Abstract
DNA methylation is an epigenetic mechanism that plays important roles in gene regulation and transposon silencing. Active DNA demethylation has evolved to counterbalance DNA methylation at many endogenous loci. Here, we report that active DNA demethylation also targets viral DNAs, tomato yellow leaf curl China virus (TYLCCNV) and its satellite tomato yellow leaf curl China betasatellite (TYLCCNB), to promote their virulence. We demonstrate that the βC1 protein, encoded by TYLCCNB, interacts with a ROS1-like DNA glycosylase in Nicotiana benthamiana and with the DEMETER (DME) DNA glycosylase in Arabidopsis thaliana. The interaction between βC1 and DME facilitates the DNA glycosylase activity to decrease viral DNA methylation and promote viral virulence. These findings reveal that active DNA demethylation can be regulated by a viral protein to subvert DNA methylation-mediated defense.
Subject terms: Plant molecular biology, Plant immunology, Plant cell biology
Plants tightly regulate DNA methylation to regulate gene expression. Here, the authors show that the tomato yellow leaf curl China virus βC1 protein interacts with plant DNA glycosylases to promote demethylation of viral DNA and promote virulence.
Introduction
Cytosine DNA methylation plays important roles in transposon silencing and gene regulation1. In plants, DNA methylation occurs in CG, CHG, and CHH (where H represents A, T, or C) sequence contexts2–4. CG, CHG, and CHH methylation are established by Domains Rearranged Methyltransferase 2 through the RNA-directed DNA methylation (RdDM) pathway5 and maintained by different mechanisms. CG and CHG methylation are maintained by METHYLTRANSFERASE 16 and CHROMOMETHYLASE 37, respectively, whereas CHH methylation is maintained by CMT2 and RdDM8. All DNA methylation reactions use S-adenosyl-l-methionine (SAM) as the donor of methyl groups9.
DNA methylation can be passively or actively removed, and the balance between methylation and demethylation determines the final methylation pattern1. In Arabidopsis, active DNA demethylation involves the removal of 5-methylcytosine (5mC) by the DME family of DNA glycosylases, which include REPRESSOR OF SILENCING 1 (ROS1)10, DEMETER (DME)11, DEMETER-LIKE PROTEIN 2 (DML2), and DML312. ROS1, DML2, and DML3 are expressed in vegetative tissues, whereas DME is preferentially expressed in the gametes11,13. ROS1, DML2, and DML3 target thousands of endogenous loci2,4,14 and are important for the development of xylem tracheary elements and leaf stomatal cells15,16. DME targets thousands of endogenous loci in vegetative cells, the companion cells of sperms17, and initiates genome-wide active DNA demethylation in central cells, the companion cells of egg cells18, thereby regulating pollen germination13 and embryo development11.
Plant viruses infect a wide range of plant species and cause devastating crop losses all over the world. The family Geminiviridae comprises circular single-stranded DNA viruses. This family is currently divided into 14 genera on the basis of their genome organization, insect vector, and host range, with the genus Begomovirus being the largest19. The begomovirus tomato yellow leaf curl China virus (TYLCCNV), which can infect Nicotiana benthamiana, is associated with a betasatellite DNA (TYLCCNB)20. TYLCCNB encodes βC1, which determines the pathogenicity of TYLCCNV20. The multifunctional protein βC1 can block Mitogen-Activated Protein Kinase signaling21, interfere with post-transcriptional and transcriptional gene silencing22,23 and manipulate jasmonic acid signaling24–26 to subvert host defense.
DNA methylation can act as a defense mechanism against DNA viruses, including geminiviruses. Viral genomic DNA can be methylated27–29, and therefore its replication is suppressed30,31. To counteract this inhibition, viruses encode suppressor proteins to interfere with DNA methylation32–36 or the production of SAM22,37–39. It has been shown that βC1 can interact with and inactivate S-adenosyl homocysteine hydrolase (SAHH), an enzyme needed for SAM production, to suppress DNA methylation and enhance TYLCCNV virulence22. Thus far, whether active DNA demethylation plays a role in plant-virus interactions and whether the active DNA demethylation machinery is targeted by virus-encoded proteins remain unknown.
In this study, we investigated the role of active DNA demethylation in plant-virus interactions. We used TYLCCNV and its betasatellite TYLCCNB as a model system20. We found that active DNA demethylation promotes viral DNA hypomethylation and TYLCCNV/TYLCCNB virulence. βC1 interacts with NbROS1L in Nicotiana benthamiana and with DME in Arabidopsis. The interaction between βC1 and DME enhances the DNA glycosylase activity to counteract viral DNA methylation and increase viral virulence.
Results
Active DNA demethylation promotes viral DNA hypomethylation and TYLCCNV/TYLCCNB virulence in N. benthamiana
By searching for ROS1 orthologs, we identified six DME-like (DML) DNA glycosylases in N. benthamiana, namely NbROS1, NbROS1L, NbDML3, NbDML4, NbDML5 and NbDML6 (Supplementary Fig. 1a). Except NbDML6, they appeared to be ubiquitously expressed in N. benthamiana plants (Supplementary Fig. 1b). We tested their role in the plant-virus interactions by knocking down their expression by using tobacco rattle virus (TRV)-induced gene silencing (VIGS) (Supplementary Figs. 2 and 3a). Plants infiltrated with pCambia1300 (TRV-free empty vector), with TRV-GUS (as a VIGS control) and with TRV-NbDMLs were mock-inoculated, inoculated with TYLCCNV alone, or inoculated with TYLCCNV plus TYLCCNB (TYLCCNV+B). pCambia1300-, TRV-GUS- or TRV-NbDMLs-infiltrated plants did not show any symptoms when inoculated with TYLCCNV alone (Fig. 1a and Supplementary Fig. 3b), consistent with the previous finding that TYLCCNV alone does not induce disease symptoms in N. benthamiana20. When inoculated with TYLCCNV+B, pCambia1300- and TRV-GUS-infiltrated plants showed dramatically curled leaves, whereas plants infiltrated with TRV-NbDMLs displayed only mild symptoms (Fig. 1a and Supplementary Fig. 3b). We next performed Southern blot and quantitative PCR (qRCR) analyses to determine the accumulation of viral DNA in infected leaves. Comparably low levels of TYLCCNV DNA accumulated in pCambia1300-, TRV-GUS- or TRV-NbDMLs-infiltrated plants when inoculated with TYLCCNV alone. High levels of TYLCCNV and TYLCCNB were accumulated in plants infiltrated with pCambia1300- or TRV-GUS then inoculated with TYLCCNV+B. However, the viral load was significantly decreased in plants infiltrated with TRV-NbDMLs (Fig. 1b, c and Supplementary Fig. 3c). These results validate that TYLCCNB promotes TYLCCNV infection20, and suggest that active DNA demethylation plays a major role in the promotion of TYLCCNV infection by TYLCCNB.
Fig. 1. Active DNA demethylation promotes TYLCCNV+B virulence and viral DNA hypomethylation in N. benthamiana.
a Symptoms of pCambia1300-, TRV-GUS-, and TRV-NbDMLs-infiltrated N. benthamiana plants mock-inoculated, or inoculated with TYLCCNV or TYLCCNV+B. Photographs were taken at 10 days post inoculation (dpi). Scale bar, 2 cm. b Accumulation of TYLCCNV and TYLCCNB in the leaves of indicated plants at 10 dpi as determined by Southern blot. Total DNA was stained with ethidium bromide (EB) as loading control. c Accumulation of TYLCCNV in the leaves of indicated plants at 10 dpi as determined by qPCR. The coat protein gene of TYLCCNV was amplified. The N. benthamiana 25S nuclear rRNA gene (Nb25SrRNA) was used as an internal control. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; **p < 0.01, ns not significant. d Percentage of methylated cytosines in the intergenic region (IR) of TYLCCNV in the leaves of indicated plants as determined by bisulfite sequencing. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; **p < 0.01, ns not significant. Uncropped pictures, blots and gels for a and b, raw data and p values for c and d are provided in the source data.
Given that active DNA demethylation counteracts DNA methylation, we wondered whether removal of DNA methylation from viral genome is associated with the high accumulation of TYLCCNV/TYLCCNB and increased symptom severity. To test this, we performed bisulfite sequencing to measure the methylation levels of TYLCCNV genomic DNA, represented by an intergenic region that contains the origin of replication and divergent viral promoters (Supplementary Fig. 3d). We found that ~23% C’s in TYLCCNV were methylated in plants inoculated with TYLCCNV alone, regardless of whether NbDMLs were knocked down (Fig. 1d). The levels of methylation decreased to ~12% in plants infiltrated with pCambia1300 or TRV-GUS then inoculated with TYLCCNV+B. Knockdown of NbDMLs greatly restored methylation of the viral DNA (Fig. 1d). These results suggest that TYLCCNB causes a decrease in viral DNA methylation, which is mainly achieved through active DNA demethylation.
βC1 interacts with NbROS1L
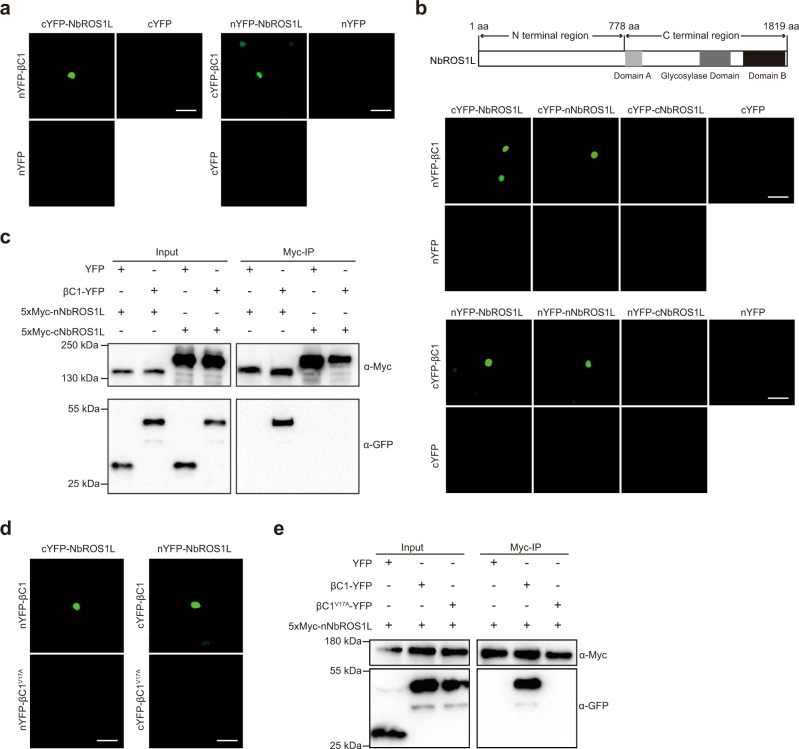
βC1 can suppress DNA methylation through inactivating SAHH22. Given our finding that the TYLCCNB-mediated decrease in viral DNA methylation is mainly dependent on NbDMLs, we reasoned that βC1 could also target the active DNA demethylation machinery. The expression levels of NbDMLs were not significantly different in mock-, TYLCCNV- or TYLCCNV+B-inoculated plants (Supplementary Fig. 4a), suggesting that βC1 does not transcriptionally regulate NbDMLs. We then tested whether βC1 can interact with NbDMLs by performing bimolecular fluorescence complementation (BiFC) assays. We found that βC1 could interact with NbROS1L in the nucleus, but not with other members of the family (Fig. 2a and Supplementary Fig. 4b, c). Further analyses revealed that the N-terminal region of NbROS1L mediated its interaction with βC1 (Fig. 2b and Supplementary Fig. 4d). The interaction between βC1 and the N-terminal region of NbROS1L was confirmed by co-immunoprecipitation (Fig. 2c). We then mapped the region of βC1 that is required for its interaction with NbROS1L, and identified a 10-amino acid motif, from residue 11 to 20, essential for the interaction (Supplementary Fig. 5a, b). Further mutational analysis showed that replacement of the Val in position 17 of βC1 with Ala (V17A) abolished the interaction with NbROS1L (Fig. 2d, e and Supplementary Fig. 5c), but did not affect its interaction with SAHH (Supplementary Fig. 5d, e), indicating that V17 is specifically required for the interaction between βC1 and NbROS1L (Fig. 2d, e).
Fig. 2. βC1 interacts with NbROS1L.
a BiFC analysis of the interaction of βC1 with NbROS1L. Scale bars, 50 μm. b BiFC analysis of the interactions of βC1 with the N-terminal region of NbROS1L (nNbROS1L) and the C-terminal region of NbROS1L (cNbROS1L). Scale bars, 50 μm. Schematic representation of NbROS1L protein structure is shown above. c Co-immunoprecipitation analysis of the interactions of βC1 with nNbROS1L and cNbROS1L. YFP was used as a negative control. d BiFC analysis of the interaction between βC1V17A and NbROS1L. Scale bars, 50 μm. e Co-immunoprecipitation analysis of the interaction between βC1V17A and nNbROS1L. YFP was used as a negative control. Images in a–e are representative of three independent experiments. Raw data for a, b and d and uncropped blots for c and e are provided in the source data.
The βC1-NbROS1L interaction is important for TYLCCNV+B virulence and viral DNA hypomethylation
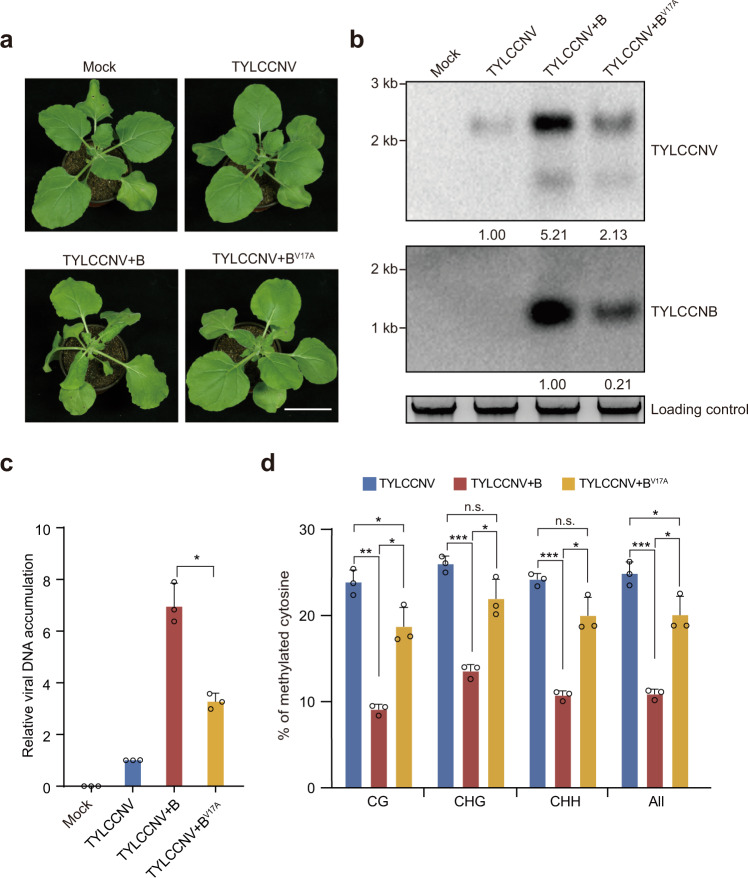
We next investigated whether the βC1-NbROS1L interaction is important for the TYLCCNB-mediated methylation decrease and promotion of virulence. For this purpose, we generated a TYLCCNB clone expressing βC1V17A. Interestingly, plants inoculated with TYLCCNV+BV17A showed very mild symptoms and lower levels of TYLCCNV/TYLCCNB accumulation (Fig. 3a–c and Supplementary Fig. 6a, b); concomitantly, higher levels of viral DNA methylation were detected (Fig. 3d). These results suggest that the interaction between βC1 and NbROS1L is important for the TYLCCNB-mediated decrease in viral DNA methylation and TYLCCNV/TYLCCNB virulence.
Fig. 3. The βC1-NbROS1L interaction is important for TYLCCNV+B virulence and viral DNA hypomethylation.
a Symptoms of N. benthamiana plants mock-inoculated or inoculated with TYLCCNV, TYLCCNV+B or TYLCCNV+BV17A at 10 dpi. Scale bar, 2 cm. b Accumulation of TYLCCNV and TYLCCNB in the leaves of indicated plants at 10 dpi as determined by Southern blot. Total DNA was stained with EB as loading control. c Accumulation of TYLCCNV in the leaves of indicated plants at 10 dpi as determined by qPCR. The coat protein gene of TYLCCNV was amplified. The N. benthamiana 25S nuclear rRNA gene (Nb25SrRNA) was used as an internal control. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; *p < 0.05. d Percentage of methylated cytosines in the intergenic region (IR) of TYLCCNV in the leaves of indicated plants at 10 dpi as determined by bisulfite sequencing. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant. Uncropped pictures, blots and gels for a and b, raw data and p values for c and d are provided in the source data.
The βC1-DME interaction promotes BSCTV virulence and viral DNA hypomethylation in Arabidopsis
To examine whether the role of βC1 in regulating active DNA demethylation and viral infection is conserved in other plant species, we tested if βC1 can interact with Arabidopsis DNA glycosylases. BiFC results revealed that βC1 could interact with DME (Supplementary Fig. 7a). Similar to what was observed for NbROS1L, we found that the N-terminal region of DME and the Val in position 17 of βC1 were required for the interaction (Fig. 4a and Supplementary Fig. 7b–e). We next determined whether the βC1-DME interaction promotes viral virulence. With this aim, we generated transgenic Arabidopsis expressing βC1 or βC1V17A, and then inoculated these plants with the Arabidopsis-infecting geminivirus beet severe curly top virus (BSCTV) (Supplementary Fig. 8a). Wild-type (Col-0) plants developed mild curly top symptoms after BSCTV infection, while the βC1-expressing plants developed dramatically more severe symptoms (Fig. 4b and Supplementary Fig. 8b). These symptoms were largely ameliorated when βC1V17A was expressed instead (Fig. 4b and Supplementary Fig. 8b). Consistent with this, a high level of BSCTV DNA was accumulated in the βC1-expressing plants, while viral accumulation was decreased in the βC1V17A-expressing plants (Fig. 4c, d and Supplementary Fig. 8c). The high accumulation of BSCTV DNA and the development of severe symptoms in βC1-expressing plants were associated with hypomethylation of BSCTV DNA, which was restored to a great extent in βC1V17A-expressing plants (Fig. 4e). These results suggest that the βC1-DME interaction is important for BSCTV DNA hypomethylation and virulence in Arabidopsis.
Fig. 4. The βC1-DME interaction promotes BSCTV virulence in Arabidopsis.
a Co-immunoprecipitation analysis of the interactions of βC1 or βC1V17A with the N-terminal region of DME (nDME). YFP was used as a negative control. Results are representative of three independent experiments. b Symptoms of Col-0, βC1-expressing lines and βC1V17A-expressing lines inoculated with BSCTV at 10 dpi. Scale bars, 2 cm. c Accumulation of BSCTV in the indicated plants at 10 dpi as determined by Southern blot. Total DNA was stained with EB as loading control. d Accumulation of BSCTV in the indicated plants at 10 dpi as determined by qPCR. The coat protein gene of BSCTV was amplified. The Arabidopsis 25S nuclear rRNA gene (At25SrRNA) was used as an internal control. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; **p < 0.01. e Percentage of methylated cytosines in the intergenic region (IR) of BSCTV in the indicated plants at 10 dpi as determined by bisulfite sequencing. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; *p < 0.05, ns not significant. Uncropped pictures, blots and gels for a, b and c, raw data and p values for d and e are provided in the source data.
βC1 promotes DME activity in vitro and in vivo
DME has both DNA glycosylase and apyrimidinic lyase activities, which mediate the removal of 5mC from double-stranded DNA and the cleavage of the DNA backbone at the abasic site to produce β- and β, δ-elimination products, respectively40,41. Because the βC1-DME interaction promotes viral DNA hypomethylation, we wondered whether the interaction can directly enhance the activity of DME. To test this, recombinant DME, GST-βC1, and GST-βC1V17A were expressed and purified (Supplementary Fig. 9a). A 5′-fluorescein-labeled oligonucleotide substrate containing 5mC was incubated with DME plus varying amounts of GST, GST-βC1, or GST-βC1V17A. β- and β, δ-elimination products were yielded when the substrate was incubated with DME. The amount of products was increased when GST-βC1 was added; however, this was abolished when GST-βC1 was replaced with GST-βC1V17A (Fig. 5a, b and Supplementary Fig. 9b). These results suggest that βC1 can directly enhance the activity of DME through protein–protein interaction.
Fig. 5. βC1 promotes DME activity in vitro and in vivo.
a In vitro 5mC excision activity of DME protein. Double-stranded oligonucleotide substrate (one strand methylated) was incubated with DME and varying amounts of GST, GST-βC1 or GST-βC1V17A. 17-nucleotide size marker and β- and δ-elimination products are indicated. b Quantification of relative DME activity. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; **p < 0.01. c Symptoms of wild-type and DME-overexpressing N. benthamiana lines mock-inoculated or inoculated with TYLCCNV, TYLCCNV+B or TYLCCNV+BV17A at 10 dpi. Scale bar, 2 cm. d Accumulation of TYLCCNV and TYLCCNB in the leaves of indicated plants at 10 dpi as determined by Southern blot. Total DNA was stained with EB as loading control. e Accumulation of TYLCCNV in the leaves of indicated plants at 10 dpi as determined by qPCR. The coat protein gene of TYLCCNV was amplified. The N. benthamiana 25S nuclear rRNA gene (Nb25SrRNA) was used as an internal control. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; *p < 0.05, ns not significant. f Percentage of methylated cytosines in the intergenic region (IR) of TYLCCNV in the leaves of indicated plants at 10 dpi as determined by bisulfite sequencing. Values are means ± SD (n = 3 independent experiments). Statistical significance was determined using two-tailed Student’s t test; **p < 0.01, ***p < 0.001, ns not significant. Uncropped pictures, blots and gels for a, c and d, raw data and p values for b, e and f are provided in the source data.
Given that DME activity was enhanced by βC1 in vitro, we asked whether this would also occur in vivo. To test this possibility, we generated transgenic N. benthamiana plants overexpressing DME (Supplementary Fig. 10a) and inoculated these plants with TYLCCNV alone, TYLCCNV+B, or TYLCCNV+BV17A. We found that DME-overexpressing plants, like wild-type plants, did not show any symptoms when inoculated with TYLCCNV alone (Fig. 5c and Supplementary Fig. 10b). However, they developed more severe symptoms than wild-type plants when inoculated with TYLCCNV+B, an effect that was abolished when the plants were inoculated with TYLCCNV+BV17A (Fig. 5c and Supplementary Fig. 10b). Consistent with this, DME-overexpressing plants inoculated with TYLCCNV+B had lower levels of viral DNA methylation and accumulated higher levels of TYLCCNV/B, compared to those inoculated with TYLCCNV+BV17A (Fig. 5d–f and Supplementary Fig. 10c). These data suggest that overexpression of DME enhances TYLCCNV infection by further decreasing the level of viral DNA methylation, and that the interaction between βC1 and DME is required for this process.
Discussion
Active DNA demethylation plays an important role in shaping genomic DNA methylation patterns and regulating transcription of many endogenous loci1. In this study, we show that active DNA demethylation targets DNA of TYLCCNV and its betasatellite TYLCCNB to promote their virulence. βC1 regulates the activity or targeting of active DNA demethylation for suppression of viral DNA methylation and enhancement of viral virulence.
DME was originally identified by mutations that cause maternal effects on seed viability11, and has been thought to be expressed and function specifically in reproductive tissues during gametogenesis for gene and transposon activation11,13,17. However, DME has been detected to be constitutively expressed in vegetative tissues by microarray, RNA-sequencing, RT-PCR and GUS reporter analyses12,42–45. DME also functions in vegetative tissues. Knockdown of DME expression leads to DNA hypermethylation at hundreds of genomic regions, which eventually leads to downregulated expression of dozens of genes under normal growth conditions45 and compromised activation of defense-related genes and increased plant susceptibility to infection when the plants are invaded by bacterial or fungal pathogens44,45. Our data suggest that DME can be activated by a viral suppressor protein to reduce DNA methylation level of the viruses in plant leaves. Our findings provide evidence that DME expression and function extend beyond reproductive tissues in Arabidopsis. In M. truncatula, DME is expressed in the root nodules, an organ formed in legumes through symbiotic interaction with nitrogen-fixing rhizobia, and DME expression is required for activation of many nodule differentiation-associated genes and normal development of nodules46. It will be interesting to investigate whether DME also targets genes of rhizobia and how its activity is regulated.
We showed that TYLCCNB is required for active DNA demethylation activity on viral DNA. We further found that βC1 interacts with DME and enhances the DNA demethylation activity of DME in vitro (Figs. 4a and 5a, b), suggesting that βC1 may increase NbROS1L and DME activity on viral DNA. This could be achieved through inducing structural or conformational changes. However, we cannot at this point exclude the possibility that βC1 may facilitate NbROS1L or DME targeting to the viral genome. Supporting this possibility, we found that loss of NbROS1L or overexpression of DME has no effect on viral DNA methylation level, viral replication, and virulence, in the absence of TYLCCNB or βC1 (Figs. 1 and 5). It is possible that access of NbROS1L/DME to the viral DNA requires assistance from another protein. Since βC1 has been shown to bind both single-stranded and double-stranded DNA in vitro, albeit lacking size or sequence specificity47, it could provide such function. Furthermore, βC1 can substitute for the DNA B component of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus, to help the DNA A component of ToLCNDV move and sustain systemic infection of tomato plants48, indicating that βC1 likely has DNA-binding ability in vivo. Nonetheless, it is possible that active DNA demethylation could also target plant endogenous genes to promote vial virulence.
We have previously found that βC1 also interacts with and inactivates SAHH to reduce viral DNA methylation22. The V17A mutation of βC1 disrupts the interaction between βC1 and NbROS1L or DME but does not affect the interaction between βC1 and SAHH, allowing us to uncouple the effects caused by βC1-mediated activation of NbROS1L or DME and those caused by βC1-mediated suppression of SAHH. We found that TYLCCNV DNA methylation level is lower when βC1 carries the V17A mutation (Fig. 3d) or when NbDMLs are silenced compared to that in TYLCCNV-inoculated control (Fig. 1d). Similarly, BSCTV DNA methylation level is lower in βC1V17A-OX plants than in Col-0 (Fig. 4e). These results suggest that βC1-mediated suppression of SAHH activity also contributes to βC1-mediated suppression of DNA methylation. Thus, DNA viruses have evolved to both suppress DNA methylation and boost active DNA demethylation as counter-defense strategies.
Methods
Plant materials and growth conditions
The 35S::RFP-H2B transgenic N. benthamiana line was previously described49. Other plant materials, including 35S::5 × Myc-DME N. benthamiana transgenic line and 35S::3 × Flag-βC1 and 35S::3 × Flag-βC1V17A Arabidopsis transgenic lines, were generated in this study. N. benthamiana plants were grown in a controlled growth chamber at 25 °C under a 16 h light/8 h dark photoperiod and Arabidopsis plants were grown in a growth room under long photoperiod conditions (16 h light, 22 °C/8 h dark, 18 °C).
Cloning
cDNAs of NbROS1, NbROS1L, NbDML3, NbDML4, NbDML5 and NbDML6 were PCR-amplified, cloned into the pLB vector (TIANGEN, VT205) and sequenced. Three cDNA fragments of NbROS1, NbDML3, and NbDML4 were fused by overlap PCR and the fusion product was cloned into the pTRV2 vector50, generating the TRV-based VIGS vector pTRV2-NbDMLs for knockdown of NbDMLs. For BiFC, the coding sequence of βC1 and cDNAs of NbROS1, NbROS1L, NbDML3, NbDML4, NbDML5, NbDML6, ROS1, DML2, DML3 and DME were PCR-amplified and cloned into p2YN and p2YC. For mapping the regions of NbROS1L and DME that mediate the interactions with βC1, cDNAs corresponding to the N-terminal and C-terminal regions of NbROS1L and DME were PCR-amplified and cloned into p2YN and p2YC. For mapping the region and residue of βC1 that mediate the interactions with NbROS1L and DME, the coding sequence of βC1 was PCR-amplified and cloned into the pLB vector. The construct was used to generate all the truncated and mutant forms of βC1 using the KOD-Plus-Mutagenesis Kit (TOYOBO, SMK-101). For co-immunoprecipitation, the coding sequences of βC1 and βC1V17A were PCR-amplified and cloned into pCambia1300-35S-YFP and pCambia1300-35S-3 × Flag to generate 35S-βC1-YFP, 35S-βC1V17A-YFP, 35S-3 × Flag-βC1, and 35S-3 × Flag-βC1V17A constructs. cDNAs corresponding to the N-terminal and C-terminal regions of NbROS1L were PCR-amplified and cloned into pCambia1300-35S-5 × Myc, generating 35S-5 × Myc-nNbROS1L and 35S-5 × Myc-cNbROS1L constructs. cDNAs corresponding to DME and the N-terminal regions of DME were PCR-amplified and cloned into pCambia1300-35S-5 × Myc, generating 35S-5 × Myc-DME and 35S-5 × Myc-nDME constructs. For the evaluation of TYLCCNV+BV17A virulence, the sequence of TYLCCNB was cloned into the pLB vector and the construct was used to introduce the βC1V17A point mutation, generating pLB-TYLCCNBV17A. The sequence of TYLCCNBV17A was PCR-amplified and cloned into pBinPLUS to generate pBinPLUS-TYLCCNBV17A using the ClonExpress II One Step Cloning kit (Vazyme, C112-02). pLB-TYLCCNBV17A was digested with KpnI and EcoRI and the product was cloned into pBinPLUS-TYLCCNBV17A, generating pBinPLUS-2TYLCCNBV17A. For the expression of recombinant proteins in E. coli, the coding sequences of βC1 and βC1V17A were PCR-amplified and cloned into the pGEX4T-3 vector and the coding sequence of DME were PCR-amplified and cloned into the pET11-6 × His-MBP vector51.
Agroinfitration and virus inoculation
All binary plasmids were individually transformed into Agrobacterium tumefaciens strain EHA105. The transformants were cultured in liquid LB with appropriate antibiotics, collected by centrifugation and resuspended in the infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.6, 100 mM acetosyringone). The suspensions were placed in dark for 2 h at room temperature before infiltration.
For TYLCCNV inoculation following VIGS, A. tumefaciens cultures carrying pCambia1300, pTRV1 plus pTRV2-GUS (TRV-GUS) or pTRV1 plus pTRV2-NbDMLs (TRV-NbDMLs) at an OD600 = 0.2 were inoculated into N. benthamiana plants at the eight-leaf stage. One week later, A. tumefaciens cultures harboring pBinPLUS (mock) or infectious clones of TYLCCNV or TYLCCNV plus TYLCCNB (TYLCCNV+B) at an OD600 = 1.0 were inoculated into pCambia1300-, TRV-GUS- or TRV-NbDMLs-infiltrated plants. For other TYLCCNV inoculation, N. benthamiana plants at the eight-leaf stage were directly agroinoculated with infectious clones of TYLCCNV, TYLCCNV+B or TYLCCNV+BV17A. For BSCTV inoculation, 4-week-old Arabidopsis plants were inoculated during bolting period and the bolts were cut at their base. The cut was applied with A. tumefaciens cultures carrying pCambia1300 (mock) or infectious clones of BSCTV and then insect pins were used to puncture the cut multiple times27. Symmetrically infected leaves were harvested at 10 days post inoculation.
RNA extraction and RT-qPCR
RNA was isolated using an Eastep Super Total RNA Extraction kit (Promega, LS1040) and 1 μg of total RNA was reversely transcribed by M-MLV reverse transcriptase (Invitrogen, 28025013). qPCR was conducted using SYBR Premix EX Taq (TaKaRa, RR420A). NbACT2 was used an internal control. Primer sequences are listed in Supplementary Table 1.
DNA extraction, Southern blot analysis, and qPCR
Total DNA was extracted from infected plant leaves using Plant Genomic DNA Kit (TIANGEN, DP305). For detection of viral DNA accumulation by Southern blot, 1 μg of total DNA was loaded onto 1.0% agarose gels and separated by electrophoresis. Agarose gels were denatured with denaturing buffer (0.5 M NaOH, 1.5 M NaCl) for 40 min and neutralized twice with neutralizing buffer (0.5 M Tris-HCl pH 7.0, 1.5 M NaCl) for 20 min. After denaturation and neutralization, total DNA was transferred to Hybond N+ nylon membranes (GE Healthcare, RPN303B). TYLCCNV+B and BSCTV DNA probes were labeled with [α-32P] dCTP using Random Primer DNA Labeling Kit Ver. 2 (TaKaRa, 6045) or with digoxigenin (DIG) using DIG High Prime DNA labeling and detection starter kit II (Roche, 11585614910). Membranes were hybridized at 65 °C overnight and washed three times with 1 × SSC, 0.1% SDS at 65 °C for 15 min. The radioactive signal was detected by Personal Molecular Imager (Bio-Rad) and the signal obtained with the DIG-labeled probes was detected by Image Lab (Bio-Rad). For detection of viral DNA accumulation by qPCR, the coat protein genes of TYLCCNV and BSCTV were amplified as described previously52. Primer sequences for generating the probes are listed in Supplementary Table 1.
DNA bisulfite sequencing analysis
Total DNA (500 ng) was subjected to bisulfite treatment using EZ DNA Methylation GoldTM Kit (Zymo Research, D5005), according to the manufacturer’s instructions. The bisulfite-treated DNA was used as templates for amplification of the IR region of TYLCCNV or BSCTV. Primer sequences are listed in Supplementary Table 1. The products were purified with AMPure XP beads (Beckman, A63880) and the purified DNA products were PCR-amplified and the adapters were added using GXL DNA polymerase (Takara, R050Q) and primer F (5′-AATGATACGGCGACCACCGAGATCTACAC-Index-TCGTCGGCAGCGTC-3′) and primer R (5′-CAAGCAGAAGACGGCATACGAGAT-Index-GTCTCGTGGGCTCGG)53. The final products were purified with AMPure XP beads for sequencing on an Illumina HiSeqX-Ten platform by Annoroad Gene Technology (Beijing). PCR-duplicated reads were removed and the unique reads with read counts above 5 were retained for cytosine methylation analysis using Kismeth (http://katahdin.mssm.edu/kismeth/revpage.pl).
BiFC assay
Equal volumes of A. tumefaciens cultures at an OD600 = 1.0. were mixed prior to infiltration. Leaves of 4-week-old 35S::RFP-H2B transgenic N. benthamiana line were infiltrated with A. tumefaciens cultures carrying pairs of BiFC constructs. Epidermal cells of the assayed leaves were examined by confocal laser scanning microscopy 980 (Zeiss, GER) 48 h post infiltration. For confocal imaging, the excitation wavelength for YFP was set at 514 nm and the emission was captured at 565–585 nm, and the excitation wavelength for RFP was set at 543 nm and the emission was captured at 590–630 nm. RFP-H2B signals were used to mark the nucleus.
Co-immunoprecipitation
Four-week-old N. benthamiana leaves were infiltrated with A. tumefaciens cultures carrying the pCambia1300 constructs. Forty-eight hours post infiltration, the leaves were collected and ground in liquid nitrogen. Total proteins were extracted from 2 g of ground powder in 4 ml of IP buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 0.5% Triton X-100, 5% glycerol, 5 mM DTT, 1 mM PMSF, and 1 Protease Inhibitor Cocktail tablet (Roche, 4693116001)). The extract was centrifuged three times at 20,000 × g for 10 min each time at 4 °C and the supernatant was collected. Twenty-five microliters of anti-c-myc-agarose affinity gel (Sigma–Aldrich, A7470) or GBP beads (homemade) were added to the supernatant and incubated at 4 °C for 2 h with rotation. After incubation, the beads were washed three times with IP buffer at 4 °C. Finally, 50 μl of 2 × SDS loading buffer were added to the sample for western blot analyses.
Western blot analysis
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare, 10600003). Membranes were incubated with primary antibodies and secondary antibodies sequentially. Primary antibodies we used include anti-GFP (Roche, 11814460001, 1:5000 dilution), anti-c-myc (Roche, 11667203001, 1:2000 dilution), anti-Flag M2 (Sigma, F1804, 1:5000 dilution), anti-HA (Roche, 11666606001, 1:5000 dilution), anti-actin (Sangon, D191048, 1:5000 dilution) and anti-α-tubulin (Sigma, T5168, 1:5000 dilution). Horseradish peroxidase-conjugated goat anti-mouse IgG (Sigma, A4416, 1:5000 dilution) was used as the secondary antibody. ECL prime western blot detection reagent (GE Healthcare, RPN2232) was added to the membranes for chemiluminescence detection using Image Lab (Bio-Rad).
Expression and purification of recombinant proteins
The constructs expressing βC1, βC1V17A and DME were transformed into E. coli BL21 (DE3) cells and transformants were grown at 37 °C until the OD600 reached 0.6. Protein expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside at 18 °C for 16 h. Cells were harvested and resuspended in lysis buffer (40 mM Tris-HCl pH 8.0, 500 mM NaCl, 10% glycerol and 1 mM PMSF). The suspension was lysed by sonication and centrifuged at 20,000 g for 1 h at 4 °C. For purification of His-tagged proteins, the supernatant was loaded onto a column packed with Ni-NTA. After washing with washing buffer (40 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole), proteins were eluted with elution buffer (40 mM Tris-HCl pH 8.0, 500 mM NaCl, and 500 mM imidazole). For purification of GST-tagged proteins, the supernatant was loaded onto a column packed with glutathione sepharose 4B agarose beads (GE Healthcare, 17-0756-01). The beads were washed with washing buffer (40 mM Tris-HCl pH 8.0, 500 mM NaCl) and proteins were eluted with elution buffer (40 mM Tris-HCl pH 8.0, 500 mM NaCl and 20 mM glutathione reduced). For purification of both His-tagged and GST-tagged proteins, gel filtration was performed on Superdex 200 increase size exclusion columns (GE Healthcare, 28-9909-44) with gel filtration buffer (40 mM Tris-HCl pH 8.0, 150 mM NaCl and 1 mM DTT) and peak fractions were collected.
DME activity assay
DME activity assay was performed as described54. A pair of Cy5 end-labeled oligonucleotides (one strand methylated) were synthesized and annealed, generating double-stranded oligonucleotide substrate. The substrate was incubated with 0.5 μM DME plus varying amounts of GST, GST-βC1 or GST-βC1V17A in a 15-μl reaction containing 40 mM Tris-HCl pH 8.0, 0.1 M KCl, 0.1 mM EDTA, 0.5 mM DTT, and 200 μg/ml BSA at 37 °C for 30 min. The reaction was terminated by the addition of 15 μl of loading buffer (95% formamide, 0.025% bromophenol blue, 0.025% SDS, 18 mM EDTA). The sample was heated at 95 °C for 10 min, and then chilled on ice. The sample was loaded onto 15% denaturing polyacrylamide gels and reaction products were separated by electrophoresis. Cy5 signal was detected using Typhoon (GE Healthcare) and quantified using ImageJ.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work is funded by grants from National Natural Science Foundation of China (31720103914, 31930089 and 31788103).
Source data
Author contributions
X.Z. and Y.Q. designed the experiments; X.G. performed the research; X.G., C.L., Y.Q., and X.Z. analyzed the data and wrote the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Bisulfite sequencing dataset generated in this study can be found in the NCBI Gene Expression Omnibus under accession number GSE188968. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yijun Qi, Email: qiyijun@tsinghua.edu.cn.
Xueping Zhou, Email: zzhou@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28262-3.
References
- 1.Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 2.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in. Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XY, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in. Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 6.Kankel MW, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 8.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roje S. S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry. 2006;67:1686–1698. doi: 10.1016/j.phytochem.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Gong ZH, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 11.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 12.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 13.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl Acad. Sci. USA. 2011;108:8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin W, et al. Active DNA demethylation regulates tracheary element differentiation in Arabidopsis. Sci. Adv. 2020;6:eaaz2963. doi: 10.1126/sciadv.aaz2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamuro C, et al. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khouider S, et al. Male fertility in Arabidopsis requires active DNA demethylation of genes that control pollen tube function. Nat. Commun. 2021;12:410. doi: 10.1038/s41467-020-20606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker PJ, et al. Changes to virus taxonomy and to the International Code of Virus Classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2021) Arch. Virol. 2021;166:2633–2648. doi: 10.1007/s00705-021-05156-1. [DOI] [PubMed] [Google Scholar]
- 20.Cui X, Tao X, Xie Y, Fauquet CM, Zhou X. A DNAbeta associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004;78:13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu T, et al. betaC1 protein encoded in geminivirus satellite concertedly targets MKK2 and MPK4 to counter host defense. PLoS Pathog. 2019;15:e1007728. doi: 10.1371/journal.ppat.1007728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, et al. Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011;7:e1002329. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Huang C, Li Z, Zhou X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014;10:e1003921. doi: 10.1371/journal.ppat.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang JY, et al. betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008;22:2564–2577. doi: 10.1101/gad.1682208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia Q, et al. CLCuMuB betaC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog. 2016;12:e1005668. doi: 10.1371/journal.ppat.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, et al. Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 2012;21:1294–1304. doi: 10.1111/j.1365-294X.2012.05457.x. [DOI] [PubMed] [Google Scholar]
- 27.Raja P, Sanville BC, Buchmann RC, Bisaro DM. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008;82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piedra-Aguilera A, et al. Integrated single-base resolution maps of transcriptome, sRNAome and methylome of tomato yellow leaf curl virus (TYLCV) in tomato. Sci. Rep. 2019;9:2863. doi: 10.1038/s41598-019-39239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deuschle K, Kepp G, Jeske H. Differential methylation of the circular DNA in geminiviral minichromosomes. Virology. 2016;499:243–258. doi: 10.1016/j.virol.2016.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Ermak G, Paszkowski U, Wohlmuth M, Mittelsten Scheid O, Paszkowski J. Cytosine methylation inhibits replication of African cassava mosaic virus by two distinct mechanisms. Nucleic Acids Res. 1993;21:3445–3450. doi: 10.1093/nar/21.15.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brough CL, et al. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 1992;18:703–712. doi: 10.1007/BF00020012. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Y. et al. Geminiviral V2 protein suppresses transcriptional gene silencing through interaction with AGO4. J. Virol.93, e01675-18 (2019). [DOI] [PMC free article] [PubMed]
- 33.Wang, L. et al. A virus-encoded protein suppresses methylation of the viral genome through its interaction with AGO4 in the Cajal body. Elife9, e55542 (2020). [DOI] [PMC free article] [PubMed]
- 34.Mei Y, Wang Y, Li F, Zhou X. The C4 protein encoded by tomato leaf curl Yunnan virus reverses transcriptional gene silencing by interacting with NbDRM2 and impairing its DNA-binding ability. PLoS Pathog. 2020;16:e1008829. doi: 10.1371/journal.ppat.1008829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Negrete E, et al. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. N. Phytol. 2013;199:464–475. doi: 10.1111/nph.12286. [DOI] [PubMed] [Google Scholar]
- 36.Wang, B., Yang, X., Wang, Y., Xie, Y. & Zhou, X. Tomato yellow leaf curl Virus V2 interacts with host histone deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 92, e00036-18 (2018). [DOI] [PMC free article] [PubMed]
- 37.Wang H, Hao L, Shung CY, Sunter G, Bisaro DM. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell. 2003;15:3020–3032. doi: 10.1105/tpc.015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ismayil A, et al. Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 2018;14:e1007282. doi: 10.1371/journal.ppat.1007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell. 2011;23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 43.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- 44.Schumann U, et al. DEMETER plays a role in DNA demethylation and disease response in somatic tissues of Arabidopsis. Epigenetics. 2019;14:1074–1087. doi: 10.1080/15592294.2019.1631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng, W. et al. Roles of DEMETER in regulating DNA methylation in vegetative tissues and pathogen resistance. J. Integr. Plant Biol.63, 691–706 (2020). [DOI] [PMC free article] [PubMed]
- 46.Satge, C. et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plants2, 16166 (2016). [DOI] [PubMed]
- 47.Cui X, Li G, Wang D, Hu D, Zhou X. A Begomovirus DNAbeta-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005;79:10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saeed M, Zafar Y, Randles JW, Rezaian MA. A monopartite begomovirus-associated DNA beta satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007;88:2881–2889. doi: 10.1099/vir.0.83049-0. [DOI] [PubMed] [Google Scholar]
- 49.Martin K, et al. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 50.Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. Virus-induced gene silencing. Methods Mol. Biol. 2003;236:287–294. doi: 10.1385/1-59259-413-1:287. [DOI] [PubMed] [Google Scholar]
- 51.Xie D, et al. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 2021;23:32–39. doi: 10.1038/s41556-020-00606-5. [DOI] [PubMed] [Google Scholar]
- 52.Zhong, X. et al. Mimic phosphorylation of a betaC1 protein encoded by TYLCCNB impairs its functions as a viral suppressor of RNA silencing and a symptom determinant. J. Virol. 91, e00300-17 (2017). [DOI] [PMC free article] [PubMed]
- 53.Jiang P, et al. 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat. Commun. 2020;11:5191. doi: 10.1038/s41467-020-19034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mok YG, et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc. Natl Acad. Sci. USA. 2010;107:19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bisulfite sequencing dataset generated in this study can be found in the NCBI Gene Expression Omnibus under accession number GSE188968. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper.