Abstract
The actin cytoskeleton and actin-dependent molecular and cellular events are responsible for the organization of eukaryotic cells and their functions. Viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), depend on host cell organelles and molecular components for cell entry and propagation. Thus, it is not surprising that they also interact at many levels with the actin cytoskeleton of the host. There have been many studies on how different viruses reconfigure and manipulate the actin cytoskeleton of the host during successive steps of their life cycle. However, we know relatively little about the interactions of SARS-CoV-2 with the actin cytoskeleton. Here, we describe how the actin cytoskeleton is involved in the strategies used by different viruses for entry, assembly, and egress from the host cell. We emphasize what is known and unknown about SARS-CoV-2 in this regard. This review should encourage further investigation of the interactions of SARS-CoV-2 with cellular components, which will eventually be helpful for developing novel antiviral therapies for mitigating the severity of COVID-19.
Introduction
In all eukaryotic cells, the actin cytoskeleton is indispensable for the anchoring and organization of the subcellular compartments and organelles. It regulates most cell functions, including cell–cell adhesion, cell movement, and cell division [1–3]. Cortical actin participates in all events related to the expression and presentation of membrane/cell surface molecules, the formation and movement of endocytic/phagocytic vesicles, virus entry, exocytosis, and virus egress. In the cell nucleus, actin regulates DNA repair, chromatin remodeling and condensation, and gene transcription [4, 5]. The two main forms of actin in eukaryotic cells are monomeric globular G-actin and polymerized filamentous F-actin. Actin polymerization and G/F actin dynamics are regulated by the small GTPase RhoA and actin-interacting proteins [2, 6–14]. Because of the pleiotropy of actin function, it is not surprising that the life cycle of viruses, from the point of cell entry to the egress of the progeny, is intimately connected with the actin cytoskeleton of the host cell and that viruses hijack various actin functions and modify or redirect them to enhance self-propagation. Viruses reconfigure and manipulate the actin cytoskeleton of the host to optimize the production of progeny. It is known that RNA viruses can promote actin polymerization and reorganization [15]. For example, the coronaviruses transmissible gastroenteritis virus (TGEV) and porcine hemagglutinating encephalomyelitis virus (PHEV) use Rac1/Cdc42 small GTPases of the RhoA family to induce actin polymerization [15–17]. The virus also manipulates cell adhesion complexes and modifies intercellular contacts, allowing directional cell-to-cell spread. The different strategies used by different viruses to manipulate the actin cytoskeleton suggest that these strategies evolved independently [18, 19]. Here, we describe strategies involving the actin cytoskeleton that are used by various viruses during entry, assembly, and egress from the host cell and point out what is already known about severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in this regard. This review should encourage further investigation of the mechanism used by SARS-Cov-2 and eventually help in the development of novel antiviral therapeutic or prevention strategies.
SARS-CoV-2 structure
SARS-CoV-2 virions are spherical particles with a diameter of ~100 nm that contain a tightly packed positive-sense, single-stranded, 29,811-nucleotide-long genomic RNA that is “translation-ready”; i.e., it serves as an mRNA that can be translated directly (using the synthetic machinery of the host) to produce a set of viral proteins that are necessary for RNA replication and formation of new copies of the virus [20–24]. The RNA genome has 10–14 open reading frames (ORFs) and codes for 24–27 proteins [20, 25, 26]. Viral transcription and replication also rely on intricate long-range RNA–RNA interactions within the viral genome as well as interactions with host RNA molecules [27]. Viral RNA is surrounded by a protective coat, called a capsid, consisting of 1000 copies of the RNA-binding nucleocapsid (N) protein (Fig. 1). The N protein is highly phosphorylated, has a molecular weight of 46.5 kDa, and through the binding to a genomic packaging signal facilitates the packaging of the viral genome within the capsid [28, 29]. The capsid is surrounded by a membrane envelope, which is derived from the host cell but has viral proteins inserted into it: ~1000 copies of the membrane (M) protein, ~20 copies of the envelope (E) protein, and ~100 copies of the spike (S) protein (Fig. 1). The M protein has a molecular weight of 25–30 kDa, and it forms a dimer that shapes the viral membrane and promotes its binding to the capsid [29, 30]. The E protein has a molecular weight of 8–12 kDa and has ion channel activity. It is crucial for assembly of the virus and its release from the host cell [29]. The S proteins form the viral spikes. Each spike contains three copies of the S protein and has a length of ~10 nm [20, 29]. The S protein has a molecular weight of 150 kDa, is N-glycosylated, and belongs to the class I family of fusion proteins, which bind to receptors and mediate membrane fusion [29, 31]. The S protein consists of two subunits: S1 and S2. The S1 subunit contains the receptor-binding domain, and the S2 subunit mediates membrane fusion [32]. The S-protein-containing spikes of SARS-CoV-2 have high affinity for angiotensin-converting enzyme 2 (ACE2), which serves as a cell-surface receptor for the virus and facilitates its entry into the cell [33–35]. The binding of the virus to the host cells involves two steps: the first is docking of the virus to ACE2, and the second is activation of the spike protein by proteolytic cleavage at two sites by host-encoded proteases [36, 37]. Proteolytic cleavage itself is also a two-step process: the first cleavage occurs at the boundary of the S1 and S2 subunits in the spike protein precursor, and the second exposes the S2 subunit. The first cleavage step can occur either before or after binding to the receptor. The second cleavage can occur before endocytosis, through the activity of the serine protease TMPRSS2, or after endocytosis, mediated by endosomal proteases. The second cleavage primes the S2 subunit to induce fusion of the viral membrane with the cell membrane. In SARS-CoV-2 and Middle East respiratory syndrome coronavirus (MERS-CoV), but not SARS-CoV-1 (called SARS-CoV before the emergence of SARS-CoV-2), the boundary between S1 and S2 subunits contains a stretch of amino acids that can be recognized and cleaved by another serine protease called furin [32, 36, 37]. The S protein also has a sialic-acid-binding domain that interacts with sialic-acid-containing molecules on the surface of the host cell and facilitates virus entry [34]. This dual feature of the S protein might be one of the reasons for the high transmissibility of SARS-CoV-2 [33, 38]. Recent studies have suggested that the recruitment of SARS-CoV-2 to the cell surface is also facilitated by heparan sulfate (HS) proteoglycan surface molecules. HS interacts with the spike protein, increasing the virus concentration and promoting engagement with the ACE2 receptor [39]. Interestingly, the human coronaviruses HCoV-OC43 and HCoV-HKU1, which, like SARS-CoV-2, belong to the genus Betacoronavirus [40], have an additional gene encoding a hemagglutinin-esterase (HE) dimer protein [29]. In these viruses, the S-protein-containing spikes are interspersed among shorter ~8-nm stubby projections composed of the HE protein. The HE protein has a dual function [41]: it has a receptor-binding lectin domain that is specific for O-acetylated sialic acid (O-Ac-Sia) and promotes attachment of the virus to sialic-acid-containing molecules on the surface of the host cell, and it has a receptor-destroying sialate-O-acetyl esterase that promotes virus release [33]. It seems that the S and HE proteins are functionally interdependent and co-evolved to optimize the balance between virus attachment and release [42]. Studies with SARS-CoV-1 have shown that the stage of binding to the receptor and virus entry requires ~10 min [20]. The process of making new virus particles within the cell lasts around 10 hours and ends with the release of ~ 1000 newly formed viruses [20]. Below, we summarize what happens inside the infected cell, from the moment of virus attachment to the receptor until the release of viral progeny, and how the actin cytoskeleton of the host is involved in these processes.
Fig. 1.

SARS-CoV-2 structure. The SARS-CoV-2 virion consists of a nucleocapsid and an envelope. The nucleocapsid contains ss(+)RNA surrounded by RNA-binding nucleocapsid (N) proteins. The nucleocapsid is surrounded by a lipid bilayer membrane that is derived from the cell membrane of the host cell but has viral proteins inserted: the membrane (M), envelope (E), and spike (S) proteins
Actin-driven “surfing” of the virus
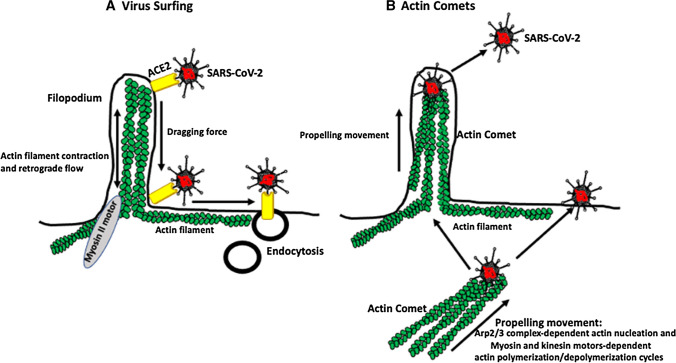
Some viruses, such as murine leukemia virus, human immunodeficiency virus, vesicular stomatitis virus, and human papillomavirus, slide (surf) on the surface of host cell protrusions called filopodia, which contain actin filaments, toward regions of the cell membrane with the highest endocytic activity [43] (Fig. 2A). Although viruses can also enter cells at other regions of the membrane, filopodium-dependent entry, due to the inherent ability of filopodia to bind and engulf various molecules, including viruses, is much more efficient [43]. Confocal time-lapse microscopy of murine leukemia virus (MLV) infecting human embryonic kidney HEK 293 cells showed that the virus, after attaching to the tip of a filopodium, moved downward toward the body of the cell [43]. The mechanism that initiates the surfing process is largely unknown, but it involves contact of the virus with its receptor(s) [43]. The surfing itself is powered by contraction (retrograde F-actin flow) of the filopodium’s actin filaments by myosin II motors located within the cortical actin at the base of the filopodium. This creates a dragging force that moves virus-receptor complexes to the base of the filopodium. Cytochalasin D, which blocks the barbed ends of actin filaments, inhibits surfing [18, 43]. Although there has been no description of SARS-CoV-2 surfing, high-resolution scanning microscopy has shown that SARS-CoV-2 adheres to the surface of filopodium‑like projections of African green monkey kidney Vero cells, which would suggest that surfing might occur [44]. Recently, Seyran et al. [34] reported that, prior to binding to the ACE2 receptor, the SARS-CoV-2 S protein interacts (through the flat sialic-acid-binding domain in the N-terminal domain of the S1 subunit) with the sialic acid layer of the epithelium, allowing the virus to move along the epithelial surface until it encounters and interacts with the ACE2 receptor. It remains to be investigated whether this movement of the virus has anything to do with the above-described surfing mediated by retrograde F-actin flow.
Fig. 2.
Hypothetical SARS-CoV-2 surfing and actin comets. A Filopodia contain bundles of actin filaments. The virus bound to its receptor surfs on the surface of the cell membrane, from the tip to the bottom of a filopodium, and to the membrane region with the highest endocytic activity for a subsequent internalization. Motor proteins located at the bottom of the filopodium cause actin filament contraction and retrograde movement, which propels the receptor-bound virus. B Viruses can also induce actin polymerization and the formation of actin filament comets (tails), which propel the virus within the cell toward the cell surface or expel it from the cell. Viral proteins induce actin nucleation via the activity of the Arp2/3 complex, and myosin- and kinesin-motor-dependent actin polymerization/depolymerization cycles propel the virus. Although these types of movement are used by many different viruses, so far, we do not know if SARS-CoV-2 can surf or use actin comets for movement
Virus entry into the cell
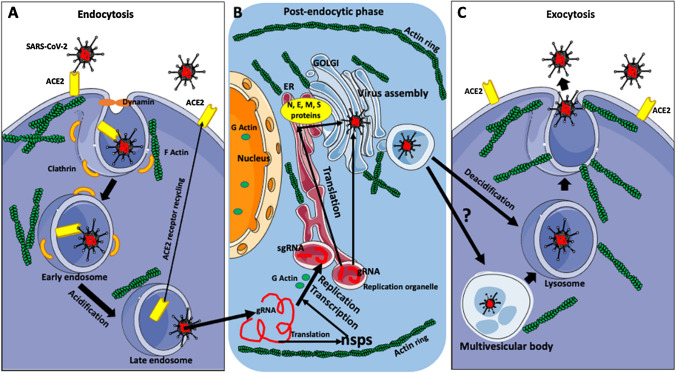
Coronaviruses can enter cells using various routes, including phagocytosis, micropinocytosis, and membrane fusion [19]. However, recent studies have shown that the preferred mode of entry of SARS-CoV-2 is through binding to the ACE2 receptor and clathrin-mediated endocytosis [45], which depends on the proper arrangement of cortical actin (Fig. 3). Owczarek et al. [46] showed that both stabilization and depolymerization of actin interfere with endocytosis and entry of the common-cold-causing human coronavirus OC43. During virus entry, the virus-receptor complex recruits clathrin, which coats the invaginating membrane pit. The initiation of membrane invagination is controlled by a multiprotein complex containing the membrane-associated protein intersectin-1, which directs interactions between endocytic vesicles and the actin cytoskeleton, the epidermal growth factor receptor substrate 15 (Eps15), which is a scaffolding adaptor protein, and the membrane-sculpting F-BAR-domain-containing Fer/Cip4 homology-domain-only proteins 1 and 2 (FCHo1/2) [46]. FCHo1/2 recruits intersectin-1 and Eps15 and induces membrane invagination [46, 47]. Eps15 [48] recruits adaptor protein 2 (AP2) [49], which recruits clathrin to form a lattice coating the membrane pit. The “neck” of the pit attracts the protein amphiphysin [50], which in turn recruits dynamin, which, via binding and hydrolysis of GTP, severs the “neck” of the pit from the cell membrane [51]. The scission results in the formation of a clathrin-coated endosomal vesicle containing the receptor and the virus (Fig. 3A) [46, 52].
Fig. 3.
SARS-CoV-2 life cycle. A Diagram summarizing virus endocytosis. The preferred mode of SARS-CoV-2 entry to the host cell is clathrin-mediated endocytosis. The binding of the virus to the ACE2 receptor, through the action of the membrane-sculpting complex (FCHo1/2, intersectin-1, and Eps15), induces a rearrangement of cortical actin, membrane bending, and recruitment of clathrin to the surface of the membrane pit. Subsequently, the “neck” of the pit recruits dynamin, which severs the “neck” of the pit from the cell membrane. The resulting clathrin-coated early endosome contains the virus and the receptor. The early endosome matures through acidification into a late endosome. In the late endosome, the virus is cleaved from the receptor by cathepsin-L, which also promotes viral and endosomal membrane fusion. The receptor recycles back to the cell membrane, and the viral genomic RNA (gRNA) is released into the cytoplasm. The movement of endocytic vesicles and their spatial distribution within the cell cytoplasm are dependent on actin/motor proteins. B Summary of the post-endocytic phase of the virus life cycle. Released viral RNA (gRNA) is translated into non-structural proteins (nsps), which are used for subsequent viral replication and transcription. First, viral gRNA (with the help of the nsps and RNA polymerases and with the involvement of the globular [G] actin) makes negative-sense RNA intermediates (not shown here) for the synthesis of gRNAs of the next generation and subgenomic RNAs (sgRNAs). The sgRNAs are translated into the viral structural proteins (N, E, M, E, and S), while the gRNAs are combined with structural proteins to form new virus particles. Viral replication and transcription occur within a specialized virus-induced, ER-derived vesicular/membrane compartment called the replication organelle. The newly synthesized viral proteins and RNAs move to the Golgi, where SARS-CoV-2 RNA coated with N proteins is enclosed by the host-cell membrane containing viral M, E, and S proteins. The maintenance and positioning of the ER and Golgi complex depend on actin filaments. Also, the ER/replication organelle and Golgi area are surrounded by a ring of actin filaments, which probably helps to concentrate the factors involved in the synthesis of the new virus within the ER/Golgi complex. Finally, the newly formed viruses are released from the cell through exocytosis. C Summary of virus exocytosis and egress. SARS-CoV-2-containing vesicles translocate through multivesicular bodies, which probably mature into lysosomes that have been deacidified, deactivating their lytic enzymes. The virus-containing lysosomes move to and fuse with the cell membrane. Virus-induced rearrangements of cortical actin and membrane bending expel the virus from the cell
The endosomal phase of the virus
The next step after the formation of the endosome containing the SARS-CoV-2-receptor complex is the acidification of the endosome through the proton-pump-dependent release of Ca2+ from the endosome via the intracellular messenger nicotinic acid adenine dinucleotide phosphate (NAADP)-sensitive two-pore channel (TPC2) [53]. The acidification of the endosome occurs during its maturation from an early endosome to the late endosome. In the late endosome, the complex of the S protein and ACE2 receptor is cleaved by cathepsin L, which promotes the fusion of the viral and endosome membranes and the release of the viral RNA into the cytoplasm (Fig. 3A) [54]. Endosomal acidification inhibitors, such as chloroquine or bafilomycin A1, have been shown to drastically reduce RNA release and replication of SARS-CoV-2 [55]. The spatial arrangement of endosomes, their trafficking within the cell, and the recycling of endosomal membranes back to the cell surface are supported by actin filaments and microtubules and are powered by various motor proteins [56].
Virus replication and translation
The released viral RNA (genomic RNA, gRNA) undergoes replication, transcription, and translation to produce viral proteins and progeny RNAs [22, 57] (Fig. 3B). First, the two large open reading frames, ORF1a and ORF1b, of the viral RNA are translated into the polyproteins pp1a and pp1ab, which, after co-translational and post-translational modification, are converted into non-structural proteins (nsps), which participate in viral replication and transcription [22, 57, 58]. In the next step, viral gRNA, with the help of the nsps, generates negative-sense RNA intermediates, which are the templates for synthesis of gRNAs of the next generation and subgenomic RNAs (sgRNAs). The sgRNAs are translated to produce the viral structural proteins (N, E, M, and S), while the gRNAs are incorporated with the structural proteins into virus particles to produce new viral progeny (Fig. 3B). A recent analysis of the SARS-CoV-2 transcriptome showed that the viral RNA also has several ORFs of unknown function [22].
The replication of the viral RNA of positive-strand (+) RNA viruses and its transcription occurs within virus-induced replication organelles situated in the vicinity of the host cell nucleus (Fig. 3B). This replication compartment, which was identified recently in cells infected with SARS-CoV-1 [59], creates a protective niche for viral replication and transcription. It is derived from the ER or other membranous structures of the host cell and consists of double-membrane vesicles (DMVs), double-membrane cups, called double-membrane spherules (DMSs), convoluted membranes (CMs), or/and tubules [57, 60–62]. We do not know if actin participates in the establishment and maintenance of the replication organelle. However, the involvement of the actin cytoskeleton in the formation and structure of other membranous organelles in eukaryotic cells [2] would imply that it does. It is known that actin regulates various transcription factors in eukaryotic cells and that binding to actin is necessary for the activity of RNA polymerases [5]. This implies that the transcription of viral RNA should also be actin-dependent. This assumption is supported by older studies showing that the actin cytoskeleton is involved in the replication and transcription of human parainfluenza virus type 3 [63]. Moreover, mutational analysis of respiratory syncytial virus (RSV) has shown that actin and profilin are necessary for viral transcription and that the divalent-cation-binding domain of actin is necessary for activation of viral RNA synthesis [64, 65]. Another study has shown that viruses that replicate in the nucleus interact with nuclear actin for replication, assembly, and export of mRNA or capsids [66]. The fact that interactions with actin are necessary for many viral functions suggests that transcription of SARS-CoV-2 RNA might also be actin-dependent. However, despite that, to our knowledge, there is no direct information available on this subject.
Virus assembly and egress
The newly synthesized viral proteins and RNAs are transported to the ER/Golgi intermediate compartment (ERGIC, also referred to as the vesicular-tubular cluster [VTC]) [67], where the SARS-CoV-2 RNA coated with N proteins is enveloped by a host-cell membrane containing viral M, E, and S proteins (Fig. 3C). From the ERGIC, the virus enters the trans-Golgi network (TGN)/Golgi complex, where the proteins undergo posttranslational modification [68]. Some time ago, a peculiar structure called “the juxtanuclear actin ring” was described in coronavirus-infected cells (Fig. 3B). The ring, composed of actin filament layers surrounding the cell nucleus and the adjacent ER and Golgi compartments, facilitates viral genome replication and protein synthesis [19, 69]. The sequestration of the molecules needed for virus replication, transcription, and translation by the actin ring may concentrate them in the vicinity of ER and Golgi for further processing. Another function of the perinuclear actin ring was described in human adenovirus infection. In this case, the adenovirus early region 4 ORF4 protein (E4orf4) induces, via the myosin II motor/Src kinase/Cdc42/N-Wasp/RhoA kinase and Rac1 pathways, actin polymerization around the nucleus. Subsequently, the actin ring, by recruiting endosomes and disrupting vesicular trafficking and membrane dynamics, activates an endo-lysosomal-based cell death pathway [70].
The egress of newly assembled virus particles from the cell occurs via exocytosis [54, 71–73] (Fig. 3C). Recent studies have shown that SARS-CoV-2 and other betacoronaviruses use a unique exit strategy. Instead of using the conventional secretory pathway used by other viruses, they use deacidified lysosomes to egress from the cell [68]. The exact mechanism of deacidification of the lysosomes, which also deactivates their lytic enzymes, is still unknown. Ghosh et al. [68] have suggested that the deacidification of lysosomes is either a direct consequence of too much cargo (viruses) and/or a dysfunctional proton pump or that some viral proteins have a viroporin function, which, by forming hydrophilic pores, modifies membrane permeability and Ca2+ homeostasis, leading to lysosome deacidification. It is also still unknown how the viruses are translocated from the TGN/Golgi to lysosomes. Ghosh et al. [68] propose two possible routes: One route is a transfer from the Golgi to multivesicular bodies (MVBs), which mature into lysosomes via the activity of the Rab7 GTPase pathway (Fig. 3C). The fact that the inhibition of Rab7 causes a reduction in both the formation of lysosomes and the egress of the virus supports this hypothesis [68]. Another, more complicated option is the retrograde transport of the virus from the Golgi back to the ER/ERGIC and then to lysosomes. A modification of the second option is the possible involvement of microphagy, with fragments of the ER containing the virus being engulfed directly by lysosomes [68]. Studies on the coronavirus avian infectious bronchitis virus have shown that budding of the virus from the cell membrane and its egress are actin-filament-dependent and involve interaction between actin and amino acids A159 and K160 of the viral membrane protein M [19, 74]. Using scanning electron microscopy and atomic force microscopy, it was shown that, just before its egress, SARS-CoV-1 induces the formation of pseudopodia and thickening of the cortical actin, which, by bending the membrane, expels the virus progeny from the host cell [75].
Actin comets
Many viruses, including vaccinia virus, Ebola virus, Marburg virus, and insect baculoviruses, use actin “tails” or so-called “actin comets” to be propelled within the cytoplasm to the cell periphery and/or to the outside of the infected cell (Fig. 2B). Viral proteins induce actin nucleation via the activity of the Arp2/3 complex. This is followed by interactions with the myosin and kinesin motors and actin polymerization/depolymerization cycles, which propel the virus. Because the comet-associated viruses were observed not only inside the cytoplasm but also in the vicinity of cell–cell junctions, it has been proposed that the propelling force of the actin comet allows the virus to pass through the actin-reinforced cellular junction and be delivered to an adjacent cell [76–78]. So far, there is no information about whether any coronaviruses, including SARS-CoV-2, use actin comets for movement inside and outside of the cell.
Actin and direct cell-to-cell transmission of the virus
Direct cell-to-cell transmission, which is faster and much more efficient than cell-free spread and allows the immune response, especially antibody-mediated immunity, to be circumvented, is used efficiently by many viruses, including SARS-CoV-2 [70, 78–80]. Below, we describe several possible routes of direct transmission of the virus between cells. All of these routes of transmission involve the actin cytoskeleton of the host cell.
a. Tunneling nanotubes (TNTs)
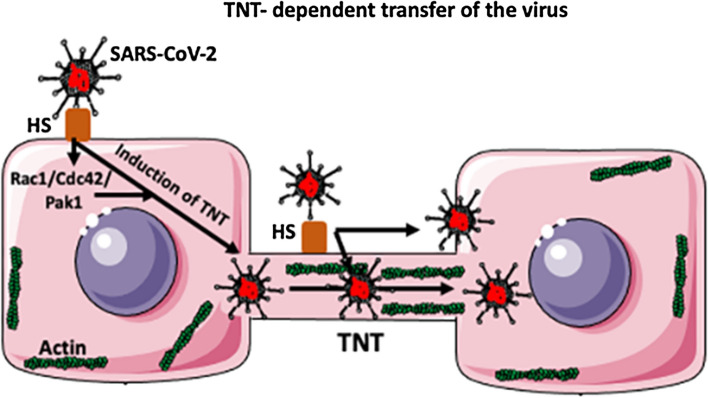
Tunneling nanotubes (TNTs) are long, thin (50–1500 nm in diameter), F-actin-based open-ended channels used by neighboring cells to exchange large cargo, such as protein complexes, lipids, nucleic acids, vesicles, pathogens, and organelles (Golgi, ER, lysosomes, endosomes, and mitochondria) at the distance of hundreds of micrometers [78, 79, 81–85] (Fig. 4). Thicker TNTs, which are used for the transport of very large cargo, in addition to F-actin, also contain microtubules. The traveling speed of the cargo through the TNTs is very fast, reaching ~7 nm/s [85]. Viruses can also “surf”, docking to heparan sulfate (HS) receptors on the surface of TNTs for long distances (Fig. 4). The same receptors can also induce the fusion of the virus with the TNT membrane and induce endocytic internalization of the virus into the TNT lumen [85] (Fig. 4). Actin polymerization required for the formation of TNTs is regulated by Rho GTPase signaling, including the Rac1/Cdc42/ Pak1 pathways [86]. Many viruses, such as HIV-1, influenza virus, and coronaviruses, induce the formation of TNTs to spread between cells [85, 86]. Interestingly, TNTs formed by Mycobacterium tuberculosis (which is known to enhance HIV-1 pathology) are used by the coinfecting HIV for cell-to-cell transfer [86]. We do not know if SARS-CoV-2 uses TNTs to spread between cells. However, the observations that the coronaviruses murine hepatitis virus (MHV) and SARS-CoV-1 promote the formation of filopodia and membrane ruffling using the Rac1/Cdc42/Pak1pathways and that SARS-CoV-2 is able to bind to the surface HS suggest that it can also induce the formation of TNTs. This should certainly be studied further in the context of SARS-CoV-2, as it may offer novel therapeutic targets for preventing or treating COVID-19.
Fig. 4.
Hypothetical TNT-dependent transfer of SARS-CoV-2. Binding of the S protein of SARS-CoV-2 to a heparan sulfate (HS) proteoglycan molecule (which acts as a co-receptor for ACE2 binding) present on the cell surface, induces the formation of a tunneling nanotube (TNT) between the infected cell and a neighboring uninfected cell. The virus is transferred within the lumen of the TNT, which contains actin filaments. Rho GTPase signaling including the Rac1/Cdc42/Pak1 pathways regulates actin polymerization, which is required for the formation of the TNT. The virus can also, after binding to HS, surf on the surface of the TNT. The virus on the surface of the TNT can also be internalized into the TNT interior. Although there are no data showing that SARS-CoV-2 spreads through TNTs, the fact that its S protein binds to HS and that the closely related SARS-CoV-1 can induce various types of cytoplasmic extensions, suggests that it does
b. Virological synapses
Many viruses, including the retroviruses human T cell leukemia virus type 1 (HTLV-1), human immunodeficiency virus type 1 (HIV-l), and simian immunodeficiency virus (SIV), spread from cell to cell through a virus-induced structure called the virological synapse (VS) or the infectious synapse, which shares similarities with neurological, immunological, and plant synapses [87–90]. During synapse formation, pre-synaptic and post-synaptic cells adhere to each other, but their membranes do not fuse. The virological synapse (VS) transmits viruses directly from the infected (pre-synaptic) to the uninfected (post-synaptic) cell [87]. Also, HIV-1, after being captured and internalized by dendritic cells, is transferred through the immunological synapse to interacting T cells. However, studies have indicated that not only internalized virus is transmitted but also virus particles attached to the surface of dendritic cells. This type of viral transmission is called transinfection [91].
The formation of any synapse, including the VS, involves the polarization of F-actin filaments and Rho GTPase signaling [92–95]. The actin network at the synapse not only plays a structural role but is a scaffold for signaling molecules and the clustering of virus particles [92, 93]. It seems that, at least in the case of HIV-1, the synapse contains a local, F-actin-depleted zone that facilitates virus entry by removing the physical cortical actin barrier [96]. Like retroviruses, coronaviruses also use the VS for direct cell-to-cell transmission. Yang et al. [97] have described VS transmission of SARS-CoV-1 between dendritic cells (DC) and target cells. Recently, additional studies have suggested that the ability of the SARS-CoV-2 to invade the nervous system involves the transmission of the virus through neurological synapses [98–100].
c. Virus interaction with the cellular junctions
A direct transfer of viruses between cells can also occur through intercellular junctions. Many viruses have evolved various strategies to alter different types of cell junctions to allow viral penetration. There are three main categories of cell junctions: Tight (occluding) junctions support the exchange of small hydrophilic molecules. Anchoring junctions (adherens junctions, desmosomes, focal adhesions, and hemidesmosomes) attach cells to each other or the extracellular matrix. Adherens junctions and focal adhesions are connected to bundles of actin filaments, while desmosomes and hemidesmosomes are connected to intermediate filaments. Gap (communicating) junctions transfer small (below 1000 Da) proteins, amino acids, nucleotides, small metabolites, ions, regulatory molecules, and secondary messengers [82, 101, 102]. Studies have shown that hepatitis C virus and retroviruses may enter through tight junctions, human papilloma virus (HPV) may enter through the adherens junctions, and HIV may modify gap junctions for entry [103]. Retroviruses modify the distribution of the tight-junction proteins occludin and claudin-1 and destroy tight junctions via the RhoA/ROCK /MLC signaling pathways [103–105]. Hepatitis C virus also affects occludin and claudin-1, and these proteins may also promote endocytosis of the virus [103, 106, 107]. HPV enters through adherens junctions. It modifies the distribution of adherens junction proteins such as β-catenin, disrupting their association with E-cadherin and the actin cytoskeleton [103, 108]. HIV hijacks gap junctions to spread apoptosis-inducing factors to uninfected cells and destroys, via the mitogen-activated protein kinase (MAPK) pathway, adherens and tight junctions [103, 109]. Studies have shown that the intermediate filament protein vimentin, connected to adherence junctions, is essential for several phases of the viral life cycle, including entry, transport, replication, assembly, and egress [110]. Intermediate filaments, actin, and microtubules are also crucial for the entry and propagation of dengue virus, Zika virus, and West Nile virus [111]. Recent studies have shown that alterations in cell junctions induced by SARS-CoV-2 are highly damaging for patients [112, 113]. Humans have over 100 proteins containing PDZ (post-synaptic density protein) domains, some of which are components of cell junctions. The envelope (E) proteins of SARS-CoV-2 and SARS-CoV-1 have a PDZ-binding motif (PBM). The binding of the viral PBM to PALS1 (protein associated with Lin seven 1) present in human cell junctions relocates PALS1 from the junctions to the ERGIC compartment, where the virus is assembled. The envelope protein also binds and dislocates other PDZ-containing junctional proteins, including the adherens junction protein syntenin and the tight junction ZO-113 (zonula occludens-113) protein, which regulates the actin-relevant RhoA/ROCK pathway [114]. Disruption of the junctional proteins in the inter-epithelial junctions of the lungs causes vascular leakage, damages the alveoli, and induces acute respiratory distress syndrome (ARDS) in COVID-19 patients [112, 115].
Virus-induced syncytia
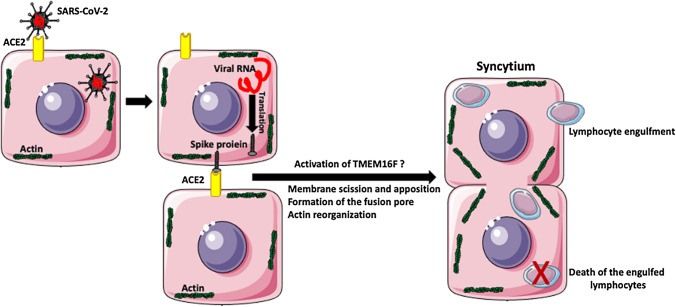
Pathological examination of the lungs of the patients with severe cases of COVID-19 infection shows the presence of SARS-CoV-2-infected, multinucleated syncytia of pneumocytes [116, 117]. These patients are also often severely lymphopenic; i.e., they have a deficit of lymphocytes, which decreases their ability to fight the infection [118, 119]. It is known that many viruses, including poxviruses, paramyxoviruses, herpesviruses, and retroviruses, induce the fusion of infected cells into multinuclear syncytia, which eventually die [120]. Recent studies have shown that SARS-CoV-2 infection causes the formation of giant syncytia and the destruction of tight junctions in the lung airway epithelium [113]. There are two main types of syncytia: (1) homotypic syncytia consisting of cells of the same type and (2) heterotypic syncytia, which result from the fusion of cells of different types. Recently, Zhang et al. [119] described another type of syncytia in the lungs of COVID-19 patients. These syncytia consist of lung cells containing internalized lymphocytes (Fig. 5). The cell fusion leading to the formation of syncytia requires the rupture of the membranes of the fusing partners, followed by membrane rejoining. The whole process is facilitated by fusogenic proteins and involves the restructuring of the supportive platform of the actin cytoskeleton underlying the cell membranes. The spike protein of SARS-CoV-2 is a fusogen with homology to mammalian syncytin-1, which is involved in the formation of the syncytial layers of the placenta [31]. The model and the details of the fusion process are described in references [31, 119]. Virus-induced syncytia not only enhance virus spread but may also serve as a tool for the elimination of immune cells that fight the virus. Studies of COVID-19 patients showed that SARS-CoV-2 induces both homotypic syncytia of pneumocytes, and heterotypic cell structures containing fused lung cells with the engulfed lymphocytes inside [36, 119]. These studies suggest that SARS-CoV-2-infected cells express the viral S (spike) protein on their surface. The interaction of the spike protein with the ACE2 receptor of the neighboring cell activates the calcium-activated lipid scramblase TMEM16F (anoctamin 6), which that exposes phosphatidylserine on the cell surface and triggers the formation of a syncytium, which subsequently engulfs the immune cells [116, 122]. These studies suggest that syncytium-mediated lymphocyte internalization and elimination may be one of the causes of lymphopenia in COVID-19 patients [119–121]. Recent studies indicate the antihelminthic drug niclosamide suppresses the activity of TMEM16F and inhibits spike-induced syncytium formation by SARS-CoV-2 [123].
Fig. 5.
SARS-CoV-2-induced syncytia. After infection of the cell and release of viral RNA, the infected cell synthesizes the viral spike (S) protein. The S protein expressed on the surface of the infected cell interacts with an ACE2 receptor on an uninfected cell. Because the viral S protein is fusogenic, this interaction initiates a cascade of events that eventually lead to the apposition of neighboring cell membranes, membrane scission, and cell fusion. The interaction of the spike protein with the ACE2 receptor of the neighboring cell activates the calcium-activated lipid scramblase TMEM16F (anoctamin 6), which exposes phosphatidylserine on the cell surface and triggers the formation of a syncytium. SARS-CoV-2 induces not only the formation of pneumocyte syncytia but also the formation of heterotypic syncytia when pneumocyte-derived syncytia engulf the lymphocytes. The internalized lymphocytes undergo apoptosis and die. SARS-CoV-2 uses this mechanism to eliminate virus-fighting lymphocytes from the lungs
Conclusions
The information presented here indicates that interactions of viruses, including SARS-CoV-2, with the actin cytoskeleton and actin-related cell functions and signaling pathways, are of crucial importance for viral functions, infectivity, and pathogenicity. It is also clear that our knowledge about the interactions of SARS-CoV-2 with actin and other components of the cytoskeleton is still very limited and requires further studies. A better understanding of these interactions and processes and the participating viral and cellular components involved can help in the design of novel antiviral therapies and/or strategies to mitigate the severity of COVID-19 and its damaging, long-term effects. The conventional approaches to fighting viral infections include vaccinations, antiviral drugs, and preventing overactivation of the immune response, the last of which is especially important in COVID-19, in which the overproduction of inflammatory factors can cause a cytokine storm in the patient’s lungs [36]. A recent analysis of gene expression profiles in the SARS-CoV-1/SARS-CoV-2 infections using Gene Ontology (GO) and MetaCore showed that the upregulation of actin/cytoskeleton remodeling via the RhoA pathway is crucial for infection [124]. Thus, we propose that interference with actin-dependent functions of the virus and/or host immune cells using RhoA pathway inhibitors, such as the multiple sclerosis drugs fingolimod and siponimod, might be a novel therapeutic approach to fighting viral diseases [125].
Acknowledgments
Some of the drawings used to make figures were from Servier Medical ART: SMART, smart.servier.com.
Declarations
Conflict of interest
None of the authors has a conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kloc M, Uosef A, Wosik J, Kubiak JZ, Ghobrial RM. RhoA pathway and actin regulation of the golgi/centriole complex. Results Probl Cell Differ. 2019;67:81–93. doi: 10.1007/978-3-030-23173-6_5. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelpsch DJ, Tootle TL. Nuclear actin: from discovery to function. Anat Rec (Hoboken) 2018;301(12):1999–2013. doi: 10.1002/ar.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloc M, Chanana P, Vaughn N, Uosef A, Kubiak JZ, Ghobrial RM. New insights into cellular functions of nuclear actin. Biology (Basel). 2021;10(4):304. doi: 10.3390/biology10040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molinie N, Gautreau A. The Arp2/3 regulatory system and its deregulation in cancer. Physiol Rev. 2018;98(1):215–238. doi: 10.1152/physrev.00006.2017. [DOI] [PubMed] [Google Scholar]
- 7.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol. 2000;2(7):441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 8.Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. doi: 10.1016/S0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- 9.Romero S, Le Clainche C, Gautreau AM. Actin polymerization downstream of integrins: signaling pathways and mechanotransduction. Biochem J. 2020;477(1):1–21. doi: 10.1042/BCJ20170719. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe N. Inside view of cell locomotion through single-molecule: fast F-/G-actin cycle and G-actin regulation of polymer restoration. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(1):62–83. doi: 10.2183/pjab.86.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borovac J, Bosch M, Okamoto K. Regulation of actin dynamics during structural plasticity of dendritic spines: signaling messengers and actin-binding proteins. Mol Cell Neurosci. 2018;91:122–130. doi: 10.1016/j.mcn.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Lee M, Kang EH. Molecular dynamics study of interactions between polymorphic actin filaments and gelsolin segment-1. Proteins. 2020;88(2):385–392. doi: 10.1002/prot.25813. [DOI] [PubMed] [Google Scholar]
- 13.Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12(21):1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards RA, Bryan J. Fascins, a family of actin bundling proteins. Cell Motil Cytoskeleton. 1995;32(1):1–9. doi: 10.1002/cm.970320102. [DOI] [PubMed] [Google Scholar]
- 15.Swaine T, Dittmar MT. CDC42 use in viral cell entry processes by RNA viruses. Viruses. 2015;7:6526–6536. doi: 10.3390/v7122955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W, Zhu L, Yang X, Lin J, Yang Q. The epidermal growth factor receptor regulates cofilin activity and promotes transmissible gastroenteritis virus entry into intestinal epithelial cells. Oncotarget. 2016;7:12206–12221. doi: 10.18632/oncotarget.7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv X, Li Z, Guan J, Hu S, Zhang J, Lan Y, Zhao K, Lu H, Song D, He H, Gao F, He W. Porcine hemagglutinating encephalomyelitis virus activation of the integrin _5_1-FAK-cofilin pathway causes cytoskeletal rearrangement to promote its invasion of N2a cells. J Virol. 2019;93:e01736–e1818. doi: 10.1128/JVI.01736-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol. 2011;9(6):427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Z, Zhang Y, Lin Z, Shi K, Jiu Y. Cytoskeleton-a crucial key in host cell for coronavirus infection. J Mol Cell Biol. 2020;12(12):968–979. doi: 10.1093/jmcb/mjaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haque SM, Ashwaq O, Sarief A, Azad John Mohamed AK. A comprehensive review about SARS-CoV-2. Future Virol. 2020;15(9):625–648. doi: 10.2217/fvl-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt N, Lareau CA, Keshishian H, et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat Microbiol. 2021;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdout H, Stein D, Israeli O, Beth-Din A, Melamed S, Weiss S, Israely T, Paran N, Schwartz M, Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 26.Katsnelson A (2020) What do we know about the novel coronavirus’s 29 proteins? https://cen.acs.orginfectious-diseaseweb2020/04
- 27.Ziv O, Price J, Shalamova L, Kamenova T, Goodfellow I, Weber F, Miska EA. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol Cell. 2020;80(6):1067–1077.e5. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molenkamp R, Spaan WJ. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- 30.Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloc M, Uosef A, Kubiak JZ, Ghobrial RM. Exaptation of retroviral syncytin for development of syncytialized placenta, its limited homology to the SARS-CoV-2 spike protein and arguments against disturbing narrative in the context of COVID-19 vaccination. Biology (Basel) 2021;10(3):238. doi: 10.3390/biology10030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim CH. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor O-acetyl sialylation in virus-host interaction. Int J Mol Sci. 2020;21(12):4549. doi: 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seyran M, Takayama K, Uversky VN, Lundstrom K, Palù G, Sherchan SP, Attrish D, Rezaei N, Aljabali AAA, Ghosh S, Pizzol D, Chauhan G, Adadi P, Mohamed Abd El-Aziz T, Soares AG, Kandimalla R, Tambuwala M, Hassan SS, Azad GK, Pal Choudhury P, Baetas-da-Cruz W, Serrano-Aroca Á, Brufsky AM, Uhal BD (2020) The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS J. 10.1111/febs.15651
- 35.Zhang H, Zhang H. Entry, egress and vertical transmission of SARS-CoV-2. J Mol Cell Biol. 2021;13(3):168–174. doi: 10.1093/jmcb/mjab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, Penn R, Jimenez-Guardeño JM, Ortega-Prieto AM, Bussani R, Cannatà A, Rizzari G, Collesi C, Schneider E, Arosio D, Shah AM, Barclay WS, Malim MH, Burrone J, Giacca M. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594(7861):88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M, Hoffmann-Winkler H, Pohlmann, S (2018) Priming time: how cellular proteases arm coronavirus spike proteins. Activation Viruses Host Proteases. 10.1007/978-3-319-75474-1_4
- 38.Sun XL (2021) The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology cwab032. 10.1093/glycob/cwab032 [DOI] [PMC free article] [PubMed]
- 39.Zhang Q, Chen CZ, Swaroop M, Xu M, Wang L, Lee J, Wang AQ, Pradhan M, Hagen N, Chen L, Shen M, Luo Z, Xu X, Xu Y, Huang W, Zheng W, Ye Y. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;6(1):80. doi: 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu DX, Liang JQ, Fung TS (2021) Human coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encyclopedia Virol 428–440. 10.1016/B978-0-12-809633-8.21501-X
- 41.Soltani S. The hemagglutinin-esterase gene in human coronaviruses SARS-CoV-2, HKU1 and OC43. Eur Rev Med Pharmacol Sci. 2020;24(12):6484–6485. doi: 10.26355/eurrev_202006_21630. [DOI] [PubMed] [Google Scholar]
- 42.Lang Y, Li W, Li Z, Koerhuis D, van den Burg ACS, Rozemuller E, Bosch BJ, van Kuppeveld FJM, Boons GJ, Huizinga EG, van der Schaar HM, de Groot RJ. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc Natl Acad Sci USA. 2020;117(41):25759–25770. doi: 10.1073/pnas.2006299117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170(2):317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caldas LA, Carneiro FA, Higa LM, Monteiro FL, da Silva GP, da Costa LJ, Durigon EL, Tanuri A, de Souza W. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci Rep. 2020;10(1):16099. doi: 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayati A, Kumar R, Francis V, McPherson PS. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owczarek K, Szczepanski A, Milewska A, Baster Z, Rajfur Z, Sarna M, Pyrc K. Early events during human coronavirus OC43 entry to the cell. Sci Rep. 2018;8(1):7124. doi: 10.1038/s41598-018-25640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328(5983):1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Bergen En Henegouwen PM. Eps15: a multifunctional adaptor protein regulating intracellular trafficking. Cell Commun Signal. 2009;7:24. doi: 10.1186/1478-811X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 50.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303(5657):495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 51.Hill E, van Der Kaay J, Downes CP, Smythe E. The role of dynamin and its binding partners in coated pit invagination and scission. J Cell Biol. 2001;152(2):309–323. doi: 10.1083/jcb.152.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 53.Petersen OH, Gerasimenko OV, Gerasimenko JV. Endocytic uptake of SARS-CoV-2: the critical roles of pH, Ca2+, and NAADP. Function. 2020;1(1):zqaa003. doi: 10.1093/function/zqaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blaess M, Kaiser L, Sauer M, Csuk R, Deigner HP. COVID-19/SARS-CoV-2 infection: lysosomes and lysosomotropism implicate new treatment strategies and personal risks. Int J Mol Sci. 2020;21(14):4953. doi: 10.3390/ijms21144953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang C, Zhuang X, Zhang H, et al. Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virol J. 2021;18:46. doi: 10.1186/s12985-021-01515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31(100):20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandersen S, Chamings A, Bhatta TR. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat Commun. 2020;11(1):6059. doi: 10.1038/s41467-020-19883-7.PMID:33247099;PMCID:PMC7695715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FFGA, Koster AJ, Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18(6):e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagy PD, Strating JR, van Kuppeveld FJ. Building viral replication organelles: close encounters of the membrane types. PLoS Pathog. 2016;12(10):e1005912. doi: 10.1371/journal.ppat.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tenorio R, Fernández de Castro I, Knowlton JJ, Zamora PF, Lee CH, Mainou BA, Dermody TS, Risco C. Reovirus σNS and μNS proteins remodel the endoplasmic reticulum to build replication neo-organelles. mBio. 2018;9(4):e01253-18. doi: 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sachse M, Fernández de Castro I, Tenorio R, Risco C. The viral replication organelles within cells studied by electron microscopy. Adv Virus Res. 2019;105:1–33. doi: 10.1016/bs.aivir.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De BP, Banerjee AK. Involvement of actin microfilaments in the transcription/replication of human parainfluenza virus type 3: possible role of actin in other viruses. Microsc Res Tech. 1999;47(2):114–123. doi: 10.1002/(SICI)1097-0029(19991015)47:2<114::AID-JEMT4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 64.Burke E, Mahoney NM, Almo SC, Barik S. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol. 2000;74(2):669–675. doi: 10.1128/jvi.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harpen M, Barik T, Musiyenko A, Barik S. Mutational analysis reveals a noncontractile but interactive role of actin and profilin in viral RNA-dependent RNA synthesis. J Virol. 2009;83(21):10869–10876. doi: 10.1128/JVI.01271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cibulka J, Fraiberk M, Forstova J. Nuclear actin and lamins in viral infections. Viruses. 2012;4(3):325–347. doi: 10.3390/v4030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Appenzeller-Herzog C, Hauri HP. The ER-golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119(Pt 11):2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- 68.Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu VW, Fehr AR, Perlman S, Achar SR, Straus MR, Whittaker GR, de Haan CAM, Kehrl J, Altan-Bonnet G, Altan-Bonnet N. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183(6):1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao S, Gao J, Zhu L, Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robert A, Smadja-Lamère N, Landry MC, Champagne C, Petrie R, Lamarche-Vane N, Hosoya H, Lavoie JN. Adenovirus E4orf4 hijacks rho GTPase-dependent actin dynamics to kill cells: a role for endosome-associated actin assembly. Mol Biol Cell. 2006;17(7):3329–3344. doi: 10.1091/mbc.e05-12-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen JR, Lin LD, Machamer CE. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J Virol. 2011;85(12):5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McBride CE, Li J, Machamer CE. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J Virol. 2007;81(5):2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perrier A, Bonnin A, Desmarets L, Danneels A, Goffard A, Rouillé Y, Dubuisson J, Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294(39):14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Fang S, Xiao H, Chen B, Tam JP, Liu DX. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS ONE. 2009;4(3):e4908. doi: 10.1371/journal.pone.0004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng ML, Lee JW, Leong ML, Ling AE, Tan HC, Ooi EE. Topographic changes in SARS coronavirus-infected cells at late stages of infection. Emerg Infect Dis. 2004;10(11):1907–1914. doi: 10.3201/eid1011.040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strauss EJ. A virus joins the movement. Intracellular pathogens. Curr Biol. 1996;6(5):504–507. doi: 10.1016/s0960-9822(02)00527-4. [DOI] [PubMed] [Google Scholar]
- 77.Rietdorf J, Ploubidou A, Reckmann I, Holmström A, Frischknecht F, Zettl M, Zimmermann T, Way M. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat Cell Biol. 2001;3(11):992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 78.Labudová M. Cell-to-cell transport in viral families: faster than usual. Acta Virol. 2020;64(2):154–166. doi: 10.4149/av_2020_205. [DOI] [PubMed] [Google Scholar]
- 79.Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. J Virol. 2010;84(17):8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng C, Evans JP, King T, Zheng YM, Oltz EM, Whelan SPJ, Saif L, Peeples ME, Liu SL (2021) SARS-CoV-2 spreads through cell-to-cell transmission. bioRxiv [Preprint]. 2021 Jun 1:2021.06.01.446579. 10.1101/2021.06.01.446579 [DOI] [PMC free article] [PubMed]
- 81.Dupont M, Souriant S, Lugo-Villarino G, Maridonneau-Parini I, Vérollet C. Tunneling nanotubes: intimate communication between myeloid cells. Front Immunol. 2018;9:43. doi: 10.3389/fimmu.2018.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kloc M, Kubiak JZ. Exogenous molecule and organelle delivery in oogenesis. Results Probl Cell Differ. 2017;63:3–16. doi: 10.1007/978-3-319-60855-6_1. [DOI] [PubMed] [Google Scholar]
- 83.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 84.McCoy-Simandle K, Hanna SJ, Cox D. Exosomes and nanotubes: control of immune cell communication. Int J Biochem Cell Biol. 2016;71:44–54. doi: 10.1016/j.biocel.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tiwari V, Koganti R, Russell G, Sharma A, Shukla D. Role of tunneling nanotubes in viral infection, neurodegenerative disease, and cancer. Front Immunol. 2021;12:680891. doi: 10.3389/fimmu.2021.680891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang S, Kazanietz MG, Cooke M. Rho GTPases and the emerging role of tunneling nanotubes in physiology and disease. Am J Physiol Cell Physiol. 2020;319(5):C877–C884. doi: 10.1152/ajpcell.00351.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia E, Piguet V (2006) Virological synapse for cell-cell spread of viruses. Cell-Cell Channels 288–297. 10.1007/978-0-387-46957-7_22
- 88.Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol. 2010;84(7):3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piguet V, Sattentau Q. Dangerous liaisons at the virological synapse. J Clin Invest. 2004;114(5):605–610. doi: 10.1172/JCI22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sewald X, Gonzalez D, Haberman A, et al. In vivo imaging of virological synapses. Nat Commun. 2012;3:1320. doi: 10.1038/ncomms2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cavrois M, Neidleman J, Kreisberg JF, Greene WC. In vitro derived dendritic cells trans-infect CD4 T cells primarily with surface-bound HIV-1 virions. PLoS Pathog. 2007;3(1):e4. doi: 10.1371/journal.ppat.0030004.PMID:17238285;PMCID:PMC1779297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dustin M, Cooper J. The immunological synapse and the actin cytoskeleton: molecular hardware for T cell signaling. Nat Immunol. 2000;1:23–29. doi: 10.1038/76877. [DOI] [PubMed] [Google Scholar]
- 93.Felts RL, Narayan K, Estes JD, Shi D, Trubey CM, Fu J, Hartnell LM, Ruthel GT, Schneider DK, Nagashima K, Bess JW, Jr, Bavari S, Lowekamp BC, Bliss D, Lifson JD, Subramaniam S. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci USA. 2010;107(30):13336–13341. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haller C, Tibroni N, Rudolph JM, Grosse R, Fackler OT. Nef does not inhibit F-actin remodelling and HIV-1 cell-cell transmission at the T lymphocyte virological synapse. Eur J Cell Biol. 2011;90(11):913–921. doi: 10.1016/j.ejcb.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 95.Kloc M, Kubiak JZ, Li XC, Ghobrial RM. The newly found functions of MTOC in immunological response. J Leukoc Biol. 2014;95(3):417–430. doi: 10.1189/jlb.0813468. [DOI] [PubMed] [Google Scholar]
- 96.Vasiliver-Shamis G, Cho MW, Hioe CE, Dustin ML. Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J Virol. 2009;83(21):11341–11355. doi: 10.1128/JVI.01440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78(11):5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92(6):552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reza-Zaldívar EE, Hernández-Sapiéns MA, Minjarez B, Gómez-Pinedo U, Márquez-Aguirre AL, Mateos-Díaz JC, Matias-Guiu J, Canales-Aguirre AA. Infection mechanism of SARS-COV-2 and its implication on the nervous system. Front Immunol. 2021;11:621735. doi: 10.3389/fimmu.2020.621735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yashavantha Rao HC, Jayabaskaran C. The emergence of a novel coronavirus (SARS-CoV-2) disease and their neuroinvasive propensity may affect in COVID-19 patients. J Med Virol. 2020;92(7):786–790. doi: 10.1002/jmv.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alberts B, Johnson A, Lewis J et al (2002) Molecular biology of the cell, 4th edn. Garland Science, New York. Cell junctions. Available from https://www.ncbi.nlm.nih.gov/books/NBK26857/
- 102.Evans WH. Cell communication across gap junctions: a historical perspective and current developments. Biochem Soc Trans. 2015;43:450–459. doi: 10.1042/BST20150056. [DOI] [PubMed] [Google Scholar]
- 103.Dong D, Xie W, Liu M. Alteration of cell junctions during viral infection. Thorac Cancer. 2020;11(3):519–525. doi: 10.1111/1759-7714.13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lopez S, Arias CF. Multistep entry of rotavirus into cells: a versaillesque dance. Trends Microbiol. 2004;12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 105.Nava P, Lopez S, Arias CF, Islas S, Gonzalez-Mariscal L. The rotavirus surface protein VP8 modulates the gate and fence function of tight junctions in epithelial cells. J Cell Sci. 2004;117:5509–5519. doi: 10.1242/jcs.01425. [DOI] [PubMed] [Google Scholar]
- 106.Harris HJ, Davis C, Mullins JG, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mailly L, Baumert TF. Hepatitis C virus infection and tight junction proteins: The ties that bind. Biochim Biophys Acta Biomembr. 2020;1862(7):183296. doi: 10.1016/j.bbamem.2020.183296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heuser S, Hufbauer M, Marx B, et al. The levels of epithelial anchor proteins beta-catenin and zona occludens-1 are altered by E7 of human papillomaviruses 5 and 8. J Gen Virol. 2016;97:463–472. doi: 10.1099/jgv.0.000363. [DOI] [PubMed] [Google Scholar]
- 109.Sufiawati I, Tugizov SM. HIV-associated disruption of tight and adherens junctions of oral epithelial cells facilitates HSV-1 infection and spread. PLOS One. 2014;9:e88803. doi: 10.1371/journal.pone.0088803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y, Wen Z, Shi X, Liu YJ, Eriksson JE, Jiu Y. The diverse roles and dynamic rearrangement of vimentin during viral infection. J Cell Sci. 2020;134(5):jcs.250597. doi: 10.1242/jcs.250597. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Y, Gao W, Li J, Wu W, Jiu Y. The Role of Host Cytoskeleton in Flavivirus Infection. Virol Sin. 2019;34(1):30–41. doi: 10.1007/s12250-019-00086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chai J, Cai Y, Pang C, et al. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat Commun. 2021;12:3433. doi: 10.1038/s41467-021-23533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu N, Wang W, Liu Z, et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raya-Sandino A, Castillo-Kauil A, Domínguez-Calderón A, Alarcón L, Flores-Benitez D, Cuellar-Perez F, López-Bayghen B, Chávez-Munguía B, Vázquez-Prado J, González-Mariscal L. Zonula occludens-2 regulates Rho proteins activity and the development of epithelial cytoarchitecture and barrier function. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1714–1733. doi: 10.1016/j.bbamcr.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 115.Teoh KT, Siu YL, Chan WL, Schlüter MA, Liu CJ, Peiris JS, Bruzzone R, Margolis B, Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21(22):3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39(23):e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin L, Li Q, Wang Y, Shi Y. Syncytia formation during SARS-CoV-2 lung infection: a disastrous unity to eliminate lymphocytes. Cell Death Differ. 2021;28(6):2019–2021. doi: 10.1038/s41418-021-00795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fathi N, Rezaei N. Lymphopenia in COVID-19: therapeutic opportunities. Cell Biol Int. 2020;44(9):1792–1797. doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Z, Zheng Y, Niu Z, Zhang B, Wang C, Yao X, Peng H, Franca DN, Wang Y, Zhu Y, Su Y, Tang M, Jiang X, Ren H, He M, Wang Y, Gao L, Zhao P, Shi H, Chen Z, Wang X, Piacentini M, Bian X, Melino G, Liu L, Huang H, Sun Q. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 2021;28:2765–2777. doi: 10.1038/s41418-021-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chan KMC, Son S, Schmid EM, Fletcher DA. A viral fusogen hijacks the actin cytoskeleton to drive cell-cell fusion. Elife. 2020;9:e51358. doi: 10.7554/eLife.51358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou X, Platt JL. Molecular and cellular mechanisms of mammalian cell fusion. Adv Exp Med Biol. 2011;713:33–64. doi: 10.1007/978-94-007-0763-4_4. [DOI] [PubMed] [Google Scholar]
- 122.Wu N, Cernysiov V, Davidson D, Song H, Tang J, Luo S, Lu Y, Qian J, Gyurova IE, Waggoner SN, Trinh VQ, Cayrol R, Sugiura A, McBride HM, Daudelin JF, Labrecque N, Veillette A. Critical role of lipid scramblase TMEM16F in phosphatidylserine exposure and repair of plasma membrane after pore formation. Cell Rep. 2020;30(4):1129–1140.e5. doi: 10.1016/j.celrep.2019.12.066.PMID:31995754;PMCID:PMC7104872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kloc M, Ghobrial RM, Kubiak JZ. How nicotine can inhibit cytokine storm in the lungs and prevent or lessen the severity of COVID-19 infection? Immunol Lett. 2020;224:28–29. doi: 10.1016/j.imlet.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu H-L, Yeh I-J, Phan NN, Wu Y-H, Yen M-C, Hung J-H, Chiao C-C, Chen C-F, Sun Z, Jiang J-Z, Hsu H-P, Wang C-Y, Lai M-D. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect Genet Evol. 2020 doi: 10.1016/j.meegid.2020.104438.Preprint.PII:S1567-1348(20)30269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kloc M, Ghobrial RM. The multiple sclerosis (MS) drugs as a potential treatment of ARDS in COVID-19 patients. Mult Scler Relat Disord. 2020;45:102437. doi: 10.1016/j.msard.2020.102437. [DOI] [PMC free article] [PubMed] [Google Scholar]