Abstract
Evidence highlights the comorbidity between emotional distress and irritable bowel syndrome (IBS) through the gut-brain axis. However, the underlying mechanism is largely unknown. Thus, the present study aimed to evaluate the associations among neurotransmitter levels and the gut microbiome profiles in persons with IBS and emotional distress. In this nested case-controlled study, emotional symptoms, including anxiety and depressive symptoms, were evaluated in 40 persons with IBS and 20 healthy controls (HC). Plasma neurotransmitters levels (serotonin and norepinephrine) and the gut microbiome profile of the collected fecal samples were examined. Emotional distress and microbiome profile were significantly different between IBS and HC groups. Lower but not significant neurotransmitters’ levels (serotonin and norepinephrine) were observed in the IBS group compared to the HC. A negative correlation was found between norepinephrine levels and alpha diversity (Shannon and Simpson indices) in the IBS group. Moreover, serotonin levels were positively associated with the abundance of Proteobacteria, and norepinephrine were positively correlated with Bacteroidetes, but negatively associated with Firmicutes phylum. The present study demonstrated alteration in the gut microbiome between persons with IBS and emotional distress compared to HC. The correlations between plasma neurotransmitters and the gut microbiome suggest that the gut microbiome may impact the regulation of neurotransmitters.
Subject terms: Medical research, Outcomes research
Introduction
Irritable bowel syndrome (IBS), the most common functional gastrointestinal disorder, is characterized by chronic abdominal pain and alteration in bowel habits1–3. IBS affects 10–15% of adults worldwide and is associated with a broad spectrum of emotional distress comorbidities4. Epidemiological data report that 40–90% of persons with IBS experience some degree of anxiety and depressive symptoms in their life5,6. Emotional distress may influence IBS symptom perception and illness behavior in patients, contribute to poor quality of life as well as increase health care costs7–9. A comorbid condition may suggest a biological association between emotional distress and IBS5.
The relationship between IBS and emotional distress is supported by neuroimaging and psychophysiological studies10,11. This association may be related to the bidirectional communication between the central nervous system (CNS) and the gut neuroendocrine system known as the brain-gut axis4,12. Emotional distress may lead to homeostasis shifts along the brain-gut axis and alteration in the gut microbiome pattern in IBS13. Several studies reported similarities in the fecal microbiome of persons with IBS and persons with depression (e.g., high abundance of Proteobacteria and low abundance of Bifidobacteria), which highlights the bidirectional interaction between the brain and the gut in comorbidity between emotional distress and IBS13,14.
Neurotransmitter imbalance is one of the mechanisms attributed to emotional distress15,16. It has been postulated that insufficient levels of the monoamine neurotransmitters (e.g., serotonin, dopamine and norepinephrine) may lead to depression17. Serotonin, as a key neurotransmitter of the brain-gut axis, plays an important role in the pathogenesis of both emotional distress and IBS6. More than 90% of the body’s serotonin is synthesized in the gut18,19. Various bacteria such as Streptococcus spp., Enterococcus spp., Escherichia spp., Lactobacillus plantarum, Klebsiella pneumonia, and Morganella morganii have the ability to produce serotonin20–23. The literature also supports that other neurotransmitters such as dopamine and norepinephrine can be produced by bacteria such as Lactobacillus, Serratia, Bacillus, Morganella and Klebsiella. Interestingly, dopamine and norepinephrine can stimulate the growth of E. coli O157: H724,25. Alteration of the gut microbiome may change the biosynthesis, release, and reuptake of neurotransmitters. The alteration may result in a disturbance in processes operating between the CNS and the enteric nervous system and may lead to emotional distress6,26. Described mutual interaction between the CNS and the gut highlights the role of the gut microbiome and neurotransmitters in emotional distress27.
Traditionally, emotional distress such as depression has been related to an imbalance in serotonin, norepinephrine, and dopamine levels. Some studies evaluated the role of serotonin in IBS, but research about the function of other involved neurotransmitters such as norepinephrine is limited28. Moreover, the gut microbiome as a cardinal part of the gut-brain axis has gained significant interest in the pathophysiology of both IBS and emotional distress in recent years. Thus, in the current study, we hypothesized a correlation between the gut microbiome and plasma neurotransmitters levels in IBS people with emotional distress. We aimed to investigate the difference in biomarkers (neurotransmitters levels and gut microbiome diversity) between IBS and healthy controls (HC), as well as the relationship between the gut microbiome and neurotransmitters in persons with IBS and emotional distress.
Results
Demographic characteristics
Demographic information of the participants, including gender, age, race, ethnicity, and marital status, is shown in Table 1. The mean age was 21 and 20 years old in the IBS and HC groups, respectively. The majority of the participants were female, White, and Non-Hispanic. The results show that the demographic characteristics were homogenous between the IBS and HC groups except in age.
Table 1.
Demographic characteristics of the participants.
| Variable | IBS (n = 40) N (%) or Mean ± SD |
HC (n = 20) N (%) or Mean ± SD |
P |
|---|---|---|---|
| Gender | |||
| Female | 26 (65%) | 10 (50%) | .402 |
| Male | 14 (35%) | 10 (50%) | |
| Age (years) | 21.67 ± 2.22 | 20.10 ± 1.41 | .007 |
| Race | |||
| White | 31 (77%) | 10 (50%) | .092 |
| Asian | 6 (15%) | 6 (30%) | |
| African-American | 3 (8%) | 4 (20%) | |
| Ethnicity | |||
| Hispanic | 5 (12%) | 4 (20%) | .391 |
| Non-Hispanic | 33 (83%) | 15 (75%) | |
| Unknown | 0 (0) | 1 (5%) | |
| Not reported | 2 (5%) | 0 (0) | |
| Marital status | |||
| Never married | 38 (95%) | 20 (100%) | .548 |
| Married | 2 (5%) | 0 (0%) | |
IBS, irritable bowel syndrome; HC, healthy control.
Levels of anxiety and depressive symptoms
The mean anxiety scores were 62.85 ± 7.67 and 54.41 ± 6.12 in the IBS and HC groups, respectively. Moreover, the mean of depressive symptoms were 56.11 ± 7.30 and 48.55 ± 6.26 in the IBS and HC groups correspondingly. The results showed that the mean of anxiety and depressive symptoms were significantly higher in the IBS group compared to the HC group. While the HC anxiety and depressive symptoms were within the normal limits, the IBS group had moderate anxiety level and mild depressive symptoms. The results can be found in Table 2.
Table 2.
Difference in anxiety and depression scores between IBS and healthy control (HC) groups.
| Variable | IBS (n = 40) Mean ± SD |
HC (n = 20) Mean ± SD |
p |
|---|---|---|---|
| Anxiety | 62.858 ± 7.678 | 54.415 ± 6.123 | < .0001 |
| Depressive symptoms | 56.110 ± 7.309 | 48.555 ± 6.268 | < .0001 |
IBS, irritable bowel syndrome; HC, healthy control.
Neurotransmitter levels
The mean plasma serotonin level was 35.47 ± 27.53 ng/ml and 42.36 ± 31.94 ng/ml and the norepinephrine level was 358.66 ± 120.25 pg/ml and 395.57 ± 169.84 pg/ml in the IBS and HC groups, respectively. While plasma serotonin and norepinephrine levels were lower in the IBS group, they were not significantly different compared to the HC group. The results are shown in Table 3.
Table 3.
Difference in plasma neurotransmitters levels between IBS and control groups.
| Variable | IBS Mean ± SD |
HC Mean ± SD |
p | ||
|---|---|---|---|---|---|
| Plasma serotonin level (ng/ml) | 35.474 ± 27.539 | N = 31 | 42.369 ± 31.914 | N = 17 | .304 |
| Plasma norepinephrine level (pg/ml) | 358.660 ± 120.256 | N = 30 | 395.579 ± 169.849 | N = 14 | .412 |
IBS, irritable bowel syndrome; HC, healthy control.
Gut microbiome profile
Gut microbiome patterns
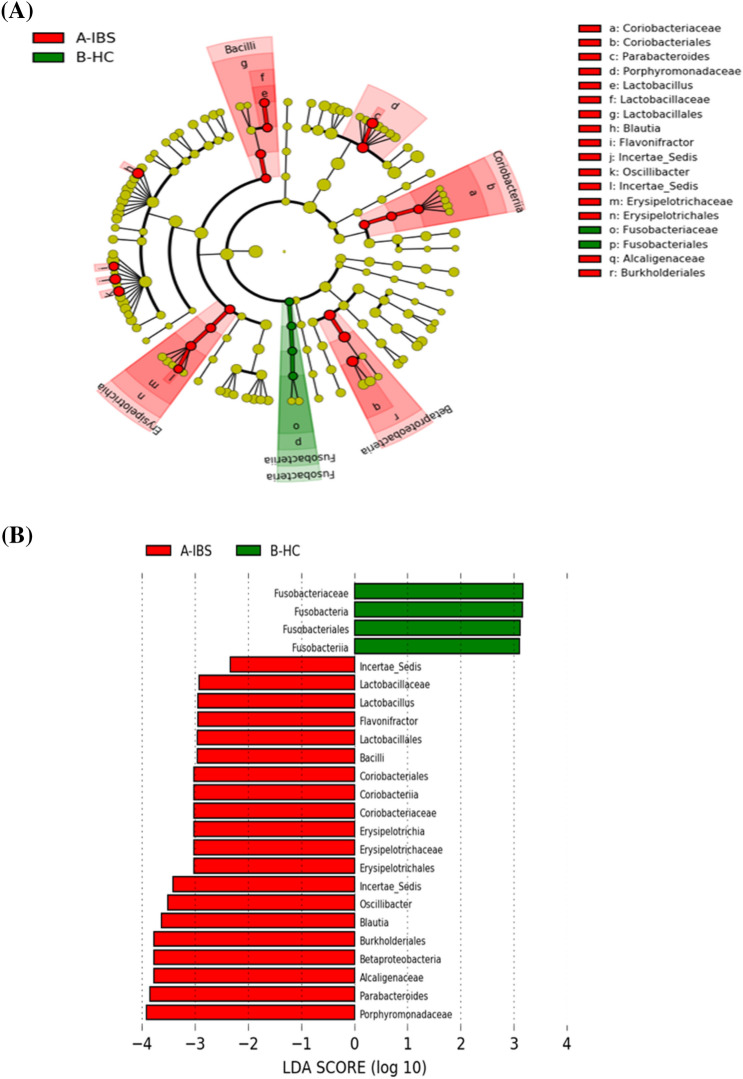
At the taxonomic level, among different OTUs, there was a difference in the abundance of bacteria between the IBS and HC groups. We observed high abundance of Fusobacteriaceae, Fusobacteria, Fusobacteriales and Fusobacales in the HC group and a high abundance of Lactobacillaceae, Lactobacillus, Flavonifractor, Lactobacillales, Bacilli, Coriobacteriales, Coriobacteriia, Coriobacteriaceae, Erysipelotrichia, Erysipelotrichaceae, Erysipelotrichales, Oscillibacter, Blautia, Burkholderiales, Betaproteobacteria, Alcaligenaceae, Parabacteroides and Porphyromonadaceae in perspons with IBS compared to HC (Fig. 1).
Figure 1.
Taxonomic differences of fecal microbiome between IBS and healthy control (HC) (Green) HC taxa; (Red) taxa enriched in people with irritable bowel syndrome (IBS). (A) Taxonomic cladogram based on the LEfSe analysis. (B) IBS-enriched taxa are indicated with a positive linear discriminant analysis (LDA) score (red) and taxa enriched in HC have a negative score (green).
Gut microbiome diversity
Alpha diversity (the richness and evenness of the microbial community) was measured using three different alpha diversity indices, including total observed species (sobs), Simpson (richness and evenness of the microbial community within a sample), and Shannon (evenness of the microbial community within a sample) indices. According to these indices, there was an increased bacteria alpha diversity in the IBS group compared to the HC group (sobs (p = 0.003), Simpson (p = 0.019), and Shannon (p = 0.013) (Figs. 2, 3, 4).
Figure 2.
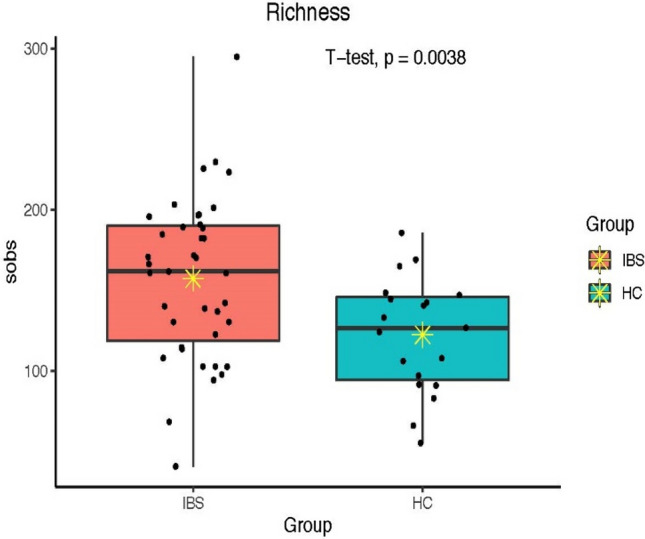
Total observed species (sobs) diversity between IBS and HC groups; * indicates mean.
Figure 3.
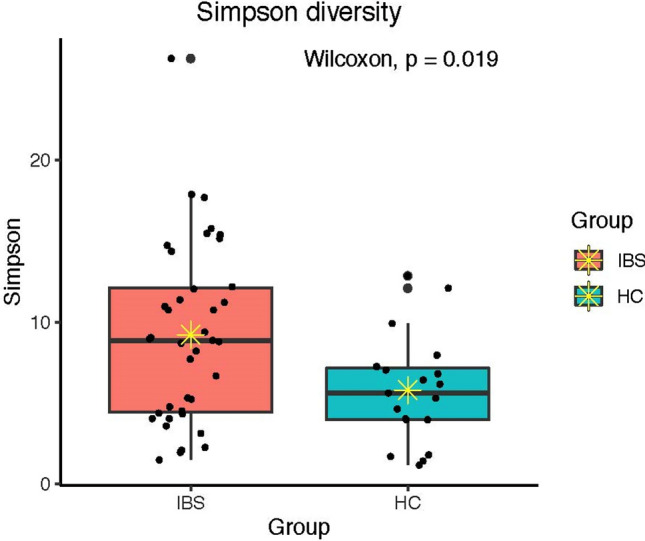
Simpson diversity between IBS and HC; * indicates mean.
Figure 4.
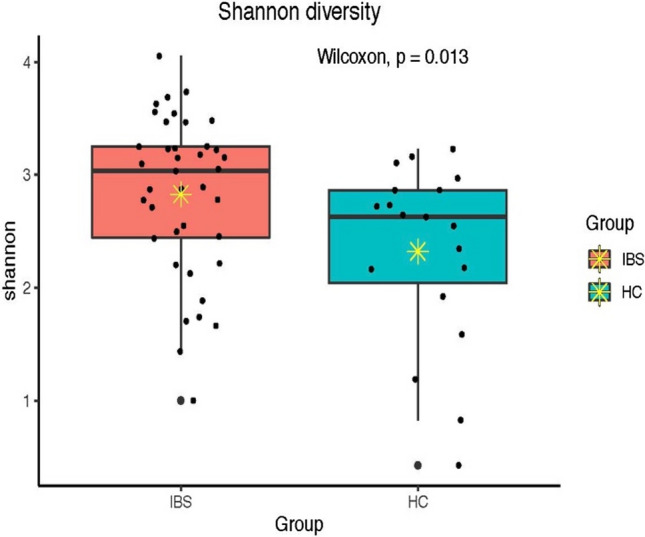
Shannon diversity between IBS and HC groups; * indicates mean.
Moreover, there was significant dissimilarity in beta diversity (the compositional dissimilarity among the microbiome community) using Bray–Curtis (abundance or read count data) and Jaccard (the presence/absence of species) dissimilarities between the two groups (Figs. 5, 6). The multivariate dispersions test of homogeneity assumption was satisfied for Bray–Curtis, but not for Jaccard dissimilarity.
Figure 5.
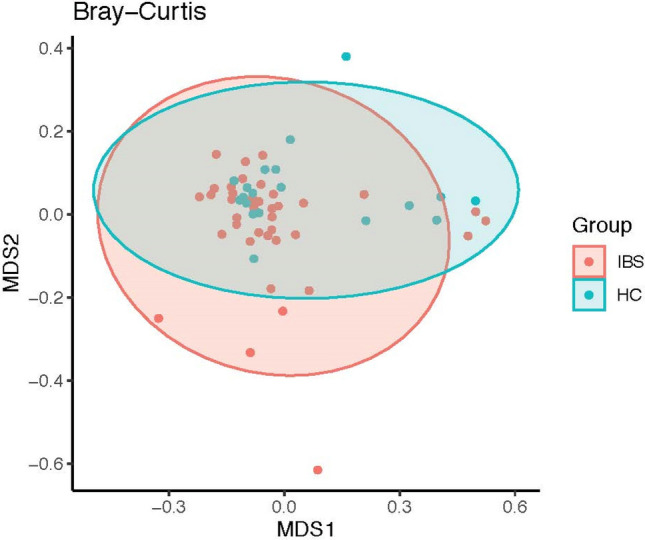
Fecal microbiome dissimilarity using Bray–Curtis distance between IBS and healthy control (HC) groups.
Figure 6.
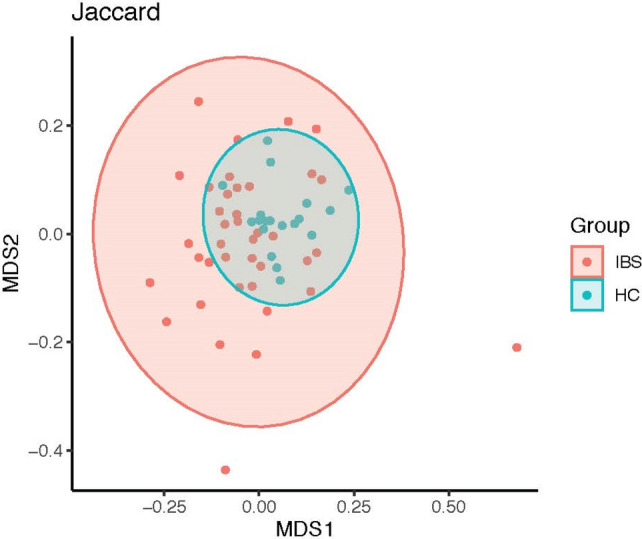
Fecal microbiome dissimilarity using Jaccard distance between IBS and healthy control (HC) groups.
Association between neurotransmitter levels and the gut microbiome in IBS people
In the analysis of associations between plasma neurotransmitter levels and alpha diversity indices in the IBS group, there were no correlations between serotonin levels and alpha diversities, but norepinephrine levels were found to be negatively correlated to Shannon and Simpson indices (p = 0.029 and p = 0.015; respectively) (Table 4).
Table 4.
Correlation between plasma neurotransmitter levels and bacterial diversity indices in IBS people.
| Neurotransmitter | Diversity index | Spearman’s r | p |
|---|---|---|---|
| Serotonin | Sobs | .068 | .717 |
| Shannon | .167 | .374 | |
| Simpson | .169 | .369 | |
| Norepinephrine | Sobs | − .262 | .168 |
| Shannon | − .403 | .029 | |
| Simpson | − .444 | .015 |
In terms of associations between plasma neurotransmitter levels and bacteria phylum, we observed a significant positive correlation between serotonin levels and Proteobacteria (p = 0.021), a significant negative correlation between norepinephrine and Firmicutes (p = 0.020), and a significant positive correlation between norepinephrine and Bacteroidetes phylum (p = 0.043) (Table 5).
Table 5.
Correlation between plasma neurotransmitter levels and bacteria phylum in IBS people.
| Neurotransmitter | Phylum | Spearman’s r | p |
|---|---|---|---|
| Serotonin | Firmicutes | .072 | .703 |
| Bacteroidetes | − .198 | .292 | |
| Actinobacteria | .116 | .541 | |
| Proteobacteria | .421 | .021 | |
| Norepinephrine | Firmicutes | − .431 | .020 |
| Bacteroidetes | .379 | .043 | |
| Actinobacteria | − .103 | .591 | |
| Proteobacteria | − .185 | .334 |
Discussion
The current study assessed the difference in the plasma neurotransmitter levels and the gut microbiome profile in persons with IBS who experienced anxiety and depressive symptoms compared to the HC. Also, we evaluated the correlation between neurotransmitter levels and the gut microbiome diversities and microbial abundance at the phylum level. Our findings revealed that gut microbiome patterns in persons with IBS who suffer from emotional distress are different from HC. Although there was no difference in neurotransmitter levels between the two groups, we observed a correlation between bacterial profile (bacterial taxa at phylum level and alpha diversity) and neurotransmitters’ levels in the IBS group.
It has been hypothesized that the imbalance in monoamine neurotransmitter levels (i.e., serotonin and norepinephrine) is a key factor in the pathophysiology of depressive symptoms and anxiety disorders29. Thus, high comorbidity between IBS and emotional distress, including anxiety and depressive symptoms, may be a consequence of an alteration in serotonin and norepinephrine30,31. Both the CNS and the enteric nervous system have the capability to produce neurotransmitters28. A decrease in the amount of serotonin and norepinephrine released and deterioration of neurotransmitters binding to receptors may lead to emotional distress32. In the current study, no significant difference was observed in either serotonin or norepinephrine levels between IBS and HC. Our results are in contrast to the findings of Zhao et al. (2014) in which IBS patients with depressive symptoms had lower plasma serotonin33. While serotonin has drawn a tremendous amount of attention regarding the pathogenesis of comorbidity of depressive symptoms and IBS, some studies have reported a lack of significant correlation between plasma serotonin levels and depressive symptoms in IBS patients compared to healthy controls34–36. Although alteration in serotonin levels has been highlighted in the comorbidity of IBS and depressive symptoms33, there are limited studies to investigate the role of other neurotransmitters such as norepinephrine in comorbidity between IBS and emotional distress. Thus, further research is needed to assess the role of norepinephrine in this comorbidity.
It has been postulated that the gut microbiome contributes to the pathogenesis of IBS and emotional distress, such as depressive symptoms and anxiety18. The gut microbiome may influence basal circuits of emotion processing via the vagal pathway from the gut37,38. Interoceptive stimuli may impede conscious and subconscious emotional and cognitive processing39. Research has shown that the dysbiosis in the gut microbiome is a shared abnormality in the pathophysiology of both IBS and depressive symptoms26,40. We observed higher alpha diversity in terms of total observed species, Simpson and Shannon indices in persons with IBS compared to HC. Our results contrast with the findings of Peter (2018), who reported non-significant lower alpha diversity in IBS patients with psychological distress13. Similarly, Hugerth (2019) found no association between bacterial diversity and depressive symptoms in IBS patients41. While we observed higher alpha diversity, Liu (2016) reported low alpha diversity (Shannon index) in persons with IBS and depressive symptoms14. Sundin (2015) also showed a negative correlation between fecal microbial diversity and anxiety and depressive symptoms in patients with IBS42. While pre-clinical studies have supported the hypothesis of lower alpha diversity in animal models after exposure to stress43,44, the results in clinical studies are contradicted and require further investigations. Regarding beta diversity, similar to our findings, Peter (2018) found that microbial beta diversity was associated with depressive symptoms13. Beta diversity as an indicator of taxa similarity within samples may clarify how emotional distress segregated the gut microbiome features among different subgroups of individuals that require further investigation45.
We also found a high abundance of several bacteria at different taxonomic levels that belong primarily to the three phyla Bacteroidetes, Firmicutes, and Verrucomicrobia in persons with IBS who suffer from emotional distress. Similar to our findings, elevated Lachnospiraceae were reported in cases of both anxiety and depressive symptoms13. However, studies have shown mixed results concerning the abundance of various gut microbiomes at different taxonomic levels among persons with IBS who had emotional distress, and thus, additional studies are needed41,46.
Recent evidence has highlighted the ability of the gut microbiome to produce neurotransmitters, which may lead to the development/maintenance of emotional distress such as anxiety and depression. While we did not observe any association between various alpha diversity indices and serotonin, there was a negative correlation between alpha diversity (Shannon and Simpson indices) and norepinephrine levels. Moreover, there was a positive association between serotonin and Proteobacteria phylum as well as norepinephrine and Bacteroidetes phylum. However, Firmicutes had a negative association with norepinephrine.
Research shows that more than 90% of the serotonin in the human body is synthesized by the gut19. For instance, Candida, Escherichia coli, Streptococcus, and Enterococcus may secrete serotonin and Escherichia coli, Bacillus and Saccharomyces can produce norepinephrine47,48. The production of neurotransmitters by various microbiota may lead to the imbalance of neurotransmitter levels and the development of pathogenic bacteria. For example, the presence of norepinephrine may stimulate the growth of Escherichia coli25. Additionally, studies show that the in vitro exposure of the Klebsiella pneumoniae, Enterobacter cloacae, Pseudomonas aeruginosa, Shigella sonnei, and Staphylococcus aureus to norepinephrine may lead to the growth of these pathogens49. So, alteration of the gut microbiome due to IBS may be attributed to the imbalance in neurotransmitters levels and subsequently emotional distress. Accumulated preclinical studies highlight the role of the gut microbiome in regulating neurotransmitters and depressive-like behavior in the animal models50. There is a strong need for future clinical studies to understand the biological mechanism of comorbidity of emotional distress with IBS by shedding light on the role of the gut microbiome in the production of neurotransmitters.
Limitations
There are limitations in the study that need to be considered when interpreting the results. The small sample did not allow the categorization of persons with IBS based on the severity of the emotional distress. Therefore, the interpretation of the biomarkers based on the severity of emotional distress is limited. Moreover, a small sample size and unbalanced design might increase the likelihood of a type II error skewing the results. Therefore, caution is needed when interpreting the results of the study, which needs to be replicated in a larger sample. The majority of the participants in our study were mixed type (IBS-M) according to the self-reported symptoms. Further studies with a larger sample size by considering history of diet, IBS subtype and other IBS symptoms (e.g., pain, bowel habits) are recommended to determine the associations between neurotransmitters and the gut microbiome in persons with IBS and emotional distress.
Conclusion
The study’s findings highlight that persons with IBS and emotional distress may be differentiated from healthy persons based on their gut microbiome profile. There were lower but not significant neurotransmitter levels in the IBS group compared to the HC. Also, we found correlation between the gut microbiome (at both diversity and operational taxonomic unit) and neurotransmitters in persons with IBS and emotional distress. Future research in a larger sample is required to correlate neurotransmitter levels with observed gut microbiome alterations. Identification of biomarkers may help advance IBS management with comorbid emotional distress.
Methods
Study design and subjects
The present cross-sectional comparative study is part of a parent randomized clinical trial titled “Precision Pain Self-Management in Young Adults with IBS” (NCT03332537)51. In the parent study, 80 persons with IBS diagnosed by a gastroenterologist were enrolled from the general community as well as two large public university campuses and two gastrointestinal (GI) clinics in the northeastern region of the United States. The participants completed emotional distress questionnaires, and their blood and fecal samples were collected at three different time points: baseline, six weeks, and 12 weeks. The data from the parent study's baseline session was utilized and 20 healthy subjects were recruited as controls in the present study.
The inclusion criteria for enrollment of people with IBS in the published protocol of the parent study were: (1) men and women 18–29 years of age, (2) with a diagnosis of IBS from a healthcare provider using the Rome IV criteria, and (3) able to read and speak in English. The exclusion criteria were: (1) having other chronic painful conditions including but not limited to fibromyalgia, chronic pelvic pain or chronic intestinal cystitis, infectious diseases (hepatitis, HIV, MRSA), celiac disease or inflammatory bowel disease, and/or diabetes mellitus, (2) serious mental health conditions (e.g., bipolar disorder, schizophrenia, mania), (3) women pregnant or post-partum within 3 months of delivery, or, (4) regular use of opioids, iron supplements, prebiotics/probiotics or antibiotics, and/or substance use. The criteria for recruitment of HC were the same as those for the IBS group, except that the HC group did not have a history of IBS or any chronic disease52.
Data collection
The IBS and HC groups completed questionnaires via a Research Electronic Data Capture (REDCap) software/system. Peripheral blood samples were collected into EDTA tubes using a standardized venipuncture procedure. The collected blood samples were diluted using 2% fetal bovine serum and centrifuged at 1000×g for 20 min at 4 °C, and then plasma was aliquoted into eppendorf tubes and stored at − 80 °C until further analysis.
The participants were requested to collect their fecal samples at the time of enrollment in the study using a stool sample collection kit (OMNIgene•GUT, DNA Genotek Inc., Canada) per manufacturer instructions. Samples were mailed back to the research laboratory and frozen and maintained at − 80 °C until processing and 16S rRNA gene sequencing analysis.
Outcome measures
Emotional distress measurement
Anxiety and depressive symptoms were assessed using the Patient-Reported Outcomes Measurement Information System (PROMIS) v1.0 questionnaires from the recommended National Institute of Nursing Research (NINR) common data elements. PROMIS instruments measure clinically important outcomes, including physical, mental, and social health. PROMIS is scored by using the Health Measures Scoring Service53,54.
Anxiety
The PROMIS questionnaire, the “Emotional Distress-Anxiety” section, which includes 6 questions, was used for measuring anxiety. Responders selected answers based on the category provided by the questionnaire (never, rarely, sometimes, often, and always). The researcher scored the responder's answers (never = 1, rarely = 2, sometimes = 3, often = 4, and always = 5). After calculating the raw score by summing responses, the total raw score was translated to a T-score which is a standardized T-score with a mean of 50 and a standard deviation of 10. According to the PROMIS measurements, a score below 55 is within the normal limit, the score of 55–60 refers to mild, and a score above 60 indicates moderate and severe anxiety symptoms55.
Depressive symptoms
For measuring depressive symptoms, the PROMIS questionnaire, “Emotional Distress-Depression” section, which includes questions with five different answer categories (never = 1, rarely = 2, sometimes = 3, often = 4, and always = 5) was used. The same approach was employed to calculate depressive symptoms according to the PROMIS measurement guidelines55.
Plasma levels of neurotransmitters measurement using ELISA
Serotonin
This study used the serotonin enzyme-linked immunosorbent assay (ELISA) assay kit named ADI-900-175 (Enzo Life Sciences, New York, New York) for the quantitative determination of serotonin in plasma56. For this purpose, the steps mentioned in the guideline of Serotonin ELISA Assay Kit, including preparation of reagents (e.g., microtiter strips, wash buffer and Equalizing Reagent), acylation of samples (20 µl) and ELISA test procedures were followed as per the manufacturer's instructions. The competitive Serotonin ELISA Assay kit uses the microtiter plate format, in which serotonin is bound to the solid phase of the plate. Antirabbit/peroxidase detects the antibody bound to the solid phase serotonin. The level of serotonin in the sample is inversely proportional to the amount of antibody bound to the solid phase serotonin. The sensitivity of the kit was 0.293 ng/ml and the range of detection was 0.49–500 ng/ml. The samples were run in duplicate, per batch and the mean inter- and intra-assay coefficients of variation for control samples was ≤ 20%.
Norepinephrine
For the quantitative determination of norepinephrine in plasma, the norepinephrine ELISA assay kit named KA1891 (Abnova, Taipei, Taiwan) was used57. For this purpose, 300 µl of plasma was processed according to the guideline of Norepinephrine ELISA Assay Kit, including preparation of reagents (e.g., wash buffer and enzyme mix), preparation of samples, and ELISA test procedure per manufacture’s directions. The kit's sensitivity was 5 pg/ml and the range of detection was 0–32 pg/ml. The samples were run in duplicate, per batch and the mean inter- and intra-assay coefficients of variation for control samples was ≤ 20%.
Microbiome sequencing and analysis
The fecal samples DNA extraction, sequencing, and analysis were carried out at the University of Connecticut, Center of Microbial Analysis, Resources, and Services (UConn MARS). Bacterial DNA was extracted from 0.25 g of stool sample using the MoBio PowerSoil isolation kit (Mo Bio Laboratories, Inc, CA, USA) in line with the manufacturer’s instruction for the Eppendorf epMotion 5076 Vac liquid handling robot58. Then the V4 region of the 16S rRNA genes of the microbial community were sequenced using the MiSeq Series Illumina platform.
The sequences produced by PCR were normalized based on the concentration of DNA in the 350–400 bp region and pooled using the QIAgility liquid handling robot (Qiagen). Then, the pooled PCR was purified using GeneRead Size Selection kit (Qiagen, Hilden, Germany, cat. Cat. ID: 180514) and paired-end sequencing (2 × 250) was performed on an Illumina MiSeq system.
The raw sequence data were processed by Mothur 1.42.3 Pipeline59 following the analysis pipeline of Miseq SOP (http://www.mothur.org/wiki/MiSeq_SOP) and the UConn MARS mothur batch file (https://github.com/krmaas/bioinformatics/blob/master/mothur.bash). Paired-end sequences were combined into contigs and aligned against to the SILVA 119 V4 16 S rRNA gene reference alignment database60. Operational taxonomic units (OTUs) were determined at a 97% identify, and then performing de novo OTU clustering on reads that failed to cluster to a reference. Taxonomic annotation was also determined by the SILVA 16S rRNA reference dataset. Sample were removed if the yielded reads were less than 10,000. For the microbiome analysis, the Mothur software59 was used. Alpha diversity, including Simpson, Shannon, and total observed species (sobs) indices, were used to evaluate the complexity of the whole microbial community. Beta diversity represented by Bray–Curtis and Jaccard was used to indicate the inter-subjects’ dissimilarity in the bacterial communities.
Statistical analysis
For data analysis, the SPSS (statistical package for social sciences) version 26 and R version 4.0.2 were used. The demographic characteristics were presented with mean and standard deviation for continuous variables and frequency and percentage for continuous variables. The statistical approaches to assess the difference in emotional distress, and neurotransmitter levels between IBS and HC groups were Independent Sample T-test and Mann–Whitney. For analysis of microbiota composition, the linear discriminant analysis effect size (LEfSe) method was performed61. For comparing alpha diversity between the IBS and HC groups, a Wilcoxon rank-sum test was used. Also, based on Bray–Curtis and Jaccard dissimilarity, non-metric multidimensional scaling (NMDS) ordination for beta diversity was performed and multivariate dispersions (PERMDIST) test was conducted to examine the assumption of homogeneity. Spearman rank correlation was used to examine the association between neurotransmitter levels and alpha diversity indices (Simpson, Shannon and sobs) as well as bacterial phylum in the IBS group.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the University of Connecticut and was conducted in accordance with the ethical guidelines of the Declaration of Helsinki. All the participants provided written informed consent.
Acknowledgements
The authors would like to acknowledge people who participated in the current study. The authors would also like to acknowledge the support from the Bio-Behavioral Lab (BBL), the Microbial Analysis, Resources and Services (MARS), the Center of Advancement in Managing Pain (CAMP), American Nurses Foundation (ANF) and NIH funded P20 Center for Accelerating Precision Pain Self-Management at the University of Connecticut School of Nursing.
Author contributions
Conceptualization, Z.A.B., X.S.C., A.R.S., and W.A.H.; formal analysis, Z.A.B., J. L., X.S.C., and M.R.; funding acquisition, Z.A.B., X.S.C., and A.R.S.; methodology, Z.A.B., X.S.C., A.R.S., and W.A.H.; project administration, Z.A.B., X.S.C., A.R.S. and JC; writing-original draft, Z.A.B., X.S.C., A.R.S., and W.A.H.; writing- review and editing, Z.A.B., X.S.C., A.R.S., W.A.H., J.L., M.R., and J.C. All authors have reviewed the manuscript and agreed to submit this version.
Funding
This study was supported by American Nurses Foundation and National Institute of Nursing Research of the National Institutes of Health (NIH-NINR) under award number: NIH-NINR P20NR016605 (PI: A.R.S; Pilot PI: X.S.C).
Data availability
The datasets generated analyzed in the current study are available from the NCBI dBGaP (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB8914789/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shahabi L, Naliboff BD, Shapiro D. Self-regulation evaluation of therapeutic yoga and walking for patients with irritable bowel syndrome: a pilot study. Psychol. Health Med. 2016;21:176–188. doi: 10.1080/13548506.2015.1051557. [DOI] [PubMed] [Google Scholar]
- 2.Barandouzi ZA, Lee J, Maas K, Starkweather AR, Cong XS. Altered gut microbiota in irritable bowel syndrome and its association with food components. J. Personal. Med. 2021;11:35. doi: 10.3390/jpm11010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.IBS, N. Nature IBS (2016).
- 4.Fadgyas-Stanculete M, Buga A-M, Popa-Wagner A, Dumitrascu DL. The relationship between irritable bowel syndrome and psychiatric disorders: From molecular changes to clinical manifestations. J. Mol. Psychiatry. 2014;2:4. doi: 10.1186/2049-9256-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y-T, et al. Risk of psychiatric disorders following irritable bowel syndrome: a nationwide population-based cohort study. PLoS ONE. 2015;10:e0133283. doi: 10.1371/journal.pone.0133283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grzesiak M, et al. Serotonin-related gene variants in patients with irritable bowel syndrome and depressive or anxiety disorders. Gastroenterol. Res. Pract. 2017;2017:4290430. doi: 10.1155/2017/4290430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C, et al. The increased level of depression and anxiety in irritable bowel syndrome patients compared with healthy controls: systematic review and meta-analysis. J. Neurogastroenterol. Motil. 2017;23:349. doi: 10.5056/jnm16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surdea-Blaga T, Băban A, Dumitrascu DL. Psychosocial determinants of irritable bowel syndrome. World J. Gastroenterol. 2012;18:616. doi: 10.3748/wjg.v18.i7.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich M, Grady SE, Wall GC. Effects of antidepressants in patients with irritable bowel syndrome and comorbid depression. Clin. Ther. 2010;32:1221–1233. doi: 10.1016/j.clinthera.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Jones MP, Crowell MD, Olden KW, Creed F. Functional gastrointestinal disorders: an update for the psychiatrist. Psychosomatics. 2007;48:93–102. doi: 10.1176/appi.psy.48.2.93. [DOI] [PubMed] [Google Scholar]
- 11.Van Oudenhove L, Coen SJ, Aziz Q. Functional brain imaging of gastrointestinal sensation in health and disease. World J. Gastroenterol. 2007;13:3438. doi: 10.3748/wjg.v13.i25.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q-E, et al. Depressive symptoms in patients with irritable bowel syndrome: a meta-analysis of comparative studies. Int. J. Biol. Sci. 2018;14:1504. doi: 10.7150/ijbs.25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peter J, et al. A microbial signature of psychological distress in irritable bowel syndrome. Psychosom. Med. 2018;80:698. doi: 10.1097/PSY.0000000000000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clinical Gastroenterol. Hepatol. 2016;14:1602–1611.e1605. doi: 10.1016/j.cgh.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 15.Kaur S, Singh R. Role of different neurotransmitters in anxiety: a systemic review. Int. J. Pharm. Sci. Res. 2017;8:411. [Google Scholar]
- 16.Pan J-X, et al. Diagnosis of major depressive disorder based on changes in multiple plasma neurotransmitters: A targeted metabolomics study. Transl. Psychiatry. 2018;8:1–10. doi: 10.1038/s41398-018-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain-gut-microbiome axis. Cell. Mol. Gastroenterol. Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Özoğul F. Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur. Food Res. Technol. 2004;219:465–469. [Google Scholar]
- 21.Shishov V, Kirovskaya T, Kudrin V, Oleskin A. Amine neuromediators, their precursors, and oxidation products in the culture of Escherichia coli K-12. Appl. Biochem. Microbiol. 2009;45:494–497. [PubMed] [Google Scholar]
- 22.Özoğul F, Kuley E, ÖZOĞUL, Y. & ÖZOĞUL, İ. The function of lactic acid bacteria on biogenic amines production by food-borne pathogens in arginine decarboxylase broth. Food Sci. Technol. Res. 2012;18:795–804. [Google Scholar]
- 23.Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: how gut microbes shape human behavior. J. Psychiatr. Res. 2015;63:1–9. doi: 10.1016/j.jpsychires.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Averina O, Danilenko V. Human intestinal microbiota: Role in development and functioning of the nervous system. Microbiology. 2017;86:1–18. [PubMed] [Google Scholar]
- 25.Freestone PP, et al. Growth stimulation of intestinal commensal Escherichia coli by catecholamines: A possible contributory factor in trauma-induced sepsis. Shock. 2002;18:465–470. doi: 10.1097/00024382-200211000-00014. [DOI] [PubMed] [Google Scholar]
- 26.Barandouzi ZA, Starkweather AR, Henderson WA, Gyamfi A, Cong XS. Altered composition of gut microbiota in depression: A systematic review. Front. Psych. 2020;11:541. doi: 10.3389/fpsyt.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Averina OV, et al. Bacterial metabolites of human gut microbiota correlating with depression. Int. J. Mol. Sci. 2020;21:9234. doi: 10.3390/ijms21239234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mudyanadzo TA, Hauzaree C, Yerokhina O, Architha NN, Ashqar HM. Irritable bowel syndrome and depression: A shared pathogenesis. Cureus. 2018;10:e3178. doi: 10.7759/cureus.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chávez-Castillo M, et al. Depression as a neuroendocrine disorder: Emerging neuropsychopharmacological approaches beyond monoamines. Adv. Pharmacol. Pharm. Sci. 2019;2019:7943481. doi: 10.1155/2019/7943481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nutt DJ. Relationship of neurotransmitters to the symptoms of major depressive disorder. J. Clin. Psychiatry. 2008;69:4–7. [PubMed] [Google Scholar]
- 31.Fond G, et al. Anxiety and depression comorbidities in irritable bowel syndrome (IBS): A systematic review and meta-analysis. Eur. Arch. Psychiatry Clin. Neurosci. 2014;264:651–660. doi: 10.1007/s00406-014-0502-z. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Zhao J, Guo W. Emotional roles of mono-aminergic neurotransmitters in major depressive disorder and anxiety disorders. Front. Psychol. 2018;9:2201. doi: 10.3389/fpsyg.2018.02201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Song W, Zhu P, Zhang Y, Bu P. A correlation study between diarrhea-predominant irritable bowel syndrome complicated functional dyspepsia patients of Gan-stagnation Pi-deficiency syndrome and gastrointestinal hormones. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2014;34:1168–1172. [PubMed] [Google Scholar]
- 34.Thijssen A, et al. Alterations in serotonin metabolism in the irritable bowel syndrome. Aliment. Pharmacol. Ther. 2016;43:272–282. doi: 10.1111/apt.13459. [DOI] [PubMed] [Google Scholar]
- 35.Park S-Y, et al. Plasma 5-hydroxytryptamine concentration and its correlation with psychopathology in patients with irritable bowel syndrome. Gut and liver. 2009;3:26. doi: 10.5009/gnl.2009.3.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin D-C, et al. Regulation of the serotonin transporter in the pathogenesis of irritable bowel syndrome. World J. Gastroenterol. 2016;22:8137. doi: 10.3748/wjg.v22.i36.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowan CS, et al. Gutsy moves: The amygdala as a critical node in microbiota to brain signaling. BioEssays. 2018;40:1700172. doi: 10.1002/bies.201700172. [DOI] [PubMed] [Google Scholar]
- 38.Bercik P, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut–brain communication. Neurogastroenterol. Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz A, Vögele C. Interoception and stress. Front. Psychol. 2015;6:993. doi: 10.3389/fpsyg.2015.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menees S, Chey W. The gut microbiome and irritable bowel syndrome. F1000Res. 2018;7:F1000. doi: 10.12688/f1000research.14592.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hugerth LW, et al. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2019;69:1076–1084. doi: 10.1136/gutjnl-2019-318717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundin J, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment. Pharmacol. Ther. 2015;41:342–351. doi: 10.1111/apt.13055. [DOI] [PubMed] [Google Scholar]
- 43.Bharwani A, et al. Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology. 2016;63:217–227. doi: 10.1016/j.psyneuen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Galley JD, et al. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 2014;14:189. doi: 10.1186/1471-2180-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan XC, Huttenhower C. Human microbiome analysis. PLoS Comput. Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeffery IB, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 47.Liu L, Zhu G. Gut–brain axis and mood disorder. Front. Psych. 2018;9:223. doi: 10.3389/fpsyt.2018.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evrensel A, Ceylan ME. The gut-brain axis: the missing link in depression. Clin. Psychopharmacol. Neurosci. 2015;13:239. doi: 10.9758/cpn.2015.13.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Donnell PM, Aviles H, Lyte M, Sonnenfeld G. Enhancement of in vitro growth of pathogenic bacteria by norepinephrine: Importance of inoculum density and role of transferrin. Appl. Environ. Microbiol. 2006;72:5097–5099. doi: 10.1128/AEM.00075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang F, Wu X. Brain neurotransmitter modulation by gut microbiota in anxiety and depression. Front. Cell Dev. Biol. 2021;9:472. doi: 10.3389/fcell.2021.649103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong X, et al. Pain self-management plus nurse-led support in young adults with irritable bowel syndrome: Study protocol for a pilot randomized control trial. Res. Nurs. Health. 2018;41:121–130. doi: 10.1002/nur.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cong X, et al. Pain self-management plus nurse-led support in young adults with irritable bowel syndrome: Study protocol for a pilot randomized control trial. Res Nurs Health. 2018;41:121–130. doi: 10.1002/nur.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A brief guide to the PROMIS© Global Health instruments.http://www.healthmeasures.net/images/PROMIS/manuals/PROMIS_Global_Scoring_Manual.pdf (2017).
- 54.NINR. Patient-Reported Outcomes Measurement Information System (PROMIS).http://www.healthmeasures.net/explore-measurement-systems/promis (2020).
- 55.Measures, H. Health Measures (PROMIS). https://www.healthmeasures.net/score-and-interpret/interpret-scores/promis/promis-score-cut-points (2021).
- 56.Sciences, E. L. ENZO. https://www.enzolifesciences.com (2020).
- 57.Abnova. Abnova. http://www.abnova.com (2020).
- 58.Inc, M. B. L. Mo BIO Laboratories Inc. https://www.selectscience.net/suppliers/mo-bio-laboratories-inc/?compID=425 (2019).
- 59.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Server, H. l. G. Huttenhower lab Galaxy server. https://huttenhower.sph.harvard.edu/galaxy (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated analyzed in the current study are available from the NCBI dBGaP (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB8914789/).