water and inoculated into MIC plates prepared at CDC according to the recommendations of the NCCLS (11, 12). Inoculation was performed with the MIC 2000 (Dynatech Laboratories, Chantilly, Va.). The final inoculum concentration was approximately 5 × 105 CFU/ml. The plates were incubated in ambient air for 24 h at 35°C. The following organisms were used as controls: Escherichia coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853. There were no quality control failures.
Nucleotide sequence accession numbers.
The 16S sequences for the nine WO-2 strains were submitted to GenBank and assigned the accession numbers AF247691 to AF247699.
RESULTS
The sources of all strains in the study are listed in Table 1. WO-2 isolates were received from seven different state public health laboratories in the United States between 1989 and 1998. No single geographic area predominated. Four of the isolates were obtained from blood, three were from sputum, and one each was from bronchial wash and maxillary sinus. Clinical information was available on seven patients and no common underlying syndrome was indicated, although significant conditions, including chronic obstructive pulmonary disease and cystic fibrosis, were reported for three of the patients. The age of the patients ranged from 35 to 76 years; five of the patients were female and four were male.
Results of DNA relatedness studies are given in Table 2. Using the established molecular criteria for species-level relatedness (strains whose DNAs are 70% or more related at optimal conditions and whose related sequences show 5% or less divergence) (20), five species-level hybridization groups were identified among the WO-2 strains. The first group contained strains G3307, G3308, G9278, and the type strain of P. apista. The relatedness within this group was greater than 83%, and the divergence was less than 3.5%. The second group consisted of G5056, G7835, G8107, and the type strain of P. pnomenusa. This group was slightly more diverse, with relatedness and divergence greater than 79% and less than 5.5%, respectively. Strains G5084, G9805, and H652 are not related at the species level with any of the other WO-2 or reference strains included in this study; however, their relatedness with P. apista, P. norimbergensis, P. pnomenusa, P. pulmonicola, and the unnamed Pandoraea species strain H1766 was substantially higher (>23%) than with P. sputorum and the other genera included in the study (<16%).
TABLE 2.
DNA relatedness of WO-2 strains, Pandoraea species, A. temperans, B. cepacia genomovar III bv. c, and Trotter's bacillusa
| Source of unlabeled DNA | Results of reaction with labeled DNA from strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G3307
|
G5056
|
G5084
|
G9805
|
H652
|
||||||
| RBR | D | RBR | D | RBR | D | RBR | D | RBR | D | |
| WO-2 G3307 | 100 | 0.0 | 45 | 10.5 | 37 | 9.5 | 35 | 8.5 | ||
| WO-2 G3308 | 99 | 0.0 | 43 | 10.5 | ||||||
| WO-2 G9278 | 90 | 3.0 | 34 | 12.0 | ||||||
| WO-2 G5056 | 57 | 12.5 | 100 | 0.0 | 34 | 9.5 | 27 | 9.0 | ||
| WO-2 G7835 | 42 | 13.5 | 87 | 5.0 | ||||||
| WO-2 G8107 | 42 | 12.5 | 80 | 3.0 | ||||||
| WO-2 G5084 | 56 | 10.5 | 47 | 10.0 | 100 | 0.0 | 38 | 7.5 | ||
| WO-2 G9805 | 49 | 11.0 | 38 | 11.0 | 42 | 9.5 | 100 | 0.0 | ||
| WO-2 H652 | 56 | 8.0 | 35 | 10.5 | 25 | 9.5 | 29 | 6.5 | 100 | 0.0 |
| Pandoraea sp. strain H1766 | 38 | 7.4 | 30 | 7.6 | 24 | 7.8 | 22 | 4.8 | 49 | 9.0 |
| P. apista H1767 | 83 | 0.3 | 34 | 7.8 | 28 | 8.2 | 27 | 5.9 | 59 | 8.3 |
| P. norimbergensis H1768 | 35 | 8.2 | 29 | 8.5 | 24 | 8.8 | 24 | 6.0 | 41 | 10.4 |
| P. pnomenusa H1769 | 34 | 8.6 | 83 | 0.2 | 27 | 8.5 | 26 | 5.5 | 46 | 10.0 |
| P. pulmonicola H1770 | 33 | 8.4 | 33 | 7.5 | 26 | 8.4 | 24 | 5.1 | 43 | 10.5 |
| P. sputorum H1771 | 8 | 8.5 | 4 | 9.9 | 4 | 9.8 | 4 | 6.7 | 5 | 13.8 |
| A. temperans G9808 | 6 | 15.5 | 2 | 14.5 | 0 | 16.5 | 1 | 11.0 | ||
| B. cepacia H857b | 10 | 16.5 | 6 | 15.5 | 1 | 15.5 | 8 | 9.5 | ||
| Trotter's bacillus G1563 | 15 | 15.0 | 10 | 15.5 | 3 | 15.5 | 5 | 11.0 | ||
RBR, relative binding ratio; D, percent divergence. Reactions were performed at 70°C.
H857, B. cepacia genomovar III bv. c.
The percent G+C was determined for a representative WO-2 strain of each DNA relatedness group, and values ranged from 65.2% for strain G5084 to 70.7% for strain G5056 (P. pnomenusa). The G+C content for G3307 (P. apista) was 69.2%, 67.1% for G9805, and 68.6% for H652. These values are slightly higher than those previously reported for Pandoraea (61.2 to 64.3%) (3).
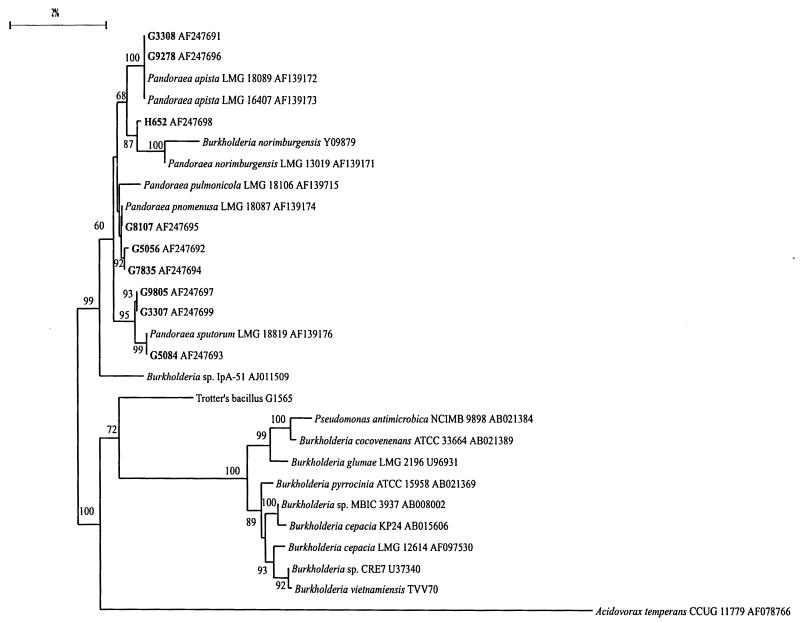
The 16S rRNA gene sequences of nine members of the WO-2 group, Trotter's bacillus, and B. cepacia genomovar III bv. c were determined and aligned with sequences of several Burkholderia and Pandoraea species from GenBank. The resulting phylogenetic tree (Fig. 1) demonstrates the close relationships between WO-2 strains and Pandoraea species. The WO-2 strains were 98.8 to 100% homologous to the reference Pandoraea strains across 1,300 bp of their 16S rRNA gene sequences. The 16S data correlated well with the DNA-DNA hybridization results. The only exception was seen with G3307. By DNA hybridization, G3307 was classified as P. apista, along with G3308 and G9278. However, the 16S rRNA gene sequence of this strain was identical to G9805, a unique genomospecies. The strains of the Pandoraea cluster were 98.4 to 98.8% similar to the sequence of an unnamed Burkholderia species, strain IpA-51 (GenBank accession number AJ011509) and 96.7 to 97.3% similar to the sequence from Trotter's bacillus. The low bootstrap value of the Trotter's bacillus branch indicates that the Trotter's bacillus sequence often was grouped with the Pandoraea strains when the tree was generated 100 separate times. The high bootstrap values for the Pandoraea branch and the Burkholderia branch indicate that in 100 trees there was little deviation from this representative tree. The Pandoraea strains and Burkholderia sp. strain IpA-51 formed a separate group from the Burkholderia species in 99% of the trees. The Pandoraea cluster, Burkholderia sp. strain IpA-51, and Trotter's bacillus were 95.4 to 96% similar to B. cepacia. The 16S ribosomal DNA sequence from Pseudomonas antimicrobica was 99.51% similar to that of Burkholderia cocovenenans, indicating a very close relationship of P. antimicrobica to the genus Burkholderia.
FIG. 1.
Phylogenetic tree including WO-2 strains and closest neighboring taxa.
The CFA compositions of the WO-2 strains, along with the Pandoraea reference strains, other phenotypically similar strains (Trotter's bacillus, B. cepacia genomovar III bv. c, and A. temperans), and a group of Burkholderia reference strains representing B. cepacia, B. gladioli, B. mallei, and B. pseudomallei (B. cepacia CFA group) are presented in Table 3. The B. cepacia CFA group and A. temperans profiles were previously published by our group after using the same methodology (22). All nine WO-2 strains shared a unique profile characterized by relatively large amounts (7 to 22%) of 16:1ω7c, 16:0, 17:0cyc, 18:1ω7c, and 19:0cyc11-12; small amounts (1 to 3%) of 12:0, 14:0 and 18:0; and eight hydroxy acids, 2-OH-12:0 (4%), 2-OH-14:0 (trace), 3-OH-14:0 (12%), 2-OH-16:1 (1%), 2-OH-16:0 (3%), 3-OH-16:0 (4%), 2-OH-18:1 (2%), and 2-OH-19:0cyc (3%). Upon acid hydrolysis of one representative strain, additional amounts of 2-OH-12:0 (2%), 3-OH-14:0 (5%), 2-OH-16:1 (1%), 2-OH-16:0 (1%), 3-OH-16:0 (9%), and 2-OH-19:0cyc (2%) were released, indicating that these acids are both ester and amide linked. No additional hydroxy acids were released upon acid hydrolysis.
TABLE 3.
Summary of CFA compositions of WO-2 strains, Pandoraea species, Trotter's bacillus, B. cepacia CFA group, B. cepacia genomovar III bv. c, and A. temperans
| Fatty acida | Mean % of total CFAb
|
|||||
|---|---|---|---|---|---|---|
| WO-2 | Pandoraea speciesc | Trotter's bacillusd | B. cepacia CFA groupc | B. cepacia genomovar III bv. cd | A. temperans | |
| 3-OH-10:0 | — | — | — | — | — | 3 |
| 12:0 | 3 | 3 | 3 | — | — | 4 |
| 13:0 | — | — | — | — | — | 1 |
| 2-OH-12:0 | 4 | 2e | — | — | — | — |
| 14:0 | 1 | 1 | T | 3 | 12 | 1 |
| 15:1ω6c | — | — | — | — | — | 6 |
| 15:0 | T | T | — | T | 1 | 6 |
| 2-OH-14:0 | T | — | — | T | T | — |
| 3-OH-14:0 | 12 | 6 | 6 | 5 | 9 | — |
| 16:1ω7c | 7 | 9 | 11 | 7 | 5 | 35 |
| 16:1ω5c | — | — | — | — | 1 | — |
| 16:0 | 22 | 29 | 27 | 25 | 21 | 23 |
| 17:0cyc | 16 | 14 | 6 | 14 | 21 | — |
| 17:0 | — | 1 | — | — | — | 8 |
| 2-OH-16:1 | 1 | T | T | 1 | 1 | — |
| 2-OH-16:0 | 3 | 1 | T | 3 | 6 | — |
| 3-OH-16:0 | 4 | 3 | 4 | 4 | 6 | — |
| 18:2 | — | — | — | — | 1 | — |
| 18:1ω9c | — | — | — | — | 1 | — |
| 18:1ω7c | 9 | 14 | 24 | 18 | 6 | 8 |
| 18:0 | 1 | 1 | 2 | 1 | 1 | 1 |
| 19:0cyc11-12 | 10 | 13 | 10 | 10 | 5 | — |
| 2-OH-18:1 | 2 | 1 | 4 | 2 | 1 | — |
| 2-OH-19:0cyc | 3 | T | 1 | 3 | 1 | — |
The number before the colon indicates the number of carbons; the number after the colon is the number of double bonds; ω is the position of the double bond counting from the hydrocarbon end of the carbon chain; OH indicates a hydroxy group at the 2(α) or 3(β) position from the carboxyl end; c, cis isomer; cyc, a cyclopropane ring structure.
T, trace (0.4 to 0.8%); —, not detected.
Pandoraea species includes a reference strain from each of P. apista, P. norimbergensis, P. pnomenusa, P. pulmonicola, P. sputorum, and other Pandoraea species. B. cepacia CFA group (22) includes type strains, reference strains, and phenotypically similar strains of Burkholderia (formerly Pseudomonas) cepacia, B. gladioli, B. mallei, and B. pseudomallei.
A strain phenotypically similar to CDC group WO-2.
2% present in all strains except CCUG 39680, in which none was detected.
The overall CFA profiles of WO-2 strains, the Pandoraea reference strains, Trotter's bacillus, the B. cepacia CFA group, and B. cepacia genomovar III bv. c are similar, in that 3-OH-14:0 (5 to 12%), 16:0 (21 to 29%), 17:0cyc (6 to 21%), 18:1ω7c (6 to 24%), and 19:0cyc11-12 (5 to 13%) predominate. Within this group, the WO-2 strains are most similar to the Pandoraea strains. The WO-2 and Pandoraea profiles can be differentiated from Trotter's bacillus by the presence of 2-OH-12:0 (4%) and the inversion of the 17:0cyc/18:1ω7c ratio. The major characteristics that differentiate the WO-2 and Pandoraea profiles from the B. cepacia CFA group and B. cepacia genomovar III bv. c are the presence of 12:0 (3%) and 2-OH-12:0 (2 to 4%). The A. temperans profile, characterized by major amounts of 16:1ω7c and 16:0 (23 to 35%), is easily differentiated from the others.
HPLC and MS data for the quinone extracts of a representative WO-2 strain from each Pandoraea species confirmed ubiquinone-8 (Q-8) as the major component. Although not all Pandoraea species were analyzed, our findings from P. apista, P. pnomenusa, and three additional genomospecies indicate that the quinone content of Pandoraea is similar to that of Burkholderia. The quinone content of Pandoraea is most similar to that of B. cepacia and B. gladioli, in that Q-8 is also the only quinone present in the latter two organisms (13). Therefore, the presence of Q-8, in combination with the described CFA profile, provides a genus-level chemical identification of Pandoraea.
All Pandoraea strains shared a common general phenotypic profile, regardless of species. Cells grown on heart infusion agar at 35°C for 18 to 24 h were short-to-medium-length straight gram-negative rods that were of medium width. Some cells produced a vacuolated or bipolar appearance. All grew heavily on RBA that was incubated either aerobically or in a candle jar atmosphere for 18 to 24 h. Five strains were also tested for growth on TSAS, and all grew at a rate and with a morphology similar to that obtained on RBA. Isolated colonies were circular, convex, semiopaque to opaque, entire, smooth, and 0.5 to 1 mm in diameter. Hemolysis was variable. No hemolytic reaction was observed after overnight incubation at 35°C with five strains; however, one P. pnomenusa strain and G9805 produced weak hemolysis, one P. apista strain produced a faint greenish discoloration, and another P. apista strain produced a partial beta-like hemolytic action underneath areas of confluent growth. All strains were positive for growth on MacConkey agar, production of catalase, alkalinization of citrate and litmus milk, growth at 25 and 35°C, and growth in nutrient broth. All strains were motile with polar flagella, ranging from one to as many as eight per cell, which were observed by using the Ryu flagellum stain (22). All strains were negative for hydrolysis of esculin and gelatin, indole, lysine and ornithine decarboxylase, and arginine dihydrolase. No acid production was observed in King's oxidation-fermentation (OF) base from mannitol, lactose, sucrose, maltose, salicin, l-arabinose, adonitol, dulcitol, d-galactose, fructose, mannose, rhamnose, trehalose, raffinose, sorbitol, inositol, cellobiose, inulin, dextrin, glycogen, erythritol, melibiose, melezitose, and starch. With one exception, all strains grew well on B. cepacia-selective medium at 35°C. One strain, assigned to P. apista (G9278), grew lightly at 35°C and the agar did not turn pink; however, at 28°C there was moderate growth and the agar turned pink.
Table 4 lists phenotypic characteristics that are useful in differentiating between the different Pandoraea species. Results are given for reference strains and the genetically identified WO-2 isolates. The reference strains of P. pulmonicola, P. sputorum, P. norimbergensis, and Pandoraea genomospecies 1 and 3 could be definitively identified using the tests listed. A nitrate-negative, cetrimide-negative P. pnomenusa strain might be confused with P. apista or Pandorea genomospecies 4. Likewise, a nitrate-negative P. pnomenusa strain that failed to grow on salmonella-shigella (SS) agar and cetrimide agar might be confused with Pandoraea genomospecies 2.
TABLE 4.
Differential phenotypic characteristics of Pandoraea strains included in this studya
| Test performedb | P. apista(n = 4) | P. pnomenusa(n = 4) | P. pulmonicola(n = 1) | P. sputorum(n = 1) | P. norimbergensis(n = 1) |
Pandoraea unnamed genomospecies
|
|||
|---|---|---|---|---|---|---|---|---|---|
| 1 (n = 1) | 2 (n = 1) | 3 (n = 1) | 4 (n = 1) | ||||||
| Acid from (OF base): | |||||||||
| d-glucose | (3w)/4 | (3w)/4 | − | +w | +w | − | (+w) | (+w) | (+w)b |
| Glycerol | (3w)/3 | (3w)/3 | +w | − | +w | +w | (+w) | (+w) | (+w) |
| d-xylose | 0/4 | 0/4 | − | +w | − | − | − | − | − |
| Oxidasec | 4/4 | 4/4 | + | + | + | + | + | − | + |
| Growth on SS agar | 1(1w)/4 | 1(1w)/4 | + | + | − | − | − | (w+) | (+) |
| Growth on cetrimide agar | 2(1w)/4 | 2/4 | + | + | − | − | − | − | − |
| Urea, Christensen's | 1(3)/4 | 4/4 | − | − | − | − | (+) | (+) | (+) |
| Nitrate | 0/4 | 3/4 | − | + | − | + | − | − | − |
| Growth at 42°C | 4/4 | 4/4 | + | + | − | − | + | + | + |
Fractions indicate number of strains positive within 48 h per total number of strains tested. Fractions in parentheses indicate number of strains positive within 7 days per total strains tested. Abbreviations: n, number of strains; +, positive; −, negative; w, weak reaction.
All strains were positive for catalase, growth on MacConkey agar and in nutrient broth, alkalinization of Simmons citrate and litmus milk, and growth at 25°C and 35°C. All strains were negative for reduction of nitrite, denitrification, indole production, gelatin and esculin hydrolysis, production of growth pigments, growth in nutrient broth with 6% NaCl, ornithine and lysine decarboxylase, arginine dihydrolase, and acid from salicin, l-arabinose, adonitol, dulcitol, d-galactose, fructose, d-mannose, l-rhamnose, trehalose, raffinose, d-sorbitol, inositol, cellobiose, inulin, dextrin, glycogen, erythritol, melibiose, melezitose, and starch.
Kovács method.
Table 5 lists the antimicrobial resistance profiles for the WO-2 isolates as classified into their Pandoraea designations. These isolates exhibited diverse antimicrobial susceptibility patterns. All of the organisms were resistant to ampicillin and cefazolin except for one strain of P. apista, G9278, and most were resistant to extended-spectrum cephalosporins, aztreonam, and piperacillin. The organisms also tended to be resistant to aminoglycosides, including gentamicin, tobramycin, and amikacin, but varied in their susceptibility to fluoroquinolones.
TABLE 5.
Antimicrobial resistance profiles of WO-2 strains classified as Pandoraeaa
| Antimicrobial agent | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
|
P. apista(n = 3)
|
P. pnomenusa(n = 3)
|
P. genomospecies 2 (n = 1) | P. genomospecies 3 (n = 1) | P. genomospecies 4 (n = 1) | |||
| Range | Mode | Range | Mode | ||||
| Ampicillin | 32–>64 | >64 | >64 | >64 | >64 | >64 | >64 |
| Amoxicillin-clavulanate | 32–>32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Amikacin | 16 | 16 | >64 | >64 | >64 | >64 | >64 |
| Cefazolin | 2–>32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Cefotaxime | 4–>64 | 4, 32, >64 NA | 32–>64 | >64 | >64 | 32 | >64 |
| Cefoxitin | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Chloramphenicol | 8–16 | 16 | 16 | 16 | 32 | 32 | 16 |
| Ciprofloxacin | 0.5–2 | 0.5, 1, 2, NA | 2–4 | 4 | 8 | 4 | >8 |
| Gentamicin | 8–16 | 16 | >16 | >16 | >16 | >16 | >16 |
| Imipenem | <1–4 | 4 | <1–2 | <1 | 2 | 2 | 32 |
| Meropenem | 1–>32 | >32 | >32 | >32 | >32 | >32 | >32 |
| Sparfloxacin | 0.12–0.5 | 0.5 | 0.5–1 | 1 | >2 | 1 | >2 |
| Tetracycline | 2–4 | 2 | 4–8 | 4 | 16 | 8 | 8 |
| Tobramycin | 4–8 | 8 | >16 | >16 | >16 | >16 | >16 |
n, number of strains; NA, not applicable. Values are given as micrograms per milliliter.
DISCUSSION
The genus Pandoraea represents a recently described group of nonfermentative gram-negative rods isolated predominately from soil and from sputa of individuals with cystic fibrosis (3). The original description of this genus identified four new species, all consisting of primarily sputum isolates (P. apista, P. pulmonicola, P. pnomenusa, P. sputorum), and reclassified Burkholderia norimbergensis as a new Pandoraea species, P. norimbergensis. One unnamed genomospecies, consisting of one sputum isolate (referred to in this study as Pandoraea unnamed genomospecies 1), was also described. The closest phylogenetic relative to Pandoraea is Burkholderia.
The findings of this study support the findings of the original Pandoraea study and assign the nonfermenters referred to as CDC WO-2 to this genus. The WO-2 group includes nine similar clinical isolates received by the CDC Special Bacteriology Reference Laboratory since 1989. Six of the nine WO-2 strains were isolated either from a respiratory site or from a patient with underlying respiratory disease, i.e., cystic fibrosis or chronic obstructive pulmonary disease. Using DNA-DNA hybridization analysis with Pandoraea type strains, three WO-2 isolates were found to be P. apista, three were found to be P. pnomenusa, and three others were found to represent unique Pandoraea genomospecies.
The assignment of the WO-2 group to the genus Pandoraea suggests a broader pathogenic potential for this genus than was previously recognized. Only two of the nine patients from which these organisms were isolated had cystic fibrosis listed as an underlying disease. Additionally, the most common site of isolation of the WO-2 isolates was blood (four isolates), demonstrating a potential for these organisms to cause invasive infections in individuals without cystic fibrosis. All three of the WO-2 isolates identified as P. pnomenusa were isolated from blood, suggesting that this particular species may have an increased potential for invasive disease. Additional pathogenesis studies will be required to better understand this association.
The great majority of confirmed Pandoraea strains have been isolated from clinical specimens. Only three nonclinical isolates have been described: a Pandoraea genomospecies 1 isolate from garden soil and one P. norimbergensis isolate each from environmental water and powdered milk (3). Our in vitro experience with the WO-2 strains indicates that Pandoraea can survive and grow in a variety of culture conditions, with adequate growth temperatures ranging at least from 25°C through 42°C. The geographic information received with the WO-2 isolates suggests that these organisms can be encountered in a variety of locations, ranging from temperate environments, such as Hawaii, to areas such as Utah and Ohio that may experience significant variations in temperature and humidity. Taken together, this information suggests that the natural habitat of Pandoraea may be soil or water. The ability of these organisms to grow on B. cepacia-selective medium should greatly enhance their isolation from environmental samples.
Pandoraea species share many phenotypic similarities with Ralstonia pickettii and Ralstonia paucula (formerly CDC group IVc-2) and with other less commonly encountered nonfermenters, including Trotter's bacillus, A. temperans, and nonsaccharolytic strains of B. cepacia (genomovar III biovar c). Trotter's bacillus was first isolated in 1990 from pleural fluid and pulmonary decortication tissue of a 5-year-old child with chronic granulomatous disease (17). A. temperans was also described in 1990 by Willems et al. (23). These organisms have been primarily isolated from clinical sources, although at least one isolate from sludge has been reported (23). Nonsaccharolytic strains of B. cepacia referred to as genomovar III biovar c were described in 1997 by Vandamme et al. (18); however, recognition of nonsaccharolytic B. cepacia-like strains was reported earlier in 1996 by Pitt et al. (14). Genus-level differentiation of Pandoraea from these other organisms can be achieved with a variety of approaches. The 16S rRNA gene sequences of the Pandoraea strains form a highly related group (greater than 98.8% homologous) that clusters most closely with a separate clade comprising several Burkholderia species. The presence of both lauric (12:0) and α-hydroxylauric (2-OH-12:0) acids in the Pandoraea CFA profile is also a useful differential characteristic. Phenotypic tests useful in differentiating between these organisms include flagellar morphology, Simmons citrate alkalinization, nitrate reduction, gas from nitrate, urease activity, and acidification of disaccharides.
The in vitro antibiotic susceptibility profile of the Pandoraea strains is similar to that observed with Burkholderia clinical isolates (6). These strains tended to be multiresistant, producing elevated MICs of most antibiotics, including most of the β-lactams and aminoglycosides tested. Additional clinical experience will be required to evaluate or recommend treatment approaches to patients with Pandoraea infections.
Although the great majority of our results with the Pandoraea type strains are in agreement with previously published findings, we did note some differences. Coenye et al. described these organisms as motile with a single polar flagellum (3). However, when we examined the type strains using the Ryu stain (22), two or more polar flagella were attached to most of the cells. This flagellar morphology is similar to the morphology of Pandoraea's closest phylogenetic relative, the genus Burkholderia. The original descriptions of P. apista, P. pnomenusa, P. sputorum, and Pandoraea genomospecies 1 indicated that they failed to grow in OF medium with d-glucose (3). We found, however, that all strains tested grew adequately in OF medium made with the King base (22) and that most P. apista, P. pnomenusa, and P. sputorum strains produced a weak acid reaction in King's OF medium with d-glucose. These laboratory-to-laboratory discrepancies show the importance of obtaining and testing valid reference strains of new taxa to verify their reactions with in-house procedures.
In their original description of this genus, Coenye et al. chose the term Pandoreae, in recognition of the potential Pandora's box of genetic diversity associated with these organisms (3). Our findings with the WO-2 isolates seem to confirm this prophecy. Although six of our nine isolates could be assigned to a specific Pandoraea species, three others represent distinct genetic entities. Until additional strains are identified and studied to better define the phenotypic characteristics of these organisms, we propose that the provisional epithet “genomospecies” be applied to the groups containing one strain only. With this provision, the genus Pandoraea contains the five previously described species and four genomospecies. Genomospecies 1 is the strain CCUG 39680, originally described as “Pandoraea species” (3). The phenotypic tests listed in Table 4 will allow for the discrimination of most of the Pandoraea species and genomospecies; however, a nitrate-negative P. pnomenusa strain might be confused with P. apista. Descriptions of the new Pandoraea genomospecies 2, 3, and 4 are given below.
(i) Pandoraea genomospecies 2.
Gram-negative, nonsporulating straight rod. Motile by means of two or more polar flagella. Weak acid production is observed in King's OF medium with d-glucose and glycerol; oxidase positive; negative for growth on centrimide and SS agars; weak and delayed urease activity is observed; nitrate is not reduced. Other phenotypic characteristics are listed in Table 4. The CFA profile is as described for the genus (3). The only known strain is CDC G5084 (ATCC BAA-108), which was isolated in 1990 from the maxillary sinus of a woman. The G+C content is 65.2%. The 16S rRNA gene sequence has been deposited at GenBank (accession no. AF247693).
(ii) Pandoraea genomospecies 3.
Gram-negative, nonsporulating straight rod. Motile by means of two or more polar flagella. Weak acid production is observed in King's OF medium with d-glucose and glycerol; oxidase negative; negative for growth on centrimide agar; weak and delayed growth is observed on SS agar; weak and delayed urease activity is observed; nitrate is not reduced. Other phenotypic characteristics are listed in Table 4. The CFA profile is as described for the genus (3). The only known strain is CDC G9805 (ATCC BAA-109), isolated in 1996 from the sputum of a 71-year-old woman. The G+C content is 67.1%. The 16S rRNA gene sequence has been deposited at GenBank (accession no. AF247697).
(iii) Pandoraea genomospecies 4.
Gram-negative, nonsporulating straight rod. Motile by means of two or more polar flagella. Weak acid production is observed in King's OF medium with d-glucose and glycerol; oxidase positive; negative for growth on centrimide agar; delayed growth is observed on SS agar; weak and delayed urease activity is observed; nitrate is not reduced. Other phenotypic characteristics are listed in Table 4. The CFA profile is as described for the genus (3). The only known strain is CDC H652 (ATCC BAA-110), isolated in 1998 from the sputum of a woman with cystic fibrosis. The G+C content is 68.6%. The 16s rRNA gene sequence has been deposited in GenBank (accession no. AF247698).
ACKNOWLEDGMENTS
We give special thanks to our colleagues at the California, Texas, Georgia, Hawaii, Louisiana, Ohio, Utah, and Oklahoma state public health laboratories for recognizing the uniqueness of these strains and referring them to us for additional study.
REFERENCES
- 1.Bonner T I, Brenner D J, Neufeld B R, Britten R J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973;31:123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- 2.Brenner D J, McWhorter A C, Leete-Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan J R W, Gillis M, Vandamme P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol. 2000;50:887–899. doi: 10.1099/00207713-50-2-887. [DOI] [PubMed] [Google Scholar]
- 4.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daneshvar M I, Hill B, Hollis D G, Moss C W, Jordan J G, Macgregor J P, Tenover F, Weyant R S. CDC group O-3: phenotypic characteristics, fatty acid composition, isoprenoid quinone content, and in vitro antimicrobic susceptibilities of an unusual gram-negative bacterium isolated from clinical specimens. J Clin Microbiol. 1998;36:1674–1678. doi: 10.1128/jcm.36.6.1674-1678.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilligan P H, Whittier S. Burkholderia, Stenotrophomonas, Ralstonia, Brevundimonas, Comamonas, and Acidovorax. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 526–538. [Google Scholar]
- 7.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 8.Mandel M, Igambi L, Bergendahl J, Dodson M L, Jr, Scheltgen E. Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J Bacteriol. 1970;101:333–338. doi: 10.1128/jb.101.2.333-338.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss C W, Guerrant G O. Separation of bacterial ubiquinones by reverse-phase high-pressure liquid chromatography. J Clin Microbiol. 1983;18:15–17. doi: 10.1128/jcm.18.1.15-17.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss C W, Kai A, Lambert M A, Patton C. Isoprenoid quinone content and cellular fatty acid composition of Campylobacter species. J Clin Microbiol. 1984;19:772–776. doi: 10.1128/jcm.19.6.772-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Vol. 17. 1997. , no. 2. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational suppl. Vol. 18 1998. , no. 1. M100-S8. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 13.Oyaizu H, Komagata K. Grouping of Pseudomonas species on the basis of cellular fatty acid composition and the quinone system with special reference to the existence of 3-hydroxy fatty acids. J Gen Appl Microbiol. 1983;29:17–40. [Google Scholar]
- 14.Pitt T L, Kaufmann M E, Patel P S, Benge L C A, Gaskin S, Livermore D M. Type characterization and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J Med Microbiol. 1996;44:203–210. doi: 10.1099/00222615-44-3-203. [DOI] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Stackebrandt E, Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990;136:37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- 17.Trotter J A, Kuhls T L, Pickett D A, Reyes de la Rocha S, Welch D F. Pneumonia caused by a newly recognized pseudomonad in a child with chronic granulomatous disease. J Clin Microbiol. 1990;28:1120–1124. doi: 10.1128/jcm.28.6.1120-1124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 19.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 20.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Truper H G. Report of the Ad Hoc Committee on Reconciliation of the Approaches to Bacterial Systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 21.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jordan J J, Cook E C, Daneshvar M I. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria. 2nd ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. [Google Scholar]
- 23.Willems A, Falsen E, Pot B, Jantzen E, Hoste B, Vandamme P, Gillis M, Kersters K, DeLey J. Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Falsen (EF) group 13, EF group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int J Syst Bacteriol. 1990;40:384–398. doi: 10.1099/00207713-40-4-384. [DOI] [PubMed] [Google Scholar]
- 24.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Pallerone and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]