Abstract
Fludioxonil and iprodione are effective fungicides widely used for crop protection and are essential for controlling plant pathogenic fungi. The emergence of fungicide-resistant strains of targeted pathogens is regularly monitored, and several cases have been reported. Non-targeted fungi may also be exposed to the fungicide residues in agricultural fields. However, there are no comprehensive reports on fungicide-resistant strains of non-targeted fungi. Here, we surveyed 99 strains, representing 12 Penicillium species, that were isolated from a variety of environments, including foods, dead bodies, and clinical samples. Among the Penicillium strains, including non-pathogenic P. chrysogenum and P. camembertii, as well as postharvest pathogens P. expansum and P. digitatum, 14 and 20 showed resistance to fludioxonil and iprodione, respectively, and 6 showed multi-drug resistance to the fungicides. Sequence analyses revealed that some strains of P. chrysogenum and Penicillium oxalicum had mutations in NikA, a group III histidine kinase of the high-osmolarity glycerol pathway, which is the mode of action for fludioxonil and iprodione. The single nucleotide polymorphisms of G693D and T1318P in P. chrysogenum and T960S in P. oxalicum were only present in the fludioxonil- or iprodione-resistant strains. These strains also exhibited resistance to pyrrolnitrin, which is the lead compound in fludioxonil and is naturally produced by some Pseudomonas species. This study demonstrated that non-targeted Penicillium strains distributed throughout the environment possess fungicide resistance.
Introduction
Fludioxonil is a member of the phenylpyrrole class of fungicides that acts on a broad spectrum of plant pathogenic fungi [1]. It is a derivative of pyrrolnitrin, a secondary metabolite produced by certain bacteria, including Pseudomonas species [2] ここをクリックまたはタップしてテキストを入力してください。. In many countries, fludioxonil is widely used for crop, as well as post-harvest, protection of pom fruits from fungal pathogens. Fludioxonil’s mode of action consists of a fungal two-component system in the high-osmolarity glycerol (HOG) pathway, which is involved in major cellular responses to external stimuli, such as osmotic shock, UV irradiation, oxidative and heavy metal stresses, and high temperature [3] ここをクリックまたはタップしてテキストを入力してください。. Treatment with fludioxonil leads to an abnormal hyphal morphology, including swelling and balloon-shapes, as well as the hyperaccumulation of glycerol, and these changes have been observed in several fungal species [4, 5]. High-doses of fludioxonil produce fungicidal effects on a wide range of fungi. Thus, fludioxonil is the first choice for controlling plant pathogenic fungi in fields and for preserving harvested crops.
However, repeated applications of fungicides have resulted in the occurrence of resistant strains of pathogenic fungi [6]. Indeed, strains of Alternaria brassicola and Alternaria alternata resistant to fludioxonil have been isolated from the fields in which fludioxonil was applied [7, 8]. Molecular analyses using laboratory-derived fungicide-resistant strains have identified that mutations in a group III histidine kinase (HHK) of the HOG pathway are responsible for fludioxonil resistance [9, 10]. The resistance mechanisms of fludioxonil have been extensively studied in several fungi, including Neurospora crassa, Magnaporthe oryzae, Botrytis cinerea, A. brassicicola, and Aspergillus nidulans [11–15]. Most of the fludioxonil-resistant strains showed multi-drug resistance to iprodione, a dicarboximide fungicide, which indicates that the fungicides’ modes of action share the same target molecule.
Some mutations conferring resistance to fludioxonil and iprodione in field isolates of plant pathogens were found in group III HHKs [15–18]. Fungal HHKs are typically classified into 11 groups. The group III HHKs have a unique structure, characterized by five to seven tandem repeats of histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases (HAMP) domains at the N-termini [19]. Owing to its essential role in many aspects of stress responses, including pathogenicity, loss-of-function mutations of the group III HHKs are thought to be maintained at a low prevalence in the field [20]. Indeed, deleting the HHK gene results in growth retardation, morphological alterations, developmental defects, and osmosensitivity [21–23], which result in higher fitness costs compared with the parental strains.
Because of the practical importance, fungicide resistance in targeted plant pathogens has been intensively investigated. However, the effects of fungicides on non-targeted fungi remain unstudied, and the ecological impact underestimated. The objective of this study was to determine whether and how non-targeted fungi acquire resistance to fludioxonil and iprodione. Thus, we searched for fludioxonil/iprodione-resistant Penicillium strains isolated from outside the fields and investigated whether resistant strains possessed mutations in the group III HHKs, while some targeted Penicillium species have been reported to show resistance to these fungicides [24, 25]. Furthermore, we examined multi-drug resistance to pyrrolnitrin among the strains and the competition among pyrrolnitrin-producing Pseudomonas strains.
Materials and methods
Strains, culture conditions, and reagents
In total, 80 Penicillium strains were provided through the National Bio-Resource Project, Japan (http://www.nbrp.jp/) and are preserved at the Medical Mycology Research Center, Chiba University, and 19 Penicillium strains were obtained from Biological Resource Center, National Institute of Technology and Evaluation (NBRC). Penicillium strains were cultured on potato dextrose agar (PDA) or in potato dextrose broth at 25°C for 5 days. Conidial suspension were prepared by scraping colony surfaces with a spreader and 0.05% Tween 20. The amount of conidia retrieved was counted using a hemocytometer. Pyrrolnitrin (from Pseudomonas cepacia) was commercially obtained (Sigma-Aldrich Co., St. Louis, MO). Fludioxonil and iprodione were obtained from the Abe laboratory at Tohoku University, Sendai, Miyagi, Japan. Oligonucleotides were synthesized by Eurofins Genomics (Tokyo, Japan).
Antifungal susceptibility assay
Sensitivity to fungicides was determined by measuring colony growth on PDA plates in the presence of fungicides. Approximately 10,000 spores of each Penicillium strain were inoculated onto both PDA and PDA supplemented with 1 μg/mL fludioxonil, 10 μg/mL iprodione, or 0.05 μg/mL pyrrolnitrin and incubated at 25°C for 5 days. The diameter of each fungal colony on PDA amended with fungicides was measured and compared with that on PDA alone. Strains having a growth rate of 50% or more were defined as “fungicide-resistant”, whereas those having a growth rate of less than 50% were defined as “fungicide-sensitive”.
Extraction of genomic DNA
Mycelia cultured in potato dextrose broth were frozen in liquid nitrogen and ground to a fine powder using a mortar and pestle. Total genomic DNA was extracted using a NucleoSpin Plant II Kit (Takara Bio, Ohtsu, Japan).
DNA sequencing
The genes encoding NikA (500 bp-upstream of the open reading frame to 500 bp-downstream of the open reading frame) in Penicillium chrysogenum and Penicillium oxalicum were amplified by PCR using genomic DNA as the template and specific primers (S1 Table). The PCR conditions were as follows: 30 cycles of 98°C for 10 s, 53°C for 5 s, and 68°C for 1 min with KOD One PCR Master Mix (Toyobo, Osaka, Japan). The PCR product was subject to agarose gel electrophoresis and purified using a Gel/PCR Extraction Kit (NIPPON Genetics, Tokyo, Japan). The purified PCR products were subjected to DNA sequencing (Eurofins Genomics). The sequences were compared with those of the nikA genes from the reference genomes of P. chrysogenum P2niaD18 (GCA_000710275) and P. oxalicum 114–2 (GCA_000346795), which were retrieved from the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/).
Genome sequencing
Whole-genome sequencing using next-generation methods was performed as described previously [26]. Briefly, we prepared a fragmented DNA library from the genomic DNA of P. roqueforti using NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England BioLabs) and NEBNext Multiplex Oligos for Illumina (New England BioLabs). Paired-end sequencing was carried out by Novogene.
Single nucleotide variant detection
To search for single nucleotide polymorphisms in nikA of P. roqueforti, we performed read mapping using CLC Genomics Workbench (CLC bio, Aarhus, Denmark). The reads from each isolate were trimmed and mapped to the nikA (PROQFM164_S03g000214) of P. roqueforti FM164 (GCA_000513255).
Results
Fludioxonil- and/or iprodione-resistant Penicillium
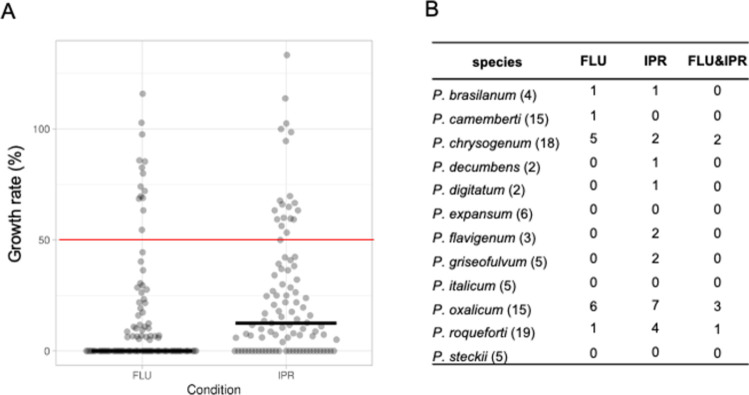
To understand the distribution of fludioxonil and iprodione resistance in non-targeted fungal species, we prepared a set of 99 Penicillium strains representing 12 species. This set contained 4 P. brasilanum, 15 P. camemberti, 18 P. chrysogenum, 2 P. decumbens, 2 P. digitatum, 6 P. expansum, 3 P. flavigenum, 5 P. griseofulvum, 5 P. italicum, 15 P. oxalicum, 19 P. roqueforti, and 5 P. steckii strains (Table 1). The isolation source was registered as unknown for 17, cheese for 25, dead body for 10, patient for 13, fruit for 6, other foods for 4, soil for 6, other creatures for 3, and other environments for 15 strains (Table 1). These strains were subjected to antifungal susceptibility assays using fludioxonil and iprodione. Colony growth on PDA containing 1 μg/mL fludioxonil or 10 μg/mL iprodione was examined (S1 and S2 Figs). The concentrations of the fungicides were determined with reference to other papers describing fungicide-resistant strains. A total of 14 strains (P. brasilanum, 1/4; P. camemberti, 1/15; P. chrysogenum, 5/18; P. oxalicum, 6/15; and P. roqueforti, 1/19) and 20 strains (P. brasilanum, 1/4; P. chrysogenum, 2/18; P. decumbens, 1/2; P. digitatum, 1/2; P. flavigenum, 2/3; P. griseofulvum, 2/5; P. oxalicum, 7/15; and P. roqueforti, 4/19) showed resistance to fludioxonil and iprodione, respectively (Table 1 and Fig 1). Interestingly, six strains (two strains of P. chrysogenum, three strains of P. oxalicum, and one strain of P. roqueforti) exhibited multi-drug resistance to fludioxonil and iprodione. No strains resistant to either of the fungicides were present in P. expansum, P. italicum, or P. steckii.
Table 1. The Penicillium strains used in this study.
| No | Species | Strain ID | FLUa | IPRb | PRNc | Source |
|---|---|---|---|---|---|---|
| 1 | P. brasilanum | IFM 42067 (= IFO 6234) | S | S | R | soil |
| 2 | P. brasilanum | IFM 42077 | S | R | R | unknown |
| 3 | P. brasilanum | IFM 60071 | S | S | R | environmental isolate |
| 4 | P. brasilanum | IFM 60072 | R | S | R | environmental isolate |
| 5 | P. camemberti | IFM 49450 (= CBS 299.48) | S | S | R | French Camembert cheese |
| 6 | P. camemberti | IFM 54179 | S | S | R | environmental isolate |
| 7 | P. camemberti | IFM 61933 | S | S | R | tongue (dead body) |
| 8 | P. camemberti | NBRC 5855 | S | S | R | unknown |
| 9 | P. camemberti | NBRC 32215 | S | S | S | Commercial cheese |
| 10 | P. camemberti | NBRC 105299 | S | S | R | Camembert cheese imported from France |
| 11 | P. camemberti | NBRC 105301 | R | S | R | Camembert cheese imported from France, Japan |
| 12 | P. camemberti | NBRC 105305 | S | S | S | Lys bleu cheese imported from France |
| 13 | P. camemberti | NBRC 105306 | S | S | S | Bonifaz cheese imported from Germany, Japan |
| 14 | P. camemberti | NBRC 105307 | S | S | S | Bonifaz cheese imported from Germany |
| 15 | P. camemberti | NBRC 105308 | S | S | S | Cambozola cheese imported from Germany |
| 16 | P. camemberti | NBRC 105309 | S | S | S | Cambozola cheese imported from Germany, Japan |
| 17 | P. camemberti | NBRC 105310 | S | S | S | Bavariablu cheese imported from Germany |
| 18 | P. camemberti | NBRC 105314 | S | S | S | Natural cheese made in Hokkaido, Japan |
| 19 | P. camemberti | NBRC 105315 | S | S | S | Camembert cheese made in Japan |
| 20 | P. chrysogenum | IFM 40614 | S | S | S | unknown |
| 21 | P. chrysogenum | IFM 47464 (= CBS 349.48) | R | S | R | unknown, UK |
| 22 | P. chrysogenum | IFM 47768 | S | S | S | unknown, Japan |
| 23 | P. chrysogenum | IFM 52203 | S | S | S | bathroom, Brazil |
| 24 | P. chrysogenum | IFM 52204 | S | S | S | kitchen, Brazil |
| 25 | P. chrysogenum | IFM 56829 | S | S | S | 50 man, China |
| 26 | P. chrysogenum | IFM 57112 | S | S | S | bioresource |
| 27 | P. chrysogenum | IFM 57243 (= CBS 282.97) | R | R | R | dust from school, Denmark |
| 28 | P. chrysogenum | IFM 57244 (= CBS 798.97) | R | R | R | Apeldoorn / indoor environment, Netherland |
| 29 | P. chrysogenum | IFM 57245 (= CBS 478.84) | R | S | R | air, fruit store, Denmark |
| 30 | P. chrysogenum | IFM 59766 | S | S | S | buttock (dead body) |
| 31 | P. chrysogenum | IFM 60605 | R | S | R | skin of jaw (dead body) |
| 32 | P. chrysogenum | IFM 60953 | S | S | S | right finger (dead body) |
| 33 | P. chrysogenum | IFM 61615 | S | S | S | swab from patient’s house |
| 34 | P. chrysogenum | IFM 61632 | S | S | S | face (dead body) |
| 35 | P. chrysogenum | IFM 62336 | S | S | S | trachea (dead body) |
| 36 | P. chrysogenum | IFM 63007 | S | S | R | right leg (dead body) |
| 37 | P. chrysogenum | IFM 64696 | S | S | S | breast bone (dead body) |
| 38 | P. decumbens | IFM 46582 | S | R | R | contaminant of Sporotrichosis patient |
| 39 | P. decumbens | IFM 63512 | S | S | R | bedroom |
| 40 | P. digitatum | IFM 60598 | S | R | S | lemon |
| 41 | P. digitatum | IFM 63755 | S | S | R | 62 F, sputum, ABPM |
| 42 | P. expansum | IFM 40618 | S | S | S | unknown |
| 43 | P. expansum | IFM 47463 (= CBS 325.48) | S | S | S | fruit of Malus syvestris |
| 44 | P. expansum | IFM 47478 (= IFO 8800) | S | S | S | unknown, Patulin production |
| 45 | P. expansum | IFM 52210 | S | S | S | nursing room, Brazil |
| 46 | P. expansum | IFM 58916 | S | S | S | unknown, cyclopiazone acid production |
| 47 | P. expansum | IFM 62049 | S | S | S | refrigerator |
| 48 | P. flavigenum | IFM 54184 (= CBS 110406) | S | S | S | soil under Chrysothamnus nauseosus |
| 49 | P. flavigenum | IFM 54185 (= CBS 110407) | S | R | R | white beans |
| 50 | P. flavigenum | IFM 54186 (= CBS 419.89) | S | R | S | flour |
| 51 | P. griseofulvum | IFM 42069 (= IAM 7212) | S | S | S | unknown |
| 52 | P. griseofulvum | IFM 47730 (= IFO 7640) | S | S | S | unknown, Belgium |
| 53 | P. griseofulvum | IFM 47791 (= CBS 124.14) | S | R | R | soil, UK |
| 54 | P. griseofulvum | IFM 54187 | S | S | R | unknown |
| 55 | P. griseofulvum | IFM 54314 | S | R | S | soil |
| 56 | P. italicum | IFM 49452 (= CBS 719.73) | S | S | S | fruit of Citrus sp., Israel |
| 57 | P. italicum | IFM 49453 (= CBS 339.48) | S | S | S | fruit of Citrus sp., USA |
| 58 | P. italicum | IFM 52160 | S | S | S | orpharyngeal swab, Brazil |
| 59 | P. italicum | IFM 53256 (= NBRC 9419) | S | S | S | fruit of Satsuma orange |
| 60 | P. italicum | IFM 59474 | S | S | S | orange of NZ, isolated in Japan |
| 61 | P. oxalicum | IFM 49446 (= CBS 219.30) | S | S | S | soil, USA |
| 62 | P. oxalicum | IFM 54751 | R | S | R | enironmental isolate |
| 63 | P. oxalicum | IFM 55886 | S | S | R | soil, China |
| 64 | P. oxalicum | IFM 57073 | S | R | S | garbage |
| 65 | P. oxalicum | IFM 59246 | R | R | R | skin (Trichechus manatus) |
| 66 | P. oxalicum | IFM 60000 | S | S | S | skull (dead body) |
| 67 | P. oxalicum | IFM 61428 | S | S | S | BALF, drowning |
| 68 | P. oxalicum | IFM 62137 | S | R | R | bean sprouts |
| 69 | P. oxalicum | IFM 62827 | S | R | R | 92 F, cornea |
| 70 | P. oxalicum | IFM 62922 | R | S | R | 49 M, tracheal mucus plug |
| 71 | P. oxalicum | IFM 62931 | S | S | S | 62 M, washing solution (lung) |
| 72 | P. oxalicum | IFM 62937 | R | R | R | 72 M, left lung apex cavity, simple pulmonary aspergilloma |
| 73 | P. oxalicum | IFM 63612 | R | R | R | 68 F, BALF |
| 74 | P. oxalicum | IFM 63698 | R | S | R | 12 F, sputum |
| 75 | P. oxalicum | IFM 65074 | S | R | S | 54 F, sputum |
| 76 | P. roqueforti | IFM 47733 (= IFO 4622) | S | S | S | requefort cheese |
| 77 | P. roqueforti | IFM 48062 | S | S | S | Blue-veained cheese (Gorgonzola) |
| 78 | P. roqueforti | IFM 48063 | S | S | S | Blue-veained cheese (Cambozola) |
| 79 | P. roqueforti | IFM 48064 | S | S | S | Blue-veained cheese (Dana blue) |
| 80 | P. roqueforti | IFM 48065 | S | S | S | Blue-veained cheese (Blue-S) |
| 81 | P. roqueforti | IFM 48066 | S | S | S | Blue-veained cheese (Stilton) |
| 82 | P. roqueforti | IFM 48067 | S | S | S | Blue-veained cheese (Stilton) |
| 83 | P. roqueforti | IFM 48068 | S | R | S | Blue-veained cheese (Roquefort) |
| 84 | P. roqueforti | IFM 48069 | S | R | S | Blue-veained cheese (Roquefort) |
| 85 | P. roqueforti | IFM 48070 | S | R | S | Blue-veained cheese (Blue-S) |
| 86 | P. roqueforti | IFM 48071 | S | S | S | Blue-veained cheese (Blue-H) |
| 87 | P. roqueforti | IFM 58915 | S | S | S | unknown, cyclopiazone acid production |
| 88 | P. roqueforti | NBRC 4622 | S | S | S | Roquefort cheese |
| 89 | P. roqueforti | NBRC 5459 | S | S | S | French roquefort cheese, USA |
| 90 | P. roqueforti | NBRC 5754 | S | S | S | unknown |
| 91 | P. roqueforti | NBRC 5956 | R | R | S | unknown |
| 92 | P. roqueforti | NBRC 6400 | S | S | S | unknown |
| 93 | P. roqueforti | NBRC 7693 | S | S | S | unknown |
| 94 | P. roqueforti | NBRC 8799 | S | S | S | unknown |
| 95 | P. steckii | IFM 62327 | S | S | S | leg (dead body) |
| 96 | P. steckii | IFM 63697 | S | S | S | 82 F, eye |
| 97 | P. steckii | IFM 64403 | S | S | S | noodle soup |
| 98 | P. steckii | IFM 64663 | S | S | S | cockroach |
| 99 | P. steckii | IFM 64664 | S | S | R | cockroach |
a FLU indicates fludioxonil: S and R indicate sensitive and resistant, respectively.
b IPR indicates iprodione.
c PRN indicates pyrrolnitrin.
Fig 1. Multi-drug resistance to fludioxonil and iprodione.
(A) Box plots showing the growth rates of 99 Penicillium strains in the presence of fludioxonil (FLU) and iprodione (IPR). The plots for growth rates ≥ 50% indicate resistant strains. (B) The number of fludioxonil and/or iprodione-resistant strains. Numbers in parentheses indicate the total number of strains.
Mutations in the group III HHK, NikA
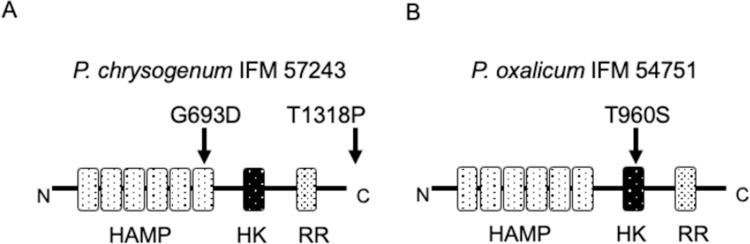
As demonstrated in several fungi, including plant pathogens, fludioxonil/iprodione-resistance might be attributed to mutations in the group III HHK, NikA, in Penicillium strains. In further analyses, we focused on P. chrysogenum, P. oxalicum, and P. roqueforti because they contained multiple multi-drug resistant strains. The nikA genes in a set of P. chrysogenum, P. oxalicum strains were sequenced and compared with those of P. chrysogenum strain P2niaD18 (GCA_000710275), P. oxalicum 114–2 (GCA_000346795), respectively. The nikA genes in a set of P. roqueforti strains were obtained by genome sequencing. There were several amino acid alterations in NikA (Fig 2), such as the combination of A404S and S1332L in P. chrysogenum IFM 56829. The S1332L mutation was also present in P. chrysogenum IFM 61615. Both strains were sensitive to fludioxonil and iprodione, suggesting that the mutations are not involved in the resistance. Conversely, in P. chrysogenum IFM 57243, which showed resistance to fludioxonil and iprodione, glycine was changed to aspartic acid at position 693 (G693D) and threonine was changed to proline at position 1318 (T1318P). The mutation at position 693 is located in the HAMP domain region, suggesting that this mutation affects the sensing of, and interactions with, the fungicides (Fig 3). For P. oxalicum, 10 of 15 strains harbored both S94F and Q151R mutations regardless of their fungicide-resistance level, indicating that these mutations are not associated with fungicide resistance. In P. oxalicum IFM 54751, which is resistant to fludioxonil but not to iprodione, threonine was changed to serine at position 960 (T960S) in addition to the abovementioned two mutations. This mutation is located in the kinase domain and potentially affects the histidine kinase function and fludioxonil resistance. In P. roqueforti, no amino acid alterations were found in nikA compared with the reference sequence.
Fig 2. Mutations in the NikA proteins and resistance to high osmotic pressure in P. chrysogenum and P. oxalicum.
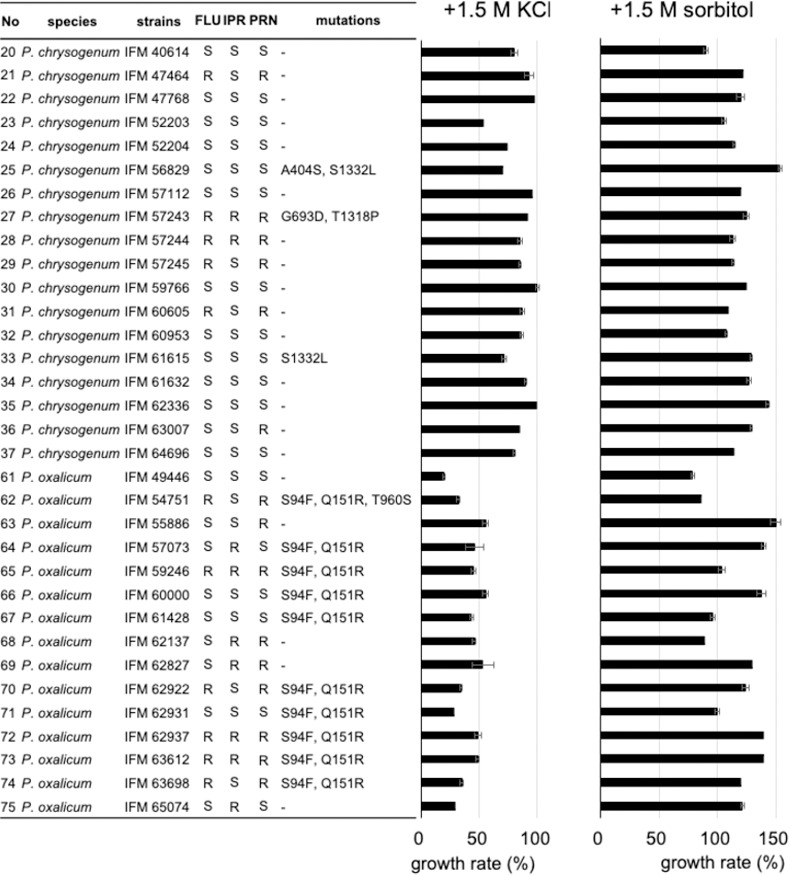
The list of mutations in the NikA proteins (left) and colony growth rates in the presence of a high concentration of KCl (middle) and sorbitol (right). The data represent the averages of triplicate individual experiments (means ± standard deviations).
Fig 3. Domain structures of fungal NikA proteins.
The group III HHK NikA is composed of six repeated HAMP, histidine kinase (HK), and response regulator (RR) domains. (A) The mutation G693D in P. chrysogenum IFM 57243 is located in the HAMP domain, whereas the mutation T1318P is located in a disordered region. (B) The mutation T960S in P. oxalicum IFM 54751 is located in the HK domain.
Resistance of Penicillium strains to high osmotic stress
As demonstrated in other species, the HOG pathway is involved in responses to fungicides and osmotic conditions. To test the link between resistance to fungicides and osmotic stress, we investigated the colony growth of P. chrysogenum and P. oxalicum strains on PDA containing high concentrations of KCl or sorbitol (1.5 M) (Fig 2). Among the 18 tested strains of P. chrysogenum, IFM 52203 showed a sensitivity to KCl (growth was less than 60% compared with under stress-free conditions). The growth rates of P. chrysogenum IFM 57243, which had G693D and T1318P mutations in NikA, in the presence of high concentrations of KCl and sorbitol were 92% and 124%, respectively. These values were comparable to those of other strains, indicating that the G693D and T1318P mutations had no effect on the strains’ sensitivity to osmotic stress. Compared with P. chrysogenum, P. oxalicum strains were relatively sensitive to KCl. Growth was less than 30% in the presence of KCl, compared with no KCl stress, in 3 of 15 strains, whereas only 1 strain showed >20% reduction in the colony growth in the presence of sorbitol. The growth rates of P. oxalicum IFM 54751, with of the T960S mutation in NikA, in high concentrations of KCl and sorbitol were 32% and 86%, respectively. The moderate sensitivity to high osmotic stresses suggests the involvement of the T960S mutation in osmotic stress adaptation.
Pyrrolnitrin-resistant Penicillium
Fludioxonil is an analog of the natural antifungal compound pyrrolnitrin, which is produced by some Pseudomonas species [27]. The mode of action for pyrrolnitrin is believed to be related to the fungal group III HKKs of the HOG pathway [28]. Therefore, we performed antifungal assays using Penicillium strains in the presence of pyrrolnitrin. Among the 99 tested strains of Penicillium, 32 (P. brasilianum, 4/4; P. camemberti, 6/15; P. chrysogenum, 6/18; P. decumbens, 2/2; P. digitatum, 1/2; P. flavigenum, 1/3; P. griseofulvum, 2/5; P. oxalicum, 9/15; and P. steckii, 1/5) showed resistance to 0.05 μg/mL pyrrolnitrin (Table 1). No strains of P. expansum, P. italicum, and P. roqueforti showed resistance to pyrrolnitrin. Notably, 13 (P. brasilanum, 1/4; P. camemberti, 1/15; P. chrysogenum, 5/18; and P. oxalicum, 6/15) and 11 (P. brasilanum, 1/4; P. chrysogenum, 2/18; P. decumbens, 1/2; P. flavigenum, 1/3; P. griseofulvum, 1/5; and P. oxalicum, 5/15) of the 32 pyrrolnitrin-resistant strains showed multi-drug resistance to fludioxonil and/or iprodione (Fig 4A). For P. chrysogenum, five and two of seven pyrrolnitrin-resistant strains showed multi-drug resistance to fludioxonil and iprodione, respectively (Fig 4B). For P. oxalicum, six and five of nine pyrrolnitrin-resistant strains showed multi-drug resistance to fludioxonil and iprodione, respectively (Fig 4C).
Fig 4. Multi-drug resistance to pyrrolnitrin, fludioxonil, and iprodione.
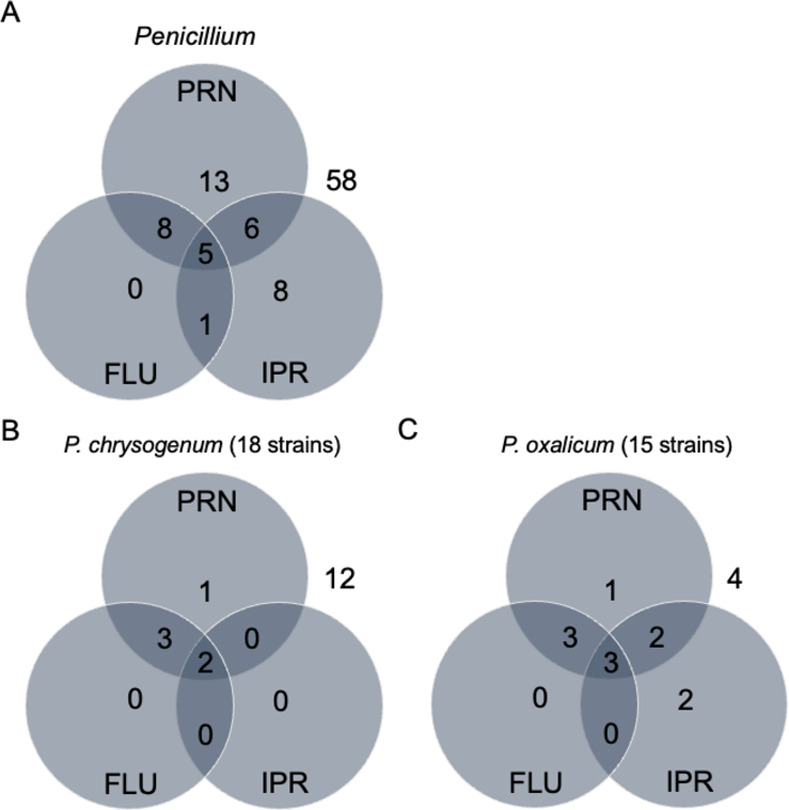
Venn diagrams of the numbers of pyrrolnitrin-, fludioxonil-, and iprodione-resistant strains in Penicillium species (A), P. chrysogenum (B), and P. oxalicum (C). PRN, pyrrolnitrin; FLU, fludioxonil; and IPR, iprodione.
Penicillium chrysogenum IFM 57243 with G693D and T1318P mutations in NikA showed resistance to pyrrolnitrin, whereas P. chrysogenum IFM 56829 and IFM 61615 were sensitive to pyrrolnitrin. Thus, the G693D and T1318P mutations may contribute to resistance against the three fungicides. Penicillium oxalicum IFM 54751 with the T960S mutation exhibited resistance to fludioxonil and pyrrolnitrin. The results of the antifungal susceptibility testing revealed that within a set of Penicillium strains isolated from various environments that are non-targeted fungi some members show resistance to an agricultural fungicide, as well as its lead compound, which is naturally produced by bacteria in the environment.
Discussion
In this study, we explored Penicillium strains resistant to the widely used fungicides fludioxonil and iprodione, as well as pyrrolnitrin. Strains of Penicillium species that cause postharvest decay of citrus and fruits, and exhibiting resistance to these fungicides, have been investigated [24, 25]. Interestingly, in the present study there were no resistant strains found in P. expansum and P. italicum, where one P. digitatum strain isolated from lemon showed resistance to iprodione. To the best of our knowledge, except for these pathogens, we are the first to demonstrate that several strains of non-targeted fungi, such as penicillin-producing P. chrysogenum, environmentally ubiquitous P. oxalicum, and cheese-producing P. roqueforti, show resistance to fludioxonil, iprodione, and pyrrolnitrin. A multi-drug resistance to these fungicides was detected in some strains. These results raised questions regarding the mechanisms and occurrence of resistance in these species.
The mechanisms underlying resistance to fludioxonil have been studied in several fungi, including plant pathogens such as B. cinerea, Cochliobolus heterostrophus, and M. oryzae [29–31]. The fungal HOG pathway involved in osmotic stress adaptation is a target of these fungicides [10, 23, 32]. Laboratory-generated mutants resistant to these fungicides provide clear perspectives on the mechanisms. Fillinger et al. showed that exposure to pyrrolnitrin or iprodione results in resistant B. cinerea mutants, most of which harbor de novo mutations in the Bos1 protein, a group III HHK of the fungus [33]. The mutations occurred in the protein’s six repeated HAMP domains. The regeneration of site-directed clones clarified that the amino acid alterations in the HAMP domains are responsible for the fungicide resistance. Some mutations resulted in fungicide resistance and hypersensitivity to osmotic stress, whereas other mutations resulted in resistance to iprodione, but not to phenylpyrroles, and sensitivity to hyperosmolarity. Here, some Penicillium strains possessed mutations in NikA that were associated with fungicide resistance. Penicillium chrysogenum IFM 57243 showed multi-drug resistance to fludioxonil and iprodione, as well as pyrrolnitrin, and had G693D and T1318P mutations in the NikA protein. Penicillium oxalicum IFM 54751 showed resistance to fludioxonil and pyrrolnitrin, but not to iprodione, and had a T960S mutation in the NikA protein. These mutations are located in the amino acid residues highly conserved among the 12 species (Fig 5). These mutations may contribute to antifungal compound resistance, while site-directed clones of the mutations need to be created in the future.
Fig 5. Sequence alignment of NikA in 12 Penicillium species.
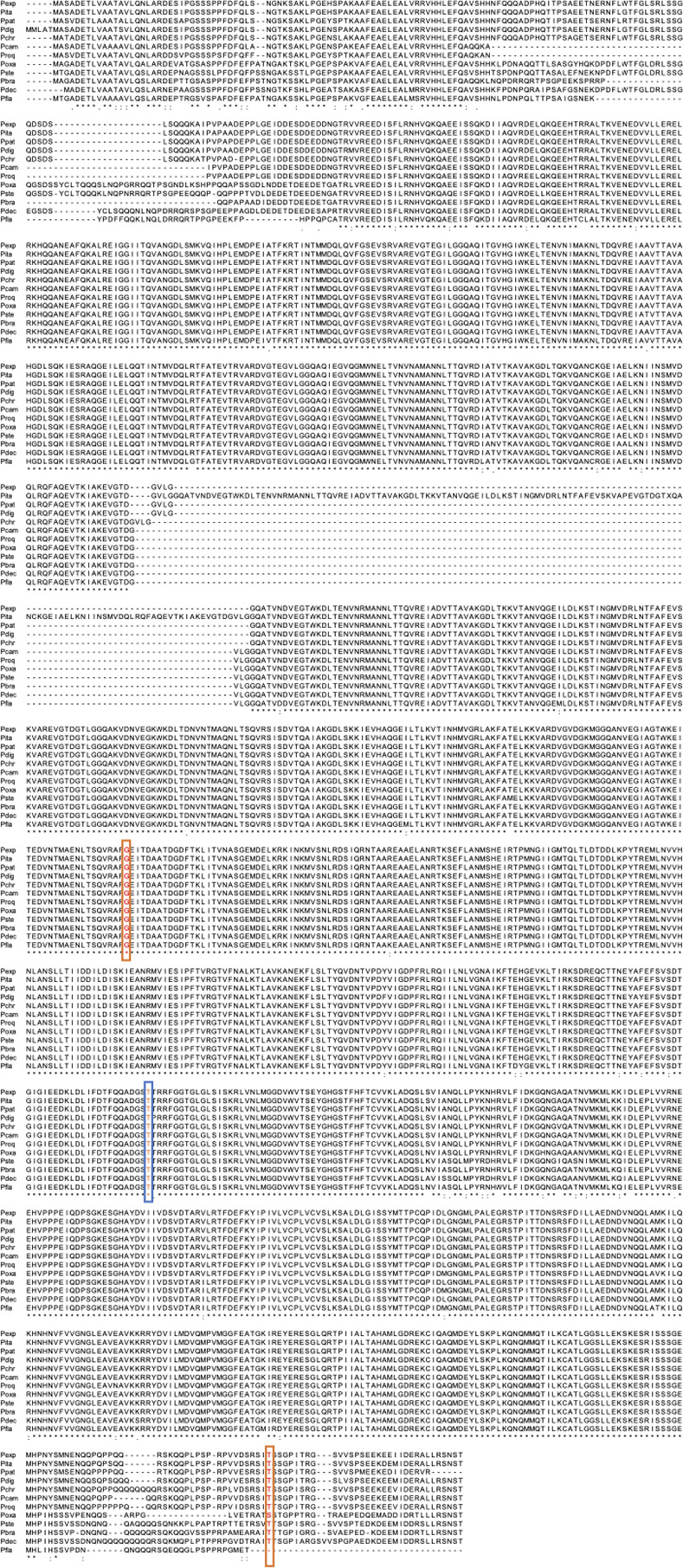
The gene IDs are as follows: P. brasilanum PMG11_02111, P. camemberti PCAMFM013_S001g000092, P. chrysogenum EN45_023640, P. decumbens PENDEC_c002G07053, P. digitatum Pdw03_4331, P. expansum PEX2_037120, P. flavigenum PENFLA_c003G00500, P. griseofulvum PGRI_040000, P. italicum PITC_092520, P. oxalicum PDE_05313, P. roqueforti PROQFM164_S03g000214, and P. steckii PENSTE_c014G10375. The amino acid residues framed by orange and blue show the mutations detected in P. chrysogenum IFM 57243 and P. oxalicum IFM 54751, respectively.
A fitness penalty has been reported in fludioxonil-resistant isolates of plant pathogens, as indicated by their relatively slower mycelial growth rates or decreased pathogenicity levels [15, 34, 35] might explain the decreased fitness levels of fludioxonil-resistant strains in the field. Interestingly, the fungicide-resistant Penicillium strains found in the present work showed no apparent growth defects on PDA compared with sensitive isolates of the same species (S1–S3 Figs). This suggested that Penicillium species pay almost no fitness costs for phenylpyrrole and dicarboximide resistance, which should be investigated further.
Here, several strains of P. chrysogenum and P. oxalicum without mutations in the NikA showed resistance to one of the antifungal compounds. In N. crassa, the components of the HOG pathway, which function downstream of the group III HKK Os1, are responsible for fungicide resistance [21, 36]. Indeed, a strain with a mutation in the os2 gene, which encodes a mitogen-activated protein (MAP) kinase in the HOG pathway, shows resistance to the fungicides. However, a mutant of the SakA MAP kinase, which is an ortholog of Os2, in A. nidulans shows only slight resistance to fludioxonil and iprodione [23]. The HOG pathway contributes to fungicide responses in different ways among fungal species. To date, only one study has investigated the HOG pathway’s role in the fungicide responses of Penicillium. Wang et al. demonstrated that the gene deletion mutant of Pdos2, which encodes an Os2 MAP kinase, constructed in P. digitatum shows only slight resistance to fludioxonil and iprodione, suggesting that its HOG pathway has a limited impact on fungicide sensitivity [37]. Thus, it is unlikely that fungicide sensitivity can be attributed to mutations in HOG pathway components, because the fungicide resistance levels identified here were relatively high. The resistance mechanisms of fludioxonil and iprodione are poorly understood, and thus uncharacterized mutations may be present in the resistant strains. More comprehensive investigations are needed to fully understand how the non-targeted fungi possess resistance to synthetic fungicides.
The Penicillium strains used in this study were collected from diverse environmental and clinical sources. According to their records, they have no history of phenylpyrrole or dicarboximide fungicide exposure. The growth test indicated that each species was naturally susceptible, and some strains acquired resistance, to the fungicides. However, it is not known where and how these Penicillium strains have become resistant. One plausible cause for resistance is exposure to fungicide residues in environmental organic matters, such as plant litter or compost. Fludioxonil and iprodione are registered as fungicides for use on a wide variety of crops, and thus, huge amounts of plant debris contaminated with residual fungicides are generated in agricultural settings. Ubiquitously present non-targeted Penicillium strains may encounter such environments, resulting in their being placed under fungicide pressure. This might lead to the natural occurrence of resistance. Another possibility is exposure to environmental pyrrolnitrin produced by certain bacteria in the environment. Pyrrolnitrin is a lead compound of phenylpyrroles, and multi-drug resistance between pyrrolnitrin and phenylpyrroles has been reported [38]. Many bacterial species that belong to the genera Burkholderia and Pseudomonas produce pyrrolnitrin [39]. Some strains have been isolated from the rhizosphere and used as biological control agents against plant pathogenic fungi in agriculture. Therefore, there might be several environmental niches having high concentrations of microbial pyrrolnitrin. Thus, non-targeted Penicillium strains may have acquired resistance to pyrrolnitrin and multi-drug resistance to fludioxonil/iprodione owing to exposure to pyrrolnitrin produced by indigenous species. This study warns of the potential risk of non-targeted fungi around the world acquiring resistance to the fungicides. This issue requires further clarification.
Supporting information
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 1 μg/mL fludioxonil (right, FLU). Colony growth rate in the presence of fludioxonil ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 10 μg/mL iprodione (right, IPR). Colony growth rate in the presence of iprodione ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 0.05 μg/mL pyrrolnitrin (right, PRN). Colony growth rate in the presence of pyrrolnitrin ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
(DOCX)
Acknowledgments
We are grateful to Yoshimi Nakazawa and Yukiko Ohata for their excellent technical support. We thank Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by a grant from the Institute for Fermentation, Osaka and partly by the National Bioresource Project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gehmann K, Nyfeler R, Leadbeater AJ, Nevill DJ, Sozzi D (1990) CGA 173506: a new phenylpyrrole fungicide for broad-spectrum disease control. Brighton Crop Protection Conference, Pests and Diseases—1990. Vol. 2., 399–406. [Google Scholar]
- 2.Chang CJ, Floss HG, Hook DJ, Manni PE, Martin LL, Schröder K, et al. (1981) The biosynthesis of the antibiotic pyrrolnitrin by Pseudomonas aureofaciens. J Antibiotics 34: 555–566. doi: 10.7164/antibiotics.34.555 [DOI] [PubMed] [Google Scholar]
- 3.Bahn YS (2008) Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell 7: 2017–2036. doi: 10.1128/EC.00323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan Y, Ge C, Liu S, Chen C, Zhou M (2013) Effect of phenylpyrrole fungicide fludioxonil on morphological and physiological characteristics of Sclerotinia sclerotiorum. Pestic Biochem Physiol 106: 61–67. doi: 10.1016/j.pestbp.2013.04.004 [DOI] [Google Scholar]
- 5.Fujimura M (2010) Mechanism of action of dicarboximide and phenylpyrrole on the stress-response signal transduction pathway. J Pestic Sci 35: 351–353. doi: 10.1584/jpestics.J10-02 [DOI] [Google Scholar]
- 6.Beever RE, Brien HMR (1983) A survey of resistance to the dicarboximide fungicides in Botrytis cinerea. New Zealand Journal of Agricultural Research 26: 391–400. doi: 10.1080/00288233.1983.10427048 [DOI] [Google Scholar]
- 7.Iacomi-Vasilescu B, Avenot H, Bataillé-Simoneau N, Laurent E, Guénard M, Simoneau P (2004) In vitro fungicide sensitivity of Alternaria species pathogenic to crucifers and identification of Alternaria brassicicola field isolates highly resistant to both dicarboximides and phenylpyrroles. Crop Prot 23: 481–488. doi: 10.1016/j.cropro.2003.10.003 [DOI] [Google Scholar]
- 8.Avenot HF, Michailides TJ (2015) Detection of isolates of Alternaria alternata with multiple-resistance to fludioxonil, cyprodinil, boscalid and pyraclostrobin in California pistachio orchards. Crop Prot 78: 214–221. doi: 10.1016/j.cropro.2015.09.012 [DOI] [Google Scholar]
- 9.Schumacher MM, Enderlin CS, Selitrennikoff CP (1997) The osmotic-1 locus of Neurospora crassa encodes a putative histidine kinase similar to osmosensors of bacteria and yeast. Curr Microbiol 34: 340–347. doi: 10.1007/s002849900193 [DOI] [PubMed] [Google Scholar]
- 10.Yoshimi A, Kojima K, Takano Y, Tanaka C (2005) Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot Cell 4: 1820–1828. doi: 10.1128/EC.4.11.1820-1828.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochiai N, Fujimura M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, et al. (2001) Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag Sci 57: 437–442. doi: 10.1002/ps.302 [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara D, Asano Y, Marui J, Yoshimi A, Mizuno T, Abe K (2009) Transcriptional profiling for Aspergillus nidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Genet Biol 46: 868–878. doi: 10.1016/j.fgb.2009.07.003 [DOI] [PubMed] [Google Scholar]
- 13.Ajouz S, Nicot PC, Bardin M (2010) Adaptation to pyrrolnitrin in Botrytis cinerea and cost of resistance. Plant Pathol 59: 556–566. doi: 10.1111/j.1365-3059.2009.02230.x [DOI] [Google Scholar]
- 14.Jacob S, Foster AJ, Yemelin A, Thines E (2015) High osmolarity glycerol (HOG) signalling in Magnaporthe oryzae: Identification of MoYPD1 and its role in osmoregulation, fungicide action, and pathogenicity. Fungal Biol 119: 580–594. doi: 10.1016/j.funbio.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 15.Ren W, Shao W, Han X, Zhou M, Chen C (2016) Molecular and biochemical characterization of laboratory and field mutants of Botrytis cinerea resistant to fludioxonil. Plant Dis 100: 1414–1423. doi: 10.1094/PDIS-11-15-1290-RE [DOI] [PubMed] [Google Scholar]
- 16.Oshima M, Banno S, Okada K, Takeuchi T, Kimura M, Ichiishi A, et al. (2006) Survey of mutations of a histidine kinase gene BcOS1 in dicarboximide-resistant field isolates of Botrytis cinerea. J Gen Plant Pathol 72: 65–73. doi: 10.1007/s10327-005-0247-7 [DOI] [Google Scholar]
- 17.Alberoni G, Collina M, Lanen C, Leroux P, Brunelli A (2010) Field strains of Stemphylium vesicarium with a resistance to dicarboximide fungicides correlated with changes in a two-component histidine kinase. Eur J Plant Pathol 128: 171–184. doi: 10.1007/s10658-010-9642-9 [DOI] [Google Scholar]
- 18.Sang H, Popko JT, Chang T, Jung G (2017) Genetics and resistance molecular mechanisms involved in qualitative and quantitative resistance to the dicarboximide fungicide iprodione in Sclerotinia homoeocarpa field isolates. Phytopathology 107: 198–207. doi: 10.1094/PHYTO-05-16-0211-R [DOI] [PubMed] [Google Scholar]
- 19.Catlett NL, Yoder OC, Turgeon BG (2003) Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2: 1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilani J, Fillinger S (2016) Phenylpyrroles: 30 years, two molecules and (nearly) no resistance. Front Microbiol 7: doi: 10.3389/fmicb.2016.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Lamm R, Pillonel C, Lam S, Xu JR (2002) Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl Environ Microbiol 68: 532–538. doi: 10.1128/AEM.68.2.532-538.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motoyama T, Ohira T, Kadokura K, Ichiishi A, Fujimura M, Yamaguchi I, et al. (2005) An Os-1 family histidine kinase from a filamentous fungus confers fungicide-sensitivity to yeast. Curr Genet 47: 298–306. doi: 10.1007/s00294-005-0572-6 [DOI] [PubMed] [Google Scholar]
- 23.Hagiwara D, Matsubayashi Y, Marui J, Furukawa K, Yamashino T, Kanamaru K, et al. (2007) Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci Biotechnol Biochem 71: 844–847. doi: 10.1271/bbb.70051 [DOI] [PubMed] [Google Scholar]
- 24.Kanetis L, Förster H, Jones CA, Borkovich KA, Adaskaveg JE (2008) Characterization of genetic and biochemical mechanisms of fludioxonil and pyrimethanil resistance in field isolates of Penicillium digitatum. Biochem Cell Biol 98: 205–214. doi: 10.1094/PHYTO-98-2-0205 [DOI] [PubMed] [Google Scholar]
- 25.Li HX, Xiao CL (2008) Characterization of fludioxonil-resistant and pyrimethanil-resistant phenotypes of Penicillium expansum from apple. Phytopathology 98: 18–28. doi: 10.1094/PHYTO-98-4-0427 [DOI] [PubMed] [Google Scholar]
- 26.Takahashi H, Oiki S, Kusuya Y, Urayama S, Hagiwara D (2021) Intimate genetic relationships and fungicide resistance in multiple strains of Aspergillus fumigatus isolated from a plant bulb. Environ Microbiol 9: 5621–5638. doi: 10.1111/1462-2920.15724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arima K, Imanaka H, Kousaka M, Fukuta A, Tamura G (1964) Pyrrolnitrin, a new antibiotic substance, produced by Pseudomonas. Agric Biol Chem 28: 575–576. doi: 10.1080/00021369.1964.10858275 [DOI] [Google Scholar]
- 28.Hagiwara D, Takahashi-Nakaguchi A, Toyotome T, Yoshimi A, Abe K, Kamei K, et al. (2013) NikA/TcsC histidine kinase is involved in conidiation, hyphal morphology, and responses to osmotic stress and antifungal chemicals in Aspergillus fumigatus. PLoS One 8: e80881-. doi: 10.1371/journal.pone.0080881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui W, Beever RE, Parkes SL, Weeds PL, Templeton MD (2002) An osmosensing histidine kinase mediates dicarboximide fungicide resistance in Botryotinia fuckeliana (Botrytis cinerea). Fungal Genet Biol 36: 187–198. doi: 10.1016/s1087-1845(02)00009-9 [DOI] [PubMed] [Google Scholar]
- 30.Yoshimi A, Tsuda AM, Tanaka AC (2004) Cloning and characterization of the histidine kinase gene Dic1 from Cochliobolus heterostrophus that confers dicarboximide resistance and osmotic adaptation. Mol Genet Genomics 271: 228–236. doi: 10.1007/s00438-003-0974-4 [DOI] [PubMed] [Google Scholar]
- 31.Motoyama T, Kadokura K, Ohira T, Ichiishi A, Fujimura M, Yamaguchi I, et al. (2005) A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fungal Genet Biol 42: 200–212. doi: 10.1016/j.fgb.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Leroux P, Fillinger S (2008) The HOG1-like MAP kinase Sak1 of Botrytis cinerea is negatively regulated by the upstream histidine kinase Bos1 and is not involved in dicarboximide- and phenylpyrrole-resistance. Fungal Genet Biol 45: 1062–1074. doi: 10.1016/j.fgb.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 33.Fillinger S, Ajouz S, Nicot PC, Leroux P, Bardin M (2012) Functional and structural comparison of pyrrolnitrin-and iprodione-induced modifications in the class III histidine-kinase Bos1 of Botrytis cinerea. PLoS One 7: e42520-. doi: 10.1371/journal.pone.0042520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan Y, Ge C, Liu S, Wang J, Zhou M (2013) A two-component histidine kinase Shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum. Mol Plant Pathol 14: 708–718. doi: 10.1111/mpp.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han X, Zhao H, Ren W, Lv CY, Chen C (2017) Resistance risk assessment for fludioxonil in Bipolaris maydis. Pestic Biochem Physiol 139: 32–39. doi: 10.1016/j.pestbp.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 36.Fujimura M, Ochiai N, Oshima M, Motoyama T, Ichiishi, Usami R, et al. (2003) Putative homologs of SSK22 MAPKK kinase and PBS2 MAPK kinase of Saccharomyces cerevisiae encoded by os-4 and os-5 genes for osmotic sensitivity and fungicide resistance in Neurospora crassa. Biosci Biotechnol Biochem 67: 186–191. doi: 10.1271/bbb.67.186 [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Chen C, Zhu C, Sun X, Ruan R, Li H (2014) Os2 MAP kinase-mediated osmostress tolerance in Penicillium digitatum is associated with its positive regulation on glycerol synthesis and negative regulation on ergosterol synthesis. Microbiol Res 169: 511–521. doi: 10.1016/j.micres.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 38.Okada A, Banno S, Ichiishi A, Kimura M, Yamaguchi I, Fujimura M (2005) Pyrrolnitrin interferes with osmotic signal transduction in Neurospora crassa. J Pestic Sci 30: 378–383. [Google Scholar]
- 39.Pawar S, Chaudhari A, Prabha R, Shukla R, Singh DP (2019) Microbial pyrrolnitrin: natural metabolite with immense practical utility. Biomolecules 9: 443-. doi: 10.3390/biom9090443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 1 μg/mL fludioxonil (right, FLU). Colony growth rate in the presence of fludioxonil ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 10 μg/mL iprodione (right, IPR). Colony growth rate in the presence of iprodione ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
99 Penicillium strains grown in PDA (left, CTRL) and PDA containing 0.05 μg/mL pyrrolnitrin (right, PRN). Colony growth rate in the presence of pyrrolnitrin ≥ 50%; “fungicide-resistant”, < 50%; “fungicide-sensitive”.
(TIFF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.