Abstract
Clinical mastitis in six Somerset dairy herds was monitored over a 12-month period. Escherichia coli was implicated in 34.7% of all clinical cases. Forty-one percent of all clinical E. coli mastitis cases occurred in just 2.2% of the population. A total of 23.9% of clinical E. coli cases occurred in quarters suffering recurrent cases of E. coli mastitis. The genotypes of strains involved in recurrent cases of clinical E. coli mastitis were compared by DNA fingerprinting with enterobacterial repetitive intergenic consensus primers. In 85.7% of cases of recurrent quarter E. coli mastitis, the same genotype was implicated as the cause of disease, suggesting persistence of the organism within the mammary environment. The same genotype as that in the original case was also implicated in 8.5% of recurrent cases occurring in different quarters of the same cow, suggesting spread between quarters. These findings challenge our current understanding of the epidemiology of E. coli mastitis and suggest that pathogen adaptation and host susceptibility may be playing a part in the changing pattern of clinical mastitis experienced in the modern dairy herd.
Mastitis remains a major cause of financial loss to the United Kingdom dairy industry and is considered to be the most economically important disease of dairy cattle, accounting for 38% of the total direct costs of all the common production diseases (21). Classically, the mastitis pathogens have been divided into contagious and environmental organisms (2) based on their proclivity to cause persistent or transient opportunistic infection, respectively. Contagious pathogens are also noted for their ability to be spread between quarters and cows during the milking process (26).
Escherichia coli has been classified as an environmental pathogen (24). Previous studies have demonstrated the rapid onset of clinical signs following inoculation (10), although these signs often present shortly before or even after elimination of viable bacteria from the gland (18). The host defense of the bovine mammary gland has been shown to be efficient in controlling and eliminating E. coli infection (15), although this ability has been shown to be less effective in early lactation due to deficiencies in neutrophil function and numbers (16, 28).
Previous studies have identified the ability of E. coli to persist in the bovine udder; however, this phenomenon has been considered to be relatively rare and of little clinical importance (14, 18, 23). We have recently demonstrated the ability of E. coli to gain access to the bovine mammary gland during the nonlactating period and then to persist, only recrudescing to cause clinical disease after the onset of lactation (4). This study demonstrated the ability of E. coli to persist within the mammary gland for in excess of 100 days and identified this phenomenon as making a significant contribution to the incidence of clinical disease.
The last 30 years has seen a dramatic change in the incidence and etiology of bovine mastitis in the United Kingdom (3). Although the incidence of clinical mastitis has fallen, the incidence of the environmental pathogens, in particular E. coli, has increased (5). Various explanations for this changing pattern of disease have been proposed and are reviewed elsewhere (11).
The change in mastitis etiology and the other recent findings are of interest and may be indicative of a shift in the behavior of E. coli. The aim of this study was to monitor clinical E. coli mastitis in a cohort of Somerset dairy herds in order to assess the importance of recurrent cases and to attempt to identify changes in the behavior of E. coli as a mastitis pathogen.
MATERIALS AND METHODS
Herd selection.
The herds were selected on the basis of location (Somerset), low bulk milk somatic cell count (3-month geometric mean of <250,000 cells/ml), nonseasonal calving pattern, and likelihood of owner compliance with the study protocol. Herds were not selected on the basis of a previous history of E. coli mastitis.
Herd details.
Clinical mastitis in six Somerset dairy herds was monitored between 1 June 1997 and 31 May 1998. The six herds totaled 810 British/Holstein Friesians and contained between 100 and 190 cows. All six herds had a nonseasonal calving pattern. Yields were 6,000 to 7,000 liters (305-day adjusted total), and mean bulk milk somatic cell counts varied between 55,000 and 230,000 cells/ml. Lactating cows in five herds were managed on grass during the summer months and in free stalls during the winter; lactating cows in the sixth herd were managed in free stalls throughout the year.
Sampling strategy.
Herdspersons were trained in the identification, grading, and sampling of clinical mastitis. Clinical mastitis was identified in quarters on the basis of the presence of abnormal secretion and/or udder changes (e.g., pain, heat, and swelling). Mastitis severity was graded according to the following system: grade 1, milk changes only (e.g., clots); grade 2, milk and udder changes (e.g., swelling and/or heat); grade 3, systemic signs (e.g., depression and/or pyrexia). Secretion samples were collected prior to institution of antibiosis by the method outlined below. The six farms were each visited weekly by the authors to reinforce initial training and to encourage compliance.
Following identification of an affected quarter, the teat was wiped to remove gross contamination (if necessary) and dipped in a solution containing 2,800 ppm of available chlorine (Agrisept; Pharmacia Animal Health). Following a minimum 30-s contact time, the teat was wiped dry. The teat was subsequently scrubbed with a cotton wool swab soaked in 70% ethanol and allowed to dry. Prior to collection of this sample, the teat end was scrubbed for a second time with 70% ethanol, and foremilk was discarded. Samples were frozen, collected by the authors, and submitted for bacteriological analysis weekly. Disposable gloves were worn throughout the sampling process.
Bacteriology.
Samples were submitted to the Langford Veterinary Investigation Centre for bacteriological analysis. Ten microliters of secretion was inoculated onto sheep blood agar and Edward's agar; 100 μl of secretion was inoculated onto MacConkey agar to enhance the detection of members of the family Enterobacteriaceae. Plates were incubated at 37°C and read at 24 and 48 h. Organisms were identified and quantified by standard laboratory techniques (25). E. coli was identified by colony morphology and oxidase and indole tests; other members of the family Enterobacteriaceae were identified with a microtube identification system (RapiD 20 E; bioMérieux). All pathogens were subject to two rounds of colony purification prior to storage in a commercial microorganism storage system (Prolab cryopreservation beads).
DNA fingerprinting.
All recurrent E. coli isolates were retrieved from the microorganism storage system and grown overnight in Luria-Bertani broth (10 g of enzymatic casein digest, 5 g of yeast extract, and 5 g of NaCl per liter) at 37°C. Chromosomal DNA was extracted with a commercially available kit (QIAamp tissue kit; Qiagen). DNA sequences were then amplified by PCR with 50 ng of template DNA, enterobacterial repetitive intergenic consensus (ERIC) sequence primers (29, 32), Taq polymerase (Qiagen), and a Techne Genie II thermal cycler. After an initial denaturation step of 2 min at 94°C, the reaction mixture was cycled 35 times for 30 s at 94°C (denaturation), 15 s at 40°C (annealing), and 5 min at 72°C (extension), which was followed by a final extension step of 10 min at 72°C. After PCR, the enterobacterial strains were discriminated by their DNA polymorphism patterns by agarose gel electrophoresis and visualization under UV light (28, 31). Enterobacterial strains exhibiting an identical DNA fingerprint were considered to be the same.
Definition of terms used for analysis. (i) Intramammary infection: cause of clinical mastitis.
If an organism was isolated in pure growth or was the predominant growth, then the organism was considered to be the cause of mastitis. A sample was called a “mixed growth” if there was growth of two or three known mastitis pathogens. A sample was labeled “contaminated” if more than three organisms were isolated.
(ii) Recurrent clinical mastitis.
A quarter was considered to be suffering a recurrent episode of clinical E. coli mastitis when there was an interval of 5 or more days between episodes. When a quarter was identified as having suffered a case of recurrent clinical E. coli mastitis, all episodes (including the first) were considered in the analysis. A recurrent cow was defined as a cow that suffered clinical E. coli mastitis in one or more quarters on one or more occasions.
(iii) Persistent infection.
An infection was considered to be persistent when the same E. coli genotype was identified in clinical mastitis samples obtained from the same quarter of the same cow.
Collation and statistical analysis of results.
Results were collated and analyzed by using Microsoft Excel, Access, and Epi-Info (Version 6.04b; Centers for Disease Control and Prevention, Atlanta, Ga.). The χ2 test was used to compare proportions. A layered Bonferroni's correction was used to compensate for multiple comparisons (6). A significance probability was set at P ≤ 0.05 for a two-tailed test.
RESULTS
Clinical mastitis incidence.
Three hundred thirty-seven cases of clinical mastitis occurred during the study period. The mean annual incidence for the six farms was 41.6 cases/100 cows/year (range, 13 to 75 cases). E. coli was the most common cause of clinical mastitis, accounting for 34.7% of all isolates. One hundred seventeen cases of clinical E. coli mastitis occurred in 101 quarters of 89 cows. Eleven percent of lactating cows suffered a case of clinical E. coli mastitis during the 12-month study period.
Recurrent E. coli mastitis results are outlined in Table 1. Thirteen cows experienced two cases of clinical E. coli mastitis, 2 cows had three cases, 2 cows had five cases, and 1 cow had six cases. Of all of the cases of clinical E. coli mastitis, 41% (48 of 117) occurred in these 18 cows, which represented just 2.2% of the population. A total of 14.5% (17 of 117) of cases of clinical E. coli mastitis occurred in quarters that had previously suffered a case of mastitis due to a different species of mastitis pathogen. At the quarter level, 10 quarters experienced two cases of clinical E. coli mastitis, and 2 quarters experienced four cases. Of all of the cases of clinical E. coli mastitis, 23.9% (28 of 117) occurred in quarters that experienced two or more cases of clinical E. coli mastitis. A total of 6.0% (7 of 117) of all cases of clinical E. coli mastitis occurred in quarters that had previously suffered a case of clinical mastitis due to a different pathogen.
TABLE 1.
Quarter and cow recurrence rates for cases of clinical E. coli mastitis in cows in this studya
| No. of times affected | No. of cows affected (n = 89) | Proportion of affected cows (%) | No. of quarters affected (n = 101) | Proportion of affected quarters (%) |
|---|---|---|---|---|
| 1 | 71 | 79.8 | 89 | 88.0 |
| 2 | 13 | 14.6 | 10 | 10.0 |
| 3 | 2 | 2.2 | 0 | 0 |
| 4 | 0 | 0 | 2 | 2.0 |
| 5 | 2 | 2.2 | 0 | 0 |
| 6 | 1 | 1.1 | 0 | 0 |
| Total experiencing a recurrent case | 18 | 20.2 | 12 | 11.9 |
n = 810.
DNA fingerprinting.
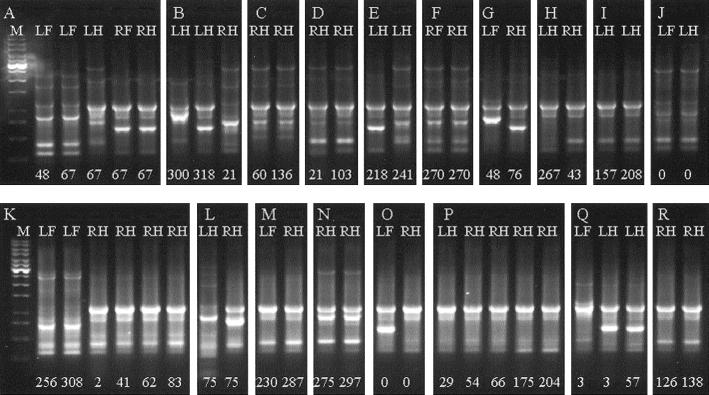
The results of the fingerprinting of all E. coli isolates from recurrent cases of clinical mastitis are illustrated in Fig. 1. In 85.7% (24 of 28) of recurrent E. coli mastitis episodes, fingerprinting implicated the same genotype as that causing the recurrent case (Fig. 1A, C, D, I, K, N, P, Q, and R). In contrast, 14.3% (4 of 28) of recurrent clinical E. coli mastitis was due to infection of the gland by a different E. coli genotype (Fig. 1B and E). A total of 20.5% (24 of 117) of all clinical E. coli mastitis cases arose in persistently infected quarters, as measured by recurrence of clinical mastitis.
FIG. 1.
DNA fingerprints of E. coli isolates from cows experiencing recurrent episodes of clinical E. coli mastitis. Lanes labeled M contain molecular weight markers (1-kb ladder; Promega). Isolates from different cows are arranged in chronological order. Lanes LF (left fore), LH (left hind), RF (right fore), and RH (right hind) denote the quarter affected. Numerical lane labels denote the day of lactation on which the clinical episode occurred.
The same E. coli genotype was identified in 10 of 26 episodes of mastitis from different quarters within the same cow (Fig. 1A, F, J, M, and P), representing 8.5% of all episodes of clinical E. coli mastitis during the study period. On three occasions, the same genotype was found in different quarters of the same cow at the same time (Fig. 1A, F, and J), whereas on two occasions, the same genotype was found in different quarters at different times (Fig. 1M and P), with 57 and 25 days, respectively, between the two episodes. The data are summarized in Table 2.
TABLE 2.
Recurrent clinical E. coli mastitis cases occurring in a cow according to genotype isolated
| Genotype | Result for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Same quarter
|
Different quarters
|
|||||||
| No. of cases | Proportion of cases (%) | No. of contributing quarters/no. of cows | Proportion of all clinical cases (%) | No. of cases | Proportion of cases (%) | No. of contributing quarters/no. of cows | Proportion of all clinical cases (%) | |
| Same | 24 | 85.7 | 10/9 | 20.5 | 10 | 38.5 | 10/5 | 8.5 |
| Different | 4 | 14.3 | 2/2 | 3.4 | 16 | 61.5 | 16/8 | 13.7 |
| Total | 28 | 23.9 | 26 | 22.2 | ||||
Mastitis severity.
Severity of E. coli mastitis is outlined in Table 3. Clinical E. coli mastitis occurring in quarters experiencing recurrent disease was significantly more likely to be mild (grade 1) (19 of 28) than disease recurring in nonrecurrent quarters (38 of 89) (P = 0.035). Recurrent clinical E. coli cases (due to persistent infection with the same genotype) tended to be more likely to be mild (grade 1) (15 of 24) than nonrecurrent cases (42 of 93) (P = 0.12); no recurrent cases resulted in clinical E. coli mastitis manifesting with systemic signs (grade 3). When compared to the expected outcome, based on nonrecurrent cases, a subsequent case of clinical E. coli mastitis in the same quarter was more likely to be of the same grade as the previous case (13 of 16) than would be expected from the study population (P < 0.05).
TABLE 3.
Severity score of clinical E. coli mastitis cases
| Gradea | Clinical E. coli mastitis severity score in cow quarters
|
||
|---|---|---|---|
| Recurrent cases
|
Nonrecurrent cases | ||
| Same genotype | Different genotype | ||
| 1 | 15 | 4 | 38 |
| 2 | 9 | 0 | 39 |
| 3 | 0 | 0 | 12 |
| Total | 24 | 4 | 89 |
Grades: 1, milk changes only (e.g., clots); 2, milk and udder changes (e.g., swelling and/or heat); 3, systemic signs (e.g., depression and/or pyrexia).
Timing of infection.
Recurrent infections became established at all stages of lactation. The mean time between calving and the first recurrent case was 100 days (range, 2 to 275 days). The mean time between calving and nonrecurrent cases was 111 days (range, 0 to 339 days). The mean time between clinical episodes caused by the same genotype within a quarter was 42.8 days (range, 12 to 109 days).
Parity of affected cows.
The mean lactation number of cows experiencing recurrent cases was 4.2 (range, 2 to 9), compared to 4.1 (range, 1 to 10) for cows experiencing nonrecurrent cases.
Between-farm variation.
Recurrent cases occurred on five of the six farms, but recurrence due to the same genotype was identified on only four farms. The same genotype was identified in different quarters of the same cow on four farms. The same genotype was identified in two different cows on one farm (Fig. 1O and P). Very similar genotypes were identified as causing recurrent disease on four of the six farms studied (Fig. 1I, K, M, N, and R).
DISCUSSION
The incidence of mastitis in these well-managed, low-bulk-milk somatic cell count herds is similar to that reported in recent national surveys (1, 22), and the finding of E. coli as the predominant etiological agent is consistent with other recent reports (30).
One major problem confronting all studies of this type is the reliance on herdspersons to identify and sample clinical cases. In this study, the herdspersons were trained and motivated on a weekly basis to identify and sample all cases; however, they were likely to be less motivated to sample recurrent cases, because they may have viewed these as being of less importance. Such a bias in the sampling strategy would have resulted in an underestimate of the importance of recurrent mastitis. It is also important to note that the herdspersons were not aware that one aim of this study was to monitor recurrent mastitis, because this may have bias towards excess sampling of cows. Another problem with studies involving milk bacteriology is in the contamination of collected samples and thus the loss of data; it is a credit to the herdspersons involved that less than 1% of samples were contaminated, none of which occurred in cows suffering recurrent E. coli mastitis.
Recurrent clinical coliform mastitis can occur via two different scenarios: either as a result of reinfection from the environmental pool or as a result of persistence of the organism within the mammary gland. In this study, we utilized DNA fingerprinting to differentiate between these two scenarios. Previous work has demonstrated the large number of strains of E. coli present in the bovine environment (24) and that these can be discriminated by DNA fingerprinting (29, 32), making reinfection from the environment with the same strain unlikely. The proportion of clinical E. coli mastitis occurring in recurrent quarters, due to the same genotype, in this study (20.2%) is much higher than those in previous reports from outside the United Kingdom (9.1% [23] and 4.77% [8]) and is also higher than those in previous reports in which fingerprinting was not employed (7.5% [18] and 5.5% [31]).
Of particular interest is the difference between the last report in the United Kingdom (5.5% [31]), generated in 1980 without the aid of fingerprinting, and the percentages generated in this study with (20.2%) and without (23.9%) the aid of fingerprinting. This apparent shift in behavior could be indicative of either a change in the susceptibility of the bovine population to persistent infection or a change in the behavior of E. coli. Despite the fact that previous studies have been unable to identify any particular virulence mechanisms involved in the pathogenesis of E. coli mastitis (20), we feel that a combination of these two scenarios is likely to be involved and that certain aspects of the data collected are suggestive of “evolution” of the E. coli strains involved. Recurrent cases of mastitis caused by the same genotype tended to be less clinically severe and were significantly more likely to elicit a similar clinical response upon recrudescence. None of the recurrent cases resulted in systemic signs of illness in affected cows, compared to 13.5% of nonrecurrent cases; it could be argued that it would be an advantage to an udder-adapted strain to elicit a less dramatic response in the manner demonstrated in this study. Additionally, recurrent cases became established at all stages of lactation and not only during the period of immunosuppression associated with parturition, when the cow is known to be less able to clear infection (28). The example of a cow being infected with two genotypes in two different quarters on 1 day, but with only one of the quarters subsequently developing recurrent clinical disease, suggests that the genotype is important and that cow factors alone (the majority of which do not vary at the quarter level) do not account for the development of persistent infections. Finally, the presence of a very similar recurrent disease-causing genotype on four of the six farms involved in the study suggests that certain strains may be more capable of causing persistent infection than others.
The proportion of clinical E. coli cases (22.2%) occurring in different quarters of the same cow is of interest. It could be argued that this is suggestive of cow susceptibility and that these cases were occurring in particularly susceptible cows; if this was the case, one would expect the genotype to be different on each occasion, because each episode should be the result of reinfection from the environmental pool, in which there are a large number of strains, all of which are assumed to be capable of causing disease (24). However, in 38% of cases, these infections were caused by a genotype previously identified in that cow, which may suggest that the organism has persisted between clinical episodes transferred between quarters and subsequently recrudesced in a manner more commonly associated with contagious pathogens such as Staphylococcus aureus (26).
E. coli strains causing recurrent clinical mastitis must have a means of persisting, such as adherence or the ability to survive intracellularly. Although well established in other mucosal systems, the ability of E. coli to exhibit such behavior in the mammary environment has not been reported. A recent study has demonstrated the ability of a limited number of E. coli strains to adhere to and invade bovine mammary epithelial cells in vitro (7). Serum resistance of E. coli has previously been associated with organism virulence in the bovine mammary environment (27), and serum-resistant strains have been shown experimentally to be capable of surviving for protracted lengths of time in the mammary gland (17). E. coli is recognized as a highly adaptive organism, existing both as a commensal organism and as a pathogen; its ability to acquire exogenous DNA and hence virulence genes is well established (9). This process may have played a role in the emergence of udder-adapted strains, such as those possibly identified in this study. Many different subsets of E. coli, such as enterohemorrhagic, enteroinvasive, and enteropathogenic, have also been demonstrated, and it is not unreasonable to expect that another such subset more adapted to the mammary environment may already be present but be as yet unidentified.
The possible contribution of cow factors in this apparent change in the behavior of E. coli should not be overlooked. The increased prevalence of intramammary infection and clinical mastitis in the periparturient period has long been recognized, and research has demonstrated that “immunosuppression” may play a pivotal role at this time. Associations between concurrent disease and mastitis are well recognized, as is the increased susceptibility of freshly calved cows to more severe coliform mastitis (19). Hill (13) demonstrated the importance of the speed of recruitment of leukocytes, during clinical coliform mastitis, for the outcome of clinical disease, and Shuster et al. (28) demonstrated more severe mastitis in early-lactation cows and that these cows had relatively lower somatic cell counts. These factors, along with as yet unknown variability in innate immune factors, such as lactoferrin and the protection that may confer, demonstrate the susceptibility of the cow in early lactation. It may be variability in these factors that results in some cows subsequently removing infections and others becoming chronically infected.
Once a chronic infection has become established, there may be some form of “triggering” event to allow recrudescence. Various such events have been hypothesized and demonstrated, such as concurrent disease and mineral and vitamin deficiencies (12). It is interesting to note that one cow suffering repeat clinical episodes in this study did so at estrus, and although she remained infected, clinical episodes ceased when she conceived.
Historically, dogma has insisted that the most significant contribution towards recurrent mastitis has come from the “contagious” pathogens. This may have been and may still be the case in herds with high somatic cell counts, although it would now appear that recurrent mastitis is potentially as big a problem in the well-managed herd with a low somatic cell count, the only difference being the pathogens involved. Undoubtedly, there are a number of factors, including both host susceptibility and pathogen virulence, that are likely to be involved in the scenario described in this paper. These challenge our current understanding of the behavior of E. coli in the mammary environment and highlight the need for further research in this area.
ACKNOWLEDGMENTS
We gratefully acknowledge the cooperation of the farmers and herdspersons involved in this study and the Langford Veterinary Investigation Centre for undertaking bacteriology.
We gratefully acknowledge financial support from Leo Animal Health. M. J. Green is supported by a BBSRC CASE studentship award. A. J. Bradley is supported by a fellowship funded by the Wellcome Trust, London, United Kingdom.
REFERENCES
- 1.Berry E. Survey of clinical mastitis incidence. Stoneleigh, England: British Mastitis Conference; 1998. pp. 78–79. [Google Scholar]
- 2.Blowey R W, Edmondson P W. Mastitis control in dairy herds. Ipswich, England: Farming Press; 1995. [Google Scholar]
- 3.Booth J M. Progress in mastitis control—an evolving problem. Stoneleigh, England: British Mastitis Conference; 1997. pp. 3–9. [Google Scholar]
- 4.Bradley A J, Green M J. A study of the incidence and significance of intramammary enterobacterial infections acquired during the dry period. J Dairy Sci. 2000;83:1957–1965. doi: 10.3168/jds.S0022-0302(00)75072-7. [DOI] [PubMed] [Google Scholar]
- 5.Bradley, A. J., and M. J. Green. The aetiology of clinical mastitis in a cohort of Somerset dairy herds. Vet. Rec., in press. [DOI] [PubMed]
- 6.Darlington R B. Multiple tests in regression and linear models. Singapore: McGraw-Hill Publishing Company; 1990. pp. 249–276. [Google Scholar]
- 7.Dopfer D, Almeida R A, Lam T J, Nederbragt H, Oliver S P, Gaastra W. Adhesion and invasion of Escherichia coli from single and recurrent clinical cases of bovine mastitis in vitro. Vet Microbiol. 2000;74:331–343. doi: 10.1016/s0378-1135(00)00191-7. [DOI] [PubMed] [Google Scholar]
- 8.Dopfer D, Barkema H W, Lam T J, Schukken Y H, Gaastra W. Recurrent clinical mastitis caused by Escherichia coli in dairy cows. J Dairy Sci. 1999;82:80–85. doi: 10.3168/jds.S0022-0302(99)75211-2. [DOI] [PubMed] [Google Scholar]
- 9.Dozois C M, Curtiss R., III Pathogenic diversity of Escherichia coli and the emergence of ‘exotic’ islands in the gene stream. Vet Res. 1999;30:157–179. [PubMed] [Google Scholar]
- 10.Erskine R J, Kirk J H, Tyler J W, DeGraves F J. Advances in the therapy for mastitis. Vet Clin N Am Food Anim Pract. 1993;9:499–517. doi: 10.1016/s0749-0720(15)30617-4. [DOI] [PubMed] [Google Scholar]
- 11.Green M J, Bradley A J. Coliform mastitis—an evolving problem. Cattle Pract. 1998;6:91–94. [Google Scholar]
- 12.Green M J, Bradley A J. Current knowledge and E. coli mastitis—what can the practitioner do? Cattle Pract. 2000;8:243–248. [Google Scholar]
- 13.Hill A W. Factors influencing the outcome of Escherichia coli mastitis in the dairy cow. Res Vet Sci. 1981;31:107–112. [PubMed] [Google Scholar]
- 14.Hill A W, Shears A L. Recurrent coliform mastitis in the dairy cow. Vet Rec. 1979;105:299–301. doi: 10.1136/vr.105.13.299. [DOI] [PubMed] [Google Scholar]
- 15.Hill A W, Shears A L, Hibbitt K G. The elimination of serum-resistant Escherichia coli from experimentally infected single mammary glands of healthy cows. Res Vet Sci. 1978;25:89–93. [PubMed] [Google Scholar]
- 16.Hill A W, Shears A L, Hibbitt K G. The pathogenesis of experimental Escherichia coli mastitis in newly calved dairy cows. Res Vet Sci. 1979;26:97–101. [PubMed] [Google Scholar]
- 17.Hill A W, Shears A L, Hibbitt K G. The survival of serum resistant Escherichia coli in the bovine mammary gland following experimental infection. Res Vet Sci. 1979;26:32–37. [PubMed] [Google Scholar]
- 18.Hogan J S, Smith K L, Hoblet K H, Schoenberger P S, Todhunter D A, Hueston W D, Pritchard D E, Bowman G L, Heider L E, Brockett B L, Conrad H R. Field survey of clinical mastitis in low somatic cell count herds. J Dairy Sci. 1989;72:1547–1556. doi: 10.3168/jds.s0022-0302(89)79266-3. [DOI] [PubMed] [Google Scholar]
- 19.Jones T O. Escherichia coli mastitis in dairy cattle—a review of the literature Vet. Bull. 1990;60:205–231. [Google Scholar]
- 20.Kaipainen T, Pohjanvirta T, Shpigel N Y, Shwimmer A, Pelkonen S, Pyörälä S. Symposium on Immunology of Ruminant Mammary Gland. Stresa, Italy: IDF; 2000. Virulence factors of Escherichia coli isolates from bovine clinical mastitis; pp. 314–318. [DOI] [PubMed] [Google Scholar]
- 21.Kossaibati M A, Esslemont R J. The costs of production diseases in dairy herds in England. Vet J. 1997;154:41–51. doi: 10.1016/s1090-0233(05)80007-3. [DOI] [PubMed] [Google Scholar]
- 22.Kossaibati M A, Hovi M, Esslemont R J. Incidence of clinical mastitis in dairy herds in England. Vet Rec. 1998;143:649–653. doi: 10.1136/vr.143.24.649. [DOI] [PubMed] [Google Scholar]
- 23.Lam T J, Lipman L J, Schukken Y H, Gaastra W, Brand A. Epidemiological characteristics of bovine clinical mastitis caused by Staphylococcus aureus and Escherichia coli studied by DNA fingerprinting. Am J Vet Res. 1996;57:39–42. [PubMed] [Google Scholar]
- 24.Nemeth J, Muckle C A, Gyles C L. In vitro comparison of bovine mastitis and fecal Escherichia coli isolates. Vet Microbiol. 1994;40:231–238. doi: 10.1016/0378-1135(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 25.Quinn P J, Carter M E, Markey B, Carter G R. Clinical veterinary microbiology. London, England: Wolfe; 1994. [Google Scholar]
- 26.Radostits O M, Leslie K E, Fetrow J. Herd health: food animal production medicine. 2nd ed. Philadelphia, Pa: Saunders; 1994. [Google Scholar]
- 27.Sanchez-Carlo V, Wilson R A, McDonald J S, Packer R A. Biochemical and serologic properties of Escherichia coli isolated from cows with acute mastitis. Am J Vet Res. 1984;45:1771–1774. [PubMed] [Google Scholar]
- 28.Shuster D E, Lee E K, Kehrli M E., Jr Bacterial growth, inflammatory cytokine production, and neutrophil recruitment during coliform mastitis in cows within ten days after calving, compared with cows at midlactation. Am J Vet Res. 1996;57:1569–1575. [PubMed] [Google Scholar]
- 29.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veterinary Laboratories Agency. Veterinary Investigation Surveillance Report. New Haw, Addleton, United Kingdom: Veterinary Laboratories Agency; 1998. [Google Scholar]
- 31.Wilesmith J W, Francis P G, Wilson C D. Incidence of clinical mastitis in a cohort of British dairy herds. Vet Rec. 1986;118:199–204. doi: 10.1136/vr.118.8.199. [DOI] [PubMed] [Google Scholar]
- 32.Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]