Abstract
To control sprouting angiogenesis, endothelial Notch signaling suppresses tip cell formation, migration, and proliferation while promoting barrier formation. Each of these responses may be regulated by distinct Notch-regulated effectors. Notch activity is highly dynamic in sprouting endothelial cells, while constitutive Notch signaling drives homeostatic endothelial polarization, indicating the need for both rapid and constitutive Notch targets. In contrast to previous screens that focus on genes regulated by constitutively active Notch, we characterized the dynamic response to Notch. We examined transcriptional changes from 1.5 to 6 h after Notch signal activation via ligand-specific or EGTA induction in cultured primary human endothelial cells and neonatal mouse brain. In each combination of endothelial type and Notch manipulation, transcriptomic analysis identified distinct but overlapping sets of rapidly regulated genes and revealed many novel Notch target genes. Among the novel Notch-regulated signaling pathways identified were effectors in GPCR signaling, notably, the constitutively active GTPase RND1. In endothelial cells, RND1 was shown to be a novel direct Notch transcriptional target and required for Notch control of sprouting angiogenesis, endothelial migration, and Ras activity. We conclude that RND1 is directly regulated by endothelial Notch signaling in a rapid fashion in order to suppress endothelial migration.
Subject terms: Cardiovascular biology, Cell migration, Cell signalling, Gene regulation, Angiogenesis
Introduction
Notch signaling regulates angiogenesis by controlling diverse endothelial cell behaviors, including migration, proliferation, metabolism, sprout formation, and barrier integrity. To control sprouting angiogenesis, Notch signaling must integrate these cellular steps in a temporal and context dependent manner1. While Notch acts as a receptor at the cell surface, activation by Notch ligands from the Delta like (DLL) or Jagged (JAG) families permit γ-secretase cleavage2,3 of the Notch Intracellular domain (ICD) which then translocates to the nucleus, forms a transcriptional complex with CSL/RBP-J and MAML, and drives transcription of genes containing RBP-J binding sites4,5. In endothelial cells, Notch transcriptional targets include those that are directly regulated, such as VEGFR3/FLT46, or those that respond to further transcriptional regulation, such as the transcriptional repressors Hairy and Enhancer-of-split family—HES and HEY genes1,7–9. Angiogenic control by Notch is driven by gene transcription and of both direct and secondary targets that control cellular behaviors, termed Notch effectors. Many Notch targets have been identified, but only in select cases has an effector function been delineated, for example NRARP controls vessel regression10,11 and EFNB2 promotes migration and proliferation12,13.
Genome-wide screens for Notch/CSL binding sites or direct Notch targets have been conducted in a variety of immortalized cell types, including T-lymphoblastic leukemia cells, myoblasts, and mouse kidney cells14–21. These screens have demonstrated that Notch targets vary widely by cell type, suggesting that targets identified in immortalized cells are not broadly applicable in endothelial cells where, for instance, Notch regulates the VEGF Receptor genes (VEGFR1, VEGFR2, and VEGFR3). Prior screens to identify Notch targets have generally utilized constitutive signaling via activating mutations or overexpression of Notch ICD. Paradoxically, examination of genes regulated by constitutive Notch ICD expression may not optimally detect direct Notch targets, due to rapid upregulation of transcriptional repressors of the Notch pathway1,7–9. In studies conducted in primary endothelial cells, microarray analysis of constitutively activated Notch signaling have identified downstream genes, such as VEGFR1, but have not been able to capture the range of rapidly regulated genes and did not distinguish between direct and indirect targets22,23.
We hypothesized that identification of genes rapidly regulated by endothelial Notch signaling in a timeframe similar to that of established direct targets would indicate novel direct Notch targets and reveal novel Notch effectors that function in angiogenesis. We therefore profiled transcriptional changes in EC in response to Notch induction and Notch repression in vitro and in vivo focusing on transcriptional events that happened within 1–6 h, rather than long-term changes in response to constitutive Notch signaling. We defined rapidly regulated Notch targets in complementary assays, including ligand-specific induction with tethered Delta-like ligand (DLL), Notch activation with EGTA, Notch inhibition by GSI in vitro and in vivo. Overlapping profiles from these different screens identified numerous previously unreported Notch regulated genes and an unappreciated mechanism of Notch signaling via GPCR and small GTPase proteins. Particularly interesting was RND1, a constitutively active Rho GTPase that is rapidly regulated by Notch, is a direct transcriptional target, controls endothelial Ras activity, and facilitates the control of endothelial migration by Notch.
Results
DLL4-activated and gamma-secretase dependent expression profiles identify endothelial Notch-regulated genes
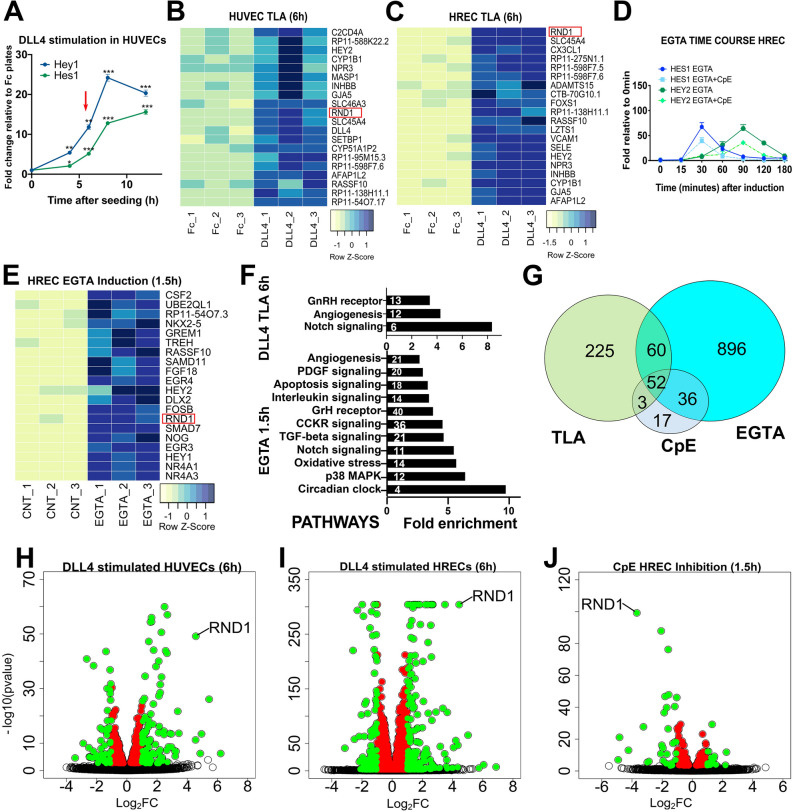
To characterize ligand-specific and rapid transcriptional changes downstream of Notch, we tethered the extracellular domain of recombinant Notch ligand DLL4 to plates on which we subsequently seeded ECs to perform a Tethered Ligand Assay (TLA) as previously described24. This approach allowed us to control the time of onset of Notch signaling (i.e. when the cells settle onto the ligand) and avoids super-physiologic and ligand non-specific activity caused by NICD25. In optimizing this assay, we documented that known Notch direct targets, such Hey1 and Hes1, could be consistently regulated with high significance at 6 h. We have therefore defined “rapidly-regulated” genes throughout this study as those which can be detected when known direct Notch targets are robustly induced. Human Umbilical Vein ECs (HUVECs) stimulated by tethered 10 ug/ml DLL4 showed robust activation of Notch canonical targets 6 h after seeding (Fig. 1A).
Figure 1.
Ligand Notch activation rapidly induces known and novel targets including RND1 in endothelial cells. (A) Time course of induction after seeding HUVEC onto DLL4-Fc coated plates as compared to Fc coated control plates (TLA assay). Robust activation of expression of known direct Notch targets was achieved as early as 6 h (red arrowhead). Single, double, and triple asterisks indicate p values of < 0.05 (*), < 0.01 (**), and < 0.001 (***), respectively, in all panels. (B–C) The 20 genes with the highest average fold induction in HUVEC (B) or HREC (C) by DLL4 TLA after 6 h. All heatmaps indicate z-score (standard deviations from the mean). RND1 is among the most significantly regulated Notch targets in both EC cell types. Red rectangle indicates RND1 in all heatmaps. (D) Induction time course of known Notch targets in HREC after 15 min of EGTA treatment or treatment with both EGTA and CpE. Robust activation of known direct Notch targets occurs between 30 and 90 min (0.5–1.5 h). (E) Heatmap of the 20 genes most highly upregulated by EGTA induction in HREC within 1.5 h shows that RND1 is rapidly significantly upregulated within the same timeframe as known direct Notch targets. (F) GO pathways significantly enriched in the 340 genes significantly upregulated by DLL4 TLA induction in both HUVEC and HREC (top) or the 1,105 genes significantly upregulated by EGTA (bottom). Inset indicates number of significantly regulated genes contributing to each pathway. The Notch signaling pathway was significantly regulated in both contexts. (G) Overlap between genes significantly upregulated by DLL4 TLA induction in both HUVEC and HREC, genes induced by EGTA, and genes repressed by CpE. (H–J) Volcano plots of gene expression changes in DLL4-stimulated HUVEC, DLL4-stimulated HREC, and CpE-inhibited EGTA-stimulated HREC (left to right, respectively). Uncolored circles indicate padj > 0.05, colored circles indicate padj < 0.05, red circles = fold change (FC) <|1.2|, green circles = FC >|1.2|.
To define the DLL4-regulated transcriptome, we selected two complementary primary endothelial cell types, HUVEC and Human Retinal ECs (HREC) (Fig. S1). HUVEC are proliferative, highly angiogenic, and the standard EC type for most assays of sprouting angiogenesis. Recent consensus among investigators studying angiogenesis suggests that results should be confirmed with a microvascular EC type26. We selected HREC as a microvascular EC of the central nervous system that is highly relevant to retinal assays of angiogenesis. We conducted triplicate DLL4-TLA assays with either HUVEC from two donor pools or HREC from a single donor, isolated RNA 6 h after cell seeding, and sequenced at a depth of 30 M single-end (SE) or 30 M paired-end (PE) reads for HUVEC and HREC, respectively. The complete RNA sequencing dataset is available in the NCBI Gene Expression Omnibus repository at https://www.ncbi.nlm.nih.gov/geo (Accession number GSE163568). DLL4-mediated Notch activation resulted in significant (padj < 0.05) changes in expression of hundreds of genes in both HUVEC (394 genes upregulated, 326 genes down-regulated) and HREC (1643 genes upregulated, 1701 genes downregulated) (Fig. 1B,C, S2A-B). 340 genes were upregulated at least 1.2 fold (Log2FC > 0.27) in both EC cell types. DLL4 TLA strongly upregulated known Notch targets HEY2, GJA5, and GUCY1A1 and GO pathways “Notch Signaling” and Angiogenesis”, consistent with Notch activation (Fig. 1F), but many of the most strongly regulated genes were not previously known to be Notch targets (Fig. 1B,C and Fig. S2G).
To detect transcriptional responses more rapidly than possible in the TLA assay and thus enrich for Notch direct targets, we screened for endothelial Notch targets using brief (15 min) exposure of HREC to EGTA, which promotes S2 cleavage of Notch within minutes of addition. Upregulation of known Notch targets HES1 and HEY2 were observed 30–90 min after HREC were treated with 10 mM EGTA and significantly repressed by 500 nM of the GSI Compound E (CpE) (Fig. 1D). This precise temporal activation of Notch permitted measurable responses within minutes and up to 2 h after Notch activation. Transcriptional profiling showed that EGTA treatment significantly upregulated 1105 genes by at least 1.2-fold in HREC after 1.5 h (Fig. 1E and S2C). To minimize conflation with other EGTA-activated pathways, we compared the transcriptional profile of EGTA-activated HREC to those pretreated with CpE to block S3 cleavage and inhibit Notch activation. 88 of the activated genes were significantly repressed by CpE (Fig. S2D-E).
Comparing transcriptional signatures from DLL4-mediated and gamma-secretase dependent Notch activation in two EC cell types identified a set of 52 commonly regulated genes likely to be direct Notch targets in endothelial cells (Fig. 1G). These included canonical Notch targets and a 55-fold enrichment for the Notch pathway signature (Fig. S2F-G). We also identified candidate Notch effectors not previously known to interact with Notch signaling, including small GTPases RND1 and RAPGEF5, and Ras-associated domain Family protein (RASSF10), highlighting potential novel roles for endothelial Notch in G protein regulation (Fig. S2G). Notably, the Rho Family GTPase 1 RND1 was clearly exceptional in both fold change and significance of response to Notch signaling (Fig. 1H–J).
RND1 and rapidly-regulated genes respond to Notch in multiple signaling contexts
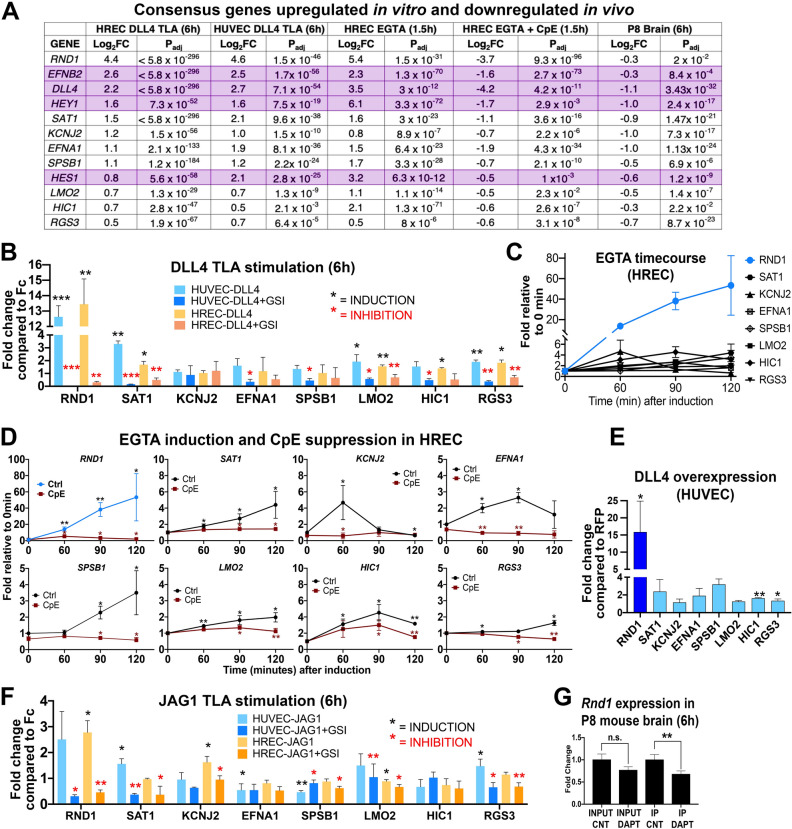
To confirm physiologic relevance in vivo, we examined endothelial transcripts from postnatal day 8 (P8) mouse brain using a RiboTag model27,28. Changes in the overall transcriptional profile of the P8 mouse brain will be described in a separate publication. Briefly, P8 pups were treated for 6 h with GSI DAPT to inhibit Notch signaling and endothelial transcripts were isolated from whole brain homogenates by immunoprecipitating endothelial-specific HA-tagged ribosomes. DAPT shows slightly different GSI activity than CpE at low concentrations, but both compounds exhibit strong inhibition of Notch cleavage and Notch signaling at the concentrations used in these experiments29. Comparison of the in vitro and in vivo rapidly regulated genes gave us a consensus set of 12 rapidly regulated genes (Fig. 2A). We examined the 12 consensus genes to determine if they are significantly regulated under a variety of in vitro and in vivo conditions. Of the consensus genes, 4 are previously established Notch targets (EFNB2, DLL4, HEY1, HES1), validating that our approach identifies Notch targets. The remaining 8 consensus genes have not previously been implicated in the Notch pathway and were not observed in analyses where constitutive Notch activation was examined in endothelial cells4,6,20,22.
Figure 2.
RND1 is the most highly regulated of the novel Notch targets in multiple contexts. (A) 12 genes were significantly regulated under all in vitro and in vivo RNA-seq screening conditions. Genes in purple are established Notch targets. (B) qPCR confirms that 7 of the 8 novel genes are regulated by DLL4 TLA in HUVEC and HREC, with RND1 showing the highest fold induction and the strongest inhibition by CpE. Black stars indicate significant induction by DLL4, red stars indicate significant inhibition of induction by CpE. (C) All novel targets are induced by EGTA; the most strongly induced gene is RND1 (blue line). (D) EGTA induction (blue or black lines) and CpE inhibition of EGTA (red lines) of the 8 novel putative endothelial Notch targets in HREC. Black stars indicate significant induction, red stars indicate significant inhibition of induction. The y axes are scaled for each gene to visualize induction of different magnitudes. (E) RND1, HIC1, and RGS3 remain induced after 48 h of DLL4 overexpression, while the other novel targets have transient expression that has returned to baseline after 48 h. (F) JAG1 TLA induces expression of most novel endothelial Notch targets at lower levels, suggesting that RND1 is a target of both DLL4 and JAG1 signaling. (G) qPCR of endothelial transcripts immunoprecipitated from the P8 mouse brain using an endothelial-specific RiboTag allele. Input = bulk brain homogenate, IP = immunoprecipitated endothelial mRNA.
We validated the 8 novel Notch target genes for response to a variety of Notch signaling activations. For all assays that had previously been analyzed by RNA-seq, we repeated experiments with independent batches of endothelial cells for qRT-PCR. Notch-responsive expression of candidate genes in HUVEC and HREC was confirmed using a DLL4 TLA assay to activate Notch signaling and the gamma-secretase inhibitor CpE to confirm Notch specificity. Of the 8 novel genes, 7 genes showed significant induction by DLL4 and/or repression by CpE after 6 h (Fig. 2B). RND1 had the highest magnitude of upregulation and was strongly repressed by GSI in both EC types, suggesting that RND1 is primarily under control of Notch signaling (Fig. 2B and S3A). Each of the 8 novel genes showed significant induction within two hours after EGTA addition when assessed by qPCR, but the magnitude and kinetics of induction varied considerably (Fig. 2C,D and Fig. S3A). KCNJ2, EFNA1, and HIC1 showed a rapid peak and then return towards baseline within 2 h, which may explain their relatively modest induction when examined after 6 h in the TLA assay. The other genes showed a steady increase in induction after EGTA addition, with RND1 again showing high magnitude induction. EGTA induction was significantly inhibited by CpE for all genes (Fig. 2D). Similar results were observed in HUVEC (Fig. S3A).
To determine whether the 8 novel endothelial Notch targets are regulated by constitutive Notch signaling, we examined expression levels in HUVEC constitutively overexpressing DLL4 for 48 h. SAT1, KCNJ2, EFNA1, SPSB1, and LMO2 were no longer significantly upregulated compared to RFP-infected controls, demonstrating that rapid response screening identifies unique Notch targets that may not be maintained under steady-state Notch signaling conditions (Fig. 2E). Induction of RND1 was maintained at approximately 15-fold above control levels under constitutive Notch signaling conditions, similar to the approximately 12-fold induction observed after 6 h in the TLA assay, suggesting that it is a target of both rapid and constitutive Notch signaling.
While DLL4 strongly activates endothelial Notch signaling and suppresses EC sprouting, the Notch ligand JAG1 is pro-angiogenic and has been variously proposed to function as weak endothelial Notch activator or a suppressor of DLL4/Notch signaling30–32. When we conducted a TLA using 40ug/ml JAG1-Fc, JAG1 significantly induced almost all of the novel Notch targets, including RND1, in at least one endothelial cell type, although the magnitude of induction was generally low (Fig. 2F). RND1 was activated 2–threefold by JAG1 signaling, in comparison to 12–14 fold activation by DLL4 signaling. These data suggest that JAG1 indeed activates endothelial Notch signaling and regulates many Notch targets that are regulated by DLL4, albeit at lower magnitude effect.
To confirm the in vivo relevance of RND1, we examined translated RNA from P8 endothelial RiboTag mouse brains treated with DAPT. Bulk brain homogenate (Input) showed no significant difference, suggesting that RND1 expression is Notch-independent in neuronal tissues, but endothelial cells showed a significant drop in RND1 mRNA, confirming that RND1 is rapidly regulated by Notch in the brain endothelium in vivo (Fig. 2G).
RND1 is a direct Notch target that facilitates Notch-mediated suppression of endothelial Ras activity
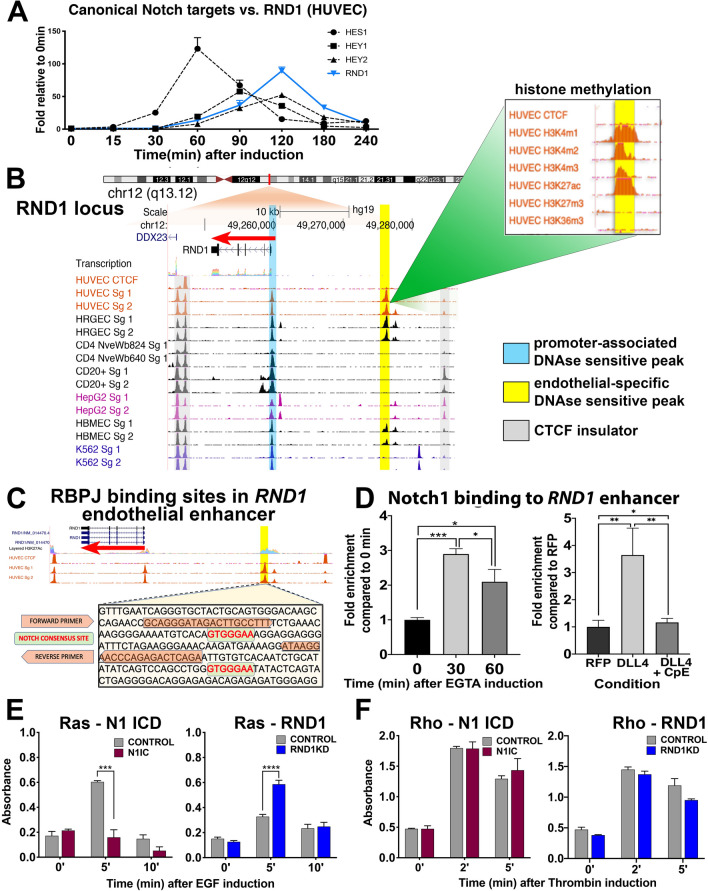
The various manipulations of Notch signaling that we tested dramatically induced or repressed RND1. We therefore determined whether RND1 is a direct or an indirect target of Notch signaling. EGTA activation of Notch signaling induced RND1 in a similar timeframe as known direct Notch targets (Fig. 3A). Examination of the ENCODE database indicated that open chromatin DNAse hypersensitivity (DHS) peaks were present near the RND1 transcriptional start site in a broad array of cell types, consistent with a promoter that facilitates expression of RND1 in diverse cell types [Fig. 3B, https://genome.ucsc.edu/encode/;33]. A separate DHS peak ~ 17 kb upstream from RND1 was observed only in endothelial cell types, suggesting that this region encodes an endothelial-specific RND1 enhancer (Fig. 3B). The putative EC enhancer exhibited histone marks typical of active enhancers34 and contains two RBPJ/CSL-binding consensus sequences (C/T)GTGGGAA, suggesting that this region may mediate Notch-responsive transcriptional regulation in EC (Fig. 3B,C)35,36.
Figure 3.
The RND1 locus contains an active endothelial specific enhancer with Notch binding sites. (A) RND1 is induced by EGTA at similar rates and magnitude as well-characterized HES and HEY Notch targets. (B) ENCODE database view of the RND1 locus shows a DNAse hypersensitivity peak at the promoter region in all cell types (blue bar) and a second peak upstream that appears only in endothelial cell types (yellow bar). The endothelial-specific peak shows histone methylation patterns consistent with active enhancers (insert box). Gray bars mark CTCF-binding insulator regions. (C) Enhancer region sequence in the RND1 locus with two Notch consensus sequence (green highlight). Also highlighted is the primer sequence for ChIP PCR (orange highlight). (D) ChIP assay of the putative endothelial-specific RND1 enhancer with antibodies against the intracellular domain of Notch1 (N1ICD). N1ICD binds to the RND1 enhancer after EGTA induction (left) or DLL4 overexpression, but binding is blocked with CpE treatment, indicating that active Notch signaling is required. (E) HUVECs transfected with siCon or siRND1 or infected with Control or Notch1IC lentiviral constructs were serum-starved for 3 h, treated with 100 ng/ml EGF, and analyzed for Ras activity by G-LISA after 0, 5, and 10 min. (F) HUVECs as in (E) were treated with 50 nM thrombin and analyzed for RhoA activity by G-LISA after 0, 2, and 5 min.
To determine whether the putative EC RND1 enhancer is bound by Notch, we conducted chromosome immunoprecipitation (ChIP) assays with antibodies against the Notch1 intracellular domain followed by PCR to detect the EC enhancer. In HUVEC, Notch1ICD bound to the EC RND1 enhancer within 30 min after EGTA treatment (Fig. 3D). Notch1ICD also bound to the EC RND1 enhancer in response to DLL4 expression in a GSI-repressible manner, demonstrating that binding depends on Notch signaling (Fig. 3D). We conclude that RND1 is a direct Notch target and regulation of RND1 may be facilitated by an endothelial enhancer found upstream of the transcriptional start site.
Examination of the remaining novel consensus genes in the ENCODE database identified putative endothelial-specific enhancers containing consensus or alternate RBPJ binding motifs in SAT1, KCNJ2, EFNA1, SPSB1, and RGS3 (Fig. S1B-C)21,37. Thus, we identified several other Notch target genes that rapidly respond to diverse means of Notch activation and whose regulatory elements include likely endothelial Notch-responsive enhancer elements, although their roles in angiogenesis are not clear in all cases. Several of the genes identified, such as KCNJ2 and LMO2 respond to VEGF-A38–41, suggesting that VEGF-induced activation of Notch signaling is critical for their regulation. SAT1 is enriched in tumor endothelial stalk cells, where Notch signaling is highly active42, and encodes a key regulator of polyamine content43, suggesting it may participate in Notch control of endothelial metabolism.
RND1 is a member of the Rho GTPase family which regulates the organization of actin cytoskeleton, cell migration, and cell adhesion44–47. Current studies of RND1 activity in HUVEC are conflicting: it has been reported to inhibit Ras activation with no effect on Rho activity, or to inhibit RhoA activation48,49. We observed that Notch activation in HUVEC significantly reduced Ras activation in response to EGF50, whereas RND1 knockdown significantly promoted Ras activation (Fig. 3E). We conclude that Notch suppression of Ras activity is facilitated, in part, by RND1. By contrast, Rho activation by thrombin51 was unaltered in HUVEC by Notch activation or siRND1, suggesting that Notch signaling does not affect this pathway (Fig. 3F).
Notch regulation of endothelial migration is mediated by RND1
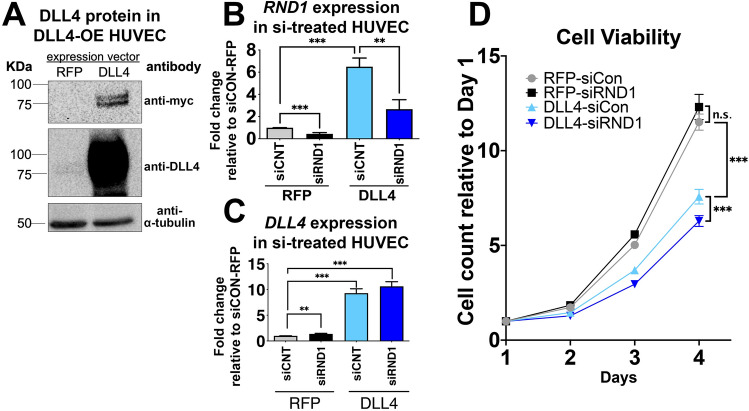
To determine if RND1 mediates Notch regulation of angiogenesis, we overexpressed DLL4 (DLL4 OE, Fig. 4A) to activate Notch and induce angiogenic responses and knocked down RND1 to establish whether this reduced the magnitude of Notch response. siRNA targeted against RND1 (siRND1) reduced RND1 transcript levels by approximately 70% at 24 and 48 h post-transfection, indicating a partial loss of RND1 function when angiogenic assays were initiated (Fig. 4B,C). DLL4 expression suppressed endothelial cell proliferation, indicative of Notch activation (Fig. 4D). RND1 knockdown did not substantively alter the growth rate of HUVEC or reverse the effects of DLL4 (Fig. 4D). We conclude that RND1 does not mediate endothelial growth suppression by Notch signaling.
Figure 4.
RND1 knockdown does not rescue Notch-mediated suppression of endothelial proliferation. (A) Transfection with a DLL4-myc expression construct induces DLL4 protein expression in HUVECs 3 days after lentivirus induction. All images are from the same blot, with uncropped version provided in Fig. S4. (B) RND1 siRNA robustly knocks down RND1 in HUVEC with normal levels of Notch signaling (RFP) or Notch signaling activated by DLL4-myc overexpression (DLL4). HUVECs were lentivirally transduced with RFP or DLL4 and transfected the next day with scrambled (siCNT) or siRNA targeting RND1 (siRND1). 3 days after lentivirus infection, HUVECs were harvested and analyzed by qPCR for RND1. (C) Treatment with siRNA1 does not grossly affect DLL4 levels in HUVEC treated as in (B). (D) Treatment with siRND1 does not restore proliferation in HUVEC where Notch signaling has been activated by DLL4. 104 HUVEC treated as in (B) were seeded onto 6-well plates cell and counted daily. Proliferation rate was normalized to counts at day 1.
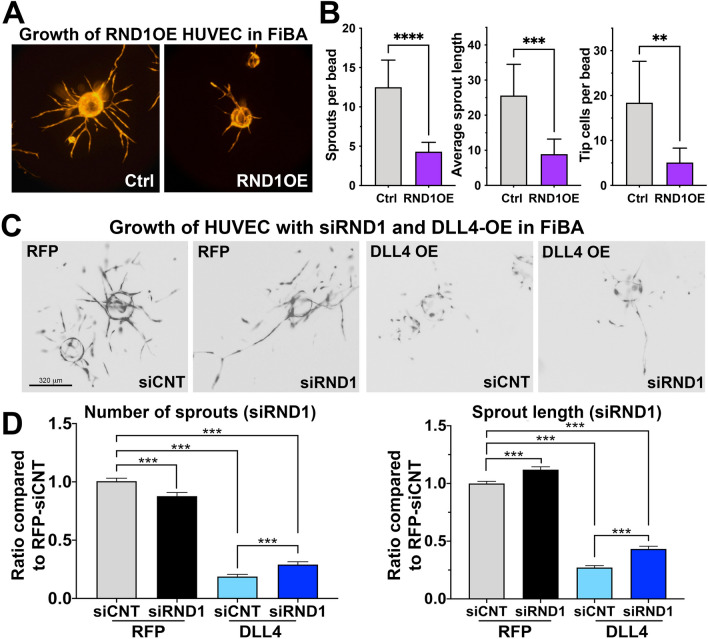
Notch signaling normally suppresses endothelial sprouting. We tested the effect of overexpression of RND1 in a 3D angiogenesis assay where endothelial cells are coated onto beads, which are then embedded in to fibrin gel (Fibrin Bead Assay or FiBA)52. Overexpression of RND1 dramatically reduced the number and length of endothelial sprouts and the number of sprouting tip cells (Fig. 5A,B). We therefore tested whether loss of RND1 can overcome Notch inhibition of sprouting. Notch signaling activation via DLL4 overexpression resulted in significant reduction of sprout numbers and length in FiBA assays, as expected (Fig. 5C,D). siRND1 resulted in a modest but significant restoration of sprout numbers and increased sprout length in Notch activated endothelial cells, demonstrating that sprout extension is suppressed in part by RND1 (Fig. 5C,D).
Figure 5.
RND1 is required for Notch-mediated suppression of sprout number and length. (A) Representative images of Fibrin bead angiogenesis (FiBA) assays using lentivirally-transduced HUVECs with full-length RND1 overexpression vectors (RND1OE) or empty vector (Ctrl). HUVEC were coated on cytodex beads, embedded in a fibrin gel with fibroblasts (D551) grown on the surface of the gel to provide growth factors, and allowed to develop for 5 days. (B) After 5 days, overexpression of RND1 reduces the number of sprouts, length of sprouts, and number of tip cells in a highly significant manner. (C) Lentivirally-transduced HUVECs with RFP or DLL4 expression constructs and were transfected with siCNT or siRND1 used for FiBA assays as above. Representative images of FiBA under each condition, scale bar indicates 320 µm. (D) After 5 days, Notch activation by DLL4 suppresses the number of sprouts from each bead, but knockdown of RND1 partially rescues sprout number (left). Notch activation by DLL4 suppresses the length of sprouts from each bead, but knockdown of RND1 partially rescues sprout length (right).
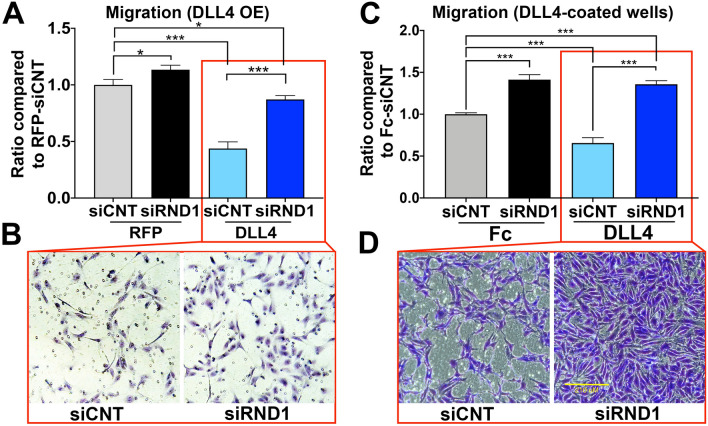
Notch activity suppresses endothelial migration and limits sprout extension via migration53. When Notch was activated via overexpression of DLL4, siRND1 knockdown substantially restored the ability of endothelial cells to migrate through transwell filters in response to VEGF (Fig. 6A,B). To ensure that DLL4 overexpression did not introduce artifacts, we repeated these experiments using DLL4-coated filters to activate Notch, which models physiologic paracrine presentation of DLL4, and confirmed that siRND1 restored the level of migration to control levels (Fig. 6C,D). These results indicate that Notch signaling requires RND1 as an effector protein to suppress VEGF-induced migration and sprout extension. We conclude that RND1 is a novel Notch effector necessary for Notch suppression of EC migration and links Notch to the Ras G-protein signaling pathway.
Figure 6.
RND1 is required for Notch-mediated suppression of EC migration. (A) Lentivirally-transduced HUVECs with RFP or DLL4 expression constructs were transfected with siCNT or siRND1 and plated in modified Boyden chambers and incubated with 50 ng/ml of VEGF for 6 h. Cells that had migrated through the pores were counted and normalized to their respective controls. DLL4 overexpression activation of Notch signaling suppresses endothelial migration (third column), but knockdown of RND1 rescues the endothelial migration to nearly control levels. (B) Representative images of cells that had migrated through Boyden chamber pores under each condition in (A). (C) Control and siRND1 transfected HUVECs were plated on a Boyden chamber inserts coating with fibronectin (5 µg/ml) and either Fc (control) or 10 µg/ml DLL4-Fc (Notch activation) and incubated with 50 ng/ml of VEGF for 6 h. Physiologic activation of Notch signaling by tethered DLL4 ligand suppresses endothelial migration (third column), but knockdown of RND1 increases migration under endogenous Notch signaling conditions (second column) or ligand-induced Notch signaling (fourth column). (D) Representative images of cells that had migrated through Boyden chamber pores under each condition in (B). The scale bar indicates 210 μm.
Discussion
Endothelial Notch signaling has distinct roles in stalk cells that must be controlled temporally; initially, Notch suppresses the tip cell phenotype, thus stimulating stalk cell attributes, this then allows Notch to subsequently promote vessel maturation. Tip and stalk cells have the ability to change places dynamically, depending on the level of Notch signaling54,55 implying that suppressing the tip cell phenotype may be coordinated with control of migration. In this study, we have identified a cohort of genes regulated by endothelial Notch within hours of signal activation or inhibition, revealing a previously underappreciated set of rapidly induced genes that are candidate effectors controlling dynamic tip/stalk regulation.
The endothelial Notch transcriptomes we identified include several GPCR regulatory proteins and GTPases, which directly control a multitude of cellular phenotypes and are excellent candidates for rapid phenotypic change but have not previously been implicated as Notch effectors. However, an overlapping but distinct set of GPCR proteins were regulated in each cell type examined, suggesting that endothelial cells from different vascular beds may employ unique GPCR effectors.
We posited that Notch signaling regulates separate effectors to control different endothelial cell phenotypes. Among the putative effector GTPases discovered, the small GTPase RND1 was consistently upregulated by Notch signaling in all cell types and signaling conditions examined, and was generally one of the most strongly regulated genes. We established that RND1 is a direct Notch target and that RND1 is required for Notch-mediated suppression of sprout extension, migration, and Ras activity, but not proliferation, confirming its role as an effector of specific Notch phenotypes.
To capture how Notch signaling carries out its diverse functions in endothelium, further examination of the rich set of novel, candidate Notch effectors identified by this study will facilitate greater understanding of Notch function in angiogenesis. RND1 was demonstrated to facilitate the inhibition of migration by Notch, but other candidates likely facilitate other cellular responses, such as proliferation, filopodia suppression, and promotion of barrier function. An appreciation of the “first wave” of rapidly regulated effectors that drive the role of Notch in sprouting angiogenesis will undoubtedly guide future design of therapeutic intervention in the Notch pathway.
Methods
Endothelial cell and tissue culture
Primary Human Umbilical Vein Endothelial Cells (HUVECs) were isolated by the Kitajewski lab from de-identified discarded human umbilical cords, which has been determined by the UIC Office for the Protection of Research Subjects (OPRS) to not meet the definition of humans subjects research. All OPRS guidelines were followed. HUVECs were isolated following established protocols56 and cultured in EGM-2 without hydrocortisone (Lonza, Cat#CC-3162) on dishes coated with rat tail type I collagen (Corning, Cat#354249). Endothelial identity was confirmed by assessing cobblestone morphology and VE-cadherin/CD31 double positivity at time of isolation (anti-CD31: Invitrogen MA5-13188, 1:100, anti-VE-cadherin: Cayman 160840, 1:100). To ensure retention of endothelial properties during culture, all HUVEC were used at passage 5 or lower, and cobblestone morphology and VE-cadherin/CD31 staining was re-assessed at passage 6 (Fig. S1). HUVEC batches A, A1, and B were used for experiments in Figs. 1, 2 and 3, batches U1 and U2 were used for experiments in Fig. 4.
Primary Human Retinal Microvascular Endothelial Cells (HREC, Cat. ACBRI 181, Passage 3) were obtained from a single vial purchased from Cell Systems, which validated these HREC for expression of CD31, VWF, and uptake of Ac-LDL (greater than 99% purity). HREC were cultured in EGM-2 with all bullet kit components on dishes coated with 0.2% gelatin (Sigma, G1393) at passage eight or lower. Retention of endothelial identity was re-assessed as above at passage 14 (Fig. S1). HEK293T cells and Detroit 551 (D551) human skin fibroblasts were purchased from ATCC and maintained DMEM (Gibco, Cat No. 11-995-073) with 10% FBS.
Tethered ligand assay (TLA)
The recombinant extracellular domains of the Notch ligands hDLL4-Fc (Cat. 10171-H02H, Sino Biologicals Inc.), hJAG1-Fc (Cat. 11648-H02H, Sino Biologicals Inc.) or IgG-Fc (Cat. 10702-HNAH, Sino Biologicals Inc.) were coated on 24-well plates (Corning, P/N 353226) in Fibronectin matrix (Cat. F1141, Sigma). Following an overnight incubation at 4 °C, primary ECs at 80% confluency were trypsinized and seeded onto the coated plates and incubated at 37 °C with 5% CO2 for indicated amount of time. Candidate targets identified by RNAseq were validated by RT-qPCR on a different frozen batch of HUVECs or HRECs.
RNA isolation and RT-qPCR
RNA from the cells or tissues were extracted using the RNeasy Mini Kit (Qiagen, Cat. 74104) as per the manufacturer’s recommendations. Reverse transcription was done using the Verso cDNA synthesis kit (Fischer Scientific, Cat. AB-1453/B) and target gene expression was assessed by Quantitative Real Time PCR (qPCR) using SYBR Green master mix (Applied Biosystems, Cat.4385612) and primers specific to genes of interest (Supplemental Table 1).
EDTA/EGTA Notch activation assay
Primary ECs at 70% confluency were treated with compound E (CpE, Cat. ALX-270–415-c250, Enzo Life Sciences) at 200 nM overnight to inhibit endogenous Notch signaling. The following day, cells were washed with PBS 2X and treated with 1XPBS with 10 mM EGTA for 15 min at 37 °C. After incubation, the PBS was replaced with fresh EGM-2. Start time was established as the moment that EGTA was added. RNA was collected as outlined above at time points from 30 min up to 4 h. For the CpE treatment group, 500 nM of CpE was added along with the EGTA at 0 h.
P8 RiboTag-based isolation of brain endothelial mRNA
All animal experiments were approved in advance by the University of Illinois Animal Care Committee and comply with the USPHS Policy on Humane Care and Use of Laboratory Animals and we report these methods in compliance with the ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines/experimental-animals). All mice were maintained on a congenic C57BL6/J background. Homozygous Rpl22tm1.1Psam; Cdh5-CreERT2 mice were mated and the day pups were first observed was assigned as postnatal day 0 (P0). Nursing females were gavaged with 250 mg/kg tamoxifen in oil at postnatal day (P) P1, P2 and P3 to induce Cre recombination. Recombination efficiency and tissue specificity of Rpl22HA expression was confirmed by sectioning the brains of P5 pups and immunostaining for HA. P8 pups (approximately 3.5 g each) of both sexes were randomized and injected subcutaneously with a single dose of 100 mg/kg DAPT (20 mg/ml in 10% ethanol 90% corn oil) or vehicle alone. Pups were humanely euthanized 6 h after injection via decapitation with a sharp blade and brains were snap frozen and homogenized. Endothelial-specific transcripts were obtained by immunoprecipitating RiboTag labeled ribosomes with anti-HA antibody (1:200, ab9118, Abcam) and purifying the resulting RNA using a Qiagen RNeasy mini kit. All genotyping was performed by Transnetyx.
Generation and validation of DLL4 and RND1 overexpressing HUVECs
To activate Notch signaling, we created a human DLL4 overexpression lentivirus vector (pCCL-PGK-DLL4) consisting of human DLL4 with C-terminal myc-6xHis-Tag in a pCCL vector and used tdTomato (RFP) expressing vector as a control. Lentivirus was generated in HEK293T cells through co-transfection of lentiviral packaging (pMDLg/pRRE and pRSV-Rev) and envelop vector (pCMV-VSV-g) with pCCL-PGK-RFP or pCCL-PGK-DLL4. For transduction with lentivirus, HUVECs were incubated with EGM-2 and the supernatant of the lentivirus infected HEK293T cells (2:1 ratio mixture) overnight and changed to fresh EGM-2 media the next day. Infection rate was estimated using RFP expression in cells. After 2 days, HUVECs were harvested with RIPA buffer (50 mM Tris (pH8.0), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% Sodium deoxycholate, 0.1% SDS) with Halt protease inhibitor cocktail (Invitrogen, Cat no. 78430). 20–40 μg of protein was loaded on 7% SDS-PAGE and DLL4 expression was evaluated with anti-myc antibody (1:1000 dilution, Sigma, Cat no. 06-549), anti-DLL4 antibody (1:1000 dilution, Cell Signaling, Cat no. 2589) and anti-α-tubulin (1:2000 dilution, Sigma, Cat No. T6074). Anti-rabbit-HRP (1:3000 dilution, Cell Signaling, Cat no. 5127) for myc and DLL4 and anti-mouse-HRP (1:3000 dilution, GE Amersham. Cat No. NA9310) were used and the specific signals were developed with ECL (GE Amersham. Cat No. RPN2209) and detected by ChemiDoc MP (Bio-Rad). To overexpress RND1, lentivirus containing pCCL-RND1, consisting of full-length unaltered human RND1, or empty pCCL vector were packaged and used to infect HUVEC as above.
Chromatin immunoprecipitation (ChIP) assay
3 × 106 HUVECs were treated with 10 mM EGTA for 15 min, then changed to new EGM-2 for 0, 30, and 60 min prior to fixation. Alternately, HUVECs were transduced with lentivirus-RFP or DLL4 and 3 × 106 cells were treated with 500 nM γ-secretase inhibitor (CpE) or vehicle control, incubated overnight, and fixed. Cells were fixed with 1% PFA at room temperature for 10 min. Cell nuclei were extracted using NP40 buffer (10 mM Tris (pH8.0), 10 mM NaCl, 0.5% NP40), lysed with SDS buffer (50 mM Tris (pH8.0), 1% SDS, 10 mM EDTA), and sonicated to an approximate DNA fragment length of 100–500 bp. Activated Notch intracellular domain (N1ICD) was precipitated by incubating the sheared genomic DNA with anti-cleaved Notch1 antibody (1:200 dilution, Cell signaling, Cat no. 4147) and Dynabeads Protein G (Invitrogen, 10003D) for 2 h at 4 °C. DNA was purified (ThermoFisher, Cat no. K0832) and probed for the RND1 enhancer with quantitative realtime PCR (qPCR). The N1ICD binding capacity was quantified by calculating the relative abundance between the experiment and control samples after normalizing to the respective inputs using the 2ΔΔCt method.
Fibrin bead assay (FiBA)
HUVECs were infected with empty vector, RND1OE, RFP or DLL4OE lentiviruses, then transfected with siRNA (control or RND1, 25 nM) using DharmaFECT 4 (Dharmacon, Cat no. T-2004-03) and trypsinized the following day. For FiBA, 6 × 104 cells were incubated (5% CO2, 37 °C) for 4 h with approximately 150 cytodex3 beads (5ul of 30 beads/ul suspension; Sigma, Cat No. C3275) suspended in EGM-2 to generate HUVEC-coated beads. HUVEC-coated beads were cultured on a 6-well plate overnight to eliminate loose HUVECs. HUVEC-coated beads were embedded in fibrin gel made with fibrinogen (3 mg/ml, Sigma, Cat No. F8630), aprotinin (1.5 TIU, Sigma, Cat No. A3428) and thrombin (312.5 mU, Sigma, Cat No. T4648). After gel formation, human dermal fibroblasts (D551, 1 × 105 cells) were seeded on top of the fibrin gel and supplemented with fresh EGM-2 media every day for 5 days. On the 5th day, the RFP positive sprouts from coated beads were assessed for the length and numbers of sprouts from 30 beads/group in three independent experiments using Image J.
VEGF induced migration
To evaluate Notch suppression of cell migration we overexpressed DLL4 or stimulated HUVECs with tethered DLL4-Fc recombinant protein. In the first approach, HUVECs transduced with lentivirus encoding RFP or DLL4 were administrated siRNA (control or RND1 siRNA, 25 nM) to knockdown RND1 expression, as described above. HUVECs were cultured overnight in the absence of added serum and VEGF in EGM-2 containing 0.2% FBS but no VEGF. 6–8 × 104 HUVECs were seeded on the upper chamber of a 0.1% gelatin coated 8 μm pore cell culture inserts (Fisher, Cat no. 08-771-21). In the second approach, 6–8 × 104 HUVECs transfected with control or RND1 siRNA as above were seeded on the upper chamber of 8 μm pore cell culture inserts coated with fibronectin (10 μg/ml, Cat. F1141, Sigma) mixed with human Fc or DLL4-Fc (10 μg/ml, Sino Biologicals Inc.) as described in the TLA assay.
In both assays, additional VEGF (50 ng/ml, R&D Cat no. 2179-VC-025/CF) was added to EGM-2 (containing 0.2% FBS but no VEGF) in bottom chamber. Cells were incubated for 6 h, fixed with 4% PFA (EMS, Cat no. 15712) for 10 min, and stained with 1% Crystal violet (Sigma, Cat no. C0775). Cells that had migrated through the pores and were adherent to the lower side of the insert were imaged to 5–7 different 10X fields per group. Three independent experiments were counted with imageJ and normalized to the corresponding control.
Detection of Ras activation and Rho activation
HUVECs were grown overnight in EBM-2 (containing 1% FBS) and changed to serum-free EBM-2 for 3 h the following day. The cells were then stimulated with 100 ng/ml hEGF for 5–10 min (Sigma-Aldrich, Cat No. E9644) for Ras activation, or with 50 nM Thrombin for 2–5 min (Enzyme Research Laboratories, Cat No. HT1002a) for RhoA activation. Assays were performed using RhoA (Cytoskeleton, Cat No. BK124), and Ras (Cytoskeleton, Cat No. BK131) G-Lisa Activation Assay Kits according to manufacturer’s recommendations and signal was measured at 490 nm absorbance.
Proliferation
HUVECs were transduced with lentivirus-RFP or DLL4 and transfected with control or RND1 siRNA (25 nM) as described above. 1 × 104 HUVECs were seeded on collagen coated 6 well plates and grown for indicated time. 30 fields per plate were imaged with 4X lens in three independent experiments. The cell growth rate was normalized to day 1 cell numbers.
RNA sequencing and data analysis
RNA quantity and integrity were measured using a Bio-analyzer (Agilent TapeStation 4200, UIC Genome Research core) prior to RNA sequencing. TLA HUVEC samples were sequenced at a ~ 30 million single-end (SE) read depth with 100-base fragments on the TruSeq platform in the Sulzberger Columbia Genome Center. TLA HREC samples and EGTA assay samples were sequenced at ~ 30 million paired-end (PE) reads with 150 base-pair fragments by Novogene (https://en.novogene.com/). Raw reads from in-vitro screens (HUVEC and HRECs) were mapped to the Human database (ENSEMBL/GRCh38) using STAR (version 2.5.0a) and processed with Samtools (version 1.4.1). The counts obtained by FeatureCounts (version 1.5.2) were analyzed by DESeq2 (version 1.18.1) to identify differentially expressed genes. The RNAseq datasets generated during this study are available in the NCBI Gene Expression Omnibus repository at https://www.ncbi.nlm.nih.gov/geo (Accession number GSE163568).
Statistics
For qPCR analysis, the ΔΔCt method57 was used to calculate the relative expression using following steps: (1) Normalization to reference gene: ΔCtGOI = CtGOI – CtBA. (2) Relative expression between conditions: ΔΔCtGOI = ΔCtEXP − ΔCtCNT. For other experiments, unless otherwise noted, t-tests or ANOVA with Bonferroni post-hoc analysis was performed on all quantified data to determine significant differences between groups using GraphPad Prism. P values less than 0.05 were considered statistically significant. p-values < 0.05 are flagged with one star (*), p values < 0.01 with two stars (**), and p-values < 0.001 with three stars (***). Unless otherwise noted, experiments were repeated at least three times.
Ethics declaration
All animal studies were done in accordance with UIC’s Institutional Animal Care and Use Committee guidelines.
Supplementary Information
Acknowledgements
We thank Dr. Reyhaan Chaudhri for help with optimizing the tethered ligand assays; James Herts for help with isolation and culturing endothelial cells; Dr. Kostandin Pajcini for technical guidance with the ChIP assay.
Author contributions
All authors have reviewed and approved this manuscript. B.S., J.D., S.W.Y., and S.R.V. designed and conducted experiments, collected data, interpreted results, and drafted the manuscript. B.S., J.B.M., H.F., C.Z., and P.S. contributed to study design and analyzed and interpreted the RNA sequencing data. B.S., S.W.Y., and L.A.N. designed and created data panels and assembled figures. R.K. contributed key insights. L.A.N. and J.K.K. guided conception of the study design, contributed to study analysis, contributed to manuscript drafting, and guided critical revisions and finalization of the manuscript.
Funding
This work was supported by NHLBI award 1R01 HL112626 (JKK), NIGMS RO1 GM55479 (RK).
Data availability
The RNAseq datasets were processed using publicly available softwares. Raw sequencing files and the processed files are deposited in NCBI Gene Expression Omnibus repository at https://www.ncbi.nlm.nih.gov/geo (Accession number GSE163568).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bhairavi Swaminathan, Seock-Won Youn and L. A. Naiche.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-05666-1.
References
- 1.Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish-Horowicz D, Pourquié O. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- 2.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 3.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 4.Borggrefe T, Oswald F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J. Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 6.Shawber CJ, et al. Notch alters VEGF responsiveness in human and murine endothelial cells by direct regulation of VEGFR-3 expression. J. Clin. Invest. 2007;117:3369–3382. doi: 10.1172/JCI24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor KL, Henderson AM, Hughes CC. Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc. Res. 2002;64:372–383. doi: 10.1006/mvre.2002.2443. [DOI] [PubMed] [Google Scholar]
- 8.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 9.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15:1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phng LK, et al. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev. Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iso T, et al. Dll4-selective Notch signaling induces ephrinB2 gene expression in endothelial cells. Biochem. Biophys. Res. Commun. 2006;341:708–714. doi: 10.1016/j.bbrc.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Zheng LC, et al. Ephrin-B2/Fc promotes proliferation and migration, and suppresses apoptosis in human umbilical vein endothelial cells. Oncotarget. 2017;8:41348–41363. doi: 10.18632/oncotarget.17298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hass MR, et al. SpDamID: Marking DNA bound by protein complexes identifies notch-dimer responsive enhancers. Mol. Cell. 2015;59:685–697. doi: 10.1016/j.molcel.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomero T, et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 2006;20:1963–1966. doi: 10.1038/sj.leu.2404409. [DOI] [PubMed] [Google Scholar]
- 16.Castel D, et al. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. U. S. A. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liefke R, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington LS, et al. Regulation of multiple angiogenic pathways by Dll4 and Notch in human umbilical vein endothelial cells. Microvasc. Res. 2008;75:144–154. doi: 10.1016/j.mvr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Lilly B, Kennard S. Differential gene expression in a coculture model of angiogenesis reveals modulation of select pathways and a role for Notch signaling. Physiol. Genom. 2009;36:69–78. doi: 10.1152/physiolgenomics.90318.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narui Y, Salaita K. Membrane tethered delta activates notch and reveals a role for spatio-mechanical regulation of the signaling pathway. Biophys. J. 2013;105:2655–2665. doi: 10.1016/j.bpj.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of Notch signaling. J. Cell Sci. 2000;113:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 26.Nowak-Sliwinska P, et al. Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis. 2018;21:425–532. doi: 10.1007/s10456-018-9613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez C, et al. Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J. Neurosci. 2014;34:10924–10936. doi: 10.1523/JNEUROSCI.0084-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T, Arslanova D, Gu Y, Augelli-Szafran C, Xia W. Quantification of gamma-secretase modulation differentiates inhibitor compound selectivity between two substrates Notch and amyloid precursor protein. Mol. Brain. 2008;1:15. doi: 10.1186/1756-6606-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedrosa A-R, et al. Endothelial Jagged1 Antagonizes Dll4 Regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler. Thromb. Vasc. Biol. 2015;35:1134–1146. doi: 10.1161/atvbaha.114.304741. [DOI] [PubMed] [Google Scholar]
- 31.Kangsamaksin T, et al. NOTCH decoys that selectively block DLL/NOTCH or JAG/NOTCH disrupt angiogenesis by unique mechanisms to inhibit tumor growth. Cancer Discov. 2015;5:182–197. doi: 10.1158/2159-8290.CD-14-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benedito R, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome browser: Update 2012. Nucleic Acids Res. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S, Mortazavi A. Integrating ChIP-seq with other functional genomics data. Brief. Funct. Genom. 2018;17:104–115. doi: 10.1093/bfgp/ely002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minoguchi S, et al. RBP-L, a transcription factor related to RBP-Jkappa. Mol. Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tun T, et al. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura M, et al. A genome-wide analysis identifies a notch-RBP-Jkappa-IL-7Ralpha axis that controls IL-17-producing gammadelta T cell homeostasis in mice. J. Immunol. 2015;194:243–251. doi: 10.4049/jimmunol.1401619. [DOI] [PubMed] [Google Scholar]
- 38.Coma S, et al. GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin-2. Angiogenesis. 2013;16:939–952. doi: 10.1007/s10456-013-9370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matrone G, et al. Lmo2 (LIM-Domain-Only 2) modulates Sphk1 (Sphingosine kinase) and promotes endothelial cell migration. Arterioscler. Thromb. Vasc. Biol. 2017;37:1860–1868. doi: 10.1161/ATVBAHA.117.309609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweighofer B, et al. The VEGF-induced transcriptional response comprises gene clusters at the crossroad of angiogenesis and inflammation. Thromb. Haemost. 2009;102:544–554. doi: 10.1160/TH08-12-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunitha P, et al. Temporal VEGFA responsive genes in HUVECs: Gene signatures and potential ligands/receptors fine-tuning angiogenesis. J. Cell Commun. Signal. 2019;13:561–571. doi: 10.1007/s12079-019-00541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Q, et al. Single-cell transcriptome analyses reveal endothelial cell heterogeneity in tumors and changes following antiangiogenic treatment. Cancer Res. 2018;78:2370–2382. doi: 10.1158/0008-5472.CAN-17-2728. [DOI] [PubMed] [Google Scholar]
- 43.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: A key metabolic regulator. Am. J. Physiol. Endocrinol. Metab. 2008;294:E995–1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 44.Gottesbuhren U, et al. Rnd3 induces stress fibres in endothelial cells through RhoB. Biol. Open. 2013;2:210–216. doi: 10.1242/bio.20123574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oinuma I, Katoh H, Harada A, Negishi M. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J. Biol. Chem. 2003;278:25671–25677. doi: 10.1074/jbc.M303047200. [DOI] [PubMed] [Google Scholar]
- 46.McCormack J, Welsh NJ, Braga VM. Cycling around cell–cell adhesion with Rho GTPase regulators. J. Cell Sci. 2013;126:379–391. doi: 10.1242/jcs.097923. [DOI] [PubMed] [Google Scholar]
- 47.Nobes CD, et al. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J. Cell Biol. 1998;141:187–197. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okada T, et al. The Rho GTPase Rnd1 suppresses mammary tumorigenesis and EMT by restraining Ras-MAPK signalling. Nat. Cell Biol. 2015;17:81–94. doi: 10.1038/ncb3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suehiro J, et al. Genome-wide approaches reveal functional vascular endothelial growth factor (VEGF)-inducible nuclear factor of activated T cells (NFAT) c1 binding to angiogenesis-related genes in the endothelium. J. Biol. Chem. 2014;289:29044–29059. doi: 10.1074/jbc.M114.555235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margolis B, Skolnik EY. Activation of Ras by receptor tyrosine kinases. J. Am. Soc. Nephrol. 1994;5:1288–1299. doi: 10.1681/ASN.V561288. [DOI] [PubMed] [Google Scholar]
- 51.van Nieuw Amerongen GP, van Delft S, Vermeer MA, Collard JG, van Hinsbergh VW. Activation of RhoA by thrombin in endothelial hyperpermeability: Role of Rho kinase and protein tyrosine kinases. Circ. Res. 2000;87:335–340. doi: 10.1161/01.res.87.4.335. [DOI] [PubMed] [Google Scholar]
- 52.Tattersall IW, et al. In vitro modeling of endothelial interaction with macrophages and pericytes demonstrates Notch signaling function in the vascular microenvironment. Angiogenesis. 2016;19:201–215. doi: 10.1007/s10456-016-9501-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leslie JD, et al. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 54.Chen W, et al. The endothelial tip-stalk cell selection and shuffling during angiogenesis. J. Cell Commun. Signal. 2019;13:291–301. doi: 10.1007/s12079-019-00511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakobsson L, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 56.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq datasets were processed using publicly available softwares. Raw sequencing files and the processed files are deposited in NCBI Gene Expression Omnibus repository at https://www.ncbi.nlm.nih.gov/geo (Accession number GSE163568).