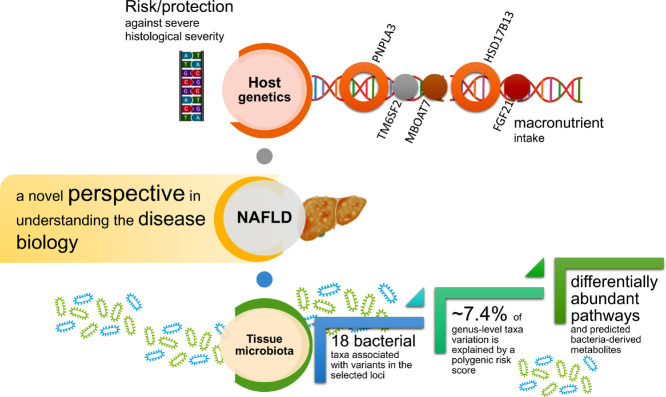
Summary
Background
Human body microbiotas are influenced by several factors, including the interaction of the host with the environment and dietary preferences. The role of host genetics in modulating the liver microbiota in the context of NAFLD remains unknown. To address this gap, we examined the interplay between the liver metataxonomic profile and host genetics.
Methods
We obtained 16S rRNA gene sequences from liver biopsies and genotypes by Taqman-assays in 116 individuals. We compared taxon abundance at the genus level across host genotypes using dominant models of inheritance. We focused the analysis on variants influencing the risk/ protection against NAFLD-histological severity (PNPLA3-rs738409, TM6SF2-rs58542926, MBOAT7-rs641738, and HSD17B13-rs72613567) and a variant influencing macronutrient intake (FGF21-rs838133). We also explored the variants' combined effect via a polygenic risk score (PRS).
Findings
We identified at least 18 bacterial taxa associated with variants in the selected loci. Members of the Gammaproteobacteria class were significantly enriched in carriers of the rs738409 and rs58542926 risk-alleles, including Enterobacter (fold change [FC]=6.2) and Pseudoalteromonas (FC=2) genera, respectively. Lawsonella (1.6-FC), Prevotella_9 (FC=1.5), and Staphylococcus (FC=1.3) genera were enriched in rs838133-minor allele carriers, which is linked to sugar consumption and carbohydrate intake. Tyzzerella abundance (FC=2.64) exhibited the strongest association (p = 0.0019) with high PRS values (>4 risk alleles). The percentage of genus-level taxa variation explained by the PRS was ∼7.4%, independently of liver steatosis score and obesity.
Interpretation
We provided evidence that genetic variation may influence the liver microbial DNA composition. These observations may represent potentially actionable mechanisms of disease.
Keywords: NASH, Genetics, PNPLA3, TM6SF2, HSD17B13, Risk score, FGF21, MBOAT7
Abbreviations: FGF21, Fibroblast growth factor 21; MBOAT7, Membrane bound O-Acyltransferase domain containing 7; NAFLD, nonalcoholic fatty liver disease; NAFL, nonalcoholic fatty liver; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13; NASH, nonalcoholic steatohepatitis; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane 6 superfamily member 2
Graphical abstract
Research in context.
Evidence before this study
NAFLD is a multifactorial disease that has become the most prevalent chronic liver disease worldwide. The disease natural history is variable among individuals. NAFLD phenotypic complexity is determined by multiple disease-triggering mechanisms, including environmental factors, genetic makeup, gut microbiota, and lifestyle factors. Pioneering studies on the liver tissue metataxonomic profile of patients with NAFLD revealed that the liver contains a diverse repertoire of bacterial DNA that may be involved in the disease severity. Nevertheless, the potential interaction between the liver microbial DNA composition and the host genetics is not entirely understood.
Added value of this study
We explored the interrelationship between the liver metataxonomic profile in patients with NAFLD across the entire spectrum of the disease severity and variants modifying either risk or protection against the disease and variants involved in carbohydrate intake and macronutrient preferences. Together, we provide evidence that the liver microbiota depends in part on the host genetic background. By combining the effects of trait-associated risk alleles, we demonstrated that ∼7.4% of genus-level taxa variation is explained by a polygenic risk score. This effect was independent of key covariate parameters, including obesity and the liver amount of fat. Furthermore, we explored the functional consequences of taxa linked to genetic variation, and we detected differentially abundant pathways and predicted bacteria-derived metabolites that may serve to understand the mechanism (s) of disease.
Implications of all the available evidence
We present a proof-of-concept study demonstrating that host genetics plays an essential role in modulating the liver microbial DNA composition. Our results show, for instance, that carriers of the PNPLA3-rs738409 risk allele may present an overabundance of DNA derived from members of the Gammaproteobacteria class, some of which are capable of metabolizing alkane-derived carbons to yield primary alcohols. Collectively, we provide a novel perspective in understanding disease biology.
Alt-text: Unlabelled box
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the leading global cause of chronic liver disease1. The disease pathogenesis involves a complex interrelationship among lifestyle (including excessive intake of energy-dense foods and diet rich in highly refined carbohydrates), systemic metabolic dysregulation associated with related comorbidities such as obesity, type 2 diabetes, and cardiovascular risk,1,2 genetic predisposition involving nuclear and mitochondrial genomes,3,4 and epigenetic mechanisms.5,6
The NAFLD natural history is characterized by not only high inter-individual variability but also phenotypic complexity, with histological stages ranging from non-alcoholic fatty liver (NAFL) to steatohepatitis (NASH), NASH-fibrosis, cirrhosis, and even hepatocellular carcinoma1.
Owing to the advances in molecular techniques, including high-throughput OMIC approaches, it has been established that a significant proportion of the variability in NAFLD-histologic traits can be explained by a highly interconnected and dynamic network of factors7 that includes the gut microbiome.8, 9, 10, 11, 12
Seminal studies examining the biogeography of bacterial communities of the human body13 revealed that tissues' microbial composition plays a vital role in maintaining not only individual organ physiology but also systemic metabolic homeostasis. We recently found that the liver metataxonomic signature may explain not only differences in the NAFLD pathogenic mechanisms and phenotypic histological complexity but also physiological functions of the host.14
Several lines of evidence suggest that the microbial composition of the body is shaped by diverse factors, including host genetics.15 Xie and co-workers demonstrated the heritability of many microbial taxa in the gut microbiome, including those associated with diseases.16 More importantly, a recent study conducted by Kolde showed that genetic attributes and single genetic variant effects on the microbial features are specific to particular body sites.17
While it is widely accepted that genetic susceptibility plays a significant role in the biology of NAFLD,4,7 the role of genetics in modulating the liver microbiota composition associated with the disease remains unknown. To explore whether the liver microbiota is shaped by host genetics, we conducted candidate gene association analyses using as a proxy of NAFLD histological severity common variants in loci that impose either risk or protection against the disease (PNPLA3-rs738409,18,19 TM6SF2-rs58542926,20, 21, 22 MBOAT7-rs641738,23 and HSD17B13-rs7261356724, 25, 26). In addition, as a proxy of major dietary modifiers, we used a variant in a locus that has been reproducibly linked to high carbohydrate intake and macronutrient preferences (FGF21-rs838133).27,28
Methods
Studied population
This study involves a secondary analysis of microbial 16S rRNA reads from the liver of 116 individuals, categorized as non-NAFLD patients (n = 19) and patients with NAFL (n = 44) and NASH (n = 53), as explained elsewhere.14 The cohort of patients with NAFLD included moderately obese or obese subjects (BMI < 40 kg/m2, n = 47) and severely/ morbidly obese patients (BMI > 40 kg/m2) elected for bariatric surgery (n = 50). By adopting a matched cohort study design, we ensured that the non-NAFLD liver samples in each group matched patients' features.14
As non-NAFLD liver tissue we included samples from patients without either NAFLD or features of the metabolic syndrome who were selected from patients attending the Liver Unit, whose age and sex matched the NAFLD patients. These patients presented near-normal liver histology in the liver specimens obtained by percutaneous liver biopsy, and the reason for performing a liver biopsy in these subjects was based on the presence of persistently mildly elevated serum liver enzymes activity. In all the non-NAFLD subjects, all causes of common liver disease were ruled out, and they were included in the study if they did not have histological evidence of fatty change; the histological diagnosis of non-NAFLD livers was minimal changes. In the population of morbid obese patients, non-NAFLD subjects were obese patients who also underwent bariatric surgery and had not features of NAFLD demonstrated in the liver biopsy.
Patients were recruited prospectively and were considered for inclusion if they had histopathologic evidence of fatty liver disease, either NAFL or NASH, on liver biopsy performed within the study period. Selection of biological samples included in the present study was based on sufficient amounts of high-quality nucleic acids (DNA) for performing metagenomic explorations. Exclusion criteria: Secondary causes of steatosis, including alcohol abuse (≥ 30 g for men and ≥ 20 g for women, of alcohol daily), total parenteral nutrition, hepatitis B and hepatitis C virus infection, and the use of drugs known to precipitate steatosis were excluded. In addition, patients with any of the following diseases were excluded: autoimmune liver disease, metabolic liver disease, Wilson's disease, and α-1-antitrypsin deficiency. Patients under treatment with antibiotics, immunosuppressive medication or proton-pump bomb inhibitors were also excluded.
Biological specimens from NAFLD-participants and non-NAFLD subjects were consecutively selected during the same study period from the same population of patients attending the participating institutions located in Argentina, ensuring that all shared the same demographic characteristics (occupation, educational level, place of residence, and ethnicity) as the matched patients.
Arterial hypertension was defined as systolic blood pressure ≥140 mm Hg and/or diastolic blood pressure ≥90 mm Hg following repeated examination and considering the average of 2 or more blood pressure measurements on at least 2 subsequent visits.29
All liver specimens were obtained by liver biopsy that was performed before any intervention (if required) with ultrasound guidance using a modified 1.4-mm-diameter Menghini needle (Hepafix, Braun, Germany) under local anaesthesia on an outpatient basis, or during bariatric surgery (surgically excised samples from the left lobe were immediately collected after the abdomen was opened and before organs were manipulated). A portion of each liver biopsy specimen was routinely fixed in 40 g/l formaldehyde (pH 7.4), embedded in paraffin, and stained with haematoxylin and eosin, Masson trichrome, and silver impregnation for reticular fibres. All biopsies were at least 3 cm in length and contained a minimum of eight portal tracts.14
Ethics
Biological specimens, including blood samples and liver biopsies from all subjects included in this study, were obtained with written, informed consent under the Institutional Review Board-approved protocols (protocol numbers: 104/HGAZ/09, 89/100, 1204/2012, and updated DI-2019-376-GCABA-HGAZ). Protocol # DI-2019-376-GCABA-HGAZ (“Genetics of steatohepatitis and its association with the liver tissue microbiome”) was approved under the Argentinean Law No. 3301 on Protection of the Rights of Subjects in Health Research, regulated by Decree No. 58/2011, which establishes the regime for health research activity with human beings. All data were de-identified prior to use. All investigations performed as a part of the present study were conducted under the guidelines of the 1975 Declaration of Helsinki, as revised in 1993.
Host genetics: selection of variants and genotyping strategy
We selected variants in genes that serve as valid instruments for assessing the interrelationship with the liver microbial DNA composition. We focused the variant selection on two axes, including variants influencing the risk and /or protection against severe NAFLD-histological phenotypes and key external factors involved in the disease pathogenesis, such as dietary preferences. Hence, variant selection involved: (i) major genetic modifiers of NAFLD and NASH natural history that have been consistently replicated across ethnicities4, including coding single nucleotide polymorphisms in PNPLA3 (rs738409), TM6SF2 (rs58542926), MBOAT7 (rs641738), and the splice donor variant in HSD17B13 (rs72613567), and (ii) the synonymous rs838133 variant in the first exon of FGF21, which is linked to food intake regulation, macronutrient preference (high carbohydrate intake), and central reward pathways.27,28
The genetic analyses were performed on genomic DNA extracted from white blood cells by a standard method.30 For rs738409 genotyping, a high-throughput genotyping method involving PCR amplification of genomic DNA with two-tailed allele-specific primers was implemented as previously described.31 Genotyping of rs58542926, rs641738, rs72613567, and rs838133 was performed using TaqMan genotyping assays (dbSNP rs58542926 assay C__89463510_10, #4351379, dbSNP rs641738 assay C___8716820_10, # 4351379, rs72613567 custom assay #4332072, and dbSNP rs838133 assay C___8832415_10 Cat. # 4351379, respectively, Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, as explained in our previous publications.22,26,32
To ensure genotyping quality, we included DNA samples as internal controls, hidden samples of known genotype, and negative controls (water). The overall genotype completion rate was 100%. To account for possible population stratification, we used a collection of 13 SNPs at different loci (located in chromosomes 4, 15, 17, 13, 1, and 3) and then analysed the data with the Structure program Version 233 as we explained elsewhere.22,26,32 We found no evidence of stratification in our sample because the cases and the non-NAFLD subjects showed similar Q values and the Structure program assigned a similar distance to clusters, with no further improvement in the fitting model by adding up to four clusters (the greatest ln of likelihood was obtained for K = 1).
In addition, we explored the combined effect/s of variants by examining a polygenic risk score (PRS) calculated as the sum of trait-associated risk alleles across the selected loci. In the case of the HSD17B13 (rs72613567) variant, we considered the deletion allele (-) as the risk allele.
Microbial DNA data collection
Using high-throughput 16S rRNA gene sequencing, we obtained pertinent liver metataxonomic information from the tissue samples of the 116 individuals as explained earlier.14 In brief, microbial DNA was isolated from fresh liver specimens by a manual protocol, after which extracted bulk DNA samples were amplified with barcoded primers, DNA libraries were constructed and high-throughput sequencing was performed on an IlluminaMiSeq platform by Macrogen Inc. (NGS Division) Seoul, South Korea. A negative (blank) control was used to test the potential presence of contaminant DNA and/or cross-contamination; the sample showed no product and did not pass the quality control analysis.14 The 16S rRNA gene hypervariable region 3 (V3) and V4 amplicons were generated via PCR amplification using primers as reported elsewhere.14 The 16S rRNA gene sequencing data were filtered, denoised, and processed on the QIIME2 (version 2018.11) platform (http://qiime2.org/index.html). To account for the generally non-normal distribution of microbial community composition data, 16S rRNA abundances underwent normalization by total sum combined with squared root transformation. High-quality sequences were assigned to operational taxonomic units (OTUs) using the QIIME pipeline34 whereby default parameters were used in the selection of OTUs for constructing the OTU table. High-quality amplicon sequence variants were classified using the vsearch algorithm with default parameters and SILVA 16S-only 99% identity database (release 137) to build our BIOM feature table of OTUs. Assessment of differences in the liver bacterial DNA composition across the entire disease severity spectrum was further confirmed by an independent molecular approach as explained earlier.14
Functional profiling of liver microbial DNA
To predict metagenome functional content from the OTU table, we utilized the bioinformatics software package PICRUSt2: Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (https://github.com/picrust/picrust2/). To additionally detect differentially abundant KEGG ortholog pathways, we used the Piphillin metagenomics inference tool.35 The normalized sequence abundance table and the weighted nearest-sequenced taxon index values per-sample were used to predict pathways. The resulting output was a list of MetaCyc pathway abundances according to the MetaCyc database (https://metacyc.org/). The STAMP software package (version 2.1.3), which relies on the concept of biological relevance in the form of confidence intervals, was employed to determine differentially enriched metabolic pathways (p < 0.05) and their effect sizes (η2) (http://kiwi.cs.dal.ca/Software/STAMP).36 In addition, we reported p-values corrected through the Benjamini-Hochberg procedure for multiple testing corrections.
Statistical analysis
We compared taxon abundances at the genus level across genotypes assuming a dominant model of inheritance and comparing YY with XY + XX, in which X is the risk allele. The use of a dominant model for all tested variants is justified as we performed pairwise comparisons of the relative abundance of each taxon. Differences between genotype groups were assessed using a pairwise Wilcoxon rank-sum test implemented in the R-base function and the default method for p-value correction using the web-application Calypso (FDR < 0.05).37
To identify the potential discriminatory power of individual bacterial DNA taxa over PRS, we used the area under the receiver operating characteristic (ROC) curve (AUC) and fold change. To adjust for relevant covariates (sex, obesity degree, and liver disease severity), we used non-parametric ROC estimation using bootstrap to obtain standard errors and confidence intervals along with linear regression to model the covariates.
We calculated the fold change in the abundance of taxon of interest as the ratio of the mean of sequencing reads assigned to the taxon normalized by the total number of sequencing reads combining with square root transformation in each group. Measurements across genotype groups were compared by non-parametric statistical tests (Wilcoxon rank test), with p < 0.05 indicating statistical significance. As indicated in some cases, pair-wise comparisons between genotype groups were performed through Student's t-test, and relative abundance was calculated as the square root of each feature read count divided by the total number of features reads in each sample, and the standard error (SE) is indicated by error bars.
The R-squared in ANOVA estimated the genetic contribution to the variance in the abundance of different genera.
We used ordinary linear regression to study the correlation between bacterial DNA abundance as dependent variable and independent variables, in particular, PRS as a continuous or dichotomic variable and relevant covariates such as sex (0=female, 1=male), steatosis score (0, 1, 2, and 3 grades), obesity degree (obese/ overweight vs. severe obesity) and liver disease severity (non-NAFLD, NAFL, and NASH). After fit the model, margins subroutine was used to estimate predicted media at fixed values of some covariates as implemented in the STATA v16.0 package (StataCorp LLC, College Station, Texas, USA).
For the assessment of the association between blood octane levels and liver fibrosis in a population-based study, we used the svyset (survey design for dataset) command (STATA 16.0). To avoid skewed distributions, continuous variables were log-transformed (Log10). Statistical analyses were performed by linear regression among blood octane concentration as the dependent variable and quantiles of the median stiffness as the independent variable. Age (years), gender (women= 0, men= 1), body mass index (kg/m2), diabetes (no = 0, yes = 1), and alcohol consumption (g per day) were also included as covariates.
For the comparison of clinical, biochemical, and histological characteristics we used Mann-Whitney U test, except for female/male proportion between studied groups that was assessed by a Chi-square test.
Sample size and power calculations indicate that a sample with ∼50 participants per genotype group has a power of ∼0.80 to detect media differences representing ∼2.0 fold-changes with a probability of <0.05 for type I error.
Role of funders
The funders had no role in the conceptualization, study design, data collection, analysis, interpretation of data, in writing the paper, or in the decision to submit the paper for publication.
Results
Host genetics and hepatic microbial DNA associations
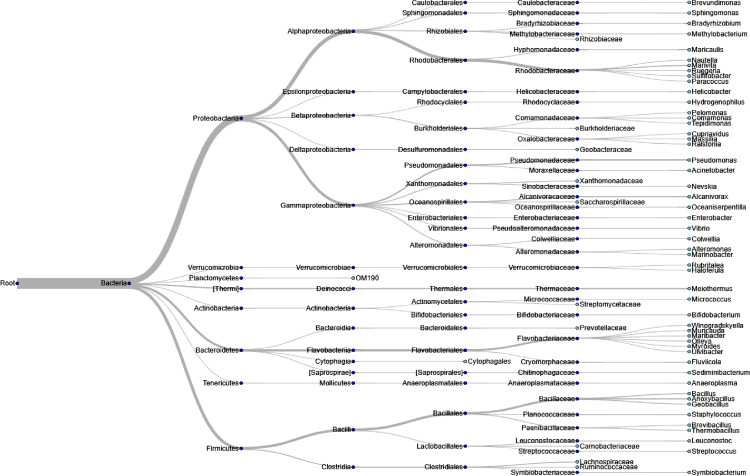
Detailed clinical, biochemical, and histological characteristics of the study participants is shown in Table 1. Briefly, the sample comprised of 55.17% women, with a mean age of 46 ± 2.3 years. Genotype frequencies were in Hardy-Weinberg equilibrium (Table 2). Visualization of the hierarchical relationship of liver microbial DNA profile in the whole population for the 950 most abundant taxa is depicted in Figure 1.
Table 1.
Baseline clinical, biochemical, and histological characteristics of the studied cohort.
| Non-NAFLD group (near normal liver histology) | ||||
|---|---|---|---|---|
| Variables | non-morbidly obese subjects | morbidly obese subjects | ||
| Number of subjects | 9 | 10 | ||
| Female/Male (n) | 5/4 | 6/4 | ||
| Age, years | 43.8±8 | 44.8±8 | ||
| BMI, kg/m2 | 24±3 | 55±14 | ||
| Type 2 diabetes (n) | 0 | 4 | ||
| Fasting plasma glucose, mg/dL | 87±10 | 105±23 | ||
| Fasting plasma insulin, μU/ml | 5±2.5 | 11±8.4 | ||
| HOMA-IR index | 1.02±0.5 | 2±1.2 | ||
| Total cholesterol, mg/dL | 188±38 | 190±36 | ||
| HDL-cholesterol, mg/dL | 60±15 | 40±10 | ||
| LDL-cholesterol, mg/dL | 110±42 | 121±36 | ||
| Triglycerides, mg/dL | 101±24 | 128±65 | ||
| ALT, U/L | 42±26 | 19.7±8 | ||
| AST, U/L | 32±11 | 19.6±8 | ||
| Patients with biopsy-confirmed diagnosis of NAFLD | ||||
|---|---|---|---|---|
| Variables | non-morbidly obese patients |
morbidly obese patients |
||
| NAFL | NASH | NAFL | NASH | |
| Number of subjects | 21 | 26 | 23 | 27 |
| Female/Male (n) | 10/11 | 16/10 | 12/11 | 15/12 |
| Age, years | 49.4±11 | 46.7±13 | 43±9 | 48±10 |
| BMI, kg/m2 | 30±5 # | 34±6 +* | 53±13 | 49±10 |
| Type 2 diabetes (n) | 6 | 15 +* | 8 | 18 * |
| Fasting plasma glucose, mg/dL | 105±26 # | 124±39 +* | 101±22 | 138±63 * |
| Fasting plasma insulin, μU/ml | 14±7 # | 19.6±12.4 + | 13±7 | 35±45 +* |
| HOMA-IR index | 3.5±1.7 # | 6.1±6 + | 3.1±1.7 | 16±40 +* |
| Total cholesterol, mg/dL | 203±39 | 196±42 | 180±37 | 179±49 |
| HDL-cholesterol, mg/dL | 58±16 | 51.2±15 | 45±10 | 37±6 * |
| LDL-cholesterol, mg/dL | 124±35 | 120±36 | 122±28 | 126±43 |
| Triglycerides, mg/dL | 143±94 | 148±67 | 154±57 | 191±102 |
| ALT, U/L | 56±42 | 52±31 +* | 28±27 # | 43±20 + |
| AST, U/L | 40±23 | 87±59 | 29±20 | 31±14 + |
| Histological Features | ||||
| Degree of steatosis (0-3) | 1.5±0.7 # | 2.2±0.44 + | 1.73±0.81 # | 2.19±0.8 + |
| Lobular inflammation (0-3) | 0.7±0.73 # | 1.24±0.8 + * | 0.35±0.6 # | 1.42±0.8 + * |
| Hepatocellular ballooning (0-2) | 0±0 # | 0.8±0.6 +* | 0.18±0.4 # | 1.07±0.62 + * |
| Fibrosis Stage | 0±0 # | 1.61±0.6 +* | 0.04±0.2 # | 1.7±0.6 +* |
| NAFLD activity score (NAS) | 2.5±1.19 # | 4.3±1 +* | 2.26±1.44 # | 4.7±2 +* |
NAFL: non-alcoholic fatty liver, NASH: non-alcoholic steatohepatitis, BMI: body mass index; HOMA: homeostatic model assessment; ALT and AST: Serum alanine and aspartate aminotransferase. Results are expressed as mean ± SD except indicated otherwise.
# p<0.001 indicates NAFL vs. controls
*p<0.001 indicates comparisons between NAFL and NASH, and + p<0.001 denotes comparisons between NASH and control subjects.
P value stands for statistical significance using Mann-Whitney U test, except for female/male proportion that p value stands for statistical significance using Chi-square test
Table 2.
Genotype distribution, variants features and genotype counts.
| Gene symbol | Alleles | Risk allele | Variant ID | Gene Consequence | Genotype counts | Dominant model (n) | HWE Chi 2 p value |
|---|---|---|---|---|---|---|---|
| PNPLA3 | C>G | G | rs738409 | missense | CC: 31 CG:54 GG:31 |
CC: 31 CG+GG: 85 |
0.45 |
| TM6SF2 | C>T | T | rs58542926 | missense | CC:89 CT:25 TT:2 |
CC: 89 CT+TT: 27 |
0.87 |
| MBOAT7 | C>T | T | rs641738 | TMC4: Missense Variant/MBOAT7: 500B Downstream Variant | CC:44 CT:51 TT:21 |
CC: 44 CT+TT: 72 |
0.36 |
| HSD17B13 | ->A | -/ | rs72613567 | Splicing donor | -/-: 83 -/A: 28 A-INS: 3 |
-/-: 83 -A+AA: 31 * |
0.73 |
| FGF21 | G>A | A | rs838133 | synonymous variant | GG: 61 AG: 43 AA: 12 |
GG: 61 GA+AA: 55 |
0.29 |
HWE: Hardy-Weinberg equilibrium; n=number of participants; PNPLA3: patatin-like phospholipase domain containing 3; TM6SF2: transmembrane 6 superfamily member 2; MBOAT7, Membrane Bound O-Acyltransferase Domain Containing 7; TMC4: Transmembrane Channel-Like Protein 4; HSD17B13: hydroxysteroid 17-beta dehydrogenase 13* two missing genotypes; FGF21: Fibroblast Growth Factor 21.
Figure 1.
Genus-level analyses of liver 16S rDNA in the entire population.
Dendrogram visualizes hierarchical structures in microbial communities, whereby the edges depict the relative abundance of the corresponding taxon.
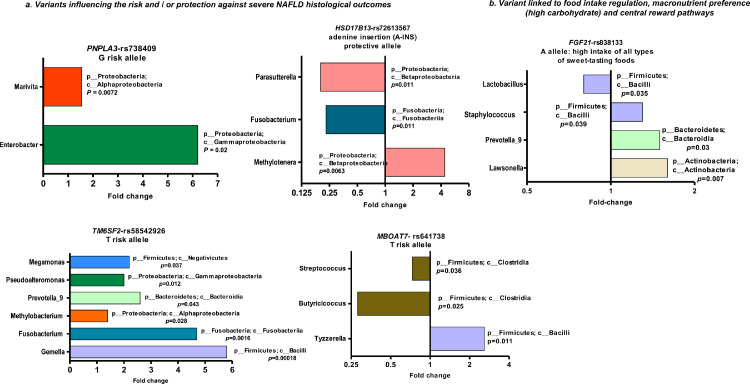
We next tested for the association between the relative abundance of bacterial DNA taxa and minor alleles of the selected variants. We found that two genera were significantly enriched in carriers of the PNPLA3-rs738409 G-risk allele, including Enterobacter and Marivita.
Gemella, Fusobacterium, Methylobacterium, Prevotella_9, Pseudoalteromonas, and Megamonas populations were significantly enriched in carriers of the TM6SF2-rs58542926 T-risk allele.
Genera Butyricicoccus and Streptococcus were significantly depleted, and Tyzzerella was increased in carriers of the MBOAT7-rs641738 T-risk allele. Among carriers of the protective A-INS allele of the HSD17B13-rs72613567 variant, the abundance of genera Fusobacterium and Parasutterella was significantly decreased while that of the Methylotenera genus was enriched.
Regarding the variant influencing dietary preferences, we found that the abundance of genera Lawsonella, Prevotella_9, and Staphylococcus was higher, and Lactobacillus quantity was diminished in carriers of the FGF21-rs838133 minor A-allele.
The direction of effects, including fold changes across all variants, is shown in Figure 2.
Figure 2.
Taxa abundance profiles according to host genetics.
Bars depict fold changes of liver bacterial DNA taxa abundance (whereby >1 indicates enrichment and <1 depletion) in carriers of variants in target loci, including NAFLD/NASH-risk loci (PNPLA3 rs738409 CC n=31, CG+GG n=85), TM6SF2 rs58542926 CC n=89, CT+TT n=27, MBOAT7 rs641738 CC n=44, CT+TT n=72, and HSD17B13 rs72613567 -/- n=83, -A+AA n=31), and a locus influencing dietary macronutrient intake (FGF21 rs838133 GG n=61, GA+AA n=55). Measurements across sample groups are compared by non-parametric statistical tests [Wilcoxon rank test], with p < 0.05 indicating statistical significance.
According to the NCBI taxonomy database, taxa are shown at the genus level (p_ stands for phyla and c_ for class). We calculated the relative abundance (fold change) of the taxon as the ratio of the media per group of sequencing reads assigned to the taxon normalized by the total number of sequencing reads combining with square root transformation.37
Combined effect of risk alleles on the liver microbial DNA composition: Taxon abundance and variance explained by the host genetics
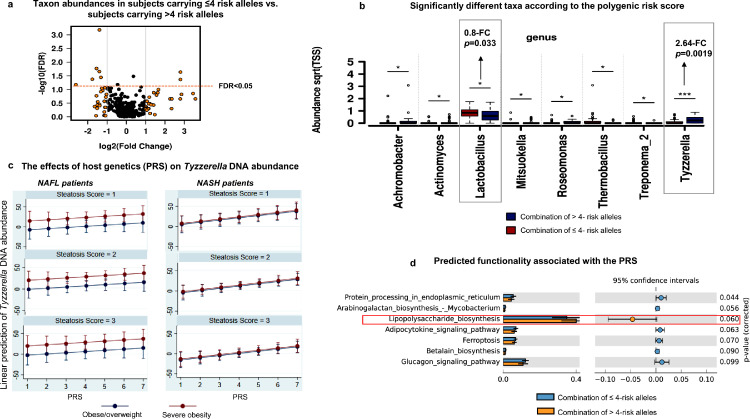
We further explored the effect of the combined effect of NAFLD-risk/protection alleles on the composition of the liver microbiota by a PRS. We first investigated whether the genetic risk of NAFLD was associated with alpha (within sample) diversity, and we found no detectable association between PRS for NAFLD and Shannon index (p = 0. 93) or Simpson index (p = 0.8).
Then, using univariate analysis, we compared the taxon abundances across two groups of individuals, including subjects carrying ≤4 risk alleles and subjects carrying >4 risk alleles, respectively. As shown in Figure 3a, among all the microbial DNA taxa present in the liver, certain taxa showed significant associations with the PRS. Specifically, the abundance of the Tyzzerella genus—a member of the Firmicutes phylum and the Clostridia class—showed the strongest association with high PRS values (>4 risk alleles), as indicated by 2.64-fold change differential abundance (p = 0.0019, FDR < 0.05) and shown in Figure 3b. Conversely, the Lactobacillus genus—a member of the Firmicutes phylum and Bacilli class—exhibited the strongest association with low PRS values (≤4 risk alleles) (0.89-fold change, p = 0.033, FDR < 0.05, Figure 3b).
Figure 3.
Combined effect of risk alleles on the liver microbial DNA composition.
a. Volcano plot shows differentially-abundant taxa at the genus level across two groups of individuals, respectively comprising of subjects carrying ≤4 risk alleles and subjects carrying >4 risk alleles. Log2-trasnformed Genus's abundance-fold changes are shown on the X-axis (vertical lines situated at -1 and 1 indicate a 2-fold change in both directions) and the negative logarithm (base 10) of the FDR is depicted on the Y-axis (the horizontal line indicates FDR = 0.05). Black or orange dots represent genus fold changes exceeding 2 in both directions.
b. Bar chart shows significantly different taxa according to the polygenic risk score (PRS) (p < 0.05, [ANOVA]). Relative abundance is calculated as the square root of each feature read count divided by the total number of features reads in each sample. The standard error is indicated by error bars. Pair-wise comparisons are performed through t-test and are annotated as *: p < 0.05, **: p < 0.01, and ***: p < 0.001.
c. Plots show the effects of host genetics (PRS, the number of risk allele carried) on Tyzzerella DNA abundance [predicted by linear regression and Margins subroutine] in NAFLD patients with steatosis scores in the 1−3 range, comparing those who are overweight/moderately obese vs. severely obese patients and after adjusting for confounding factors (age and gender, in addition to steatosis score and obesity degree); the analysis was performed in patients stratified by liver disease severity, separately (NAFL in the left panels vs. NASH in the right panels).
d. Predicted functionality associated with the PRS. The functional inference was explored based on KEGG KO pathways by applying the Piphillin metagenomics inference tool35. Bars indicate mean proportions (%), confidence intervals, and their associated p-values for the top-ranked metabolic pathways corresponding to both groups with 4 ≥ PRS > 4. STAMP was used to determine differentially enriched metabolic pathways (corrected p < 0.05) and their effect sizes (η2). The Welch's test was performed to assess statistical significance and corrected p values were calculated using Storey's FDR approach.
ROC analysis was performed to identify the potential discriminatory power of individual bacterial DNA taxa over PRS. For the genus Tyzzerella, an AUC of 0.70 (95% CI 0.59−0.80, p = 0.00067) was obtained, whereas Lactobacillus presented an AUC of 0.63 (95% CI 0.51−0.75, p = 0.034). Even after adjusting for sex, obesity degree, and liver disease severity, similar non-parametric AUCs were obtained for Tyzzerella (AUC: 0.71, bootstrap 95% CI: 0.60-0.83) and Lactobacillus (AUC: 0.62, bootstrap 95% CI: 0.50-0.75); both are significant (p<0.05) as the bootstrap 95% CIs did not include 0.
Next, we calculated the percentage of the liver microbial taxa variance that could be explained by the PRS, which was built with proxy-based variants of NAFLD histological severity and macronutrient intake. The proportion of genus-level taxa variance explained by the host genetics (PRS) was ∼7.4% (adjusted R2 = 0.0738, p < 0.005). This effect was independent of the presence of fatty liver and the obesity status of the studied population.
Figure 3c shows the predicted marginal means of Tyzzerella abundance by PRS according to subjects' liver steatosis scores in obese/overweight and severely obese groups and across the two main stages of the disease severity, namely NAFL and NASH. The Margins algorithm performed the estimation after linear regression with Tyzzerella abundance and PRS as dependent and independent continuous variables, respectively, and sex, steatosis score, and BMI-classified groups as covariates. We found that correlation between Tyzzerella abundance and the number of risk alleles (PRS) was only significant in NASH (Coefficient: 5.42, 95% CI: 0.92 - 9.93, p value = 0.02) as opposed to in NAFL (Coefficient: 2.89, 95% CI: -2.97 - 8.76, p value = 0.32) patients.
Because of previous observations in well-characterized cohorts addressing the relationship between the gut microbiota composition and cardiovascular disease (CVD) risk demonstrated a potential link between Tyzzerella and Tyzzerella 4 abundance and high CVD risk profile,38 we explored the possible association between Tyzzerella enrichment and the presence of arterial hypertension. We found a trend toward a relative increased abundance of Tyzzerella in the liver of subjects with arterial hypertension (mean: 13.85 ± SE: 2.85, n = 53) compared to non-hypertensive individuals (mean: 10.03 ± SE: 2.80, n = 60) (p value = 0.056 [Mann-Whitney test]). However, the association was not significant after adjustment for the relevant covariates.
Functional annotation enrichment shows distinctive predicted functional signatures
After demonstrating the host genetic impact on liver microbial taxa, we examined genetic contributions to liver microbial functions. We found significantly enriched specialized pathways in carriers of the PNPLA3-rs738409 G-risk allele. The most remarkable ones include octane oxidation and fatty acid salvage, also known as β-oxidation degradation de novo fatty acid biosynthesis shunt (Figure 4a). As shown in Figure 4b, among the pathways associated with the TM6SF2-rs58542926 T-risk allele, we found L-tryptophan degradation to 2-amino-3-carboxymuconate semialdehyde and NAD biosynthesis II from tryptophan (one of the essential coenzymes in redox reactions in the cell). In carriers of the HSD17B13-rs72613567 A-INS allele, we found that phosphatidylglycerol biosynthesis I and gondoate biosynthesis microbial pathways were under-represented and the super pathway of adenosylcobalamin salvage from cobinamide I was over-represented (Figure 4c). Specialized pathways observed in carriers of the MBOAT7-rs641738 T-risk allele is shown in Figure 4d, indicating that the superclass of nucleoside and nucleotide and purine biosynthesis serving as generating cellular energy are enriched. Finally, predicted functionality associated with the PRS showed a trend toward the association with lipopolysaccharide biosynthesis (Figure 3d).
Figure 4.
Functional annotation enrichment.
Functional profiling was performed by PICRUSt from the 16S rRNA gene amplicon sequencing. a.PNPLA3-rs738409 (CC n=31, CG+GG n=85); b. TM6SF2-rs58542926 (CC n=89, CT+TT n=27); c. HSD17B13-rs72613567 (-/- n=83, -A+AA n=31); d. MBOAT7-rs641738 (CC n=44, CT+TT n=72). The top metabolic pathways (MetaCyc) for each genotype (host genetics by assuming a dominant model of inheritance) are represented by effect sizes, confidence intervals, and their associated p values. Bars indicate mean proportions (%). STAMP was used to determine differentially enriched metabolic pathways (p < 0.05) and their effect sizes (η2). The Welch's test was performed to assess statistical significance and corrected p values were calculated using Storey's FDR approach.
Discussion
Our results indicate that host genetic determinants of the risk/protection for NAFLD/NASH and dietary macronutrient intake are related to the liver microbial DNA composition. The strongest associations were found between members of the Gammaproteobacteria class and NAFLD/NASH risk alleles of the rs738409 and rs58542926 variants, including Enterobacter and Pseudoalteromonas genera, respectively. These findings are consistent with our previous observation indicating that DNA derived from Gammaproteobacteria was associated with more severe forms of liver disease, including high scores of lobular and portal inflammations.14
It is also known that the body microbiome is sensitive to the host's diet and lifestyle. In this study, we found that carriers of the FGF21-rs838133-minor allele presented higher abundances of Lawsonella (a member of the Actinobacteria class), Prevotella_9, and Staphylococcus genera, and a decreased abundance of Lactobacillus. This finding suggests that dietary sugar consumption and carbohydrate intake favor certain bacterial taxa, such as Prevotella_9 that has been associated with the risk of major common diseases, including cardiovascular disease,38 colon cancer,39 and even immunologic diseases such as rheumatoid arthritis.40
Most importantly, a PRS generated to assess the combined effect of NAFLD/NASH associated variants' risk alleles and the FGF21-rs838133 minor allele revealed that the abundance of the Tyzzerella taxa showed the strongest association with high PRS values (>4 risk alleles). Findings yielded by the analyses of the gut microbiome of the participants of the Bogalusa Heart Study—a long-term epidemiologic study investigating the natural history of atherosclerosis—indicate that the genus Tyzzerella is associated with lifetime cardiovascular risk profile.38 Collectively, our findings might serve to explain, at least in part, the link between NASH/NAFLD and CVD.41,42
Although several factors influence the human (and mammalian) body microbiota composition, it appears that host genetic factors account for a considerable proportion of variance. For example, seminal studies in twins showed that microbial gut composition is ∼40% heritable.15,16 Recent evidence that leveraged 16,234 gut microbiome profiles collected over 14 years from 585 wild baboons suggests that, after controlling for diet, age, and socioecological variation, 97% of the gut microbiome phenotypes are significantly heritable.43 Of note, the percentage of genus-level taxa variation explained by our NAFLD/NASH PRS was ∼7.4%, which was independent of key covariate parameters that may affect this effect, including the steatosis score and obesity degree. Even age and gender were not contributing factors. Considering that the PRS based on selected variants in few genes was significant, it is justified to consider that the observed variance is higher than expected by chance.
It should be highlighted that host genetics is also relevant in modelling host–gut microbe interactions in health and disease44, including NAFLD in obese youth as reported recently.45
Finally, we aimed to detect differentially abundant pathways and predicted bacteria-derived metabolites from the taxa associated with the tested variants. We reasoned that exploring putative functional consequences of taxa linked to genetic variation may aid in better understanding of the disease mechanism(s). We found multiple altered microbial metabolic pathways associated with host genetics. For example, in carriers of the rs738409 G-risk allele, octane oxidation and the novo fatty acid biosynthesis shunt were over-represented. The key process in octane oxidation is the alkane hydroxylase system that introduces molecular oxygen in the C1 atom of the hydrocarbons at the expense of NADH to yield primary alcohols.46 Previous studies demonstrated that several alkane-degrading bacteria can use diverse compounds as a carbon source in addition to alkanes,47 which are further oxidated to fatty acids via bacterial β-oxidation pathway (BioCyc ID: P221-PWY). While the alkB domain has been found in many bacterial genomes, including Proteobacteria, Actinobacteria, and Bacteroidetes, it appears that the alkane hydroxylase system is particularly relevant in the Gammaproteobacteria class, including those from Pseudomonas aeruginosa.48
Volatile organic compounds, including octane, were found either in the exhalation or faeces of human subjects with diverse medical conditions associated with oxidative stress and chronic inflammation, including lung cancer,49 obstructive sleep apnoea,50 gastrointestinal diseases,51 and NAFLD.52 Notably, data analysis based on NHANES 2017−2018 population-based survey of non-occupationally exposed individuals that included liver fibrosis assessment via transient elastography (FibroScan) examination shows a significant association between blood octane concentrations and liver stiffness (beta 0.0016 ± 0.0007, p = 0.031 adjusted by age, gender, diabetes, obesity [BMI], and alcohol consumption) (Supplementary Figure 1).
Collectively, primary alcohols can be synthesized either via fatty acid or amino acid pathways in diverse bacteria. The link between the gut microbiota and alcohol (ethanol) production has been described by several authors.53,54 However, our study provides novel evidence regarding the association between PNPLA3-rs738409 and taxa capable of metabolizing alkane-derived carbons to yield primary alcohols.
Likewise, the rs738409 G-risk allele was found to be associated with a metabolic pathway involved in β-oxidation degradation, also known as de novo fatty acid biosynthesis shunt. Fatty acid biosynthesis shunt is initiated by the β-ketoacyl-[acyl-carrier-protein] synthase that catalyses the condensation of acetyl-CoA with malonyl-[acp], producing acetoacetyl-[acp] and that can be transformed into longer fatty acids via the fatty acid elongation cycle. This might be a new mechanism to explain why carriers of the PNPLA3 variant are exposed to 3.28-fold higher risk of liver fat accumulation.18,19
The comparison of functional prediction between carriers and non-carriers of the HSD17B13-72613567 protective A-INS allele yielded results that might expand our knowledge of the protective function of this variant against the most detrimental NAFLD-histological outcomes.24,26 Specifically, we found over-representation of the super pathway of adenosylcobalamin salvage from cobinamide I—a pathway involved in vitamin and cofactor biosynthesis—in A-INS allele carriers, along with a lower proportion of taxa linked to fatty acid metabolism, such as phosphatidylglycerol biosynthesis and components of the gondoate pathway, which is essential for the production of unsaturated fatty acids, including arachidonate. Arachidonate production is the rate-limiting step in the synthesis of prostaglandins and is associated with NAFLD55 and also participates in the Land's cycle.
In addition, the predicted functional consequence/s of associated taxa with the PRS suggested that carriers of >4 risk alleles might be exposed to some genera with high potential for lipopolysaccharide biosynthesis. This finding is consistent with our earlier observation that lipopolysaccharides derived from Gram-negative bacteria localize in the portal tract of patients with more severe disease.14
However, when interpreting these findings, some study limitations should be considered. In particular, our results are based on the examination of the bacterial DNA profile of liver specimens in association with a few genetic variants. Therefore, much greater (by many orders of magnitude) sample sizes will be necessary to fully explore the influence of the whole genome on the liver microbial DNA composition. Nonetheless, we were able to overcome several challenges, including explorations of microbiome profiles in liver tissue, which is very difficult to obtain and analyze for relevant data.
It is also plausible that different results may arise in other populations, as the inter-relationship between genetics and the liver microbiome can be modified by environmental factors not explored in the present study. These potential differences can be explained by the presence of population structure across individuals from different parts of the world, as well as by different biogeography of bacterial taxa in different parts of the body that are determined not only by host genetics, but are also modified by factors that impose selective pressure on the body microbiotas, such as antibiotic usage, travel, diet, microbes in the soil, vertical transmission from mothers to children, and diverse causes of horizontal transmission between humans and/or animals.56
In conclusion, we studied the intricate link between host genetics and the liver microbiome composition in patients with NAFLD. We were able to show the most significant liver bacterial DNA that may be affected by host variation in candidate genes relevant to the disease biology. The most striking result to emerge from our study is that host genetics may shape the metabolic and biological environment in which the NAFLD-associated liver tissue-microbial DNA resides.
Declaration of interests
Nothing to declare.
Acknowledgments
Contributors
All authors (CJP; AS; MFQ; GOC; MG; and SS) read and approved the final version of the manuscript. CJP: study concept and design; biological material collection, data acquisition; data analysis and interpretation; genetic studies; general study supervision; statistical analysis; manuscript drafting; and securing funding. AS: bioinformatics analysis of sequencing data and data processing and presentation. MFQ: sample processing. GOC and MG: performed liver biopsies and collected biological samples. SS: study concept and design; data acquisition; performed liver biopsies and collected biological material; histological evaluation; genetic studies; data analysis and interpretation; general study supervision; manuscript drafting; securing funding. All authors have read and approved the final version of the manuscript. In addition, CJP, GOC, and SS have verified the underlying data.
Acknowledgements
This study was partially supported by grants PICT 2018-889, PICT 2019-0528, PICT2016-0135 and PICT 2018-0620 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT), CONICET Proyectos Unidades Ejecutoras 2017, PUE 0055.
Data sharing statement
Data supporting the observations of this study, including the methodology, are available upon reasonable request from the corresponding author. Access to data concerning genetic information may be restricted due to some research participants of this study did not agree for their genetic data to be shared publicly.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103858.
Contributor Information
Carlos Jose Pirola, Email: pirola.carlos@conicet.gov.ar.
Silvia Sookoian, Email: ssookoian@intramed.net.
Appendix. Supplementary materials
References
- 1.Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sookoian S, Flichman D, Scian R, et al. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol. 2016;240(4):437–449. doi: 10.1002/path.4803. [DOI] [PubMed] [Google Scholar]
- 4.Sookoian S, Pirola CJ, Valenti L, Davidson NO. Genetic pathways in nonalcoholic fatty liver disease: Insights from systems biology. Hepatology. 2020;72(1):330–346. doi: 10.1002/hep.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirola CJ, Gianotti TF, Burgueno AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 6.Pirola CJ, Sookoian S. Epigenetics factors in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2020;1:1–16. doi: 10.1080/17474124.2020.1765772. [DOI] [PubMed] [Google Scholar]
- 7.Sookoian S, Pirola CJ. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis. 2019;39(2):124–140. doi: 10.1055/s-0039-1679920. [DOI] [PubMed] [Google Scholar]
- 8.Aron-Wisnewsky J, Vigliotti C, Witjes J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17(5):279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63(3):764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25(5):1054–1062. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwimmer JB, Johnson JS, Angeles JE, et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology. 2019;157(4):1109–1122. doi: 10.1053/j.gastro.2019.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sookoian S, Salatino A, Castano GO, et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69(8):1483–1491. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich JK, Davenport ER, Beaumont M, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie H, Guo R, Zhong H, et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 2016;3(6):572–584. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolde R, Franzosa EA, Rahnavard G, et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med. 2018;10(1):6. doi: 10.1186/s13073-018-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 20.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milano M, Aghemo A, Mancina RM, et al. Transmembrane 6 superfamily member 2 gene E167K variant impacts on steatosis and liver damage in chronic hepatitis C patients. Hepatology. 2015;62(1):111–117. doi: 10.1002/hep.27811. [DOI] [PubMed] [Google Scholar]
- 22.Sookoian S, Castano GO, Scian R, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61(2):515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 23.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of european descent. Gastroenterology. 2016;150(5):1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Belyaeva OV, Brown PM, et al. 17-Beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69(4):1504–1519. doi: 10.1002/hep.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirola CJ, Garaycoechea M, Flichman D, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60(1):176–185. doi: 10.1194/jlr.P089953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu AY, Workalemahu T, Paynter NP, et al. Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet. 2013;22(9):1895–1902. doi: 10.1093/hmg/ddt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merino J, Dashti HS, Li SX, et al. Genome-wide meta-analysis of macronutrient intake of 91,114 European ancestry participants from the cohorts for heart and aging research in genomic epidemiology consortium. Mol Psychiatry. 2019;24(12):1920–1932. doi: 10.1038/s41380-018-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unger T, Borghi C, Charchar F, et al. 2020 International society of hypertension global hypertension practice guidelines. Hypertension. 2020;75(6):1334–1357. doi: 10.1161/HYPERTENSIONAHA.120.15026. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki ES. In: PCR Protocols. A Guide to Methods and Applications. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Academic Press, INC; San diego: 1990. Sample preparation from blood, cells, and other fluids; pp. 146–152. editors. [Google Scholar]
- 31.Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50(10):2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sookoian S, Flichman D, Garaycoechea ME, et al. Lack of evidence supporting a role of TMC4-rs641738 missense variant-MBOAT7- intergenic downstream variant-in the susceptibility to nonalcoholic fatty liver disease. Sci Rep. 2018;8(1):5097. doi: 10.1038/s41598-018-23453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17(R2):R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwai S, Weinmaier T, Schmidt BL, et al. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zakrzewski M, Proietti C, Ellis JJ, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33(5):782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly TN, Bazzano LA, Ajami NJ, et al. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circ Res. 2016;119(8):956–964. doi: 10.1161/CIRCRESAHA.116.309219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 40.Wells PM, Adebayo AS, Bowyer RCE, et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: a cross-sectional study. Lancet Rheumatol. 2020;2(7):e418–e427. doi: 10.1016/S2665-9913(20)30064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sookoian S, Pirola CJ. Non-alcoholic fatty liver disease is strongly associated with carotid atherosclerosis: a systematic review. J Hepatol. 2008;49(4):600–607. doi: 10.1016/j.jhep.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Targher G, Byrne CD, Tilg H. NAFLD and increased risk of cardiovascular disease: clinical associations, pathophysiological mechanisms and pharmacological implications. Gut. 2020;69(9):1691–1705. doi: 10.1136/gutjnl-2020-320622. [DOI] [PubMed] [Google Scholar]
- 43.Grieneisen L, Dasari M, Gould TJ, et al. Gut microbiome heritability is nearly universal but environmentally contingent. Science. 2021;373(6551):181–186. doi: 10.1126/science.aba5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonder MJ, Kurilshikov A, Tigchelaar EF, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48(11):1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 45.Monga KA, Testerman T, Galuppo B, et al. Effect of gut microbiota and PNPLA3 rs738409 variant on nonalcoholic fatty liver disease (NAFLD) in obese youth. J Clin Endocrinol Metab. 2020;105(10) doi: 10.1210/clinem/dgaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, Janssen DB, Witholt B. Growth on octane alters the membrane lipid fatty acids of Pseudomonas oleovorans due to the induction of alkB and synthesis of octanol. J Bacteriol. 1995;177(23):6894–6901. doi: 10.1128/jb.177.23.6894-6901.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojo F. Degradation of alkanes by bacteria. Environ Microbiol. 2009;11(10):2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 48.Nie Y, Chi CQ, Fang H, et al. Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep. 2014;4:4968. doi: 10.1038/srep04968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poli D, Carbognani P, Corradi M, et al. Exhaled volatile organic compounds in patients with non-small cell lung cancer: cross sectional and nested short-term follow-up study. Respir Res. 2005;6:71. doi: 10.1186/1465-9921-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aoki T, Nagaoka T, Kobayashi N, et al. Editor's Highlight: prospective analyses of volatile organic compounds in obstructive sleep apnea patients. Toxicol Sci. 2017;156(2):362–374. doi: 10.1093/toxsci/kfw260. [DOI] [PubMed] [Google Scholar]
- 51.Garner CE, Smith S, de Lacy CB, et al. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007;21(8):1675–1688. doi: 10.1096/fj.06-6927com. [DOI] [PubMed] [Google Scholar]
- 52.Raman M, Ahmed I, Gillevet PM, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(7):868–875. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 53.Cope K, Risby T, Diehl AM. Increased gastrointestinal ethanol production in obese mice: implications for fatty liver disease pathogenesis. Gastroenterology. 2000;119(5):1340–1347. doi: 10.1053/gast.2000.19267. [DOI] [PubMed] [Google Scholar]
- 54.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 55.Pirola CJ, Sookoian S. The lipidome in nonalcoholic fatty liver disease: actionable targets. J Lipid Res. 2021;62 doi: 10.1016/j.jlr.2021.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garud NR, Pollard KS. Population genetics in the human microbiome. Trends Genet. 2020;36(1):53–67. doi: 10.1016/j.tig.2019.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.