Abstract
Hypertrophic/dilated cardiomyopathy, often a prequel to heart failure, is accompanied by maladaptive transcriptional changes that contribute to arrythmias and contractile misfunction. Transgenic mice constitutively expressing high levels of calcineurin are known to develop extreme heart hypertrophy, which progresses to dilated cardiomyopathy, and to die several weeks after birth. Here, we characterized aberrant transcriptional and epigenetic pathways in this mouse model and established a pharmacological approach to treat established cardiomyopathy. We found that H3K4me3 (trimethyl histone 3 lysine 4) and H3K9me3 (trimethyl histone 3 lysine 9) Jumonji histone demethylases are markedly increased at the protein level and show enhanced enzymatic activity in diseased hearts. These epigenetic regulators continued to increase with time, further affecting cardiac gene expression. Our findings parallel the lower H3K4me3 and H3K9me3 levels seen in human patients. Inhibition of Jumonji demethylase activities in vivo results in lower histone demethylase enzymatic function in the heart and higher histone methylation levels and leads to partial reduction of heart size, reversal of maladaptive transcriptional programs, improved heart function, and prolonged survival. At the molecular level, target genes of transcription factor myocyte enhancer factor 2 are specifically regulated in response to pharmacological or genetic inhibition of Jumonji demethylases. Similar transcriptional reversal of disease-associated genes is seen in a second disease model based on cardiac mechanical overload. Our findings validate pharmacological inhibitors of Jumonji demethylases as potential therapeutics for the treatment of cardiomyopathies across disease models and provide evidence of the reversal of maladaptive transcriptional reprogramming leading to partial restoration of cardiac function. In addition, this study defines pathways of therapeutic resistance upregulated with disease progression.
Keywords: cardiomyopathy, Jumonji enzymes, small-molecule inhibitor, JIB-04, transcriptional reprogramming, calcineurin
Abbreviations: cDNA, complementary DNA; CnA, calcineurin A; EMT, epithelial-to-mesenchymal transition; GEO, Gene Expression Omnibus; GO, Gene Ontology; GSEA, gene set enrichment analysis; HDAC, histone deacetylase; H3K4me3, trimethylation of histone 3 lysine 4; H3K9me3, trimethylation of histone 3 lysine 9; HRP, horseradish peroxidase; IgG, immunoglobulin G; KDM, histone lysine demethylase; KO, knock-out; LVAD, left ventricular assist device; MEF2, myocyte enhancer factor 2; MHC, myosin heavy chain; mTORC1, mammalian target of rapamycin complex 1; NFAT, nuclear factor of activated T-cell; TAC, transaortic constriction; Tg, transgenic; TSA, trichostatin A; UTSW, UT Southwestern
Transcriptionally driven heart remodeling occurs in response to multiple physiological and pathological conditions such as mechanical overload or hypertension, resulting in left ventricular hypertrophy, cardiomyopathy, and other cardiac abnormalities. Although physiological remodeling is reversible, such as occurs during pregnancy, pathological remodeling becomes established and leads to heart enlargement, fibrosis, electrical conductance abnormalities, and heart failure (1, 2, 3). Treatment with inhibitors that block angiotensin or calcium signaling, or with diuretics, can improve symptoms, prevent worsening of the condition, and partly normalize heart function; yet pathological cardiac remodeling itself neither is reversed by these measures nor are they preventive (1, 2).
Over the last several years, we and others have uncovered a role for epigenetic mechanisms in the transcriptional remodeling of the heart that occurs under pathological conditions and establishes heart hypertrophy and stiffening. Multiple transcriptional programs, including hypertrophic and profibrotic gene expression patterns, are now known to be controlled by histone-modifying enzymes including histone deacetylases (HDACs) and histone demethylases (4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15). Indeed, in human patients, it has been noted that heart disease tissue has abnormal levels of certain histone marks. In comparison to normal healthy tissue, for example, heart tissue from cardiomyopathies shows lower levels of methylation on histone 3 lysines 4 and 9 (lower H3K4 and H3K9 methylation), whereas other methylation marks remain unchanged (16). Consistent with these findings, the balance between acetylation and deacetylation of histones on genomic loci containing prohypertrophy genes, as well as the levels and sites of histone methylation on genes controlling calcium signaling and heart muscle contractility is altered in samples of human heart failure patients (8, 9, 16, 17, 18). Thus, epigenetic drivers of the remodeling process in the heart may be targetable key players in the development and progression of cardiac hypertrophy, cardiac myopathies, and cardiac dysfunction.
Multiple studies have established that high levels of calcineurin activity are found in patients with congestive heart failure and more recently, also in those with left ventricular hypertrophy (19, 20, 21, 22, 23). Given this, here we characterized the transcriptional and epigenetic pathways at play in a model of severe dilated cardiomyopathy driven by constitutively active calcineurin, and found wide deregulation of cardiac transcriptional programs and increased levels and activity of a subset of histone-demethylating enzymes not yet described to be involved in heart pathology. A subset of these changes continues to worsen with time. We related these findings to other mouse models of disease including mechanical overload models and genetic overexpression and knock-outs (KOs) of histone demethylases. We show that pharmacological intervention blocking Jumonji demethylase activity mimics genetic KOs and reverses gene expression patterns that drive disease, essentially partly reversing transcriptional changes of the remodeling process.
Results
The functional transcriptome of myosin heavy chain-calcineurin A transgenic mice is deregulated
As has been reported, the hearts of myosin heavy chain (MHC)-calcineurin A (CnA) transgenic (Tg) mice are 2 to 2.5 times larger than Wt mice of the same age and demonstrate gross anatomical abnormalities with clear dilated cardiomyopathy (Fig. S1A) (24). Overall, heart over body weights are nearly tripled already by 1 month of age (Fig. S1B). Heart function at distinct time points using echocardiography confirmed that the ability of the heart to contract is significantly reduced in Tg animals at 28 days and remains reduced at 77 days of age consistent with prior reports (25, 26) (Fig. S1C). The increase of end-systolic volume and the reduction of stroke volume and ejection fraction seen here are similar to the phenotype of heart failure models mediated by the combination of transverse aortic constriction and distal left anterior coronary ligation (27). In addition, disease appears to be fully established by 28 days (compare 28 versus 77 days, Fig. S1C) by these measurements. We next examined in detail the expression of key functional and structural genes in these hearts. Consistent with the pathological cardiac dilation/enlargement phenotype, quantitative RT–PCR showed that the expression of multiple hypertrophy genes was significantly upregulated as were fibrosis genes (Fig. S1, D and E). Some genes in both categories, including Rcan1, showed higher expression at 77 days compared with earlier time points (Fig. S1G).
Dong et al. (28) have reported that MHC-CnA mice suffer from arrhythmias, although ion channel gene expression has not been comprehensibly described in this mouse model. We therefore measured the expression of ion channel, accessory/regulatory, and other related genes contributing to action potential in heart myocytes (such as calcium pumps, calsequestrins, and sodium/calcium exchangers and their stabilizers) of mice with fully established disease (77 days old). As shown in Figure S1F, there was robust deregulation of these genes. We observed, for example, reduced levels of the sodium channel Scn5a, responsible for sodium ion influx in phase 0 (29), as well as lower expression of multiple potassium channel genes responsible for the K+ current in phases 1, 2, and 3 of the myocyte action potential (30), including Kcnd2, Kcna5, and Kcnq1. In contrast, Kcnc1 shows a likely compensatory increase, and the channels responsible for K+ current in phase 4, Kcnj2 (30) and Hcn4 (29), are increased. In addition, multiple accessory/regulatory genes are deregulated. The expression of calcium channels responsible for calcium influx into the cytoplasm during phase 2 (29), Cacna1c and Cacna1g, is increased, whereas expression of Ryr2, which releases calcium from the sarcoplasmic reticulum (31), is down. Calsequestrin mRNA levels are increased, also potentially indicating greater retention of calcium in the sarcoplasmic reticulum. In addition, expression of calcium pump genes that take calcium from the cytoplasm back into sarcoplasmic reticulum such as sarcoplasmic/endoplasmic reticulum calcium ATPase 1 is decreased, but the sodium/calcium exchanger Slc8a1/NCx1 is increased in Tg hearts, suggesting an overall imbalance in calcium regulation and activation of compensatory mechanisms. Consistent with the published induction of the Fgf21 gene upon cardiac stress (32, 33), we also observed increased levels in these Tg hearts, whereas other stress response genes were not significantly altered (Fig. S1H).
Epigenetic erasers controlling histone methylation are upregulated in CnATg hearts
Given the extensive changes in functional gene expression observed in CnATg hearts, and the known observation that H3K4 and H3K9 trimethyl marks are decreased genome wide in heart disease patients (16), we wondered if epigenetic regulation was aberrant in CnTg hearts. We and others have previously established the contribution of members of the Jumonji histone demethylase family in models of mechanical overload by transaortic constriction (TAC) (6, 11, 14, 15, 34). We therefore examined mRNA and protein levels of multiple Jumonji demethylases including those that remove H3K4 or H3K9 methylation. Tg hearts with established disease (42 days) did not show increased mRNA expression for any of the many Jumonji demethylases measured versus aged-matched Wt littermates (Fig. S1I). At the protein level, however, we found higher levels of two enzymes that remove H3K4 trimethylation, namely Kdm5a and Kdm2b (histone lysine demethylase [KDM]), as well as increased levels of the H3K9me3 (trimethyl histone 3 lysine 9)/H3K36me3 (trimethyl histone 3 lysine 36) demethylase Kdm4c (Fig. 1, A and B), whereas other Jumonji enzyme levels were unaltered (Fig. S2, A and B).
Figure 1.
Jumonji KDMexpression and activity are increased in CnA transgenic (Tg) hearts.A, Jumonji proteins increase in Tg hearts. Western blots of H3K4me3 and H3K9me3 Jumonji KDM demethylases in hearts from Wt versus CnATg mice. Protein extracts for Western blots of Kdm5a (whole cell), Kdm2b (whole cell), and Kdm4c (nuclear extract) are lysates from ventricle powders of Wt and Tg mice, between 35 and 42 days of age. B, quantification of Jumonji protein levels. Left panels, H3K4me3 demethylases. Kdm5a: Wt n = 6, Tg n = 6; Kdm2b: Wt n = 5, Tg n = 6. Right panel, H3K9me3 demethylase. Kdm4c: Wt n = 4, Tg n = 4. Data are average ± SEM. p Values are from Mann–Whitney's test. C, H3K4me3 and H3K9me3 Jumonji demethylase activity in heart nuclear extracts (left ventricle) from Wt versus CnATg mice between 84 and 103 days of age on exogenous trimethylated substrates. Left panel, H3K4me3 demethylase activity assays; Wt n = 4, Tg n = 4. Right panel, H3K9me3 demethylase activity assays, Wt n = 3, Tg n = 3. Data are average ± SEM. p Values are from t test, two-tailed, unequal variance. B and C, ∗p ≤ 0.05 and ∗∗p ≤ 0.01. CnA, calcineurin A; H3K4me3, trimethylation of histone 3 lysine 4; H3K9me3, trimethylation of histone 9 lysine 4; KDM, histone lysine demethylase.
We then evaluated Jumonji histone demethylase activity in CnATg hearts with fully established dilated cardiomyopathy. We found that the levels of both H3K4me3 (trimethyl histone 3 lysine 4) demethylase activity and H3K9me3 demethylase activity were doubled compared with Wt (Fig. 1C). The enhanced protein abundance and activity of Jumonji demethylases acting on H3K4me3 has not been described in other heart disease models to our knowledge, and it parallels the clinical observation that patients with cardiomyopathies have lower levels of cardiac H3K4me3 (16). Consistent with the increased levels of Kdm4c enzyme and its H3K9me3 demethylase activity, expression of a target gene regulated by H3K9 methylation in the heart (14), four and a half LIM domains 1 (Fhl1), was robustly increased in these animals (Fig. S2C) in line with other studies showing Fhl1 is induced in hypertrophic hearts (14, 35). The Fhl1 protein was likewise upregulated (Fig. S2D) in the hearts of these Tg mice.
Inhibition of Jumonji demethylase activity decreases heart size, improves heart function, and extends the survival of MHC-CnA Tg mice
Because of the observed increased Jumonji demethylase protein levels and activities on H3K4me3 and H3K9me3, we used a pharmacological approach to block these functions. We have previously published that the small-molecule JIB-04 inhibits H3K4me3 and H3K9me3 demethylase activities in vivo in other model systems and importantly that it does not negatively impact healthy hearts (15, 36, 37, 38). We therefore evaluated if JIB-04 could target Jumonji KDM activity in MHC-CnA Tg hearts. Treatment of 4-week-old to 5-week-old Tg animals with JIB-04 significantly reduced both H3K4me3 and H3K9me3 Jumonji demethylase activity in these hearts after 2 or 7 weeks of treatment (Fig. 2A) while not significantly affecting total HDAC activity in the same extracts (Fig. S3A). Thus, JIB-04 can block Jumonji enzyme activity in the heart in vivo. To evaluate the consequences of this treatment, we measured heart size in vehicle versus JIB-04-treated MHC-CnA animals after 6 weeks of treatment. JIB-04-treated mice showed a ∼15% modest yet significant reduction in heart size in response to JIB-04 compared with no change in heart size in Wt controls (Figs. 2B and S3B). Together, this indicates on-target activity of JIB-04 in these diseased hearts. Indeed, JIB-04-treated mice showed partly improved heart function already after 2 weeks of treatment as measured by echocardiographs (Figs. 2C and S3C).
Figure 2.
Pharmacological inhibition of Jumonji KDMs in the heart improves cardiac function and prolongs survival.A, H3K4me3 and H3K9me3 demethylase activity in nuclear extracts from two pairs of transgenic (Tg) mice treated with vehicle (Ve) or JIB-04 for 2 weeks (orange symbols) and two pairs treated for 7 weeks (green symbols), all starting at 4 weeks of age. p Values are t test, two-tailed, and paired. B, percent changes of Wt or Tg hearts treated with JIB-04 for 6 weeks, starting from 5 weeks of age, normalized to Ve control of the same genotype. Wt n = 10, Tg n = 12, p value is t test, two-tailed, and of unequal variance. C, echocardiography data of mice treated with JIB-04 in the same age range as mice in panel A (orange symbols). Stroke volume and cardiac output are shown. p Values are from t test, two-tailed, and of unequal variance. Wt Ve n = 12, Tg Ve n = 9, Tg JIB-04 = 14. D, percent survival of Tg mice treated with Ve or JIB-04, starting from 4 weeks of age. Ve n = 25, JIB-04 n = 19. p Value is Gehan–Breslow–Wilcoxon test. 75% median and 25% survival are shown by horizontal lines. E, body weight changes of Tg animals treated with Ve or JIB-04 over the duration of the survival experiment shown in D. F, quantification of heart rate and ST interval from ECG measurements of Wt mice treated with Ve and of CnATg mice treated with Ve or JIB-04 starting at 4 weeks of age. Ages of mice used for ECG range from 88 to 149 days old. Wt Ve n = 7, Tg Ve n = 5, and Tg JIB-04 n = 6. p Values are from t test, two-tailed, and of unequal variance. G, echocardiography parameters of Wt Ve-treated, Tg Ve-treated, and Tg JIB-04-treated mice in the same age range used for ECG measurements in F. Animals in F and G were treated for 9–13 weeks. Wt Ve n = 2, Tg Ve n = 4, and Tg JIB-04 n = 3. p Values are t test, one-tailed, and of unequal variance. A, B, C, E, F, and G, data are average ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, or p values are shown. H3K4me3, trimethylation of histone 3 lysine 4; H3K9me3, trimethylation of histone 9 lysine 4; KDM, histone lysine demethylase.
The severely enlarged fibrotic hearts of MHC-CnATg mice lead to congestive heart failure and sudden death triggered by arrythmia as early as 8 weeks of age (24, 28). We therefore examined next if JIB-04 would extend the survival of these mice by improving heart function. We treated the Tg mice with vehicle or JIB-04 starting at 4 weeks of age when the Tg phenotype is already robust and followed them until they succumbed to the disease. JIB-04 extended the survival of MHC-CnA mice, increasing 75% survival from 57 to 130 days, median survival from ∼110 to 140 to 177 days, and 25% survival from 156 to 195 days (Figs. 2D and S3D). Animals tolerated the treatment well with no decreases in body weight or changes in overall health performance including in pregnant females who gave birth to pups normally while under treatment (Figs. 2E and S3E).
Of note, vehicle-treated mice showed two distinct patterns, with one population succumbing to disease early (around day 45) and a second population surviving this threshold and succumbing around day 160 (Figs. 2D and S3D). The two subpopulations did neither significantly differ in gender nor in levels of transgene expression. Although both populations benefited by JIB-04 treatment, the major extension of survival corresponded to the treated population that dies around day 45 when on vehicle. In addition to a survival benefit, however, JIB-04 treatment also prevented older animals from the weight loss seen in the vehicle-treated cohort of the same age (Fig. 2E).
To further understand inhibitor action during prolonged treatment, we followed heart function in these animals by ECG and echocardiographs. In anesthetized mice undergoing ECG, the basal heart rate of Tg mice is lower than that of Wt mice, whereas the basal PR and ST intervals are significantly increased in the Tg mice under these experimental conditions (Figs. 2F and S3F). Treatment with JIB-04 did not significantly change the PR or QRS interval. Remarkably, however, the length of the ST intervals was significantly reduced toward the levels of the Wt mice in JIB-04-treated Tg animals, and the heart rates under anesthesia were partly normalized toward the Wt heart rates (Figs. 2F, both panels, and S3, F and G). Thus, treatment with JIB-04 inhibits cardiac H3K4me3 and H3K9me3 demethylase activity, reduces heart size, improves ST interval timing, normalizes heart rate under anesthesia, and prolongs survival in Tg animals overexpressing CnA.
To further confirm improved heart performance, we also carried out echocardiographs on vehicle versus JIB-04-treated Tg mice of the same age cohort as the mice studied by ECG treated for prolonged periods. Multiple parameters of myocyte function showed improved trends, including fraction shortening, stroke volume, cardiac output, and ejection fraction (Figs. 2G and S3H).
Jumonji enzyme inhibition reprograms heart gene expression toward normal myogenesis and contractile function
To understand the impact of pharmacological inhibition of Jumonji demethylases on the transcriptome of the heart, we performed RNA-Seq analysis of cardiac tissue (left ventricle to enable direct comparison to models of left ventricular hypertrophy) from vehicle versus JIB-04-treated animals and compared observed changes in gene expression to genes known to be deregulated in heart disease mouse models including several TAC versus sham datasets and MHC-CnA Tg versus Wt mice (14, 15, 39), as described in detail in the Experimental procedures section. Gene set enrichment analysis (GSEA) of gene expression levels revealed that while mouse hearts that undergo mechanical overload by TAC and MHC-CnA Tg mouse hearts show enrichment in genes that contribute to myogenesis when compared with their respective sham/Wt control counterparts, JIB-04 treatment depleted this gene set in the diseased hearts (Table S1 and Figs. 3, A and B and S4A). Indeed, of 66 myogenesis genes that were upregulated by the disease process, JIB-04 downregulated 51 of these or 77% (Fig. 3C). Similarly, genes involved in left ventricular assist device (LVAD) support of failing heart function were upregulated in disease models versus healthy normal controls while being downregulated by JIB-04 in disease (Figs. 3, D and E and S4B) as were gene sets involved in muscle contraction (Figs. 3, G and H and S4C). Indeed, JIB-04 depleted 73% of the genes enriched by disease involved in LVAD support of failing hearts and 70% of disease genes involved in muscle contraction (Fig. 3, F and I). In addition, fatty acid metabolism genes were robustly depleted in disease models compared with healthy controls, and JIB-04 treatment markedly upregulated these genes in disease hearts restoring their levels toward normal (Fig. S4, D and E). Tables S2–S4 contain a detailed list of these genes.
Figure 3.
Jumonji enzyme inhibition reprograms gene expression in the heart toward normal myogenesis and contractile function.A, D, G, and J, GSEA curves of disease models showing the enrichment of genes in Hallmark myogenesis, Chen LVAD support of failing heart, muscle contraction genes, and MEF2 motifs, respectively. B, E, H, and K, GSEA curves showing that JIB-04 treatment depletes disease-enriched gene sets. C, F, I, and L, Venn diagrams showing genes that are upregulated in disease and reversed by JIB-04 treatment in either CnATg or TAC models. Genes used to make Venn diagram in I for muscle contraction are the sum of following GSEA terms: GO actin-mediated cell contraction, GO contractile fiber, GO muscle contraction, and GO heart process. Genes used to make Venn diagram in L for MEF2 motifs are the sum of genes with the following motifs: MEF2_02, MEF2_03, MEF2_Q6_01, CTAWWWATA_RSRFC4_Q2, and RSRFC4_Q2, in a region spanning up to 4 kb around the transcription start sites according to GSEA. CnA, calcineurin A; GO, Gene Ontology; GSEA, gene set enrichment analysis; LVAD, left ventricular assist device; MEF2, myocyte enhancer factor 2; TAC, transaortic constriction.
JIB-04 downregulates myocyte enhancer factor 2 target genes involved in heart disease
Further analysis of RNA-Seq datasets revealed that myocyte enhancer factor 2 (MEF2) motif–containing genes were enriched in diseased hearts in both mechanical overload and CnA genetic models (Figs. 3J and S4F). JIB-04 downregulated 67% of these genes, partly normalizing MEF2-driven gene expression (Fig. 3, K and L and Table S5). Since MEF2 is known to act in conjunction with nuclear factor of activated T-cell (NFAT) in the regulation of cardiac gene expression during disease (40, 41, 42), we bioinformatically examined if MEF2 target genes upregulated in disease and downregulated by JIB-04 included genes coregulated by NFAT (Fig. S5, A and B and Table S6). We found that of the 54 genes that are MEF2 target genes upregulated in disease and downregulated by JIB-04 treatment in the diseased heart, about a third (17 of 54 or 32%) contain NFAT binding motifs (Fig. S5C), whereas genes containing GATA4 motifs were not significantly represented (Table S6). This suggests that JIB-04 treatment directly or indirectly leads to downregulation of MEF2 targets including MEF2–NFAT coregulated genes in the diseased heart.
Transcriptional reprogramming by JIB-04 reverses KDM target genes deregulated in cardiac hypertrophy mouse models
Since Jumonji demethylases have different biological roles and the use of a pan inhibitor does not delineate the complex roles of individual Jumonji enzymes, we assessed if JIB-04 in vivo action was partly through modulation of Jumonji KDM targets. In addition to measuring enzymatic activity in hearts from treated animals (done in Fig. 2A), we also analyzed Kdm3a and Kdm4a Tg and KO genetic mouse models and compared their cardiac transcriptomes to those seen in the TAC and CnA disease models basally and after JIB-04 treatment. First, we performed GSEA analysis on the Jumonji Kdm3a and Kdm4a Tg models, which show an exacerbated phenotype in response to TAC as published previously (14, 15). We also did GSEA analysis in Kdm3a and Kdm4a KO models, which are protective against TAC-induced cardiac hypertrophy (14, 15). We found that the Jumonji Kdm3a and/or Kdm4a Tg animals showed overlapping gene set enrichments with the MHC-CnA and TAC models and that Kdm3a and/or Kdm4a KO animals showed depletion of these, including myogenesis, LVAD support of failing heart, and muscle contraction gene sets (Figs. 4, A–C and S6A). In addition, fatty acid metabolism gene sets found depleted in the TAC and CnA disease models (Fig. S4, D and E, left GSEA plots) were enriched in the hearts of the KDM KO animals (Fig. 4D), similar to what we observed in JIB-04-treated TAC and CnA mice (Fig. S4, D and E, right GSEA plots). Furthermore, MEF2 motif–containing gene sets were enriched in the Kdm4a Tg animals (Fig. 4E), as seen also in the TAC and MHC-CnA animals (Figs. 3J and S4F).
Figure 4.
KDM transgenic (Tg) mouse hearts show similar deregulation of transcriptional pathways as TAC and MHC-CnA animals,and KDM KO in the heart partly mimics JIB-04 treatment.A, GSEA enrichment curve of Kdm3a Tg/Wt of Hallmark myogenesis. B, GSEA depletion curve of Kdm4a heart-specific (hs) KO/Wt of gene sest “Chen LVAD support of failing heart”. C, GSEA enrichment curves of Kdm4a Tg/Wt of muscle contraction genes, including GO muscle contraction and GO contractile fiber. D, GSEA enrichment curves of Kdm4a general (g) KO/Wt for fatty acid metabolism, including reactome fatty acid metabolism and Hallmark fatty acid metabolism. E, GSEA enrichment curves of Kdm4a Tg/Wt of MEF2 motifs, including MEF2_Q6_01 and RSRFC4_Q2. F, overlap of JIB-04-upregulated and KDM-regulated genes. Upper panel, Venn diagram of JIB-04-upregulated and Kdm3a- or Kdm4a KO-upregulated genes (JIB-04/Ve ≥1.5, Kdm3a or Kdm4a KO/Wt ≥1.3). Lower panel, Venn diagram of JIB-04-upregulated and Kdm3a- or Kdm4a Tg-downregulated genes (JIB-04/Ve ≥1.5; Kdm3a or Kdm4a Tg/Wt ≤0.77). G, overlap of JIB-04-downregulated and KDM-regulated genes. Upper panel, Venn diagram of JIB-04-downregulated and Kdm3a- or Kdm4a KO-downregulated genes (JIB-04/Ve ≤0.67, Kdm3a or Kdm4a KO/Wt ≤0.77). Lower panel, Venn diagram of JIB-04-downregulated and Kdm3a- or Kdm4a Tg-upregulated genes (JIB-04/Ve ≤0.67; Kdm3a or Kdm4a Tg/Wt ≥1.3). CnA, calcineurin A; GSEA, gene set enrichment analysis; KDM, histone lysine demethylase; LVAD, left ventricular assist device; MEF2, myocyte enhancer factor 2; MHC, myosin heavy chain; TAC, transaortic constriction.
Given these parallels in the biology across disease models, and the commonalities between KDM KO and JIB-04-altered pathways, we next determined if genes whose expression is altered by JIB-04 treatment in the diseased heart actually overlapped with KDM-regulated genes, as would be expected from the on-target action of this compound that inhibits Kdm3a and Kdm4a as well as other KDM enzymes including Kdm5a (15, 36). This analysis revealed that 22% (178 of 799) of genes upregulated in the diseased heart by JIB-04 treatment are similarly regulated by Kdm3a or Kdm4a (Fig. 4F), and that 36% (446 of 1243) of genes downregulated by JIB-04 treatment are similarly regulated by Kdm3a or Kdm4a in the heart (Fig. 4G). Thus, JIB-04 acts on Kdm3a and Kdm4a target genes in the diseased heart (Table S7), among other targets.
Finally, we determined if genes regulated by JIB-04 that are Kdm3a or Kdm4a targets are relevant to disease. We first defined KDM target genes (genes oppositely regulated in the KDM Tg versus KO animals, see the Experimental procedures section). We then overlapped disease genes commonly altered in the TAC and CnA models with JIB-04-regulated genes and KDM target genes, yielding 88 genes (Fig. S6B and Table S8). Of these, 59 genes were upregulated in disease, and within this set, 57 genes were downregulated by JIB-04 and 20 of these were Kdm3a target genes, whereas 40 were Kdm4a target genes (Fig. S6, C and D and Table S8). Similarly, of the 29 genes downregulated in disease, 21 genes were upregulated by JIB-04 with seven of these being Kdm3a targets and 14 being Kdm4a target genes (Fig. 6, E and F and Table S8). Overall, this indicates that JIB-04 acts on KDM target genes to reverse gene expression patterns that contribute to heart dysfunction.
The transcriptome of CnATg animals becomes less responsive to Jumonji inhibitors over time
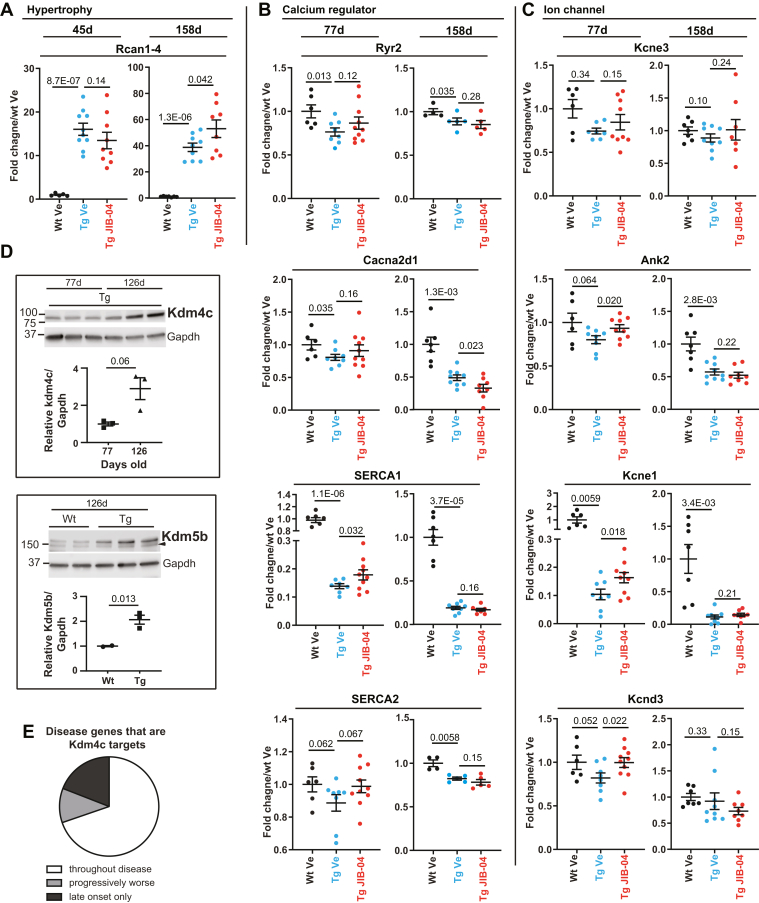
To understand how these overall gene expression changes may translate into the increased survival seen in the younger versus the older population of CnATg mice, we measured gene expression at early versus later time points in animals being treated with JIB-04. At the earlier time points (45–77-day-old mice, treated since week 4–5 years of age), we observed reversal of pathological gene expression. For example, some hypertrophy genes and multiple ion channels deregulated in disease, such as calcium regulators Ryr2, sarcoplasmic/endoplasmic reticulum calcium ATPase 1 and sarcoplasmic/endoplasmic reticulum calcium ATPase 2, and potassium channels Kcne3, Kcne1, and Kcnd3 were partly normalized by JIB-04 treatment in the younger cohorts after a few weeks of treatment (Figs. 5, A–C and S7A, “early” panels). In contrast, by the later time points (158 days), the reversal of pathological gene expression was lost. The older animals no longer showed partly normalized expression of ion channel genes or hypertrophy genes (Figs. 5, A–C and S7A, “late” panels), indicating that the transcriptome is more refractory to changes later in disease progression and/or as animals adapt to JIB-04 treatment over time. Indeed, when we quantified Jumonji demethylase activity on H3K4me3 or H3K9me3 substrates in hearts from 158-day-old littermates treated since 4 weeks of age with vehicle versus JIB-04, we found no measurable inhibition by the drug (Fig. S7B), in contrast with the effect of JIB-04 seen in the younger animals (Fig. 2A). Furthermore, the levels of Jumonji demethylase gene expression trended upward over time in Tg animals overall and significantly increased for multiple KDMs as animals aged (Fig. S7C). Strikingly, among the KDMs with higher gene expression levels over time, at least two also showed increased protein levels in the older Tg animals. Kdm4c protein, seen upregulated at the earlier time points compared with Wt age-matched controls (Fig. 1, A and B), for example, continued to increase with time (Fig. 5D, top panel) with protein levels tripling in Tg hearts between 77 and 126 days of age. In addition, Kdm5b protein, which had not shown higher levels than Wt age-matched controls in younger animals (Fig. S2, A and B), did increase markedly in the older Tg mice (Fig. 5D, lower panel). Furthermore, the increased Kdm4c protein in older Tg mice coincided with progressively increased or newly increased expression of disease genes that are Kdm4c gene targets (43) (Fig. 5E and Table S9). These results indicate that CnA-driven disease is progressive, and its transcriptome worsens with time in part driven by increased levels of Jumonji KDMs with age. Overall, these observations suggest that the animals that benefited less from JIB-04 treatment in terms of extended survival coincide with those whose transcriptomes did not respond to treatment or became resistant to it over time, consistent with the lack of demethylase inhibition by JIB-04 and increased enzyme expression in the older and longer-treated population versus the younger-treated and shorter-treated population.
Figure 5.
Reduced transcriptional responsiveness of CnATg animals to Jumonji inhibitor coincides with increased levels of Jumonji proteins in older animals during disease progression. Relative mRNA levels in response to JIB-04 treatment of hypertrophy (A), calcium regulator (B), and ion channel genes (C) at early (45 or 77 days old) versus late (125–158 days old) time points of CnATg mice. In all cases, RNA was extracted from left ventricle from mice treated starting at 4 to 5 weeks of age and measured by quantitative RT–PCR except for older animal time points for Ryr2 and SERCA2 (RNA-Seq). About 45-day-old mice: Wt vehicle (Ve) n = 5, Tg Ve n = 10, Tg JIB-04 n = 9; 77-day-old mice: Wt Ve n = 5–6, Tg Ve n = 7–8, Tg JIB-04 n = 9–10; 158-day-old mice: Wt Ve n = 7, Tg Ve n = 8–9, Tg JIB-04 n = 7–8; Ryr2 and SERCA2 158 days: Wt Ve n = 4, Tg Ve n = 5, Tg JIB-04 n = 5. D, Western blot analysis and quantification of Kdm4c (upper panel) and Kdm5b (lower panel). Mice used in Kdm4c Western blot are Tg mice at 77 (n = 3) or 126 (n = 3) days old. Mice used in Kdm5b Western blot are Wt or Tg, as indicated, at 126 days old. Wt n = 2; Tg n = 3. Data are average ± SEM, p values are from t test, one-tailed, and of unequal variance. Arrowhead: nonspecific band. Samples are from ventricle powders. E, pie chart of disease genes that are Kdm4c targets modulated throughout the disease, progressively modulated, or modulated at late time points only when Kdm4c protein is further increased. See text for details and Table S9 for a list of genes and their expression over time in Tg hearts. CnA, calcineurin A; SERCA2, sarcoplasmic/endoplasmic reticulum calcium ATPase 2; Tg, transgenic.
To identify potential pathways leading to this resistance/loss of response over time, we performed RNA-Seq analysis on left ventricle tissue at early (45–77 days old) versus late time points (125–158 days old) from CnATg mice continuously treated with vehicle or JIB-04 since 4 to 5 weeks of age. We identified several gene sets whose response to JIB-04 reversed or was lost over time (Fig. S7D and Table S10). For example, while at early time points during treatment, JIB-04 triggered significant depletion of E2F target genes, G2M checkpoint genes, glycolysis genes, and mammalian target of rapamycin complex 1 (mTORC1) signaling genes; at later time points, these gene sets became enriched by JIB-04 compared with vehicle cohorts. EMT genes and interferon gamma and alpha response gene sets behaved similarly. Given the roles of these pathways in promoting heart pathologies (44, 45, 46, 47, 48, 49, 50, 51, 52, 53), their increased expression at late time points either renders JIB-04 ineffective at the given dose and/or indicates that JIB-04 resistance has developed and expression goes back up. In contrast, fatty acid metabolism genes were enriched by JIB-04 at both early and later time points suggesting this transcriptional pathway is not a main contributor to changes seen in the survival benefit.
We also then evaluated Wt versus CnATg animals overtime to assess disease progression and define if significant changes in the disease transcriptome were occurring over time, in addition to the increase in Kdm4c target genes, which could account for weaker response to JIB-04 and decreased survival benefit. We found that indeed there was increased fibrotic and hypertrophy gene expression and lower expression of adult Mhy6 in the late versus early time points when comparing CnATg animals to age-matched Wt controls (Fig. S7E and Table S10). Thus, not only does JIB-04 resistance develop overtime through upregulation of mTORC1 signaling genes, EMT genes, and similar pathways but also the disease itself transcriptionally progresses with more pronounced expression of Jumonji KDM enzymes and of fibrotic, hypertrophy, and other disease-specific genes, likely contributing to lowering the effects of JIB-04 at the later time points. In conclusion, the CnATg hearts continue to undergo transcriptional changes even after the establishment of dilated cardiomyopathy around a month of age. This progressive transcriptional dysregulation is coincident with and at least partly driven by further increased levels of Jumonji enzymes with time in CnATg hearts.
Discussion
In this study, we have shown that overlapping transcriptional programs are aberrantly regulated across multiple mouse models of cardiac hypertrophy and cardiomyopathy, including calcineurin-driven disease, mechanical overload–triggered disease, and genetic models predisposed to heart dysfunction including animals overexpressing Jumonji H3K9 demethylases Kdm4a and Kdm3a. Remarkably, genetic knock down or pharmacological inhibition of these Jumonji demethylases partly reverses disease gene expression programs, including myogenesis, heart muscle contractility, and fatty acid metabolism gene sets. In addition to increased activity of H3K9 demethylases, we find that the CnATg hearts express enhanced levels of Jumonji H3K4 demethylases Kdm5a and Kdm2b already at early stages of the disease and Kdm5b at later stages of disease and have higher levels of enzyme activity on H3K4me3 compared with Wt littermates, a discovery in this mouse model that is consistent with the lower levels of H3K4 methylation found in cardiomyopathy patients (16) and recent reports in other cardiac disorders (54, 55). The involvement of these novel Jumonji KDMs opens further venues for intervention. In fact, pharmacological inhibition with JIB-04 lowers H3K4me3 demethylase activity in CnATg hearts as well as H3K9me3 demethylase activity and partly normalizes cardiac gene expression, decreases heart weighs, improves heart function and specifically ST segments and HR, as well as fraction shortening, stroke volume, and cardiac output and extends survival in animals that would succumb to disease. These findings suggest that a wide range of patients with congenital mutations or predispositions to develop cardiac hypertrophy or related cardiomyopathies may benefit from Jumonji inhibition to block maladaptive transcriptional remodeling, partly reverse the course of the disease, or prevent its progression. Early intervention appears to be critical as disease worsens and/or treatment becomes less effective with time. Importantly, JIB-04 treatment had adverse effects neither on the survival, heart function, or transcriptome of Wt animals nor on pup development during pregnancy in drug-treated Tg moms (Fig. S3D and Table S10) (15, 36).
Multiple studies to date including our own have implicated Jumonji KDMs in cardiomyopathies (6, 11, 14, 15, 34) establishing the role of several Jumonji enzymes as generally prohypertrophic. Genetic and pharmacological blockade of this catalytic activity results in attenuation of pathological cardiac phenotypes by direct reversal of maladaptive gene expression and perhaps other indirect mechanisms as well. One gene set of particular interest that is downregulated by disease in both TAC and CnA Tg models and is restored by JIB-04 treatment in both models are genes involved in fatty acid metabolism. The heart's high energy demand relies on fatty acid oxidation for ATP production, and the expression of many of the genes responsible for fatty acid oxidation is decreased during maladaptive hypertrophy/cardiomyopathies (2). We found that both pharmacological inhibition of Jumonji KDMs as well as genetic ablation of Kdm4a specifically, enrich fatty acid metabolism gene expression likely balancing cardiac energy needs/restoring energy homeostasis, by normalizing energy production. It will be important to define the target genes of the KDM4 family in the heart to further understand their regulation of fatty acid metabolism genes, including peroxisome proliferator–activated receptor α/γ targets, under cardiac stress.
We have previously reported decreased size of phenylephrine-induced rat primary ventricular myocytes upon JIB-04 treatment or KO of Kdm3a (15). Consistent with this, here, we report a decrease in heart size in JIB-04-treated animals after the establishment of severe cardiomyopathy in the CnATg hearts. This suggests that KDM inhibition or another aspect of the action of JIB-04 may interfere with progrowth and protein synthesis pathways in the heart, such as mTOR, Hippo, or PI3K signaling, a possibility that can have mechanistic importance. Regardless, the actual beneficial effects of Jumonji inhibition appear to be partly generalizable across models of cardiac dysfunction and partly context dependent. Fibrotic and fetal gene programs, for example, are markedly reversed by JIB-04 treatment in TAC models and primary culture (15); yet, this is not the main effect of treatment in vivo in the MHC-CnTg model. In CnATg animals, we see partial normalization of electric signaling genes and other disease genes. Thus, we propose that cardiac Jumonji KDMs activated during hypertrophy, during fibrosis, and during the development of dilated cardiomyopathy have both common and unique targets depending on the initiating aberrant event that triggers disease. Importantly, these phenotypes are at least partly reversible by Jumonji inhibitor JIB-04 or equivalent interventions and may synergize with standard therapies currently used to manage disease symptoms. The beneficial effects of Jumonji inhibitor intervention in the CnTg animals become of high interest considering the continued observations that human patients suffering from heart pathologies have increased levels of calcineurin activity (19, 20, 21, 22, 23).
The present study, at the same time, points to limitations of this strategy as mice that were treated for long periods with JIB-04 had less of a survival benefit than younger mice. This seems to be due to the combined effects of both disease progression (with worsening of fibrosis/hypertrophy and increased levels of Jumonji KDM enzymes over time) as well as activation of resistance pathways that render JIB-04 less effective. It is unlikely that the difference in survival benefit between the younger and the older mice was due to developing arrythmias, since CnATg animals can succumb to arrhythmias as young as 8 to 11 weeks old (28). However, it is possible that arrhythmic episodes become stronger or more frequent with disease development. Some channel gene expression changes do get exacerbated with time. Ultimately, however, the intervention with Jumonji inhibitors does benefit animals across various models of heart pathologies and highlights the potential of this approach across multiple etiologies of human cardiac disorders.
Experimental procedures
Mouse models and animal work
MHC-CnA male mice, a gift of Dr Eric Olson (24), were crossed with C57BL6/N females (Envigo) to generate Tg mice for this study. Wt or Tg (MHC-CnA) mice were identified by genotyping using a CnA-genotype primer pair (forward: 5′-CAC TCC AGC TTG GTT CCC GAA TAG AC-3′, reverse: 5′-GAA CAA GCA GTT CCT GTG TGT ACA CG-3′) and Kapa BioSystem (KK5621) kit. Mice were housed at the certified UT Southwestern (UTSW) Medical Center animal facility. Female and male mice were treated with JIB-04 (50 mg/kg, three times per week) or vehicle (12.5% Cremophor EL [Sigma; C5135-500G] and 12.5% dimethyl sulfoxide [Sigma; D-26650-100 ML] in water) by gavage. Treatment with JIB-04 started on or around 28 to 35 days postbirth as indicated specifically in figure legends. At the indicated times, a subset of mice was subjected to echocardiography or ECGs before and during the treatment period. For imaging and molecular analysis, hearts were collected ∼4 to 5 h post treatment. All animal experiments were carried out under approved Institutional Animal Care and Use Committee protocols and followed UTSW animal care procedures.
Histology
For H&E staining, hearts were collected while beating, rinsed with PBS, and then submerged in cardiac relaxant buffer (118 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 25 mM NaHCO3, and 11 mM glucose) for 30 min. Hearts were transferred into a container with at least 20× volume of 4% paraformaldehyde in PBS and gently massaged to remove residual blood, and the relaxant buffer replaced with the fixative reagent, then incubated at room temperature with gentle agitation for 48 h. Hearts were then transferred into 50% ethanol and kept at 4 °C until paraffin processing took place at UTSW Histology Pathology Core. Hearts were cut into four-chamber view slides and stained using H&E. Hearts were imaged using Zeiss Axio Scan.Z1 scope.
Heart processing and quantitative RT–PCR
RNA was extracted from flash-frozen hearts after dissection and tissue homogenization in a tissue lyzer (Qiagen Tissuelyzer II), using Trizol (Thermo Scientific; 15596018) according to the manufacturer's instructions. See legends for specific anatomical dissection details for each figure. Complementary DNAs (cDNAs) were synthesized using High capacity cDNA reverse transcription kit (Life Technology; 4368813). Quantitative PCR was performed using Invitrogen SYBR GreenER qPCR SuperMix For ABI PRISM Instrument (11-760-500) in a QuanStudio7Flex (Life Technology) with an initial 2 min preincubation at 50 °C, followed by 10 min at 95 °C, and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Reference genes used are Gapdh or Tbp (Primer Table). Data were analyzed using the ΔΔCt method (56).
Echocardiography and ECG measurements and data analysis
Echocardiographs were performed on Vevo 2100 (VisualSonics) under conscious and unsedated conditions. For ECG measurements, mice treated from 28 days of age were anesthetized with 1% isoflurane. Mouse chest hair was shaved and then removed by topical treatment with Nair (Church & Dwight) hair remover product. Mice were restrained while anesthetized with 1% isoflurane using mouse breathing masks, and ECG probes were attached on their chest and signals recorded for ∼30 s. Once recording was completed, the ECG probes and then the breathing masks were removed. Mice recovered in warm bedding. Data were analyzed manually using LabChart5 software (ADInstruments).
Jumonji histone demethylase activity assays
Nuclear extracts were isolated from the left ventricle using EpiQuik Nuclear Extraction Kit (Epigentek; OP-0002-1) according to the manufacturer's instructions. Nuclear extracts were used as the enzyme source to measure demethylase activity on exogenous H3K4me3 or H3K9me3 biotinylated histone peptide substrates as previously described (36, 37, 38), using Epigenase JARID Demethylase Activity/Inhibition Assay Kit (Epigentek; P-3083) for H3K4me3 demethylation or Epigenase JMJD2 Demethylase Activity/Inhibition Assay Kit (Epigentek; P-3081) for H3K9me3 demethylation. The reaction was carried out in 50 μl reactions with 1 mM of α-ketoglutarate, 2 mM of ascorbate, 100 μM of FeSO4.7H2O (Sigma; F7002), 50 ng biotinylated histone peptide substrate (either H3K9me3 or H3K4me3), 0.25× of EDTA-free protease inhibitor (Roche; 05892791001), and 20 μg nuclear extract at 37 °C for 2 h. A mix of nuclear extracts that were subjected to heat inactivation at 95 °C for 10 min and then cooled to 25 °C was used as the negative control. Activity was determined by subtracting the values obtained from the heat-inactive samples from the active nuclear extracts for reactions performed at the same time. Jumonji activity of JIB-04-treated mice reported corresponds to female animals.
HDAC activity assays
Nuclear extracts were prepared as aforementioned. HDAC activity assays were performed using Millipore's HDAC Fluorometric Assay Kit (Millipore; 17-356) according to the manufacturer's protocol. Briefly, 5 μg of nuclear extracts was used as the enzyme source for HDAC reactions in the absence or the presence of 2 μM of HDAC inhibitor trichostatin A (TSA) at room temperature for 15 min. Activity of a nuclear extract is determined by subtracting the reading from the reaction in the absence of TSA to the one with TSA.
Western blots
Hearts were collected, blood flushed out, and atria removed. Ventricles were ground in the presence of liquid nitrogen to generate ventricle powders or used fresh. Nuclear extracts were prepared as described in the Jumonji histone trimethyl demethylase activity assay section, using either freshly collected left ventricle or ventricle powder as the starting material. Whole cell lysates prepared from freshly collected left ventricle or ventricle powder using T-Per (Thermo Scientific; 78510) according to the manufacturer's instructions in the presence of complete proteinase inhibitor (Roche; 11836153001). Proteins were separated in 4 to 15% (Bio-Rad; 465-1083) or 4 to 20% polyacrylamide (Bio-Rad; 465-1093) gradient gels, transferred on to polyvinylidene fluoride membranes, and probed with the indicated antibodies. The primary antibodies that were used for Western blots in this study are Kdm4c (Abcam; ab85454), Kdm4d (Cayman Chemical; 10383), Kdm5a (Cell Signaling; 3786S), Kdm5b (Abcam; ab181089), Kdm5c (Abcam; ab190181), Gapdh (Gene Tex; GTX627408), Histone H3 (Abcam; ab12079), Histone H2B (Cell Signaling; 12364S), tubulin (Sigma; T5168), Fhl1 (ProteinTech; 10991-1-AP), and Rcan1 (DSCR1) (Sigma; D6694). Secondary antibodies that were used in this study were anti-rabbit immunoglobulin G (IgG), horseradish peroxidase (HRP) linked (Cell Signaling; 7074), antimouse IgG, HRP linked (Cell Signaling; 7076), antigoat IgG, HRP linked (Santa Cruz; sc-2020), IRDye680 Donkey antigoat IgG (H + L) (LI-COR; P/N 926-68074). Membranes were imaged in a LICOR ODYSSEY Fc. Quantification was performed using Image Studio Lite, version 5.2 (Li-COR Biosciences).
RNA-Seq and gene expression data analysis
RNA was isolated using Trizol (Thermo Scientific; 15596018) from the left ventricle of Tg hearts from animals 125 to 158 days old, which were treated with either JIB-04 or vehicle, starting at 28 days of age, according to the manufacturer's instructions. cDNAs were synthesized by Novogene, Inc or in house using TruSeq Stranded mRNA Library Preparation kits (Ilumina; RS-122-2101, RS-122-2102) according to manufacturer's protocol. Library quality was validated on an Agilent 2100 Bioanalyzer prior to sequencing on an Illumina NextSeq 500 (McDermott Next Generation Sequencing Core, UTSW) at an average of 37,000,000 reads per sample. Raw reads were processed, data were normalized, low expressor genes were removed, and differential expression of protein-coding genes was then calculated. For published datasets, raw normalized data were downloaded from Gene Expression Omnibus (GEO) and similarly analyzed. For microarray data, genes that had detection p value for all samples >0.05 were removed. CnATg/Wt were obtained from (39) and from our own RNA-Seq experiments. Further details including n for each group are given in the relevant figure legends. All original datasets have been deposited with GEO (GSE169078, GSE168847, and GSE175711).
GSEA analysis and Venn and pie diagrams
Fold-change ratios (CnATg/WT [two experimental sets], Veh/JIB-04, TAC/sham [four distinct experimental sets], Kdm Tg/WT, or Kdm KO/WT) were used to perform GSEA preranked analysis of hallmarks, curated datasets, Gene Ontology (GO), and motifs. CnTg/Wt sets used included our own RNA-Seq data (CnTg n = 5; Wt n = 4 animals) and published microarray data (39). TAC/sham model 1 used Wt mice of Kdm3a KO (15); TAC/sham model 2 used WT mice treated with vehicle (15); TAC/sham model 3 used WT mice of Kdm3a Tg (15); and TAC/sham model 4 used WT mice of Kdm4a Tg (14). Gene sets reported showed significant reciprocal enrichment or depletion across groups. GEO accession numbers for data used are GSE27689, GSE169645, GSE120739, GSE120737, for KDM genetic models +/− TAC; GSE120598 for TAC models; GSE168847, GSE169078, and GSE175711 for CnATg model. The ID numbers for the gene sets reported here are as follows: Hallmark myogenesis M5909, G2 M checkpoint M5901, glycolysis M5937, interferon alpha response M5911, interfering gamma response M5913, fatty acid metabolism M5935, E2F targets M5925, mTORC1 signaling M5924, EMT M5930, angiogenesis M5944, LVAD support of failing heart M12300, reactome fatty acid metabolism M27854, muscle contraction M12628 or GO: 0006936, 2M14073 or GO: 0070252, M6298 or GO: 0043292 and M14929 or GO: 0003015. MEF2 motifs include M11407, M17361, M15929, M9937, and M16022.
Overlapping Venn diagram analyses were performed using BioVenn (http://www.biovenn.nl/) (57). Disease genes in Figure 3C contained core-enriched genes as defined by GSEA for TAC/sham plus CnATg/Wt. In Figure 3F, CnTg/Wt genes upregulated by at least twofold were considered, in addition to GSEA core-enriched genes in at least two of the TAC/sham models. In Figure 3I, genes upregulated in disease were defined by ≥1.6 across disease models. For Figure 3L, disease genes were defined by ≥1.5 for at least two TAC models plus CnTg. These cutoffs were more stringent than GSEA core enrichment cutoffs. JIB-04-depleted genes were manually defined as those whose expression changed by ≥0.67 in animals that underwent TAC and were treated with JIB-04 or by GSEA core-depleted genes for CnTg animals treated with JIB-04 in all aforementioned cases. In Figure S4D, disease downregulated genes were defined as GSEA core-depleted genes in at least two TAC/sham models and in CnATg/Wt. JIB-04/vehicle-enriched genes were GSEA core-enriched genes in both TAC and CnATg models. In Figure S4E, disease downregulated genes were defined as in Figure S4D, and JIB-04-enriched genes were defined by ≥1.3-fold increase in TAC and by GSEA enrichment in CnATg. All other GSEA analyses and related Venn diagrams are described in detail in the corresponding figures.
Disease genes at three time points (45, 77, and 125–158 days) were defined by the cutoff ≥1.3 or ≤0.77 fold expression for Tg Ve/Wt Ve; then the lists were overlapped individually with KDM4C targets from Pedersen et al. (43), yielding disease genes that are KDM4C targets at individual time points. Next, the disease genes at the three time points were overlapped to determine Kdm4c target disease genes common to early (45 and/or 77 days) and late (125–158 days) time points termed “throughout disease genes.” Genes that were gradually upregulated or downregulated over time were termed “progressive genes.” Finally, late-onset genes were defined as any gene with ≥1.2-fold or ≤0.8-fold expression in late/early time points. The proportion of throughout, progressive, and late onset genes was used to make the pie chart in Figure 5E, and the expression of those genes at individual time points is shown in Table S9.
Statistical analyses
Unless otherwise indicated in figure legends, p values, comparing groups are from t test, two-tailed, and of unequal variances, calculated in excel. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ns, not significant.
Data availability
All the data described in the article are included within the article or deposited in GEO database accession numbers GSE169078, GSE168847, and GSE175711.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are deeply indebted to Dr Eric Olson and Dr Rhonda Bassel-Duby for the gift of MHC-CnA Tg mice. We thank Dr Nik Munshi and Dr Beverly Rothermel for helpful discussions.
Author contributions
T. G., Z.-P. L., and E. D. M. conceptualization; Z.-P. L. methodology; T. A. T. and Q.-J. Z. validation; T. A. T., Q.-J. Z., L. G., and H. M. formal analysis; T. A. T., Q.-J. Z., L. W., and C. G. investigation; L. G., H. M., T. G., and Z.-P. L. resources; E. D. M. writing–original draft; Z.-P. L. and E. D. M. writing–review & editing; T. A. T. and Q.-J. Z. visualization; Z.-P. L. and E. D. M. supervision; E. D. M. project administration; Z.-P. L. and E. D. M. funding acquisition.
Funding and additional information
This work was partly funded by The Welch Foundation (I-1878 to E. D. M), the American Heart Association (19TPA34910171 to Z. P. L.), and the John P. Perkins, PhD Distinguished Professorship in Biomedical Science Endowment (to E. D. M.).
Edited by Roger Colbran
Contributor Information
Zhi-Ping Liu, Email: zhi-ping.liu@utsouthwestern.edu.
Elisabeth D. Martinez, Email: elisabeth.martinez@utsouthwestern.edu.
Supporting information
References
- 1.Dorn G.W., 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J. Clin. Invest. 2005;115:527–537. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey N., Olson E.N. Cardiac hypertrophy: The good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 3.Maillet M., van Berlo J.H., Molkentin J.D. Molecular basis of physiological heart growth: Fundamental concepts and new players. Nat. Rev. Mol. Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao D.J., Wang Z.V., Battiprolu P.K., Jiang N., Morales C.R., Kong Y., Rothermel B.A., Gillette T.G., Hill J.A. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Nachef D., Oyama K., Wu Y.Y., Freeman M., Zhang Y., MacLellan W.R. Repressive histone methylation regulates cardiac myocyte cell cycle exit. J. Mol. Cell Cardiol. 2018;121:1–12. doi: 10.1016/j.yjmcc.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Z., Lu J., Li J., Wang P., Li Z., Zhong Y., Guo K., Wang J., Ye J., Liu P. JMJD3 inhibition protects against isoproterenol-induced cardiac hypertrophy by suppressing beta-MHC expression. Mol. Cell Endocrinol. 2018;477:1–14. doi: 10.1016/j.mce.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Han K.A., Kang H.S., Lee J.W., Yoo L., Im E., Hong A., Lee Y.J., Shin W.H., Chung K.C. Histone deacetylase 3 promotes RCAN1 stability and nuclear translocation. PLoS One. 2014;9:e105416. doi: 10.1371/journal.pone.0105416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohl M., Wagner M., Reil J.C., Muller S.A., Tauchnitz M., Zimmer A.M., Lehmann L.H., Thiel G., Bohm M., Backs J., Maack C. HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 2013;123:1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C.F., Tang W.H.W. Epigenetics in cardiac hypertrophy and heart failure. JACC Basic Transl. Sci. 2019;4:976–993. doi: 10.1016/j.jacbts.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Wang D.-Z. An epigenetic “LINK(RNA)” to pathological cardiac hypertrophy. Cell Metab. 2014;20:555–557. doi: 10.1016/j.cmet.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Wang X., Bi Y., Bu P., Zhang M. The histone demethylase PHF8 represses cardiac hypertrophy upon pressure overload. Exp. Cell Res. 2015;335:123–134. doi: 10.1016/j.yexcr.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Rabkin S.W., Klassen S.S. Jumonji is a potential regulatory factor mediating nitric oxide-induced modulation of cardiac hypertrophy. J. Cardiovasc. Med. (Hagerstown) 2009;10:206–211. doi: 10.2459/JCM.0b013e3283212ecd. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C.L., McKinsey T.A., Chang S., Antos C.L., Hill J.A., Olson E.N. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q.J., Chen H.Z., Wang L., Liu D.P., Hill J.A., Liu Z.P. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J. Clin. Invest. 2011;121:2447–2456. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q.J., Tran T.A.T., Wang M., Ranek M.J., Kokkonen-Simon K.M., Gao J., Luo X., Tan W., Kyrychenko V., Liao L., Xu J., Hill J.A., Olson E.N., Kass D.A., Martinez E.D., et al. Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis. Nat. Commun. 2018;9:5230. doi: 10.1038/s41467-018-07173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneda R., Takada S., Yamashita Y., Choi Y.L., Nonaka-Sarukawa M., Soda M., Misawa Y., Isomura T., Shimada K., Mano H. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14:69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 17.Movassagh M., Choy M.K., Knowles D.A., Cordeddu L., Haider S., Down T., Siggens L., Vujic A., Simeoni I., Penkett C., Goddard M., Lio P., Bennett M.R., Foo R.S. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilsbach R., Schwaderer M., Preissl S., Gruning B.A., Kranzhofer D., Schneider P., Nuhrenberg T.G., Mulero-Navarro S., Weichenhan D., Braun C., Dressen M., Jacobs A.R., Lahm H., Doenst T., Backofen R., et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun. 2018;9:391. doi: 10.1038/s41467-017-02762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra V., Rothermel B.A. Calcineurin signaling in the heart: The importance of time and place. J. Mol. Cell Cardiol. 2017;103:121–136. doi: 10.1016/j.yjmcc.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D., Ma S., Tan Y., Li D., Tang B., Zhang X., Sun M., Yang Y. Increased expression of calpain and elevated activity of calcineurin in the myocardium of patients with congestive heart failure. Int. J. Mol. Med. 2010;26:159–164. doi: 10.3892/ijmm_00000448. [DOI] [PubMed] [Google Scholar]
- 21.Cao J.L., Yang Y.Q., Nabeel D.M., Sun Y.L., Zou H.Y., Kong X.Q., Lu X.Z. Correlation between serum calcineurin activity and left ventricular hypertrophy in hypertensive patients and its clinical significance. Cardiology. 2018;139:124–131. doi: 10.1159/000481280. [DOI] [PubMed] [Google Scholar]
- 22.Wang J.C., Zhao Y., Li X.D., Zhou N.N., Sun H., Sun Y.Y. Proteolysis by endogenous calpain I leads to the activation of calcineurin in human heart. Clin. Lab. 2012;58:1145–1152. [PubMed] [Google Scholar]
- 23.Burkard N., Becher J., Heindl C., Neyses L., Schuh K., Ritter O. Targeted proteolysis sustains calcineurin activation. Circulation. 2005;111:1045–1053. doi: 10.1161/01.CIR.0000156458.80515.F7. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin J.D., Lu J.R., Antos C.L., Markham B., Richardson J., Robbins J., Grant S.R., Olson E.N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semeniuk L.M., Severson D.L., Kryski A.J., Swirp S.L., Molkentin J.D., Duff H.J. Time-dependent systolic and diastolic function in mice overexpressing calcineurin. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H425–H430. doi: 10.1152/ajpheart.00546.2002. [DOI] [PubMed] [Google Scholar]
- 26.Bourajjaj M., Armand A.S., da Costa Martins P.A., Weijts B., van der Nagel R., Heeneman S., Wehrens X.H., De Windt L.J. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J. Biol. Chem. 2008;283:22295–22303. doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- 27.Weinheimer C.J., Lai L., Kelly D.P., Kovacs A. Novel mouse model of left ventricular pressure overload and infarction causing predictable ventricular remodelling and progression to heart failure. Clin. Exp. Pharmacol. Physiol. 2015;42:33–40. doi: 10.1111/1440-1681.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong D., Duan Y., Guo J., Roach D.E., Swirp S.L., Wang L., Lees-Miller J.P., Sheldon R.S., Molkentin J.D., Duff H.J. Overexpression of calcineurin in mouse causes sudden cardiac death associated with decreased density of K+ channels. Cardiovasc. Res. 2003;57:320–332. doi: 10.1016/s0008-6363(02)00661-2. [DOI] [PubMed] [Google Scholar]
- 29.Grant A.O. Cardiac ion channels. Circ. Arrhythm. Electrophysiol. 2009;2:185–194. doi: 10.1161/CIRCEP.108.789081. [DOI] [PubMed] [Google Scholar]
- 30.Outdit G.Y., Backx Peter H. In: Cardiac Electrophysiology: From Cell to Bedside. Douglas J.J., Zipes P., Stevenson W.G., editors. Elsevier; Amsterdam: 2018. Voltage-gated potassium channels; pp. 25–37. [Google Scholar]
- 31.Kushnir A.a.M., S O. In: Cardiac Electrophysiology. Douglas J.J., Zipes P., Stevenson W.G., editors. Elsevier; Amsterdam: 2018. Voltage-gated calcium channels; pp. 12–24. [Google Scholar]
- 32.Brahma M.K., Adam R.C., Pollak N.M., Jaeger D., Zierler K.A., Pocher N., Schreiber R., Romauch M., Moustafa T., Eder S., Ruelicke T., Preiss-Landl K., Lass A., Zechner R., Haemmerle G. Fibroblast growth factor 21 is induced upon cardiac stress and alters cardiac lipid homeostasis. J. Lipid Res. 2014;55:2229–2241. doi: 10.1194/jlr.M044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planavila A., Redondo I., Hondares E., Vinciguerra M., Munts C., Iglesias R., Gabrielli L.A., Sitges M., Giralt M., van Bilsen M., Villarroya F. Fibroblast growth factor 21 protects against cardiac hypertrophy in mice. Nat. Commun. 2013;4:2019. doi: 10.1038/ncomms3019. [DOI] [PubMed] [Google Scholar]
- 34.Yu S., Li Y., Zhao H., Wang Q., Chen P. The histone demethylase JMJD1C regulates CAMKK2-AMPK signaling to participate in cardiac hypertrophy. Front Physiol. 2020;11:539. doi: 10.3389/fphys.2020.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheikh F., Raskin A., Chu P.H., Lange S., Domenighetti A.A., Zheng M., Liang X., Zhang T., Yajima T., Gu Y., Dalton N.D., Mahata S.K., Dorn G.W., 2nd, Brown J.H., Peterson K.L., et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J. Clin. Invest. 2008;118:3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Chang J., Varghese D., Dellinger M., Kumar S., Best A.M., Ruiz J., Bruick R., Pena-Llopis S., Xu J., Babinski D.J., Frantz D.E., Brekken R.A., Quinn A.M., Simeonov A., et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayo J., Tran T.A., Wang L., Pena-Llopis S., Das A.K., Martinez E.D. Jumonji inhibitors overcome radioresistance in cancer through changes in H3K4 methylation at double-strand breaks. Cell Rep. 2018;25:1040–1050.e5. doi: 10.1016/j.celrep.2018.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalvi M.P., Wang L., Zhong R., Kollipara R.K., Park H., Bayo J., Yenerall P., Zhou Y., Timmons B.C., Rodriguez-Canales J., Behrens C., Mino B., Villalobos P., Parra E.R., Suraokar M., et al. Taxane-platin-resistant lung cancers co-develop hypersensitivity to JumonjiC demethylase inhibitors. Cell Rep. 2017;19:1669–1684. doi: 10.1016/j.celrep.2017.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bousette N., Chugh S., Fong V., Isserlin R., Kim K.H., Volchuk A., Backx P.H., Liu P., Kislinger T., MacLennan D.H., Emili A., Gramolini A.O. Constitutively active calcineurin induces cardiac endoplasmic reticulum stress and protects against apoptosis that is mediated by alpha-crystallin-B. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18481–18486. doi: 10.1073/pnas.1013555107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada-Aviles R., Rodriguez G., Zarain-Herzberg A. The cardiac calsequestrin gene transcription is modulated at the promoter by NFAT and MEF-2 transcription factors. PLoS One. 2017;12:e0184724. doi: 10.1371/journal.pone.0184724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putt M.E., Hannenhalli S., Lu Y., Haines P., Chandrupatla H.R., Morrisey E.E., Margulies K.B., Cappola T.P. Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ. Cardiovasc. Genet. 2009;2:212–219. doi: 10.1161/CIRCGENETICS.108.816686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao A., Luo C., Hogan P.G. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen M.T., Agger K., Laugesen A., Johansen J.V., Cloos P.A., Christensen J., Helin K. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 2014;34:1031–1045. doi: 10.1128/MCB.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovacic J.C., Mercader N., Torres M., Boehm M., Fuster V. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: From cardiovascular development to disease. Circulation. 2012;125:1795–1808. doi: 10.1161/CIRCULATIONAHA.111.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovacic J.C., Dimmeler S., Harvey R.P., Finkel T., Aikawa E., Krenning G., Baker A.H. Endothelial to mesenchymal transition in cardiovascular disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019;73:190–209. doi: 10.1016/j.jacc.2018.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hlaing M., Spitz P., Padmanabhan K., Cabezas B., Barker C.S., Bernstein H.S. E2F-1 regulates the expression of a subset of target genes during skeletal myoblast hypertrophy. J. Biol. Chem. 2004;279:43625–43633. doi: 10.1074/jbc.M408391200. [DOI] [PubMed] [Google Scholar]
- 47.Sciarretta S., Volpe M., Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ. Res. 2014;114:549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Wang J., Wu Y., Li W., Gong K., Zhao P. Identification of SLED1 as a potential predictive biomarker and therapeutic target of post-infarct heart failure by bioinformatics analyses. Int. Heart J. 2021;62:23–32. doi: 10.1536/ihj.20-439. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z., Dudek J., Maack C., Hofmann U. Pharmacological inhibition of GLUT1 as a new immunotherapeutic approach after myocardial infarction. Biochem. Pharmacol. 2021;190:114597. doi: 10.1016/j.bcp.2021.114597. [DOI] [PubMed] [Google Scholar]
- 50.Honka H., Solis-Herrera C., Triplitt C., Norton L., Butler J., DeFronzo R.A. Therapeutic manipulation of myocardial metabolism: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2021;77:2022–2039. doi: 10.1016/S0735-1097(21)03378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rai A., Narisawa M., Li P., Piao L., Li Y., Yang G., Cheng X.W. Adaptive immune disorders in hypertension and heart failure: Focusing on T-cell subset activation and clinical implications. J. Hypertens. 2020;38:1878–1889. doi: 10.1097/HJH.0000000000002456. [DOI] [PubMed] [Google Scholar]
- 52.Bansal S.S., Ismahil M.A., Goel M., Zhou G., Rokosh G., Hamid T., Prabhu S.D. Dysfunctional and proinflammatory regulatory T-lymphocytes are essential for adverse cardiac remodeling in ischemic cardiomyopathy. Circulation. 2019;139:206–221. doi: 10.1161/CIRCULATIONAHA.118.036065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang L., Ellims A.H., Beale A.L., Taylor A.J., Murphy A., Dart A.M. Systemic inflammation is associated with myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy in patients with hypertrophic cardiomyopathy. Am. J. Transl. Res. 2017;9:5063–5073. [PMC free article] [PubMed] [Google Scholar]
- 54.Coste Pradas J., Auguste G., Matkovich S.J., Lombardi R., Nee Chen S., Garnett T., Chamberlain K., Mahmud Riyad J., Weber T., Singh S.K., Robertson M.J., Coarfa C., Marian A.J., Gurha P. Identification of genes and pathways regulated by lamin A in heart. J. Am. Heart Assoc. 2020;9:e015690. doi: 10.1161/JAHA.119.015690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S., Frisk C., Eriksson M.J., Walentinsson A., Corbascio M., Hage C., Kumar C., Asp M., Lundeberg J., Maret E., Persson H., Linde C., Persson B. Transcriptomics of cardiac biopsies reveals differences in patients with or without diagnostic parameters for heart failure with preserved ejection fraction. Sci. Rep. 2019;9:3179. doi: 10.1038/s41598-019-39445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bookout A.L., Cummins C.L., Mangelsdorf D.J., Pesola J.M., Kramer M.F. High-throughput real-time quantitative reverse transcription PCR. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb1508s73. Chapter 15:Unit 15.8. [DOI] [PubMed] [Google Scholar]
- 57.Hulsen T., de Vlieg J., Alkema W. BioVenn - A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described in the article are included within the article or deposited in GEO database accession numbers GSE169078, GSE168847, and GSE175711.