Abstract
We conducted a study to examine the clonal distribution of invasive serotype 1 and 5 isolates as representatives of serotypes that are rarely carried by healthy individuals compared to that of invasive serotype 6B and 23F isolates as representatives of serotypes often carried by young children for prolonged periods. All invasive serotype 1, 5, 6B, and 23F isolates recovered from blood cultures during January 1995 to May 1999 were analyzed; these included 66 serotype 1, 30 serotype 5, 11 serotype 6B, and 15 serotype 23F isolates. One hundred thirty-three nasopharyngeal (NP) isolates of the indicated four serotypes from healthy children were also studied. The strains were characterized using serotyping, antimicrobial susceptibility testing, and pulsed-field gel electrophoresis profiling. We found that both invasive and NP serotype 1 and 5 isolates were susceptible to penicillin and that each serotype showed only one clonal type. In contrast, serotype 6B and 23F strains showed different phenotypic characteristics as well as multiple clonal types; 10 clones were identified among 6B isolates, and 11 clones were identified among 23F isolates.
Infections due to Streptococcus pneumoniae continue to cause significant morbidity and mortality in children (14). More than 90 pneumococcal types have been identified on the basis of differences in their structures and the antigenicities of their capsular polysaccharides (8). The levels of virulence of S. pneumoniae strains differ between serogroups and types, with serogroups 1, 2, 3, 5, 7, 14, 16, 25, 28, 36, 43, 46, and 47 being more virulent and more likely to cause invasive disease than serogroups 6, 18, 19, and 23, which are recovered frequently from healthy carriers (15, 17).
The frequencies of disease-causing types have been shown to differ according to geographic region (12). The serotype distribution for children in developing countries tends to be somewhat different than in developed countries. Serotypes 1 and 5 are important types in developing countries and less so in developed settings. These two serotypes are the dominant cause of disease in southern Israel (N. Porat, R. Trefler, and R. Dagan, Abstr. Second Int. Symp. Pneumococci Pneumococcal Dis., abstr. P-12, 2000), South America (15, 18, 20), Africa (3), and India (11). However, other serotypes (6B, 14, 19F, and 23F) are common causes of invasive diseases in all settings and these types are also most frequently associated with drug resistance.
In the last decade the two most common capsular types among pediatric invasive isolates in southern Israel were 1 (23%) and 5 (10%); these two serotypes were rare among asymptomatic carriers. Other serotypes, like 6B and 23F, highly prevalent in the nasopharyngeal (NP) flora of healthy children, were less frequently associated with invasive disease: 4 and 3% of 6B and 23F isolates, respectively, were associated with invasive disease during 1989 to 1999 (6; Porat et al., Abstr. Second Int. Symp. Pneumococci Pneumococcal Dis., 2000).
New subtyping methods using DNA fingerprint profiles provide additional discriminatory power compared with that of serotyping alone. These methods are extremely important in cases where phenotypic characteristics are insufficient to show the relatedness among strains, e.g., strains carrying the same capsular type and antimicrobial resistance pattern. In this study we applied one of the genotyping methods, namely, pulsed-field gel electrophoresis (PFGE), to look at the clonal distribution of strains carrying capsular types 1, 5, 6B, and 23F. Our goal was to see the clonal diversity of serotypes that are frequently found among invasive isolates and to compare it with that of serotypes that are less common among invasive strains but often carried by young children for prolonged periods. The basic hypothesis was that invasive serotypes that are not carried in the nasopharynx for prolonged periods will be represented by a more homogenous population due to their lower chance of interacting with other organisms and/or being exposed to antibiotic pressure, resulting in genetic transformation.
MATERIALS AND METHODS
Bacterial isolates.
The 255 isolates included in this study were recovered from children living in southern Israel, aged 0 to 12 years. The study population in this area consisted of Jewish children with a lifestyle resembling that of developed populations and Moslem Bedouin children with much lower standards of living, representing that of developing populations. All isolates of serotypes 1, 5, 6B, and 23F recovered from blood cultures of children aged 0 to 12 years during January 1995 through May 1999 at the Soroka University Medical Center were analyzed. These included 66 serotype 1, 30 serotype 5, 11 serotype 6B, and 15 serotype 23F isolates (total of 122 isolates). In cases of strains with identical PFGE patterns originating from an outbreak (5) or where several cultures were taken from the same child during the course of the disease episode, resulting in genetically identical strains, only one isolate per child was included in the study. The 133 NP isolates included all serotype 1 and 5 strains isolated during the above-indicated time period and organisms of serotypes 6B and 23F which were chosen randomly from a collection of 4,600 strains recovered from pediatric healthy carriers in southern Israel. Only 13 serotype 1 and 15 serotype 5 epidemiologically unrelated isolates from healthy carriers were found. The 53 6B isolates and 52 23F isolates were randomly chosen from a total of 205 6B and 444 23F isolates (Table 1). In cases where an epidemiological linkage was detected among strains with identical PFGE patterns, only one representative isolate was included in the study. This exclusion criterion refers to (i) strains originating from the same day care center during a period of 6 months or less and (ii) repeated cultures obtained from the same child during a 6-month period or less.
TABLE 1.
Survey of S. pneumoniae organisms isolated from children in southern Israel during January 1995 to May 1999
| Type of isolates and characteristic | Yr of isolation | No. (%) of isolates of serotype:
|
|||
|---|---|---|---|---|---|
| 1 (n = 79) | 5 (n = 45) | 6B (n = 64) | 23F (n = 67) | ||
| NP | 1995 | 1 (7.7) | 1 (6.7) | 9 (17) | 4 (7.7) |
| 1996 | 0 (0) | 1 (6.7) | 8 (15.1) | 3 (5.8) | |
| 1997 | 5 (38.5) | 5 (33.3) | 10 (18.9) | 8 (15.4) | |
| 1998 | 7 (53.9) | 4 (26.7) | 22 (41.5) | 30 (57.7) | |
| 1999 | 0 (0) | 4 (26.7) | 4 (7.6) | 7 (13.5) | |
| Total by serotype | 13 (100) | 15 (100) | 53 (100) | 52 (100) | |
| Total by ethnic group | |||||
| Jewish | 7 (53.9) | 9 (60) | 26 (49.1) | 37 (71.2) | |
| Bedouin | 6 (46.2) | 6 (40) | 27 (50.9) | 15 (28.9) | |
| Blood or CSF | 1995 | 17 (25.8) | 3 (10) | 4 (36.4) | 3 (20.0) |
| 1996 | 6 (9.1) | 3 (10) | 2 (18.2) | 3 (20.0) | |
| 1997 | 19 (28.8) | 11 (36.7) | 2 (18.2) | 3 (20.0) | |
| 1998 | 20 (30.3) | 8 (26.7) | 2 (18.2) | 5 (33.3) | |
| 1999 | 4 (6.1) | 5 (16.7) | 1 (9.1) | 1 (6.7) | |
| Total by serotype | 66 (100) | 30 (100) | 11 (100) | 15 (100.0) | |
| Total by ethnic group | |||||
| Jewish | 12 (18.2) | 10 (33.3) | 6 (54.6) | 8 (53.3) | |
| Bedouin | 54 (81.8) | 20 (66.7) | 5 (45.5) | 7 (46.7) | |
Isolates were confirmed as S. pneumoniae by inhibition with optochin and by a positive slide agglutination test (Phadebact; Pharmacia Diagnostics, Uppsala, Sweden). One S. pneumoniae colony per culture was subcultured, harvested, and kept frozen at −70°C for further testing.
Antibiotic susceptibility testing.
Testing of isolate susceptibility to penicillin, erythromycin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole (SXT) was performed by the disk diffusion method of Bauer and Kirby according to NCCLS recommendations (13). Isolates exhibiting an inhibition zone with a radius of ≤19 mm around a 1-μg-oxacillin disk were further tested for susceptibility to penicillin by means of the E-Test (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions (10). Antibiotic susceptibility testing was carried out on all the isolates described in Table 1.
Serogrouping and serotyping.
Serogrouping and serotyping of S. pneumoniae was done by means of the quellung reaction using antisera provided by the Statens Serum Institute of Copenhagen, Denmark (1).
PFGE.
Chromosomal DNA fragments, generated by SmaI digestion, were prepared and analyzed as described elsewhere (16). A contour-clamped homogeneous electric field DRIII apparatus (Bio-Rad Laboratories, Richmond, Calif.) was used for running the gels. Running conditions were 23 h at 11.3°C at 200 V ramped with an initial forward time of 5 s and a final forward time of 35 s. Gels were stained with ethidium bromide and photographed. Interpretation of strain relatedness on the basis of PFGE pattern was according to current consensus (19).
RESULTS
Distribution of types.
Table 1 shows the yearly distribution of the strains and their partitioning among Jewish and Bedouin children. A total of 255 S. pneumoniae strains were analyzed, 133 of which were from healthy carriers and 122 of which were from children with invasive disease. The 122 invasive strains included all serotype 1, 5, 6B, and 23F organisms that were cultured at the Soroka University Medical Center in southern Israel during January 1995 to May 1999. The 133 NP isolates included all 13 (0.28%) serotype 1 isolates, all 15 (0.33%) serotype 5 isolates, and 53 6B isolates and 52 23F isolates that were randomly chosen from our collection.
As can be seen in Table 1, the distribution of invasive S. pneumoniae strains carrying serotypes 1 and 5 differed significantly between Jews and Bedouins (P = 0.002); 81.8% of the invasive type 1 isolates and 66.7% of the invasive type 5 isolates were found in the Bedouin population, whereas no significant difference was found for the invasive isolates of serotypes 6B and 23F.
Antimicrobial susceptibility.
Table 2 summarizes the susceptibility testing results for the six antimicrobial agents evaluated in this study. All the strains carrying capsular serotype 1 or 5 were susceptible to penicillin, erythromycin, clindamycin, tetracycline, and chloramphenicol; 52% of serotype 1 isolates and 36% of serotype 5 isolates were not susceptible to SXT. Isolates carrying serotypes 6B and 23F presented a completely different resistance pattern. Of 64 isolates with serotype 6B, 33 (52%) were not susceptible to penicillin and 36 (56%) were not susceptible to three or more different antimicrobial agents. Among the 67 serotype 23F isolates, 60 (90%) were not susceptible to penicillin and 14 (21%) were not susceptible to three or more different antimicrobial agents. Different resistance rates were also noticed for organisms isolated from the nasopharynx compared to those isolated from blood or cerebrospinal fluid (CSF).
TABLE 2.
Resistance pattern of S. pneumoniae to antimicrobial agentsa
| Antimicrobial agent | No. (%) of resistant isolates of serotype:
|
|||||
|---|---|---|---|---|---|---|
| 6B
|
23F
|
|||||
| NP (n = 53) | Blood or CSF (n = 11) | Total (n = 64) | NP (n = 52) | Blood or CSF (n = 15) | Total (n = 67) | |
| Penicillin | 30 (57) | 3 (27) | 33 (52) | 48 (92) | 12 (80) | 60 (90) |
| SXT | 30 (57) | 3 (27) | 33 (52) | 46 (88) | 12 (80) | 58 (87) |
| Erythromycin | 32 (60) | 3 (27) | 35 (55) | 6 (12) | 0 | 6 (9) |
| Clindamycin | 30 (57) | 4 (36) | 34 (53) | 5 (10) | 0 | 5 (7) |
| Tetracycline | 33 (62) | 4 (36) | 37 (58) | 5 (10) | 0 | 5 (7) |
| Chloramphenicol | 15 (28) | 0 | 15 (23) | 5 (10) | 0 | 5 (7) |
| ≥3 different agents | 32 (60) | 4 (36) | 36 (56) | 13 (25) | 1 (7) | 14 (21) |
All 79 serotype 1 isolates and 45 serotype 5 isolates were susceptible to penicillin, erythromycin, clindamycin, tetracycline, and chloramphenicol; 52% of serotype 1 isolates and 36% of serotype 5 isolates were not susceptible to SXT.
Molecular typing.
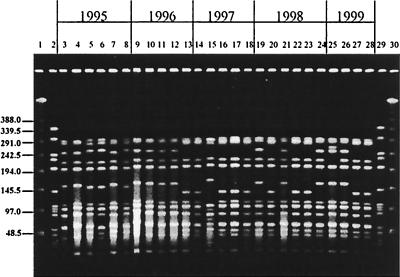
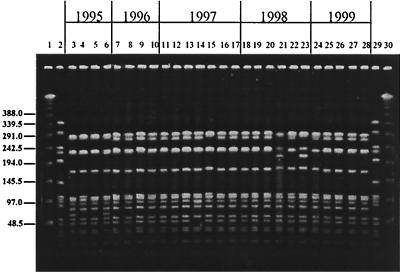
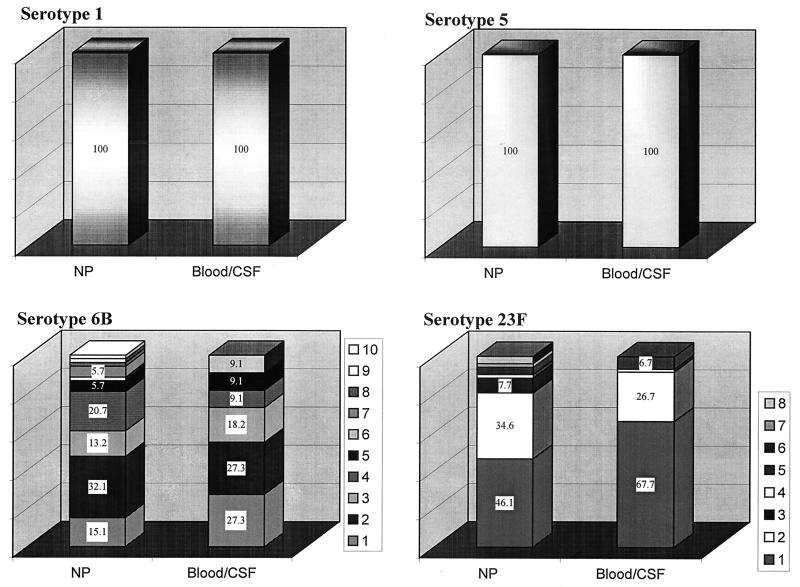
Analysis by PFGE provided data on the variability of the genetic background for each serotype. All type 1 isolates, both invasive and NP, belonged to the same clonal type (Fig. 1); three PFGE patterns differing from each other by no more than five bands represented 86% of all type 1 isolates and 92% of the invasive isolates. Type 5 isolates also shared one clonal type (Fig. 2), with 62% of the isolates showing indistinguishable PFGE patterns and the rest distributed into eight closely related profiles. The PFGE pattern of serotype 5 isolates studied here resembles that of the serotype 5 “Colombian clone” (18).
FIG. 1.
PFGE patterns generated by SmaI digestion of S. pneumoniae serotype 1 isolates recovered from children in Southern Israel during 1994 to 1999. Lanes 1 and 30 contain a lambda ladder; lanes 2 and 29 contain a reference strain, R6, used as a molecular weight marker. Numbers on the left show molecular sizes in kilobases. Lanes 3, 14, 15, and 19 to 21 contain NP isolates; lanes 4 to 13, 16, 17, and 22 to 28 contain blood isolates; lane 18 contains a CSF isolate.
FIG. 2.
PFGE patterns generated by SmaI digestion of S. pneumoniae serotype 5 isolates recovered from children in Southern Israel during 1994 to 1999. Lanes 1 and 30 contain a lambda ladder; lanes 2 and 29 contain a reference strain, R6, used as a molecular weight marker. Numbers on the left show molecular sizes in kilobases. Lanes 3, 7, 11, 12, 18, 19, and 24 contain NP isolates; lanes 4 to 6, 8 to 10, 13 to 17, 20 to 23, and 25 to 27 contain blood isolates; lane 28 contains a CSF isolate.
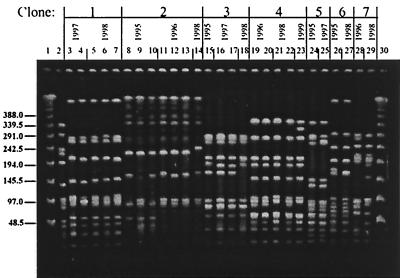
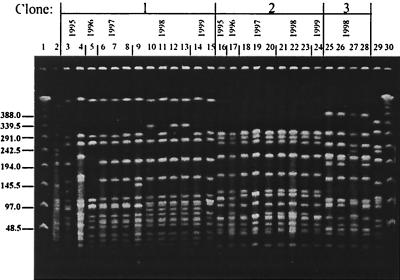
In contrast to serotypes 1 and 5, pneumococci of capsular type 6B (Fig. 3) and 23F (Fig. 4) showed a large diversity in their phenotypic and genotypic characteristics: six clonal types (arbitrarily named 6B-1 to 6B-6) were identified among the 11 invasive serotype 6B isolates; all of them plus four additional PFGE patterns (6B-7 to 6B-10) were present among the 53 serotype 6B NP isolates (Fig. 5). Clone 6B-4 resembled the “Icelandic 6B clone” (16). Multiple clonal types were also found among the 23F isolates (Fig. 4); three clonal types were found among the 15 invasive isolates (23F-1 to 23F-3), and eight types (23F-1 to 23F-8) were found among the 52 NP isolates (Fig. 5). For all capsular types examined here, the clones found among invasive isolates were also detected among the organisms recovered from the NP samples.
FIG. 3.
PFGE patterns generated by SmaI digestion of S. pneumoniae serotype 6B isolates recovered from children in Southern Israel during 1994 to 1999. Lanes 1 and 30 contain a lambda ladder; lane 2 contains a reference strain, R6, used as a molecular weight marker. Numbers on the left show molecular sizes in kilobases. Lanes 5 to 9, 12 to 13, 15 to 23, 25, and 27 to 29 contain NP isolates; lanes 3, 4, 10, 11, 14, 24, and 26 contain blood isolates.
FIG. 4.
PFGE patterns generated by SmaI digestion of S. pneumoniae serotype 23F isolates recovered from children in Southern Israel during 1994 to 1999. Lanes 1 and 30 contain a lambda ladder; lanes 2 and 29 contain a reference strain, R6, used as a molecular weight marker. Numbers on the left show molecular sizes in kilobases. Lanes 5, 7, 10 to 16, 18 to 21, and 23 to 28 contain NP isolates; lanes 3, 4, 6, 8, 17, and 22 contain blood isolates; lane 9 contains a CSF isolate.
FIG. 5.
Clonal distribution of NP and invasive (blood or CSF) S. pneumoniae isolates carrying serotypes 1, 5, 6B, and 23F recovered from children in Southern Israel during 1994 to 1999. The numbers in the keys attached to the figures of serotypes 6B and 23F refer to the clonal types shown in Fig. 3 and 4. The numbers in the bar graphs are percentages.
DISCUSSION
There is no doubt that NP carriage plays a crucial role in S. pneumoniae epidemiology and disease (4; P. Yagupsky, N. Peled, and R. Dagan, Abstr. 36th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-48, 1996). From this site the pneumococcus may spread to adjacent mucosal surfaces to cause mucosal infections, such as otitis and pneumonia, or invade the bloodstream and cause systemic infections, such as bacteremia and meningitis. However, the frequency with which certain serotypes cause an invasive disease is not necessarily reflected in their prevalence as serotypes carried in the nasopharynx.
The two most common capsular types among pediatric invasive isolates in southern Israel in the last 10 years were 1 (23%) and 5 (10%) and were most frequently associated with low-socioeconomic-level, overcrowded populations like that of the Bedouins and with outbreaks (5). These serotypes were infrequently detected among healthy carriers. In contrast, serotypes 6B and 23F were the most common serotypes found in the nasopharynx as part of the normal flora but were rare in invasive diseases.
These two groups of serotypes differed markedly in their resistance patterns. Organisms of the two invasive serotypes with capsular types 1 and 5 presented similar resistance patterns and were susceptible to all antimicrobial agents tested (penicillin, erythromycin, tetracycline, and chloramphenicol) except SXT. However, most of the strains belonging to serotypes 6B and 23F were not susceptible to penicillin, showing considerable heterogeneity in their resistance pattern to other antimicrobial agents as well. While none of the invasive serotypes were resistant to more than one antimicrobial agent, high percentages of the strains belonging to the carried serotypes were multidrug resistant: 56% of serotype 6B and 21% of serotype 23F.
The homogeneity of the invasive strains belonging to serotypes 1 and 5 compared with the diversity of 6B and 23F was further examined by looking at the genetic background of the strains. Organisms of the two invasive serotypes, 1 and 5, appeared to be genetically related, with each one of them having a single clonal origin despite their being collected during a long period of time (5.5 years). In contrast, pneumococci of capsular types 6B and 23F were represented by multiple clonal types: 10 for type 6B and 8 for 23F. For all capsular types examined here, the same clones were detected both in the nasopharynx and in blood or CSF.
Previous reports have documented genetic diversity to be higher among penicillin-susceptible than among non-penicillin-susceptible S. pneumoniae strains (7, 20, 21). Increasing penicillin MICs for organisms was associated with decreasing genetic variability, fewer serotypes, and a higher probability of multidrug resistance. While the exact mechanism of this relationship is not clear, one could hypothesize that resistance to antimicrobial agents provides an advantage which supports the survival of the organism in the normal flora. This postulate was further validated by the extensive international spread of a relatively few multidrug-resistant clones such as the serotype 23F “Spanish/USA” clone and the serotype 14/9V “French/Spanish” clone (9).
In this study we describe two penicillin-susceptible invasive clones which tend to persist in our region for a long period of time without undergoing a major genetic transformation. These serotypes are rarely found in the nasopharynxes of children and thus have fewer chances of exchanging genetic material with the environment and fewer chances of being exposed to antibiotic pressure. We speculate that the poor adaptation of these serotypes to mucosal surfaces is one of the factors controlling their low tendency to acquire genes from other NP bacterial inhabitants, resulting in DNA homogeneity. The other less pathogenic serotypes might be more prone to long-term colonization and thus more inclined to multiple antibiotic exposure. Preliminary laboratory investigations of pneumococcal infection in a mouse model carried out by Azoulay-Dupuis et al. (2) have suggested a complex relationship among capsular type, penicillin susceptibility, and virulence. Based on our data, the capsular type seems to be the dominant factor in the virulence of S. pneumoniae and in its tendency to acquire resistance genes. Further studies of the polysaccharide compositions of the invasive versus noninvasive serotypes may shed some light on the adhesion mechanism of S. pneumoniae to epithelial cells and its role in colonization. Moreover, characteristics associated with the clonality of serotypes 1 and 5 may represent an advantage for invasiveness, from a knowledge of which one can study the genetic mechanism involved in S. pneumoniae pathogenicity.
ACKNOWLEDGMENT
This study was partially supported by grant 4520 from the Ministry of Health, Jerusalem, Israel.
REFERENCES
- 1.Austrian R. The quellung reaction, a neglected microbiologic technique. Mt Sinai J Med. 1976;43:699–709. [PubMed] [Google Scholar]
- 2.Azoulay-Dupuis E, Rieux V, Muffat-Joly M, Bedos J P, Vallee E, Rivier C, Isturiz R, Carbon C, Moine P. Relationship between capsular type, penicillin susceptibility and virulence of human Streptococcus pneumoniae isolates in mice. Antimicrob Agents Chemother. 2000;44:1575–1577. doi: 10.1128/aac.44.6.1575-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogaerts J, Lepage P, Taelman H, Rouvroy D, Batungwanayo J, Kestelyn P, Hitimana D G, Van de Perre P, Vandepitte J, Verbist L, Verhaegen J. Antimicrobial susceptibility and serotype distribution of Streptococcus pneumoniae from Rwanda, 1984–1990. J Infect. 1993;27:157–168. doi: 10.1016/0163-4453(93)94728-t. [DOI] [PubMed] [Google Scholar]
- 4.Dagan R, Melamed R, Muallem M, Piglansky L, Greenberg D, Abramson O, Mendelman P M, Bohidar N, Yagupsky P. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J Infect Dis. 1996;174:1271–1278. doi: 10.1093/infdis/174.6.1271. [DOI] [PubMed] [Google Scholar]
- 5.Dagan R, Gradstein S, Belmaker I, Porat N, Siton Y, Weber G, Janco J, Yagupsky P. An outbreak of Streptococcus pneumoniae serotype 1 in a closed community in southern Israel. Clin Infect Dis. 2000;30:319–321. doi: 10.1086/313645. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, D., N. Givon-Lavi, N. Bilenko, and R. Dagan. A decade (1989–1998) of pediatric invasive pneumococcal disease in two populations residing in one geographic location: implications for vaccine choice. Clin. Infect. Dis, in press. [DOI] [PubMed]
- 7.Hall L M C, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermans P W M, Overweg K, Sluijter M, Groot R. Penicillin-resistant Streptococcus pneumoniae: an international molecular epidemiological study. In: Tomasz A, editor. Streptococcus pneumoniae: molecular biology & mechanisms of disease. Larchmont, N.Y: Marry Ann Liebert, Inc., Publishers; 2000. pp. 457–466. [Google Scholar]
- 10.Jacobs M R, Bajaksouzian S, Appelbaum P C, Bolstrom A. Evaluation of the E-Test for susceptibility testing of pneumococci. Diagn Microbiol Infect Dis. 1992;15:473–478. doi: 10.1016/0732-8893(92)90093-9. [DOI] [PubMed] [Google Scholar]
- 11.John T J, Pai R, Lalitha M K, Jesudason M V, Brahmadathan K N, Sridharan G, Steinhoff M C. Prevalence of pneumococcal serotypes in invasive diseases in southern India. Indian J Med Res. 1996;104:205–207. [PubMed] [Google Scholar]
- 12.Musher D M, Brieman R F, Tomasz A. Streptococcus pneumoniae: at the threshold of the 21st century. In: Tomasz A, editor. Streptococcus pneumoniae: molecular biology & mechanisms of disease. Larchmont, N.Y: Mary Ann Liebert, Inc., Publishers; 2000. pp. 485–466. [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: 9th informational supplement. Vol. 19 1999. , no. 1. M100–S9. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 14.Robbins J B, Austrian R, Lee C J, Rastogi S C, Schiffman G, Henrichsen J, Makela P H, Broome C V, Facklam R R, Tiesjema R H, Parker J C., Jr Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 15.Scott J A G, Hall A J, Dagan R, Dixon J M S, Eykyn S J, Fenoll A, Hortal M, Jette L P, Jorgensen J H, Lamothe F, Latorre C, Macfarlane J T, Shlaes D M, Smart L E, Taunay A. Serotype-specific epidemiology of Streptococcus pneumoniae: association with age, sex and geography in 7,000 episodes of invasive disease. Clin Infect Dis. 1996;22:973–981. doi: 10.1093/clinids/22.6.973. [DOI] [PubMed] [Google Scholar]
- 16.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 17.Takala A K, Vuopio-Varkila J, Tarkka E, Leinonen M, Musser J M. Subtyping of common pediatric pneumococcal serotypes from invasive disease and pharyngeal carriage in Finland. J Infect Dis. 1996;173:128–135. doi: 10.1093/infdis/173.1.128. [DOI] [PubMed] [Google Scholar]
- 18.Tamayo M, Sa-Leao R, Sanches I S, Castaneda E, de Lencastre H. Dissemination of a chloramphenicol- and tetracycline-resistant but penicillin-susceptible invasive clone of serotype 5 Streptococcus pneumoniae in Colombia. J Clin Microbiol. 1999;37:2337–2342. doi: 10.1128/jcm.37.7.2337-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasz A, Corso A Members of the PAHO/Rockefeller University Workshop (E. P. Severina, G., Echaniz-Aviles, M. C. de Cunto Brandileone, T. Camou, E. Castaneda, O. Figueroa A. Rossi, and J. L. di Fabio) Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American countries: an overview. Microb Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]
- 21.Vaz Pato M V, de Carvalho C B, Tomasz A the Multicenter Study Group. Antibiotic susceptibility of Streptococcus pneumoniae isolates in Portugal. A multicenter study between 1989 and 1993. Microb Drug Resist. 1995;1:59–69. doi: 10.1089/mdr.1995.1.59. [DOI] [PubMed] [Google Scholar]