Abstract
Plasma specimens collected in 1999 from 32 human immunodeficiency virus type 1 (HIV-1)-infected foreigners living in Madrid, Spain, were examined for the presence of non-B subtypes. Furthermore, plasma viremia was quantified using two different AMPLICOR HIV-1 MONITOR tests, version 1.0 and the new upgraded and automated version 1.5 (COBAS). Most patients came from Africa, where they most likely had acquired HIV-1 infection through sexual contact. HIV-1 genetic subtyping was based on the phylogenetic analysis of the protease gene. Twenty-two subtype B, six subtype G, two subtype C, one subtype A, and one D subtype infection were found. Overall, non-B subtypes represented 31.25% of the study population. Irrespective of the HIV-1 variant, viral load values above the detection limit (200 HIV RNA copies/ml) increased from 56.2 to 71.9% for results obtained using MONITOR version 1.0 and COBAS, respectively. Moreover, significant differences in viral load values (>0.5 logs) were recognized in up to 37.5% of samples. In summary, COBAS seemed to be more reliable for testing plasma viral load in HIV-infected immigrants living in Spain, one third of whom carried non-B subtypes.
Human immunodeficiency virus type 1 (HIV-1) mutates rapidly, contributing to its high degree of genetic heterogeneity in vivo (3). Methods based on nucleotide sequence analyses allow the recognition of phylogenetic relationships between different sequences. So far HIV-1 can be divided into three distinct and highly divergent groups: M (major), O (outlier), and N (new) (19, 25, 30). At least fourteen major genetic variants can be recognized within HIV-1 group M, including several subtypes (A, B, C, D, F, G, H, J, and K) and four major circulating recombinant forms (CRF01-AE, CRF02-AG, CRF03-AB, and CRF04-cpx, “complex”) (19, 33). Classification of HIV-1 into subtypes is based primarily on the analysis of genetic sequences coding for the envelope (env) and other structural (gag, pol) proteins.
In Spain, as in North America and other Western European countries, HIV-1 subtype B is the most prevalent HIV-1 variant (15). Non-B subtypes have been reported mainly in Africa, where a large diversity of HIV-1 variants has been found (18). However, the prevalence of HIV-1 non-B subtypes seems to be increasing in North America (24, 34) and Europe (6, 10, 15, 16, 24), and limitations of the current commercial HIV-1 quantitation assays examining these specimens have been pointed out recently (1, 2, 17, 32), since these tests were originally designed on the basis of HIV-1 subtype B sequences. Herein we investigate the prevalence of HIV-1 subtypes in a group of 32 infected foreigners living in Madrid, Spain, and analyze the performance of two different versions of the AMPLICOR HIV-1 MONITOR test in samples belonging to these subjects.
MATERIALS AND METHODS
Blood specimens from 32 HIV-1-infected immigrants attending one HIV unit located in Madrid were collected in 1999. Twenty-seven (84.4%) were from Africa, four (12.5%) were from South America, and one (3.1%) was from Eastern Europe. Epidemiological and clinical data are summarized in Table 1. Plasma aliquots were separated from blood cells within 4 h following phlebotomy and were frozen at −80°C until the time of analysis. The CD4+ lymphocyte count was analyzed by flow cytometry (Coulter, Barcelona, Spain).
TABLE 1.
Epidemiological features of the study population and genetic heterogeneity in amino acid positions of the protease gene product associated with resistance
| Sample code | Gendera | Age | Clinical status | Country of birthb | Country of diagnosis | Transmission routec | CD4 count (cells/μl) | Antiretroviral therapy (months)d | Amino acid changes of the protease gene producte | Subtype | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1158 | M | 24 | A3 | Morocco | Spain | Htsex | 232 | Naive | L63T | B | AF247007 |
| M432 | M | 32 | A2 | Bulgaria | Spain | Htsex | 531 | Naive | None | B | AF247008 |
| M2426 | F | 8 | C2 | EG | EG | Vertical | 100 | Naive | V77I | B | AF247009 |
| M2131 | F | 24 | A1 | Nigeria | Spain | Htsex | 988 | Naive | V77I | B | AF247010 |
| M513 | M | 33 | A2 | Peru | Spain | Htsex | 478 | Naive | None | B | AF247011 |
| M2385 | F | 31 | A2 | EG | Spain | Htsex | 487 | Naive | None | B | AF247012 |
| M1240 | M | 6 | B2 | Honduras | Honduras | Htsex | 606 | Naive | L101, L63T | B | AF247013 |
| M927 | F | 28 | A1 | EG | EG | Htsex | 785 | Naive | L63S, V77I | B | AF247014 |
| M1642 | M | 33 | A3 | Morocco | Spain | Htsex | 198 | Naive | L10I, L63P | B | AF247015 |
| M963 | M | 37 | B3 | Ecuador | Spain | Homo | 95 | Naive | M36I | B | AF247016 |
| M2067 | F | 23 | A1 | EG | Spain | Htsex | 737 | Naive | M36L, L63P | B | AF247017 |
| M2133 | F | 39 | A1 | EG | Spain | Htsex | 847 | Naive | M36I | A | AF247018 |
| M959 | F | 34 | A2 | EG | Spain | Htsex | 309 | Naive | K20R, M36I, L63A | C | AF247019 |
| M2489 | F | 20 | A1 | EG | EG | Htsex | 916 | Naive | M36I, L63A | C | AF247020 |
| M1296 | F | 31 | A3 | EG | EG | Htsex | 94 | Naive | M36I, L63S | D | AF247021 |
| M1911 | F | 33 | A3 | Zaire | Spain | Htsex | 247 | Naive | L10I, K20I, M36I, V77I | G | AF247022 |
| M2444 | F | 35 | A2 | EG | EG | Htsex | 471 | Naive | L10S, K20I, M36I, L63S, V77I | G | AF247023 |
| M916 | F | 30 | A2 | EG | Spain | Htsex | 406 | Naive | K20I, M36I | G | AF247024 |
| M635 | F | 43 | C3 | EG | Spain | Htsex | 142 | AZT + 3TC (11) | M46L, L63P | B | AF247025 |
| M2671 | F | 32 | A3 | Nigeria | Spain | Htsex | 374 | AZT + 3TC (29) | M36I, V77I | B | AF247026 |
| M2389 | F | 64 | A2 | EG | Spain | Htsex | 399 | AZT + ddI (24) | L10I, K20M, M36I, M46L, L63P, A71V, I84V | B | AF247027 |
| M2558 | F | 30 | A2 | EG | Spain | Htsex | 343 | AZT + ddC (30) | K20I, M36I, L63P | G | AF247028 |
| M787 | M | 48 | A2 | Nigeria | Spain | Htsex | 467 | 3TC + d4T (13) | L10I, K20M, M36I, M46L, L63P, A71V, L90M | B | AF247029 |
| M2814 | F | 39 | A2 | EG | Spain | Htsex | 504 | AZT + 3TC + NVP (9) | L10I, K20M, M36I, M46L, L63P, A71V, I84V, L90M | B | AF247030 |
| M1303 | F | 9 | A1 | Zaire | Spain | Vertical | 450 | 3TC + d4T + RTV (5) | M36I | B | AF247031 |
| M430 | F | 27 | A3 | Nigeria | Spain | Htsex | 140 | 3TC + d4T + RTV (7) | L10I, M36I, M46L, L63P, A71V, L90M | B | AF247032 |
| M2346 | F | 28 | B3 | EG | Spain | Htsex | 28 | 3TC + d4T + IDV (16) | L63P, V77I | B | AF247033 |
| M2725 | F | 33 | B3 | Zaire | Spain | Htsex | 10 | AZT + 3TC + IDV (16) | L10I, K20M, M36I, M46L, L63P, A71V, L90M | B | AF247034 |
| M2686 | M | 26 | A3 | Ecuador | Spain | Htsex | 427 | AZT + 3TC + SQV (30) | L10I, K20M, M36I, M46L, L63P, A71V | B | AF247035 |
| M2388 | F | 32 | A2 | Cape Verde | Spain | Htsex | 535 | AZT + ddI + NVP (7) | L10S, K20I, M36I, L63P, V82I | G | AF247036 |
| M2773 | M | 36 | C3 | Cameroon | Cameroon | Htsex | 161 | D4T + NVP + NFV (8) | K20I, M36I | G | AF247037 |
| M1743 | M | 27 | C3 | Nigeria | Spain | Htsex | 134 | ddI + NVP + SQV + NFV (15) | M36I, L63P | B | AF247038 |
F, female; M, male.
EG, Equatorial Guinea.
Htsex, heterosexual; Homo, homosexual.
AZT, zidovudine; 3TC, lamivudine; d4T, stavudine; ddI, didanosine; NVP, nevirapine; RTV, ritonavir; IDV, indinavir; SQV, saquinavir; NFV, nelfinavir.
Amino acid codons at positions 10, 20, 30, 36, 46, 48, 50, 54, 63, 71, 77, 82, 84, and 90 were analyzed.
The characterization of HIV-1 subtypes was performed by phylogenetic analysis of the protease gene as previously described (16). We used 21 HIV-1 reference sequences belonging to HIV-1 groups M and N having full-length genomes available at GenBank. The tree topology was obtained using the neighbor-joining program (27). Alignment of DNA sequences was performed using the CLUSTAL W method (31). Pairwise distance matrices were estimated using the Kimura two-parameter model with the DNADIST program, as implemented in the PHYLIP software package (13). Bootstrap resampling (1,000 data sets) of the multiple alignment was done to test the statistical robustness of the tree.
Plasma viremia was quantified using the two different versions of the AMPLICOR HIV-1 MONITOR test (Roche Diagnostics, Barcelona, Spain), a reverse transcription-PCR-based assay designed for quantifying HIV-1 RNA in plasma (22). Version 1.0 was the original commercial test. The other one was a prototype automated procedure of version 1.5 (COBAS), in which HIV-1 RNA amplification and detection take place on the COBAS AMPLICOR instrument (9). Both methods differ in the primers used for reverse transcription and PCR, the composition of the reverse transcription-PCR mixture, the thermal cycling parameters, and the internal quantification standard RNA (21, 32). Hypothetically, COBAS provides more reliable viral load data, since it is substantially less influenced by viral subtype (21).
Nucleotide sequence accession numbers.
Protease sequences have been submitted to the GenBank database with the accession numbers AF247007 to AF247038.
RESULTS
HIV-1 genetic subtype characterization and main epidemiological features.
The presence of HIV-1 non-B subtypes in Spain has been reported previously (15, 16). All patients enrolled in this study were HIV-1-infected foreigners living in Madrid, mostly coming from African countries (84.4%) where most HIV-1 subtypes cocirculate (18). Interestingly, although all 32 subjects had probably acquired HIV-1 infection in their country of origin, in 78% of the cases their first diagnosis was in Spain.
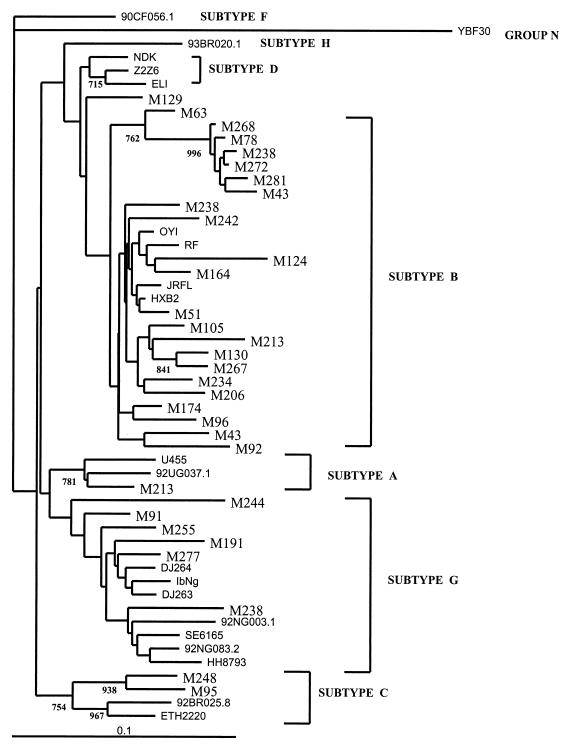
The epidemiological features of the study population and the assignment of its genetic HIV-1 subtype are summarized in Table 1. Twenty-two subtype B, six subtype G, two subtype C, one subtype A, and one D subtype infection were found. Phylogenetic tree topology was supported by high bootstrap values (Fig. 1). Although sample no. M1296 could not be assigned initially to a specific known subtype, when the tree was performed exclusively with this sample and the reference strains, this specimen clustered within subtype D variants (data not shown).
FIG. 1.
Phylogenetic tree of the HIV-1 protease coding region from 32 foreigners living in Spain (bold). Bootstrap resampling values (1,000 sets) are indicated. The lengths of the branches are proportional to the relative evolutionary distances.
Overall, nearly one third (31.25%) of the HIV-1-infected foreigners living in Madrid carried HIV-1 non-B subtypes. All of them were African. All but one were women, and seven of them admitted to being engaged in prostitution. The origins of individuals harboring HIV-1 subtype G were Equatorial Guinea (n = 3), Zaire (n = 1), Cape Verde (n = 1), and Cameroon (n = 1). All subjects carrying subtypes A, C, and D came from Equatorial Guinea, a former Spanish colony.
Drug resistance mutations in non-B subtypes.
Genetic changes associated with resistance to protease inhibitors were recognized in both naive and pretreated individuals carrying HIV-1 non-B subtypes (Table 1). For example, the secondary substitution Met36→Ile was found in all seven naive subjects infected with non-B subtypes, while the change Val77→Ile was seen in two subtype G specimens. These changes have been associated with resistance to nelfinavir and ritonavir, respectively (29). On the other hand, drug resistance mutations in subjects with non-B subtypes under antiretroviral therapy appeared at the same positions as those that were reported for subtype B (29) (Table 1).
Performance of viral load tests.
Regardless of the HIV-1 variant, positive quantitative values above the detection limit (200 HIV-RNA copies/ml) increased from 56.2% for MONITOR version 1.0 to 71.9% for COBAS (Student's t test, P < 0.05) (Table 2). On average, the newer method outperformed version 1.0 in all aspects of HIV-1 testing. The differences between the geometric mean titers of version 1.0 (341,401) and COBAS (539,009) were found to be statistically significant (Fisher exact test, P = 0.05).
TABLE 2.
Differences between HIV-1 plasma viral load results provided by AMPLICOR HIV-1 MONITOR version 1.0 and COBAS
| Sample code | Genetic subtype | Viral load results (titers) for:
|
Difference of COBAS vs. version 1.0 (log) | |
|---|---|---|---|---|
| Version 1.0 | COBAS | |||
| M2133 | A | <200 | 3,650 | >1.26 |
| M2489 | C | 32,700 | 6,720 | <0.68 |
| M959 | C | 44,600 | 19,600 | <0.36 |
| M1296 | D | 59,300 | 337,000 | >0.75 |
| M916 | G | <200 | 99,500 | >2.69 |
| M2444 | G | 4,300 | 4,230 | <0.0004 |
| M2388 | G | 5,800 | 10,300 | >0.25 |
| M1911 | G | 28,700 | 222,000 | >0.88 |
| M2558 | G | 146,000 | 82,200 | <0.25 |
| M2773 | G | 2,349,500 | 9,160,000 | >0.59 |
| M2131 | B | <200 | <200 | 0 |
| M2067 | B | <200 | <200 | 0 |
| M2671 | B | <200 | <200 | 0 |
| M2389 | B | <200 | <200 | 0 |
| M787 | B | <200 | <200 | 0 |
| M2814 | B | <200 | <200 | 0 |
| M2346 | B | <200 | <200 | 0 |
| M2725 | B | <200 | <200 | 0 |
| M2686 | B | <200 | <200 | 0 |
| M430 | B | <200 | 667 | >0.52 |
| M635 | B | <200 | 8,100 | >1.61 |
| M1303 | B | <200 | 20,500 | >2.01 |
| M927 | B | 8,900 | 4,330 | <0.31 |
| M432 | B | 36,700 | 53,700 | >0.15 |
| M2426 | B | 47,100 | 138,000 | >0.46 |
| M2385 | B | 60,600 | 176,000 | >0.45 |
| M513 | B | 90,300 | 1,530,000 | >1.23 |
| M963 | B | 95,350 | 543,000 | >0.75 |
| M1240 | B | 198,300 | 201,000 | >0.03 |
| M1158 | B | 213,200 | 182,000 | <0.07 |
| M1642 | B | 2,340,000 | 56,000 | <1.61 |
| M1743 | B | 5,160,700 | 4,260,000 | <0.08 |
Differences in viral load values above 0.5 logs were considered significant and were recognized in up to 37.5% of cases. They occurred more frequently in non-B rather than subtype B specimens (60 versus 27.3%) (Table 2). One specimen belonging to subtype G (M916) yielded repeated discrepancies in plasma viremia above 2.5 logs. However, viral load values were significantly higher using version 1.0 in 4 of 10 HIV-1 non-B subtype specimens (Table 2). With respect to subtype B specimens, 41% showed significantly higher values using COBAS, and 18.2% showed higher values using version 1.0.
DISCUSSION
Performance of two different versions of the AMPLICOR HIV-1 MONITOR test.
Quantitative values of plasma viremia using the currently available viral load assays can be unreliable for testing non-B subtypes or recombinant forms of HIV-1 (1, 5, 7, 11, 17). The AMPLICOR HIV-1 MONITOR test, version 1.0, was developed when little sequence information on HIV-1 subtypes was available (22). The primers used (SK431 and SK462) were designed on the basis of a subtype B consensus sequence (22). This fact explains the low performance of this assay for testing non-B subtypes. It is estimated that HIV-1 subtypes A, E, F, and G were underestimated by 10-fold or more. The use of COBAS, which was developed to minimize subtype-related variation (21), seemed to allow an equivalent quantitation of HIV-1 RNA regardless of the subtype. It uses a set of primers (SK145 and SKCC1B) which are based on a non-B subtype consensus sequence (21, 32). The lower viremia values provided by COBAS in 40% of samples from subjects carrying non-B subtypes and in 18.2% of those with subtype B found in our study was an unexpected finding. It could be due to the difference in how the amplicons are captured (microwells versus microbeads) by the detection reagents used in the prototype automated test or the presence of mismatches in sample primer binding sites. The recognition of higher levels of HIV-1 plasma viremia by either assay in two subjects carrying subtypes G (M2773) and B (M1743) and being under triple or quadruple antiretroviral combinations was also unexpected, but it might be explained by noncompliance with the prescribed treatment noticed in these subjects.
The high number of specimens showing significant differences (>0.5 log) in viral load values between the AMPLICOR quantification versions strongly reinforces the importance of always monitoring HIV-1-infected patients with the same version of any viral load quantitation technique (17).
Epidemiological implications of the spreading of non-B subtypes.
The presence of HIV-1 non-B subtypes in Spain has been reported previously (15, 16). However, in our study nearly one third of the population (31.25%) carried non-B subtypes. All 10 subjects infected with non-B subtypes came from Africa, where a large variety of HIV-1 subtypes and recombinant forms are circulating (18). Seven of those 10 patients with HIV-1 non-B subtypes were identified as prostitutes. The reported promiscuity of the three other individuals or their sexual partners should remain in question. Spreading of these minor HIV variants among native individuals in Spain is of particular concern, as it seems to happen in other European countries (10) and the United States (34). The primary diagnosis of more than three-fourths of these patients occurred in Spain. This reinforces the fact that HIV testing should be offered to all persons belonging to high-risk groups and/or emigrating from regions of high endemicity where testing is not available.
The spread of different HIV-1 subtypes in a single geographic region coupled with intersubtype recombination (16, 26) has serious implications for the efforts to control the AIDS pandemic. The impacts of the different genetic subtypes on pathogenesis, the course of HIV infection, transmissibility, vaccine efficacy, and diagnosis based on serologic (20) or PCR assays (2) are not yet well known and must be further studied (12). It has been previously reported that susceptibility to antiretroviral drugs might differ between distinct subtypes (8, 23). In this study we have shown that mutations associated with resistance to protease inhibitors appear in HIV-1 non-B subtypes at the same positions that they do in subtype B strains. This observation suggests that the distinct HIV-1 subtypes evolve convergently at the genetic level when antiretroviral drugs act as selective forces (28).
Studies designed to monitor the spread of HIV-1 subtypes should be encouraged. In areas where non-B subtypes represent a significant proportion of infections, viral load quantitation tests able to appropriately recognize the different viral variants should be implemented.
ACKNOWLEDGMENTS
This work was funded in part by grants from Instituto de Salud Carlos III, Comunidad Autónoma de Madrid (CAM), and Asociación Investigación y Educación en SIDA (AIES).
REFERENCES
- 1.Alaeus A, Lidman K, Sönnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Arnold C, Barlow K, Kaye S, Loveday C, Balfe P, Clewley J. HIV type 1 sequence subtype G transmission from mother to infant: failure of variant sequence species to amplify in the Roche Amplicor test. AIDS Res Hum Retrovir. 1995;11:999–1001. doi: 10.1089/aid.1995.11.999. [DOI] [PubMed] [Google Scholar]
- 3.Coffin J. HIV population dynamic in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 4.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J The UNAIDS Network for HIV Isolation and Characterization. HIV-1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J-P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of HIV-1 RNA in plasma. J Med Virol. 1996;50:293–302. doi: 10.1002/(SICI)1096-9071(199612)50:4<293::AID-JMV3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Couturier E, Damon F, Roques P, Flevry H, Barin F, Brunet J, Brun-Vézinet F, Simon F the AC II Laboratory Network. HIV-1 diversity in France, 1996–1998. AIDS. 2000;14:289–296. doi: 10.1097/00002030-200002180-00011. [DOI] [PubMed] [Google Scholar]
- 7.Debyser Z, Van Wijngaerden E, Laethem K, Beuselinck K, Reynders M, De Clercq E, Desmyter J, Vandamme A. Failure to quantify viral load with two of the three commercial methods in a pregnant woman harboring an HIV-1 type 1 subtype G strain. AIDS Res Hum Retrovir. 1998;14:453–459. doi: 10.1089/aid.1998.14.453. [DOI] [PubMed] [Google Scholar]
- 8.Descamps D, Apetrei C, Collin G, Damond F, Simon F, Brun-Vézinet F. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS. 1998;12:1109–1111. [PubMed] [Google Scholar]
- 9.Di Domenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z, Rosentraus M. COBAS Amplicor: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chemother. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 10.Dietrich U, Ruppach H, Gehring S, Knechten H, Knickmann M, Jäger H, Wolf E, Husak R, Orfanos C, Brede H, Rübsamen-Waigmann H, Von Briesen H. Large proportion of non-B subtypes and presence of zidovudine resistance mutations among German seroconverters. AIDS. 1997;11:1532–1533. [PubMed] [Google Scholar]
- 11.Dunne A, Crowe S. Comparison of branched DNA and reverse transcriptase PCR for quantifying six different HIV-1 subtypes in plasma. AIDS. 1997;11:126–127. [PubMed] [Google Scholar]
- 12.Expert Group of the Joint United Nations Programme on HIV/AIDS. Implications of HIV variability for transmission: science and politic issues. AIDS. 1997;11:1–15. [PubMed] [Google Scholar]
- 13.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1992. [Google Scholar]
- 14.Gobbers E, Fransen K, Oosterlaken T, Janssens W, Heyndrickx L, Ivens T, Vereecken K, van de Wiel P, van der Groen G. Reactivity and amplification system with regard to different HIV-1 subtypes. J Virol Methods. 1997;66:293–301. doi: 10.1016/s0166-0934(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 15.Holguín A, Rodés B, Dietrich U, Soriano V. Human immunodeficiency viruses type 1 subtypes circulating in Spain. J Med Virol. 1999;53:856–860. [PubMed] [Google Scholar]
- 16.Holguín A, Rodés B, Soriano V. Recombinant human immunodeficiency viruses type 1 circulating in Spain. AIDS Res Hum Retrovir. 2000;16:501–511. doi: 10.1089/088922200309179. [DOI] [PubMed] [Google Scholar]
- 17.Holguín A, de Mendoza C, Soriano V. Comparison of three commercial methods for quantification of plasma viraemia in clinical specimens belonging to HIV-1 non-B subtypes. Eur J Clin Microbiol Infect Dis. 1999;18:256–259. doi: 10.1007/s100960050273. [DOI] [PubMed] [Google Scholar]
- 18.Janssens W, Buvé A, Nkengasong J. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Korber B, Kuiken C, Foley B, Hanh B, McCutchan F, Mellors J, Sodroski J. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1998. [Google Scholar]
- 20.Loussert-Ajaka I, Ly T, Chaix M, Ingrand D, Saragosti S, Courouce A, Brun-Vézinet F, Simon F. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet. 1994;343:1393–1394. doi: 10.1016/s0140-6736(94)92524-0. [DOI] [PubMed] [Google Scholar]
- 21.Michael N, Herman S, Kwok S, Dreyer K, Wang J, Christopherson C, Spadoro J, Young K, Polonis V, McCutchan F, Carr J, Mascola J, Jagodzinski L, Robb M. Development of calibrated viral load standards for group M subtypes of HIV-1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of HIV-1 RNA: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer S, Alaeus A, Albert J, Cox S. Drug susceptibility of subtypes A, B, C, D, and E HIV-1 primary isolates. AIDS Res Hum Retrovir. 1998;14:157–162. doi: 10.1089/aid.1998.14.157. [DOI] [PubMed] [Google Scholar]
- 24.Paraskevis D, Hatzakis A. Molecular epidemiology of HIV-1 infection. AIDS Reviews. 1999;1:238–249. [Google Scholar]
- 25.Peeters M, Liegeois F, Torimiro N, Bourgeois A, Mpoudi E, Vergne L, Saman E, Delaporte E, Saragosti S. Characterization of a highly replicative intergroup M/O HIV-1 recombinant isolated from a Cameroonian patient. J Virol. 1999;73:7368–7375. doi: 10.1128/jvi.73.9.7368-7375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quiñones-Mateu M, Arts E. Recombination in HIV-1: update and implications. AIDS Reviews. 1999;1:89–100. [Google Scholar]
- 27.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Sato H, Tomita Y, Shibamura K, Shiino T, Miyakuni T, Takebe Y. Convergent evolution of RT genes of HIV-1 subtype E and B following nucleoside analogue RT inhibitor therapies. J Virol. 2000;74:5357–5362. doi: 10.1128/jvi.74.11.5357-5362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schinazi R, Larder B, Mellors J. Mutations in retroviral genes associated with drug resistance: 1999-2000 update. Int Antivir News. 1999;7:46–69. [Google Scholar]
- 30.Simon F, Mauclère P, Roques P, Loussert-Ajaka I, Müller-Trutwin M, Saragosti S, Georges-Courbot M, Barré-Sinoussi F, Brun-Vézinet F. Identification of a new HIV-1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triques K, Coste J, Perret J, Segarra C, Mpoudi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of HIV-1. J Clin Microbiol. 1999;37:110–116. doi: 10.1128/jcm.37.1.110-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triques K, Bourgeois A, Vidal N, Mpoudi-Ngole E, Mulanga-Kabeya C, Nzilambi N, Torimiro N, Saman E, Delaporte E, Peeters M. Near-full-length genome sequencing of divergent African HIV type 1 subtype F viruses leads to the identification of a new HIV type 1 subtype designated K. AIDS Res Hum Retrovir. 2000;16:139–151. doi: 10.1089/088922200309485. [DOI] [PubMed] [Google Scholar]
- 34.Weidle P, Ganea C, Irwin K, Pieniazek D, McGowan J, Olivo N, Ramos A, Schable C, Lal R, Holmberg S, Ernst J. Presence of HIV-1 group M, non-B subtypes in Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J Infect Dis. 2000;181:470–475. doi: 10.1086/315253. [DOI] [PubMed] [Google Scholar]