An end-to-end pipeline for deep learning–assisted computational 3D histology analysis of whole prostate biopsies shows that nondestructive 3D pathology has the potential to enable superior prognostic stratification of patients with prostate cancer.
Abstract
Prostate cancer treatment planning is largely dependent upon examination of core-needle biopsies. The microscopic architecture of the prostate glands forms the basis for prognostic grading by pathologists. Interpretation of these convoluted three-dimensional (3D) glandular structures via visual inspection of a limited number of two-dimensional (2D) histology sections is often unreliable, which contributes to the under- and overtreatment of patients. To improve risk assessment and treatment decisions, we have developed a workflow for nondestructive 3D pathology and computational analysis of whole prostate biopsies labeled with a rapid and inexpensive fluorescent analogue of standard hematoxylin and eosin (H&E) staining. This analysis is based on interpretable glandular features and is facilitated by the development of image translation–assisted segmentation in 3D (ITAS3D). ITAS3D is a generalizable deep learning–based strategy that enables tissue microstructures to be volumetrically segmented in an annotation-free and objective (biomarker-based) manner without requiring immunolabeling. As a preliminary demonstration of the translational value of a computational 3D versus a computational 2D pathology approach, we imaged 300 ex vivo biopsies extracted from 50 archived radical prostatectomy specimens, of which, 118 biopsies contained cancer. The 3D glandular features in cancer biopsies were superior to corresponding 2D features for risk stratification of patients with low- to intermediate-risk prostate cancer based on their clinical biochemical recurrence outcomes. The results of this study support the use of computational 3D pathology for guiding the clinical management of prostate cancer.
Significance:
An end-to-end pipeline for deep learning–assisted computational 3D histology analysis of whole prostate biopsies shows that nondestructive 3D pathology has the potential to enable superior prognostic stratification of patients with prostate cancer.
Introduction
Prostate cancer is the most common cancer in men and the second leading cause of cancer-related death for men in the United States (1). Currently, prostate cancer management is largely dependent upon examination of prostate biopsies via two-dimensional (2D) histopathology (2), in which a set of core-needle biopsies is formalin-fixed and paraffin-embedded (FFPE) to allow thin sections to be cut, mounted on glass slides, and stained for microscopic analysis. To quantify the aggressiveness of the cancer, the Gleason grading system is used, which relies entirely upon visual interpretation of prostate gland morphology as seen on a few histology slides (thin 2D tissue sections) that only “sample” approximately 1% of the whole biopsy. Gleason grading of prostate cancer is associated with high levels of interobserver variability (3, 4) and is only moderately correlated with outcomes, especially for patients with intermediate-grade prostate cancer (5). This contributes to the undertreatment of patients with aggressive cancer (e.g., with active surveillance; ref. 6), leading to preventable metastasis and death (7), and the overtreatment of patients with indolent cancer (e.g., with surgery or radiotherapy; ref. 8), which can lead to serious side effects, such as incontinence and impotence (9).
Motivated by recent technological advances in optical clearing to render tissue specimens transparent to light [i.e., iDISCO (10), CUBIC (11) etc.] in conjunction with high-throughput three-dimensional (3D) light-sheet microscopy, a number of groups have been exploring the value of nondestructive 3D pathology of clinical specimens for diagnostic pathology (12–17). Compared with conventional slide–based histology, nondestructive 3D pathology can achieve vastly greater sampling of large specimens along with volumetric visualization and quantification of diagnostically significant microstructures, all while maintaining intact specimens for downstream molecular assays (18). Like others before us (12, 19, 20), we hypothesized that 3D versus 2D pathology datasets could allow for improved characterization of the convoluted glandular structures that pathologists currently rely on for prostate cancer risk stratification. However, since the associated information content of a 3D pathology dataset of a biopsy is >100× larger than a 2D whole-slide image representation (in terms of total number of pixels), computational tools are necessary to analyze these large datasets efficiently and reproducibly for diagnostic and prognostic determinations.
For computational analysis of 3D pathology datasets, a multistage pipeline was chosen for classifying patient outcomes based on interpretable “hand-crafted” features (i.e., glandular features; refs. 21–23) rather than an end-to-end deep learning (DL) strategy for risk classification based directly on the imaging data (24–26). This was motivated by: (i) the attractiveness of an intuitive feature-based approach as an initial strategy to facilitate hypothesis testing and clinical adoption of an emerging modality in which datasets are currently limited and poorly understood (18, 27), and (ii) the observation that when case numbers are limited, a hand-crafted feature-based approach can be more reliable than an end-to-end DL classifier (28, 29).
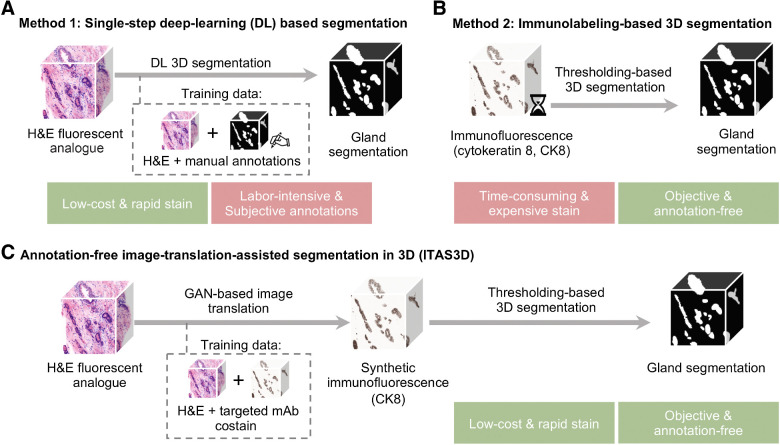
Multistage feature-based classification approaches rely on the accurate segmentation of morphologic structures such as nuclei (30–32), collagen fibers (33, 34), vessels (14, 35), or in our case, prostate glands (21, 36). This is typically achieved in one of two ways: (i) direct DL-based segmentation methods (37–40) that require manually annotated training datasets, which are especially tedious and difficult to obtain in 3D (Fig. 1A; ref. 41); or (ii) traditional computer vision (CV) approaches based on intensity and morphology, provided that tissue structures of interest can be stained/labeled with high specificity (Fig. 1B; refs. 19, 42, 43). While immunolabeling can confer a high degree of specificity for traditional CV-based segmentation, it is not an attractive strategy for clinical 3D pathology assays due to the high cost of antibodies to stain large tissue volumes, and the slow diffusion times of antibodies in thick tissues (up to several weeks; refs. 10, 44).
Figure 1.
General methods for 3D gland segmentation. A, A single-step DL segmentation model can be trained with imaging datasets of tissues labeled with a fluorescent analogue of H&E paired with manually annotated ground-truth segmentation masks. While H&E analogue staining is low-cost and rapid, manual annotations are labor-intensive (especially in 3D) and based on subjective human judgements. B, By immunolabeling a tissue microstructure with high specificity, 3D segmentations can be achieved with traditional CV methods without the need for manual annotations. While this is an objective segmentation method based on a chemical biomarker, immunolabeling large intact specimens is expensive and time-consuming due to the slow diffusion of antibodies in thick tissues. C, With ITAS3D, H&E-analogue datasets are computationally transformed in appearance to mimic immunofluorescence datasets, which enables the synthetically labeled tissue structures to be segmented with traditional CV methods. The image-sequence translation model is trained with a GAN based on paired H&E-analogue and immunofluorescence datasets. ITAS3D is rapid and low-cost (in terms of staining) as well as annotation-free and objective (i.e., biomarker-based).
To address these challenges, we developed a generalizable annotation-free 3D segmentation method, hereafter referred to as “image-translation-assisted segmentation in 3D (ITAS3D).” In our specific implementation of ITAS3D (Fig. 1C), 3D hematoxylin and eosin (H&E) analogue images of prostate tissues are synthetically converted in appearance to mimic 3D immunofluorescence (IF) images of cytokeratin 8 (CK8) – a low molecular weight keratin expressed by the luminal epithelial cells of all prostate glands – thereby facilitating the objective (biomarker-based) segmentation of the glandular epithelium and lumen spaces using traditional CV tools. The DL image translation algorithm is trained with a generative adversarial network (GAN), which has been previously used for 2D virtual staining applications (45–47). However, unlike those prior 2D image translation efforts, we developed a “2.5D” virtual-staining approach based on a specialized GAN that was originally designed to achieve video translation with high spatial continuity between frames, but which has been adapted within our ITAS3D framework to ensure high spatial continuity as a function of depth (see Results; ref. 48).
As a preliminary study to investigate the value of a computational 3D pathology workflow versus a computational 2D pathology workflow, 300 ex vivo biopsies were extracted from archived radical prostatectomy (RP) specimens obtained from 50 patients who underwent surgery over a decade ago. We stained the biopsies with an inexpensive small-molecule (i.e., rapidly diffusing) fluorescent analogue of H&E, optically cleared the biopsies with a simple dehydration and solvent–immersion protocol to render them transparent to light, and then used an open-top light-sheet (OTLS) microscopy platform to obtain whole-biopsy 3D pathology datasets. The prostate glandular network was then segmented using ITAS3D, from which 3D glandular features (i.e., histomorphometric parameters) and corresponding 2D features were extracted from the 118 biopsies that contained prostate cancer. These 3D and 2D features were evaluated for their ability to stratify patients based on clinical biochemical recurrence (BCR) outcomes, which serve as a proxy endpoint for aggressive versus indolent prostate cancer.
Materials and Methods
Key methodologic information is provided below for our preliminary clinical demonstration. The Supplementary Methods contain technical details on the training and validation of ITAS3D, as well as computational details for the clinical study, such as a description of gland features.
Collection and processing of archived tissue to obtain simulated biopsies for the clinical study
The following study was approved by the institutional review board (IRB) of the University of Washington (Seattle, WA; Study 00004980), where research specimens were previously obtained from patients with informed consent. Archived FFPE prostatectomy specimens were collected from 50 patients with prostate cancer (see Supplementary Table S1 for clinical data), of which, 46 cases were initially graded during post-RP histopathology as having Gleason scores of 3+3, 3+4 or 4+3 (Grade Group 1–3). All patients were followed up for at least 5 years post-RP as part of a prior study (Canary TMA; ref. 49). FFPE tissue blocks were identified from each case corresponding to the six regions of the prostate targeted by urologists when performing standard sextant and 12-core (2 cores per sextant region) biopsy procedures. The identified FFPE blocks were first deparaffinized by heating them at 75°C for 1 hour until the outer paraffin wax was melted, and then placing them in 65°C xylene for 48 hours. Next, one simulated core-needle biopsy (∼1-mm in width) was cut from each of the six deparaffinized blocks (per patient case), resulting in a total of n = 300 biopsy cores. All simulated biopsies were then fluorescently labeled with the To-PRO-3 and eosin (T&E) version of our H&E analogue staining protocol (Supplementary Fig. S1).
The T&E staining protocol (H&E analogue) for the clinical study
Biopsies were first washed in 100% ethanol twice for 1 h each to remove any excess xylene, then treated in 70% ethanol for 1 hour to partially rehydrate the biopsies. Each biopsy was then placed in an individual 0.5 mL Eppendorf tube (catalog no. 14–282–300, Thermo Fisher Scientific), stained for 48 hours in 70% ethanol at pH 4 with a 1:200 dilution of Eosin-Y (catalog no. 3801615, Leica Biosystems) and a 1:500 dilution of To-PRO-3 Iodide (catalog no. T3605, Thermo Fisher Scientific) at room temperature with gentle agitation. The biopsies were then dehydrated twice in 100% ethanol for 2 hours. Finally, the biopsies were optically cleared (n = 1.56) by placing them in ethyl cinnamate (catalog no. 112372, Sigma-Aldrich) for 8 hours before imaging them with open-top light-sheet (OTLS) microscopy.
OTLS microscopy and preprocessing
We utilized a previously developed OTLS microscope (15) to image tissues slices (for training data) and simulated biopsies (for the clinical study). For this study, ethyl cinnamate (n = 1.56) was used as the immersion medium, and a custom-machined HIVEX plate (n = 1.55) was used as a multibiopsy sample holder (12 biopsies per holder). Multichannel illumination was provided by a four-channel digitally controlled laser package (Skyra, Cobolt Lasers). Tissues were imaged at near-Nyquist sampling of approximately 0.44 μm/pixel. The volumetric imaging time was approximately 0.5 minute per mm3 of tissue for each wavelength channel. This allowed each biopsy (∼1 × 1 × 20 mm), stained with two fluorophores (T&E), to be imaged in approximately 20 minutes.
Statistical analysis of the correlation between glandular features and BCR outcomes
Patient-level glandular features were obtained by averaging the biopsy-level features from all cancer-containing biopsies from a single patient. Patients who experienced BCR within 5 years post-RP are denoted as the “BCR” group, and all other patients are denoted as “non-BCR.” BCR was defined here as a rise in serum levels of prostate-specific antigen (PSA) to 0.2 ng/mL after 8 weeks post-RP (49). The box plots indicate median values along with interquartile ranges (25%–75% of the distribution). The whiskers extend to the furthest data points excluding outliers defined as points beyond 1.5× the interquartile range. The P values for the BCR group versus non-BCR group are calculated using the two-sided Mann–Whitney U test (50). To assess the ability of different 3D and 2D glandular features to distinguish between BCR versus non-BCR groups, we applied ROC curve analysis, from which an area-under-the-curve (AUC) value could be extracted. The t-SNE (51) analyses were performed with 1000 iterations at a learning rate of 100.
To develop multiparameter classifiers to stratify patients based on 5-year BCR outcomes, a least absolute shrinkage and selection operator (LASSO) logistic regression model was developed (22) using the binary 5-year BCR category as the outcome endpoint. LASSO is a regression model that includes a L1 regularization term to avoid overfitting and to identify a subset of features that are most predictive. Here, the optimal LASSO tuning parameter, λ, was determined with 3-fold cross validation (CV), where the dataset was randomly partitioned into three equal-sized groups: two groups to train the model with a specific λ, and one group to test the performance of the model. Along the LASSO regularization path, the λ with the highest R2 (coefficient of determination) was defined as the optimal λ. Because of the lack of an external validation set, a nested CV schema was used to evaluate the performance of the multivariable models without any bias and data leakage between parameter estimation and validation steps (52). The aforementioned CV used for hyperparameter tuning was performed in each iteration of the outer CV. LASSO regression was applied on the training set of the outer CV once an optimal λ was identified in the inner CV. AUC values were then calculated from the testing group of the outer CV (Supplementary Fig. S2). This nested CV was performed 200 times to determine an AUC (average and SD). The exact same pipeline was used to develop multiparameter classifiers based on 3D and 2D features.
Kaplan–Meier (KM) analysis was carried out to compare BCR-free survival rates for high-risk versus low-risk groups of patients. This analysis utilized a subset of 34 cases, for which time-to-recurrence data is available (see Supplementary Table S1). The performance of the models, either based on 2D or 3D features, was quantified with P values (by log-rank test), HRs, and concordance index (C-index) metrics. For the multiparameter classification model used for KM analysis, the outer CV (3-fold) in our nested CV schema was replaced by a leave-one-out approach, where one case was left out of each iteration (50 total iterations) to calculate the probability of 5-year BCR for that patient (53). The samples were categorized as low- or high-risk by setting a posterior class probability threshold of 0.5. MATLAB was used for the KM analysis and all other statistical analysis was performed in Python with the “Scipy” and “Scikit-learn” packages.
Data availability
Relevant clinical data for this study are provided in Supplementary Table S1 and Supplementary Table S5. Example prostate images for testing ITAS3D codes and models are available in a GitHub repository at https://github.com/WeisiX/ITAS3D. Full 3D prostate imaging datasets, “simulated 2D whole-slide images” extracted from those datasets (three levels per biopsy), as well as other clinical data are available upon reasonable request and with the establishment of a material-transfer or data-transfer agreement.
Code availability
The Python code for the deep-learning models, and for 3D glandular segmentations based on synthetic-CK8 datasets, are available on GitHub at https://github.com/WeisiX/ITAS3D.
Results
Annotation-free 3D gland segmentation
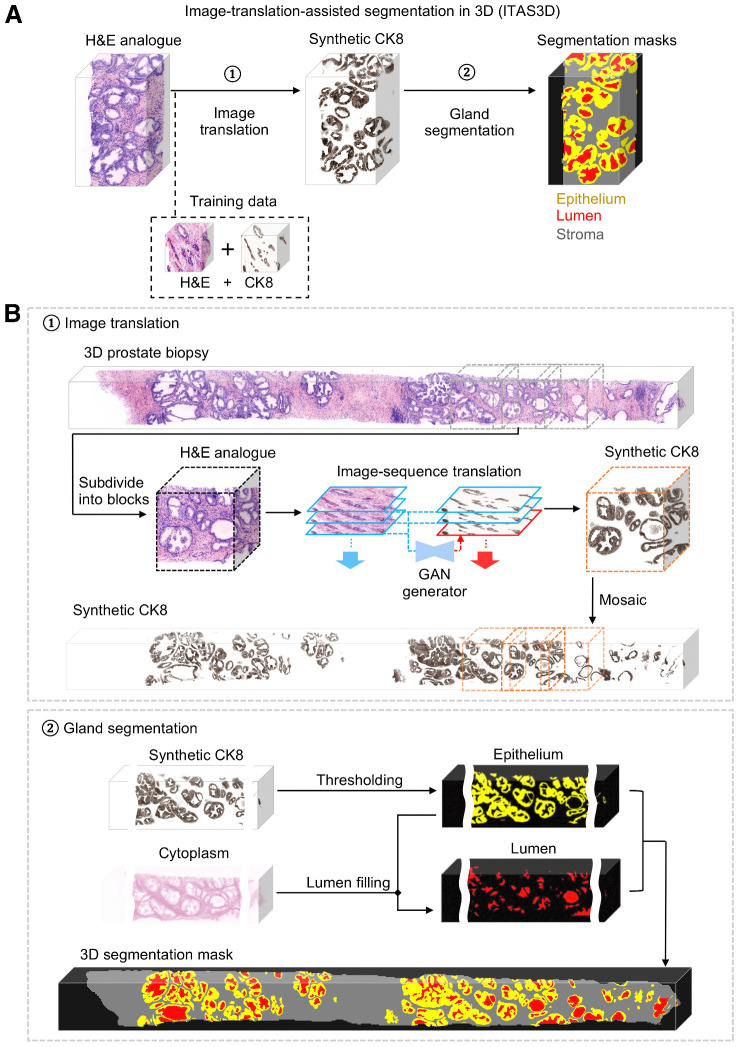
To segment the 3D glandular network within prostate biopsies, we first trained a GAN-based image-sequence translation model (see Supplementary Methods) to convert 3D H&E analogue images into synthetic CK8 IF images, which can be false colored to resemble chromogenic IHC (Fig. 2A). As mentioned, the CK8 biomarker is expressed by the luminal epithelial cells of all prostate glands. The image translation model is trained in a supervised manner with images from prostate tissues that are fluorescently tri-labeled with our H&E analogue and a CK8-targeted mAb (Supplementary Table S2).
Figure 2.
ITAS3D: a two-step pipeline for annotation-free 3D segmentation of prostate glands. A, In step 1, a 3D microscopy dataset of a specimen, stained with a rapid and inexpensive fluorescent analogue of H&E, is converted into a synthetic CK8 immunofluorescence dataset by using an image-sequence translation model that is trained with paired H&E analogue and real-CK8 immunofluorescence datasets (tri-labeled tissues). The CK8 biomarker, which is utilized in standard-of-care genitourinary pathology practice, is ubiquitously expressed by the luminal epithelial cells of all prostate glands. In step 2, traditional computer-vision algorithms are applied to the synthetic-CK8 datasets for semantic segmentation of the gland epithelium, lumen, and surrounding stromal regions. B, In step 1, a 3D prostate biopsy is subdivided into overlapping blocks that are each regarded as depth-wise sequences of 2D images. A GAN-trained generator performs image translation sequentially on each 2D level of an image block. The image translation at each level is based on the H&E analogue input at that level while leveraging the H&E analogue and CK8 images from two previous levels to enforce spatial continuity between levels (i.e., a “2.5D” translation method). The synthetic-CK8 image-block outputs are then mosaicked to generate a 3D CK8 dataset of the whole biopsy to assist with gland segmentation. In step 2, the epithelial cell layer (epithelium) is segmented from the synthetic-CK8 dataset with a thresholding-based algorithm. Gland lumen spaces are segmented by filling in the regions enclosed by the epithelia with refinements based on the cytoplasm channel (eosin fluorescence). See Supplementary Methods for details.
As shown in step 1 of Fig. 2B, for whole-biopsy H&E-to-CK8 conversion, we first sub-divide the 3D biopsy (∼ 1 mm × 0.7 mm × 20 mm) datasets into ∼1 mm × 0.7 mm × 1 mm (∼ 1024 × 700 × 1024 pixel) blocks. Each 3D image block is treated as a 2D image sequence as a function of depth. At each depth level, a synthetic CK8 image is inferred from the H&E analogue image at that level while simultaneously utilizing the images (H&E analogue and CK8) from two previous levels to enforce spatial continuity as a function of depth. This “2.5D” image translation method is based on a previously reported “vid2vid” method for video translation (time sequences rather than depth sequences; ref. 48; see Supplementary Fig. S3; Supplementary Methods). However, our modified model omits the “coarse-to-fine” training strategy implemented in the original vid2vid method because this enables training times to be minimized with negligible performance loss (see Supplementary Note S1; Supplementary Fig. S4; Supplementary Video S1). Once the synthetic CK8 image blocks are generated, they are mosaicked to generate a whole-biopsy CK8 IHC dataset for gland segmentation. In step 2 of Fig. 2B, the synthetic CK8 dataset is used to segment the luminal epithelial cell layer (“Epithelium” in Fig. 2B) via a thresholding algorithm. The gland-lumen space, which is enclosed by the epithelium layer, can then be segmented by utilizing both the epithelium segmentation mask and the cytoplasmic channel (eosin-analogue images). Algorithmic details are provided in the Supplementary Methods and Supplementary Fig. S5.
Evaluation of image translation and segmentation
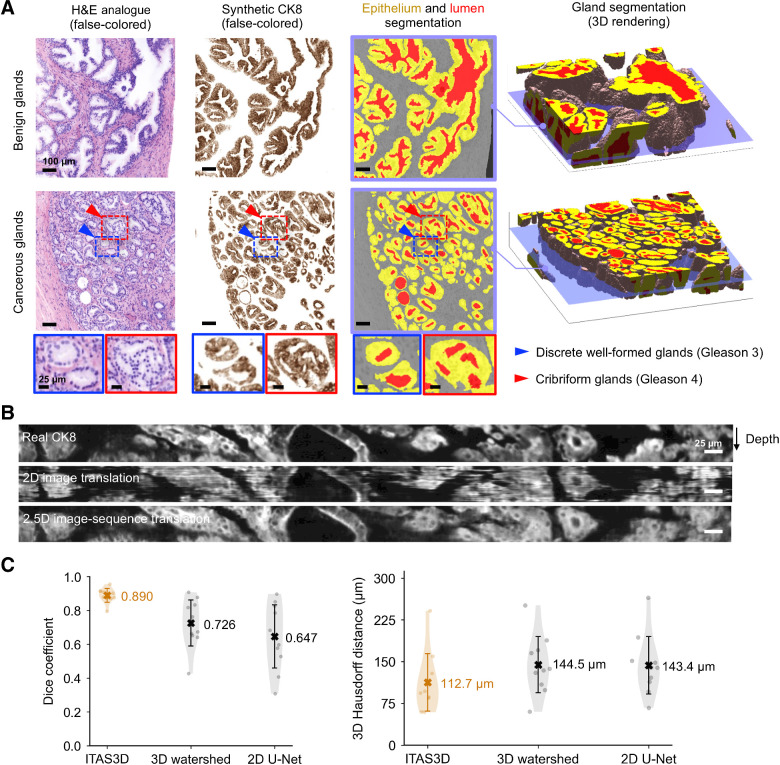
Example 3D prostate gland-segmentation results are shown for benign and cancerous regions in Fig. 3A. While the glands can be delineated on the H&E analogue images by a trained observer, automated computational segmentation of the glands remains challenging (54, 55). Here we demonstrate that 3D image translation based on H&E analogue inputs results in synthetic CK8 outputs in which the luminal epithelial cells are labeled with high contrast and spatial precision. We further show that these synthetic-CK8 datasets allow for relatively straightforward segmentation of the gland epithelium, lumen, and surrounding stromal tissue compartments (Fig. 3A). Glands from various prostate cancer subtypes are successfully segmented as shown in Fig. 3A, including two glandular patterns that are typically associated with low and intermediate risk, respectively: small discrete well-formed glands (Gleason 3) and cribriform glands consisting of epithelial cells interrupted by multiple punched-out lumina (Gleason 4). Supplementary Video S2 shows depth sequences of an H&E analogue dataset, a synthetic-CK8 dataset, and a segmentation mask of the two volumetric regions shown in Fig. 3A. A whole-biopsy 3D segmentation is also depicted in Supplementary Video S3.
Figure 3.
Segmentation results with ITAS3D. A, 2D cross-sections are shown (from left to right) of false-colored H&E analogue images, synthetic-CK8 IHC images generated by image-sequence translation, and gland-segmentation masks based on the synthetic-CK8 images (yellow, epithelium; red, lumen; gray, stroma). The example images are from large 3D datasets containing benign glands (first row) and cancerous glands (second row). Zoom-in views show small discrete well-formed glands (Gleason pattern 3, blue box) and cribriform glands (Gleason pattern 4, red box) in the cancerous region. Three-dimensional renderings of gland segmentations for a benign and cancerous region are shown on the far right. Scale bar, 100 μm. B, Side views of the image sequences (with the depth direction oriented down) of real- and synthetic-CK8 immunofluorescence images. The 2.5D image translation results exhibit substantially improved depth-wise continuity compared with the 2D image translation results. Scale bar, 25 μm. C, For quantitative benchmarking, Dice coefficients (larger is better) and 3D Hausdorff distances (smaller is better) are plotted for ITAS3D-based gland segmentations along with two benchmark methods (3D watershed and 2D U-net), as calculated from 10 randomly selected test regions. Violin plots are shown with mean values denoted by a center cross and SDs denoted by error bars. For the 3D Hausdorff distance, the vertical axis denotes physical distance (in microns) within the tissue.
To demonstrate improved depth-wise continuity with our 2.5D image-translation strategy versus a similar 2D image-translation method (based on the “pix2pix” GAN), vertical cross-sectional views of a synthetic-CK8 dataset are shown in Fig. 3B. While obvious distortions and discontinuities are seen as a function of depth with 2D image translation, the results of our 2.5D image-sequence translation exhibit optimal continuity with depth. To further illustrate this improved continuity with depth, Supplementary Video S4 shows a depth sequence of en face images (z stack). Abrupt morphologic discontinuities between levels are again obvious with 2D translation but absent with the 2.5D translation approach. To quantify the performance of our image-translation method, a 3D structural similarity (SSIM) metric was calculated in which real CK8 IF datasets were used as ground truth. For images generated with 2.5D versus 2D image translation, the 3D SSIM (averaged over 58 test volumes that were 0.2-mm3 each) was 0.41 vs. 0.37, reflecting a 12% improvement at a P value of 7.8 × 10–6 (two-sided paired t test). This enhanced image-translation performance facilitates accurate 3D gland segmentations in subsequent steps of our computational pipeline.
To assess segmentation performance, ground-truth gland-segmentation datasets were first generated under the guidance of board-certified genitourinary pathologists (L.D. True and N.P. Reder). A total of 10 tissue volumes from different patients (512 × 512 × 100 pixels each, representing 0.2-mm3 of tissue) were manually annotated. We then compared the accuracy of ITAS3D with that of two common methods: 3D watershed (as a 3D non-DL benchmark; ref. 56) and 2D U-Net (as a 2D DL benchmark; ref. 57). ITAS3D outperforms the two benchmark methods in terms of Dice coefficient (58) and 3D Hausdorff distance (Fig. 3C; ref. 59). As a visual comparison between ITAS3D and the two benchmark methods, Supplementary Video S5 displays image-stack sequences from three orthogonal perspectives of a representative segmented dataset, where the higher segmentation accuracy of ITAS3D can be appreciated. Note that a 3D DL-based benchmark method is not provided since there are currently insufficient 3D-annotated prostate gland datasets to train an end-to-end 3D DL segmentation model (40, 60, 61); again, this is one of the main motivations for developing the annotation-free ITAS3D method.
Clinical validation study: glandular feature extraction and correlation with BCR outcomes
Due to the slow rate of progression for most prostate cancer cases, an initial clinical study to assess the prognostic value of 3D versus 2D glandular features was performed with archived prostatectomy specimens. Our study consisted of N = 50 patients with prostate cancer who were followed up for a minimum of 5 years post-RP as part of the Canary TMA case-cohort study (primarily low- to intermediate-risk patients; ref. 49). The Canary TMA study was based on a well-curated cohort of patients with prostate cancer in which the primary study endpoints were 5-year BCR outcomes and time to recurrence, which are also used as endpoints for our validation study. In the original Canary TMA study, approximately half of the patients experienced BCR within 5 years of RP, making it an ideal cohort for our study. We randomly selected a subset of 25 cases that had BCR within 5 years of RP (“BCR” group), and 25 cases that did not have BCR within 5 years of RP (“non-BCR” group).
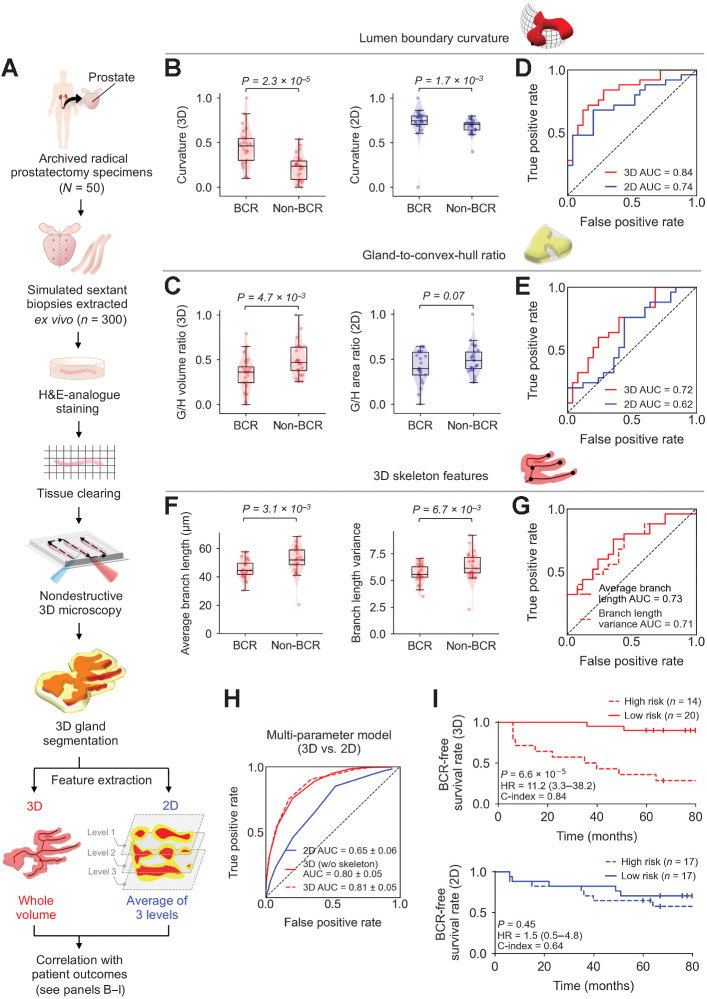
FFPE tissue blocks were identified from each case corresponding to the six regions of the prostate targeted by urologists when performing standard sextant and 12-core (2 cores per sextant region) biopsy procedures (Fig. 4A). Next, a simulated core-needle biopsy was extracted from each of the six FFPE tissue blocks for each patient (n = 300 total biopsy cores). The biopsies were deparaffinized, labeled with a fluorescent analogue of H&E, optically cleared, and imaged nondestructively with a recently developed OTLS microscope (see Materials and Methods; ref. 15). Note that we have previously shown that there is no difference in the quality/appearance of our H&E-analogue 3D pathology datasets whether the tissue is formalin-fixed only versus FFPE (where de-paraffinization is necessary; refs. 15–17). Review of the 3D pathology datasets by pathologists (L.D. True and N.P. Reder) revealed that 118 out of the 300 biopsy cores contained cancer (1–5 biopsies per case). The ITAS3D pipeline was applied to all cancer-containing biopsies (see Supplementary Methods and Supplementary Fig. S6 for cancer-region annotation). We then calculated histomorphometric features from the 3D gland segmentations, and from individual 2D levels from the center region of the biopsy cores, which were then analyzed in terms of their association with BCR outcomes. For 2D analysis, average values from a total of 3 levels were calculated, in which the three levels were separated by 20 μm (mimicking clinical practice at many institutions) as shown in Fig. 4A.
Figure 4.
Clinical study comparing the performance of 3D versus 2D glandular features for risk stratification. A, Archived (FFPE) RP specimens were obtained from a well-curated cohort of 50 patients, from which, 300 simulated (ex vivo) needle biopsies were extracted (6 biopsies per case, per sextant-biopsy protocol). The biopsies were labeled with a fluorescent analogue of H&E staining, optically cleared to render the tissues transparent to light, and then comprehensively imaged in 3D with OTLS microscopy. Prostate glands were computationally segmented from the resultant 3D biopsy images using the ITAS3D pipeline. Three-dimensional glandular features were extracted from tissue volumes containing prostate cancer. Two-dimensional glandular features were extracted from three levels per volume and averaged. B and C, Violin and box plots are shown for two examples of 3D glandular features, along with analogous 2D features, for cases in which BCR was observed within 5 years of RP (“BCR”) and for cases with no BCR within 5 years of RP (“non-BCR”). For both sets of example features, “lumen boundary curvature” in B and “gland-to-convex hull ratio” (G/H) in C, the 3D version of the feature shows improved stratification between BCR and non-BCR groups. D and E, ROC curves also show improved risk stratification with the 3D features versus corresponding 2D features, with considerably higher AUC values. F, Violin and box plots are shown of representative gland-skeleton features (average branch length and branch length variance), which can only be accurately derived from the 3D pathology datasets, showing significant stratification between BCR and non-BCR groups. G, ROC curves are shown, along with AUC values, for average branch length and branch length variance. H, ROC curves are shown of various multiparameter models, including those trained with 2D glandular features, 3D glandular features excluding skeleton features, and 3D glandular features including skeleton features. I, KM curves are shown for BCR-free survival, showing that a multiparameter model based on 3D glandular features is better able to stratify patients into low-risk and high-risk groups with significantly different recurrence trajectories (P = 6.6 × 10–5, HR = 11.2, C-index = 0.84).
We compared multiple 3D and 2D glandular histomorphometric features (see Supplementary Table S3 for a detailed list). For example, the curvature of the boundary between the lumen and epithelium is a feature that increases as glands become smaller or more irregular, as is often seen with aggressive prostate cancer (21). This can be quantified in the form of the average surface curvature of the object in 3D, or the curvature of the object's cross-sectional circumference in 2D (Fig. 4B). As another example (Fig. 4C), the gland-to-convex-hull ratio (G/H) is defined as the volume ratio (in 3D) or the area ratio (in 2D) of the gland mask (epithelium + lumen) divided by the convex hull that circumscribes the gland. This G/H feature is inversely related to the irregularity or “waviness” of the periphery of the gland (at the scale of the gland itself rather than fine surface texture), which is generally expected to increase with aggressive prostate cancer (21). For various 3D and 2D features (Fig. 4D and E; Supplementary Table S3), ROC curves were generated to quantify the ability of the features to stratify patients based on 5-year BCR outcomes. When comparing analogous 3D and 2D glandular features, the 3D features largely exhibit an improved correlation with 5-year BCR outcomes in comparison with their 2D counterparts. This is exemplified by the significant P values for the 3D features showcased in Fig. 4B and C (between BCR and non-BCR groups) and higher area-under-the-ROC-curve (AUC) values (Fig. 4D and E).
We also extracted the 3D skeleton of the lumen network and quantified its branching parameters (skeleton-derived features). Here, a “gland skeleton” is defined as a collection of lines that approximate the center axes of various glands as they propagate in 3D space (similar to a line representation of a vessel network). Example skeleton networks for benign and cancerous glands are shown in Supplementary Video S6. Due to the complex 3D branching-tree architecture of the gland-lumen network, there are no straightforward 2D analogues for these skeleton-derived features. In Fig. 4F, we show two examples of skeleton-derived features: the average branch length and the variance of the branch lengths. Both features are correlated with BCR outcomes based on P values and AUC values (Fig. 4F and G). Our analysis reveals that aggressive cancers (BCR cases) have shorter branch lengths and a smaller variance in branch lengths, which agrees with prior observations from 2D histology that glandular structures in higher grade prostate cancer are smaller and more abundant (i.e., less differentiated and varied in size). A histogram of branch lengths (Supplementary Fig. S7) demonstrates that the vast majority of branches are < 200-μm long, which suggests that the diameter of standard prostate biopsies (∼1-mm) is sufficient for whole-biopsy 3D pathology to quantify prostate cancer branch lengths with reasonable accuracy.
To explore the prognostic value of combining multiple glandular features, we used logistic regression models for feature selection and classification based on 3D versus 2D features (see Materials and Methods). Brief descriptions and AUC values for the 3D and 2D glandular features involved in training the multiparameter models are shown in Supplementary Table S3. The ROC curve of a model that combined 12 non-skeleton 3D features (“3D nonskeleton model”) yielded an AUC value of 0.80 ± 0.05 (average ± SD; Fig. 4H), which is considerably higher than the AUC value (0.65 ± 0.06) of the model trained with 12 analogous 2D features (2D model). By adding five skeleton-derived features to the 12 non-skeleton 3D features, a retrained 3D multiparameter model (3D model) yielded a slightly higher AUC value of 0.81 ± 0.05. The distribution of the 50 cases, based on their glandular features, can be visualized using t-distributed stochastic neighbor embedding (t-SNE), where a clearer separation between BCR and non-BCR cases is evident based on 3D versus 2D glandular features (Supplementary Fig. S8). Multiparameter classification models based on 3D features alone (nonskeleton) or 2D features alone were used to divide patients into high- and low-risk groups based on 5-year BCR outcomes (Supplementary Table S1), from which KM curves of BCR-free survival were constructed for a subset of cases in which time-to-recurrence (BCR) data are available (Fig. 4I). Compared with the 2D model, the 3D model is associated with a higher HR and C-index, along with a significant P value (P < 0.05), suggesting superior prognostic stratification.
Discussion
As high-resolution biomedical imaging technologies continue to evolve and generate increasingly larger datasets, computational techniques are needed to derive clinically actionable information, ideally through explainable approaches that generate new insights and hypotheses. Interpretable feature-based analysis strategies in digital pathology generally hinge upon obtaining high-quality segmentations of key structural primitives (e.g., nuclei, glands, cells, collagen; refs. 18, 29). However, a common bottleneck to achieving accurate segmentations is the need for large amounts of manually annotated datasets (41). In addition to being tedious and difficult to obtain (especially in 3D), such annotations are often performed by one or more individuals who are not representative of all pathologists, thereby introducing an early source of bias. The use of simulated data has been explored to alleviate the need for manual annotations, and has been reported to be effective for training DL-based segmentation models for highly conserved and predictable morphologies [e.g., ellipsoidal nuclei (62, 63), or tubular vessel networks (63, 64)]. However, the 3D glandular networks of prostate tissues are highly irregular and variable, making it challenging to computationally generate simulated datasets. This complex and varied 3D morphology is also in part why 2D Gleason patterns may not be ideal for characterizing prostate glands.
The ITAS3D pipeline is a general approach for the volumetric segmentation of tissue structures (e.g., vasculature/endothelial cells, neurons, collagen fibers, lymphocytes) that can be immunolabeled with high specificity and that are also discernable to a deep-learning model when labeled with small-molecule stains like our H&E analogue or similar covalent stains (65). ITAS3D obviates the need for tedious and subjective manual annotations and, once trained, eliminates the requirement for slow/expensive antibody labeling of thick tissues (Fig. 1). The 2.5D segmentation approach employed in ITAS3D (i.e., image-sequence translation) offers an attractive compromise between computational speed/simplicity and accuracy for 3D objects that are relatively continuous in space (e.g., prostate glands). Details regarding 2.5D versus 3D image translation are provided in Supplementary Note S2 and Supplementary Table S4. In addition, a video summary of our ITAS3D-enabled gland-segmentation approach for prostate cancer assessment is provided in Supplementary Video S7. Note that in this specific implementation of ITAS3D, intermediate images are synthetically generated to mimic an IHC stain (CK8) that is routinely used by genitourinary pathologists. Therefore, it has the added advantage of enabling intuitive troubleshooting and facilitating clinical acceptance of our computational 3D pathology approach. A limitation of our implementation of ITAS3D is that the tissues used to train our deep-learning image-sequence translation modules were predominantly from Gleason pattern 3 and 4 regions, as well as benign regions. The reason for this was that the Canary TMA case–cohort study from which we derived our tissue specimens mainly consisted of low- to intermediate-grade (i.e., pattern 3 and 4) patients. In the future, improved ITAS3D performance over a wider range of prostate cancer grades would be facilitated by a more-diverse set of training specimens.
In this initial clinical study, we have intentionally avoided comparing our method with extant risk classifiers or nomograms that incorporate parameters based on human interpretation of 2D histopathology images [e.g., Kattan (66), CAPRA (67), and Canary-PASS (68)]. Rather, our goal has been to demonstrate the basic feasibility and value of 3D pathology by providing a direct comparison of intuitive 3D versus 2D glandular features analyzed computationally. A direct comparison of computational 3D pathology versus computational 2D pathology allows us to avoid the challenging variability and subjectivity of human interpretation. A human-observer study would require much-larger patient cohorts and a large panel of pathologist observers to account for inevitable interobserver variabilities. Nonetheless, to encourage the clinical adoption of 3D pathology methods, such large-scale studies will be necessary in the future, including prospective randomized studies on active surveillance versus curative therapies for low- to intermediate-risk patients (e.g., the PROTECT study; ref. 69), as well as studies to demonstrate the ability of computational 3D pathology to predict the response of individual patients to specific treatments such as androgen deprivation and both neoadjuvant and adjuvant chemotherapy. For completeness, in Supplementary Table S5, we provide Gleason scores for the 118 cancerous biopsies based on single-pathologist review (N.P. Reder) of whole-biopsy 3D pathology datasets with level-by-level examination of all 2D planes.
When trained with large numbers of images/cases and an optimal set of histomorphometric features, computational 2D pathology (based on whole slide images) has already been shown to be highly prognostic (21, 70–72). The goal of our study is not to suggest otherwise, but to provide early evidence of the additional prognostic value that 3D pathology can provide. The metrics presented in this study (Fig. 4) are intended to be comparative in nature (between computational 3D vs. 2D pathology) rather than regarded as definitive figures from a large prospective study. Our results show clear improvements in risk stratification based on 3D glandular features, both individually and in combination (Fig. 4B–I). As mentioned, the added prognostic value of 3D pathology is due in part to the significantly increased microscopic sampling of specimens (e.g., whole biopsies vs. sparse tissue sections). In addition, there are a number of advantages of 3D pathology datasets for computational analyses: (i) more-reliable segmentation of tissue structures due to the ability to leverage out-of-plane information (e.g., through continuity constraints) (ii); the ability to quantify tissue structures more accurately in 3D while avoiding 2D artifacts (73, 74), and (iii) the ability to extract novel prognostic features that cannot be derived from 2D tissue sections (e.g., gland-skeleton features).
As future work, ITAS3D can be used for the extraction and analysis of other 3D features (e.g., nuclear features, vascular features, and stromal features) to develop powerful classification models based on multiple morphologic primitives for a variety of tissue types. Annotation-free ITAS3D segmentation results, once available in sufficient quantities, can also be used to train end-to-end DL-based segmentation methods that bypass the image-translation step within ITAS3D. In the context of prostate cancer, studies are underway to identify additional prognostic 3D features based on our unique 3D-pathology datasets. A tiered approach to analyzing prostate cancer glandular features could be useful, such as first identifying broad classes of glandular morphologies (e.g., cribriform glands) and then analyzing class-specific features, as has been suggested in recent studies based on 2D whole-slide images of prostate cancer (70). Future studies should also aim to combine computational 3D pathology with patient metadata, such as radiomics, genomics, and electronic health records, to develop holistic decision-support algorithms (18). Nonetheless, as an initial step towards these goals, the results of this study, as enabled by the ITAS3D computational approach, provide the strongest evidence to date in support of the value of computational 3D pathology for clinical decision support, specifically for patients with low- to intermediate-risk prostate cancer.
Authors' Disclosures
W. Xie reports a patent for U.W. Ref. No. 49141.01US1, “Annotation-free segmentation of 3D digital pathology datasets with image-sequence translation” pending. N.P. Reder reports grants from Department of Defense and grants from Prostate Cancer Foundation during the conduct of the study; other support from Lightspeed Microscopy and grants from NIH outside the submitted work; in addition, N.P. Reder has a patent for “Annotation-free segmentation of 3D digital pathology datasets with image-sequence translation” pending. C.F. Koyuncu reports a patent for US20210049759A1 pending. P. Leo reports a patent for automated analysis of prostate pathology tissue pending and issued, and is presently employed by Genentech. K.W. Bishop reports grants from National Science Foundation during the conduct of the study. P. Fu reports grants from Case Western Reserve University during the conduct of the study. J.L. Wright reports grants from Merck, Altor Biosciences, Nucleix, Janssen, Movember; and other support from UpToDate outside the submitted work. A. Janowczyk reports grants from NIH during the conduct of the study; personal fees from Roche, Merck, and personal fees from Lunaphore outside the submitted work. A.K. Glaser reports other support from Lightspeed Bio outside the submitted work; in addition, A.K. Glaser has a patent for US10409052B2 issued and licensed to Lightspeed Bio, a patent for US20210033838 pending and licensed to Lightspeed Bio, a patent for US20210325656 pending and licensed to Lightspeed Bio, a patent for WO2021097300A1 pending and licensed to Lightspeed Bio, and a patent for WO2020167959A1 pending and licensed to Lightspeed Bio. A. Madabhushi reports grants from Bristol Myers-Squibb, AstraZeneca, Boehringer-Ingelheim; other support from Elucid Bioimaging and other support from Inspirata Inc; personal fees from Aiforia Inc, Caris Inc, Roche, Cernostics, and personal fees from Biohme outside the submitted work; in addition, A. Madabhushi has a patent 10,528,848 issued, a patent 10,769,783 issued, a patent 10,783,627 issued, and a patent 10,902,256 issued. L.D. True reports other support from LightSpeed Microscopy, Inc., outside the submitted work; in addition, L.D. True has a patent for patent no. 10,409,052 issued to University of Washington. J.T. Liu reports grants from NIH, US Department of Defense, and grants from National Science Foundation during the conduct of the study; other support from Lightspeed Microscopy, Inc., outside the submitted work; in addition, J.T. Liu has a patent for U.S. Patent No. 10,409,052. “Inverted light-sheet microscope,” filed on September 28, 2017, issued and licensed to Lightspeed Microscopy, a patent for U.S. Patent Application 17/356,135 “Apparatuses, Systems and Methods For Solid Immersion Meniscus Lenses,” filed 6/23/2021 pending and licensed to Lightspeed Microscopy, a patent for US Patent Application 17/310,309 “Apparatuses, Systems and Methods For Microscope Sample Holders,” filed 7/27/2021 pending and licensed to Lightspeed Microscopy, a patent for U.S. Patent Application No. 16/901,629, “Analysis of Prostate Glands Using Three-Dimensional (3D) Morphology Features of Prostate From 3D Pathology Images,” filed June 15, 2020 pending, and a patent for patent application 63/136,605 (provisional), “Annotation-free segmentation of 3D digital pathology datasets with image-sequence translation” filed on 1/12/2021 pending. No disclosures were reported by the other authors.
Disclaimer
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation, the NIH, the Department of Defense, the Department of Veterans Affairs, or the United States Government.
Supplementary Material
Supplementary methods, notes, figures, tables, video captions, and references
Acknowledgments
The authors acknowledge funding support from the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) through W81XWH-18-10358 (to J.T. Liu, L.D. True, and J.C. Vaughan), W81XWH-19-1-0589 (to N.P. Reder), W81XWH-15-1-0558 (to A. Madabhushi), and W81XWH-20-1-0851 (to A. Madabhushi and J.T. Liu). Support was also provided by the NCI through K99 CA240681 (to A.K. Glaser), R01CA244170 (to J.T. Liu), U24CA199374 (to A. Madabhushi), R01CA249992 (to A. Madabhushi), R01CA202752 (to A. Madabhushi), R01CA208236 (to A. Madabhushi), R01CA216579 (to A. Madabhushi), R01CA220581 (to A. Madabhushi), R01CA257612 (to A. Madabhushi), U01CA239055 (to A. Madabhushi), U01CA248226 (to A. Madabhushi), and U54CA254566 (to A. Madabhushi). Additional support was provided by the National Heart, Lung and Blood Institute (NHLBI) through R01HL151277 (to A. Madabhushi), the National Institute of Biomedical Imaging and Bioengineering (NIBIB) through R01EB031002 (to J.T. Liu) and R43EB028736 (to A. Madabhushi), the National Institute of Mental Health through R01MH115767 (to J.C. Vaughan), the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs (to A. Madabhushi), the National Science Foundation (NSF) 1934292 HDR: I-DIRSE-FW (to J.T. Liu), the NSF Graduate Research Fellowships DGE-1762114 (to K.W. Bishop) and DGE-1762114 (to L. Barner), the Nancy and Buster Alvord Endowment (to C.D. Keene), and the Prostate Cancer Foundation Young Investigator Award (to N.P. Reder). The training and inference of the deep learning models were facilitated by the advanced computational, storage, and networking infrastructure provided by the Hyak supercomputer system, as funded in part by the student technology fee (STF) at the University of Washington.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
W. Xie: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. N.P. Reder: Conceptualization, data curation, formal analysis, supervision, validation, investigation, methodology, project administration, writing–review and editing. C. Koyuncu: Software, formal analysis, validation, investigation, methodology, writing–review and editing. P. Leo: Software, formal analysis, validation, investigation, methodology, writing–review and editing. S. Hawley: Resources, data curation, writing–review and editing. H. Huang: Methodology, writing–review and editing. C. Mao: Resources, methodology, writing–review and editing. N. Postupna: Resources, methodology, writing–review and editing. S. Kang: Supervision, methodology, writing–review and editing. R. Serafin: Software, investigation, methodology, writing–review and editing. G. Gao: Investigation, methodology, writing–review and editing. Q. Han: Methodology. K.W. Bishop: Methodology, writing–review and editing. L.A. Barner: Methodology, writing–review and editing. P. Fu: Formal analysis, validation, methodology, writing–review and editing. J.L. Wright: Formal analysis, validation, writing–review and editing. C.D. Keene: Resources, formal analysis, supervision, methodology, writing–review and editing. J.C. Vaughan: Resources, supervision, methodology, writing–review and editing. A. Janowczyk: Formal analysis, investigation, methodology, writing–review and editing. A.K. Glaser: Software, methodology, writing–review and editing. A. Madabhushi: Resources, formal analysis, supervision, funding acquisition, validation, investigation, methodology, project administration, writing–review and editing. L.D. True: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, project administration, writing–review and editing. J.T.C. Liu: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Epstein JI. A new contemporary prostate cancer grading system. Ann Pathol 2015;35:474–6. [DOI] [PubMed] [Google Scholar]
- 3. Ozkan TA, Eruyar AT, Cebeci OO, Memik O, Ozcan L, Kuskonmaz I. Interobserver variability in Gleason histological grading of prostate cancer. Scand J Urol 2016;50:420–4. [DOI] [PubMed] [Google Scholar]
- 4. Shah RB, Leandro G, Romerocaces G, Bentley J, Yoon J, Mendrinos S, et al. Improvement of diagnostic agreement among pathologists in resolving an “atypical glands suspicious for cancer” diagnosis in prostate biopsies using a novel “Disease-Focused Diagnostic Review” quality improvement process. Hum Pathol 2016;56:155–62. [DOI] [PubMed] [Google Scholar]
- 5. Kane CJ, Eggener SE, Shindel AW, Andriole GL. Variability in outcomes for patients with intermediate-risk prostate cancer (Gleason Score 7, International Society of Urological Pathology Gleason Group 2–3) and implications for risk stratification: A systematic review. Eur Urol Focus 2017;3:487–97. [DOI] [PubMed] [Google Scholar]
- 6. Albertsen PC. Treatment of localized prostate cancer: when is active surveillance appropriate? Nat Rev Clin Oncol 2010;7:394–400. [DOI] [PubMed] [Google Scholar]
- 7. Haffner MC, De Marzo AM, Yegnasubramanian S, Epstein JI, Carter HB. Diagnostic challenges of clonal heterogeneity in prostate cancer. J Clin Oncol 2015;33:e38–40. [DOI] [PubMed] [Google Scholar]
- 8. Bill-Axelson A, Holmberg L, Garmo H, Rider JR, Taari K, Busch C, et al. Radical prostatectomy or watchful waiting in early prostate cancer. N Engl J Med 2014;370:932–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frey AU, Sonksen J, Fode M. Neglected side effects after radical prostatectomy: a systematic review. J Sex Med 2014;11:374–85. [DOI] [PubMed] [Google Scholar]
- 10. Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014;159:896–910. [DOI] [PubMed] [Google Scholar]
- 11. Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014;157:726–39. [DOI] [PubMed] [Google Scholar]
- 12. van Royen ME, Verhoef EI, Kweldam CF, van Cappellen WA, Kremers GJ, Houtsmuller AB, et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 2016;69:985–92. [DOI] [PubMed] [Google Scholar]
- 13. Olson E, Levene MJ, Torres R. Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express 2016;7:3089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka N, Kanatani S, Tomer R, Sahlgren C, Kronqvist P, Kaczynska D, et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng 2017;1:796–806. [DOI] [PubMed] [Google Scholar]
- 15. Glaser AK, Reder NP, Chen Y, Yin CB, Wei LP, Kang S, et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat Commun 2019;10:2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reder NP, Glaser AK, McCarty EF, Chen Y, True LD, Liu JTC. Open-top light-sheet microscopy image atlas of prostate core needle biopsies. Arch Pathol Lab Med 2019;143:1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barner LA, Glaser AK, Huang H, True LD, Liu JT. Multi-resolution open-top light-sheet microscopy to enable efficient 3D pathology workflows. Biomedical Optics Express 2020;11:6605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu JTC, Glaser AK, Bera K, True LD, Reder NP, Eliceiri KW, et al. Harnessing non-destructive 3D pathology. Nat Biomed Eng 2021;5:203–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tolkach Y, Thomann S, Kristiansen G. Three-dimensional reconstruction of prostate cancer architecture with serial immunohistochemical sections: hallmarks of tumour growth, tumour compartmentalisation, and implications for grading and heterogeneity. Histopathology 2018;72:1051–9. [DOI] [PubMed] [Google Scholar]
- 20. Humphrey PA. Complete histologic serial sectioning of a prostate-gland with adenocarcinoma. Am J Surg Pathol 1993;17:468–72. [DOI] [PubMed] [Google Scholar]
- 21. Lee G, Sparks R, Ali S, Shih NN, Feldman MD, Spangler E, et al. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients. PLoS One 2014;9:e97954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leo P, Janowczyk A, Elliott R, Janaki N, Bera K, Shiradkar R, et al. Computer extracted gland features from H&E predicts prostate cancer recurrence comparably to a genomic companion diagnostic test: a large multi-site study. NPJ Precis Oncol 2021;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee G, Singanamalli A, Wang H, Feldman MD, Master SR, Shih NNC, et al. Supervised multi-view canonical correlation analysis (sMVCCA): Integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging 2015;34:284–97. [DOI] [PubMed] [Google Scholar]
- 24. Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 2021;594:106–10. [DOI] [PubMed] [Google Scholar]
- 25. Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158–64. [DOI] [PubMed] [Google Scholar]
- 26. Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020;26:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leo P, Elliott R, Shih NNC, Gupta S, Feldman M, Madabhushi A. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: a multi-site study. Sci Rep 2018;8:14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin W, Hasenstab K, Cunha GM, Schwartzman A. Comparison of handcrafted features and convolutional neural networks for liver MR image adequacy assessment. Sci Rep 2020;10:20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chandramouli S, Leo P, Lee G, Elliott R, Davis C, Zhu G, et al. Computer extracted features from initial H&E Tissue biopsies predict disease progression for prostate cancer patients on active surveillance. Cancers 2020;12:2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Selective invocation of shape priors for deformable segmentation and morphologic classification of prostate cancer tissue microarrays. Comput Med Imaging Graph 2015;41:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lefebvre AEYT, Mai D, Kessenbrock K, Lawson DA, Digman MA. Automated segmentation and tracking of mitochondria in live-cell time-lapse images. Nat Methods 2021;18:1091. [DOI] [PubMed] [Google Scholar]
- 33. Bhargava HK, Leo P, Elliott R, Janowczyk A, Whitney J, Gupta S, et al. Computationally derived image signature of stromal morphology is prognostic of prostate cancer recurrence following prostatectomy in african american patients. Clin Cancer Res 2020;26:1915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiemen A, Braxton AM, Grahn MP, Han KS, Babu JM, Reichel R, et al. In situ characterization of the 3D microanatomy of the pancreas and pancreatic cancer at single cell resolution. bioRxiv 2020. [Google Scholar]
- 35. Arunkumar N, Mohammed MA, Mostafa SA, Ibrahim DA, Rodrigues JJPC, de Albuquerque VHC. Fully automatic model-based segmentation and classification approach for MRI brain tumor using artificial neural networks. Concurr Comp-Pract E 2020;32:4962. [Google Scholar]
- 36. Koyuncu CF, Janowczyk A, Lu C, Leo P, Alilou M, Glaser AK, et al. Three-dimensional histo-morphometric features from light sheet microscopy images result in improved discrimination of benign from malignant glands in prostate cancer. International Society for Optics and Photonics; 2020;113200G. [Google Scholar]
- 37. Chen H, Dou Q, Yu L, Qin J, Heng P-A. VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018;170:446–55. [DOI] [PubMed] [Google Scholar]
- 38. Bui TD, Shin J, Moon T. 3D densely convolutional networks for volumetric segmentation. arXiv2017.
- 39. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. Springer. 2016:424–32. [Google Scholar]
- 40. Zhu Z, Xia Y, Shen W, Fishman E, Yuille A. A 3D coarse-to-fine framework for volumetric medical image segmentation. IEEE. 2018:682–90. [Google Scholar]
- 41. van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med 2021;27:775–84. [DOI] [PubMed] [Google Scholar]
- 42. Di Cataldo S, Ficarra E, Acquaviva A, Macii E. Automated segmentation of tissue images for computerized IHC analysis. Comput Methods Programs Biomed 2010;100:1–15. [DOI] [PubMed] [Google Scholar]
- 43. Migliozzi D, Nguyen HT, Gijs MAM. Combining fluorescence-based image segmentation and automated microfluidics for ultrafast cell-by-cell assessment of biomarkers for HER2-type breast carcinoma. J Biomed Opt 2018;24:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee SS, Bindokas VP, Lingen MW, Kron SJ. Nondestructive, multiplex three-dimensional mapping of immune infiltrates in core needle biopsy. Lab Invest 2019;99:1400–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bayramoglu N, Kaakinen M, Eklund L, Heikkilä J. Towards virtual H&E staining of hyperspectral lung histology images using conditional generative adversarial networks. IEEE International Conference on Computer Vision Workshops, 22–29 Oct. 2017. p.64–71. [Google Scholar]
- 46. Burlingame EA, McDonnell M, Schau GF, Thibault G, Lanciault C, Morgan T, et al. SHIFT: speedy histological-to-immunofluorescent translation of a tumor signature enabled by deep learning. Sci Rep 2020;10:17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y, et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng 2019;3:466–77. [DOI] [PubMed] [Google Scholar]
- 48. Wang T-C, Liu M-Y, Zhu J-Y, Liu G, Tao A, Kautz J, et al. Video-to-video synthesis. arXiv2018.
- 49. Hawley S, Fazli L, McKenney JK, Simko J, Troyer D, Nicolas M, et al. A model for the design and construction of a resource for the validation of prognostic prostate cancer biomarkers: the Canary Prostate Cancer Tissue Microarray. Adv Anat Pathol 2013;20:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Annal Mathemat Stat 1947;18:50–60. [Google Scholar]
- 51. Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 52. Dobbin KK, Cesano A, Alvarez J, Hawtin R, Janetzki S, Kirsch I, et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume II - clinical validation and regulatory considerations. J Immunother Cancer 2016;4:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ, Dotan ZA, DiBlasio CJ, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol 2005;23:7005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singh M, Kalaw EM, Giron DM, Chong KT, Tan CL, Lee HK. Gland segmentation in prostate histopathological images. J Med Imaging 2017;4:027501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bulten W, Bándi P, Hoven J, van de Loo R, Lotz J, Weiss N, et al. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci Rep 2019;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soille PJ, Ansoult MM. Automated basin delineation from digital elevation models using mathematical morphology. Signal Process 1990;20:171–82. [Google Scholar]
- 57. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Springer. 2015:234–41. [Google Scholar]
- 58. Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297–302. [Google Scholar]
- 59. Cignoni P, Rocchini C, Scopigno R. Metro: measuring error on simplified surfaces. Wiley Online Library; 1998:167–74. [Google Scholar]
- 60. Cirillo MD, Abramian D, Eklund A. Vox2Vox: 3D-GAN for brain tumour segmentation. arXiv2020.
- 61. Haft-Javaherian M, Fang LJ, Muse V, Schaffer CB, Nishimura N, Sabuncu MR. Deep convolutional neural networks for segmenting 3D in vivo multiphoton images of vasculature in Alzheimer disease mouse models. PLoS One 2019;14:e0213539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dunn KW, Fu CC, Ho DJ, Lee S, Han S, Salama P, et al. DeepSynth: Three-dimensional nuclear segmentation of biological images using neural networks trained with synthetic data. Sci Rep 2019;9:18295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu Q, Gaeta IM, Zhao M, Deng R, Jha A, Millis BA, et al. Towards annotation-free instance segmentation and tracking with adversarial simulations. arXiv2021. [DOI] [PMC free article] [PubMed]
- 64. Tetteh G, Efremov V, Forkert ND, Schneider M, Kirschke J, Weber B, et al. DeepVesselNet: Vessel segmentation, centerline prediction, and bifurcation detection in 3-D Angiographic Volumes. Front Neurosci 2020;14:592352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mao C, Lee MY, Jhan J-R, Halpern AR, Woodworth MA, Glaser AK, et al. Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci Adv 2020;6:eaba4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol 1999;17:1499–507. [DOI] [PubMed] [Google Scholar]
- 67. Cooperberg MR, Freedland SJ, Pasta DJ, Elkin EP, Presti JC Jr, Amling CL, et al. Multiinstitutional validation of the UCSF cancer of the prostate risk assessment for prediction of recurrence after radical prostatectomy. Cancer 2006;107:2384–91. [DOI] [PubMed] [Google Scholar]
- 68. Newcomb LF, Thompson IM, Boyer HD, Brooks JD, Carroll PR, Cooperberg MR, et al. Outcomes of active surveillance for clinically localized prostate cancer in the prospective, multi-institutional Canary PASS cohort. J Urol 2016;195:313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016;375:1415–24. [DOI] [PubMed] [Google Scholar]
- 70. Leo P, Chandramouli S, Farré X, Elliott R, Janowczyk A, Bera K, et al. Computationally derived cribriform area index from prostate cancer hematoxylin and eosin images is associated with biochemical recurrence following radical prostatectomy and is most prognostic in gleason grade group 2. Eur Urol Focus 2021;7:722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wulczyn E, Nagpal K, Symonds M, Moran M, Plass M, Reihs R, et al. Predicting prostate cancer specific-mortality with artificial intelligence-based Gleason grading. Commun Med 2021;1:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nagpal K, Foote D, Liu Y, Chen PHC, Wulczyn E, Tan F, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2019;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Glaser AK, Reder NP, Chen Y, McCarty EF, Yin CB, Wei LP, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng 2017;1:0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Epstein JI, Allsbrook WC Jr, Amin MB, Egevad LL, Committee IG. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol 2005;29:1228–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, notes, figures, tables, video captions, and references
Data Availability Statement
Relevant clinical data for this study are provided in Supplementary Table S1 and Supplementary Table S5. Example prostate images for testing ITAS3D codes and models are available in a GitHub repository at https://github.com/WeisiX/ITAS3D. Full 3D prostate imaging datasets, “simulated 2D whole-slide images” extracted from those datasets (three levels per biopsy), as well as other clinical data are available upon reasonable request and with the establishment of a material-transfer or data-transfer agreement.