Abstract
This report describes a rapid detection procedure for salmonellae from chicken feces by the combination of tetrathionate primary enrichment (preenrichment [PE])-bacterial lysis-capillary PCR and capillary gel electrophoresis. Pure Salmonella enterica serovar Enteritidis 64K was reisolated and detected by capillary PCR after buffered peptone water and nutrient broth, tetrathionate broth base Hajna (TTBH), and tetrathionate broth (TTB) preenrichments. When the same culture was mixed with intestinal homogenate, bacteriological reisolation and capillary PCR detection was achieved only by TTBH and TTB preenrichments. Capillary gel electrophoresis revealed that a Salmonella genus-specific 281-bp PCR product was detected when Salmonella strains but not non-Salmonella strains were tested. The detection limit of capillary PCR with whole-cell DNA extracted from pure Salmonella enterica serovars Enteritidis 64K, Typhimurium LT2-CIP60-62, and Gallinarum 64K was 3, 3, and 9 CFU ml−1, respectively. The detection limit of capillary PCR from whole-cell DNA extracted from intestinal homogenate artificially contaminated with the same three strains was 3, 3, and 7 CFU ml−1, respectively. We compared the results of the capillary PCR and bacteriological examination from the natural samples. Thirty-five of 53 naturally contaminated samples produced a specific PCR product. In 9 of the 35 PCR-positive samples, Salmonella could not be detected bacteriologically either by PE or a primary and delayed secondary enrichment (DSE) combination. In the 18 PCR-negative samples, 4 samples were found to harbor Salmonella by both PE and DSE and 14 samples were positive after DSE. Fifty-three additional intestinal homogenate samples, which were negative by their PE and DSE in bacteriological examination, were found to be also negative by their PCRs. The total time required to detect Salmonella with the capillary PCR method we used was approximately 20 h. If samples are from clinically diseased birds, the total time for PCR and detection is reduced to 2 h since the 18-h PE is not required. These results indicate that TTB enrichment, bacterial lysis, and genus-specific capillary PCR combined with capillary gel electrophoresis constitute a sensitive and selective procedure which has the potential to rapidly identify Salmonella-infected flocks.
Salmonellae are among the major bacterial pathogens of poultry in Turkey and in the world (3, 8). Prevention of Salmonella infection is important for poultry health and for the food industry, and prevention can be achieved only by good monitoring and screening programs (10, 17, 27). Salmonella detection by bacteriological methods usually requires 5 to 11 days (10, 27), and samples with low numbers of Salmonella cells, usually seen in subclinically infected chickens, may give false-negative results (7). Efforts have been made to reduce the time required and to increase the sensitivity of methods to detect Salmonella serovars in poultry samples (16, 25). PCR with preincubation in an enrichment broth has been performed for human (5, 13, 14, 28), animal (6, 23, 24), fecal, and food (1, 2, 4, 9, 11, 19) samples. This has been found to be a useful and more rapid method because it increases the number of viable Salmonella in the sample and, therefore, increases the sensitivity of the assay (5, 9, 11, 22). PCR methods and DNA extraction procedures for Salmonella have been described for human, animal, and food samples (5, 6, 21, 23, 24). However, little information exists about these techniques used with poultry fecal samples (15, 26). These methods seem to be expensive and time-consuming (15, 26). This study describes a practical DNA extraction procedure from primary enrichment cultures of Salmonella, genus-specific capillary PCR, and capillary gel electrophoresis to detect chickens carrying the viable Salmonella in their intestines.
MATERIALS AND METHODS
Bacteria.
Salmonella enterica serovars Enteritidis 64K, Typhimurium LT2-CIP60–62, and Gallinarum 64K were obtained from M. Y. Popoff, Institut Pasteur, 28 rue du Dr Roux, 75015 Paris Cedex 15, France, and non-Salmonella strains (Citrobacter sp., Escherichia coli, Klebsiella sp., Pseudomonas aeruginosa, and Streptococcus sp.) were from the Department of Microbiology, Medical School, and Faculty of Veterinary Medicine, Uludag University, Bursa, Turkey.
Comparison of different PE broths.
Ileocecal parts of healthy chickens, which were negative when tested with Salmonella antigen (Intervet) were dissected and minced in sterile conditions. One gram from this mince was used as an intestinal homogenate in the following procedure. Pure Salmonella serovar Enteritidis cultures, with and without intestinal homogenate, were grown in buffered peptone water (BPW) (L37; Oxoid), nutrient broth (NB) (CM67; Oxoid), tetrathionate broth (TTB) (CM29; Oxoid), and tetrathionate broth base Hajna (TTBH) (0491-17; Difco). After an 18-h incubation at 37°C, 20 μl from each tetrathionate primary enrichment (preenrichment [PE]) was plated onto xylose-lysine-Tergitol 4 agar (XLT4) (0234-17-9; Difco) and 1 ml was used to prepare a template for PCR.
Cultivation of Salmonella serovars.
The purity of Salmonella strains was verified, and biochemical and antigenic identifications were performed. One colony from each strain was inoculated into 10 ml of TTB. Cultures were incubated at 37°C for 18 h. One milliliter from each culture was taken into a sterile Eppendorf tube and was stored at −20°C until used.
DNA extraction.
Crude DNA was prepared by modifying the method described by Soumet et al. (21). One milliliter of TTB culture was centrifuged for 4 min at 4,600 × g. The pellet was suspended in 0.85% saline, was centrifuged, and was resuspended in 20 μl of deionized water. This bacterial suspension was then boiled for 10 min and was centrifuged for 3 min at 18,000 × g. Five microliters of the supernatant was used as a template in PCR.
Oligonucleotide primers and capillary PCR.
Salmonella genus-specific primers 139 and 141 were described by Rahn et al. (18) and have, respectively, the following nucleotide sequences based on the invA gene of Salmonella: 5′-GTG AAA TTA TCG CCA CGT TCG GGC AA-3′ and 5′-TCA TCG CAC CGT CAA AGG AAC C-3′. Both primers were synthesized in the Expetide DNA synthesizer (Perseptive Biosystems) and were purified using reverse-phase high-pressure liquid chromatography (BioCAD700E; Perseptive Biosystems). Primer 139 (18) was synthesized as 6-FAM labeled. The 25-μl PCR mixture, which contained 0.3 μl of Taq DNA polymerase (IonTaq; 5 U/μl), 2.5 μl of 10× PCR buffer (3.5 mM MgCl2), 2.5 μl of deoxynucleoside triphosphate (dNTP) mixture (2 mM), 1 μl of each primer (5 pmol/ml), 5 μl of template DNA, 2 μl of bovine serum albumin (2.5 mg/ml), and 10.2 μl of deionized water, was taken into borosilicate capillary tubes (Idaho Technology) by capillary action. The expected product size was reported as 284 bp (18).
PCR incubations were performed using a DNA Air Thermal Cycler, model 1605 (Idaho Technologies). The cycle conditions were as follows: an initial incubation at 94°C for 15 s followed by 30 cycles of denaturation at 94°C for 0 s, primer annealing at 50°C for 0 s, and primer extension at 72°C for 15 s. Following the last cycle, there was a 5-min incubation at 72°C.
Capillary gel electrophoresis.
Capillary gel electrophoresis was performed on an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer). Experimental conditions were optimized by using PCR products from Salmonella serovars Enteritidis and Gallinarum. After capillary PCR, 2 μl from the reaction mixture was added to 12.5 μl of sampling buffer (12 μl of formamide, 0.5 μl of GeneScan 500 standard). The GeneScan 500 standard was labeled with the fluorescent dye TAMRA. The final mixture was heated at 95°C for 5 min and was chilled on ice before being loaded on the analyzer. We used Performance Optimized Polymer 6 (part no. 402837; Perkin-Elmer Applied Biosystems, ABI PRISM) for analysis. The electrophoresis conditions were as follows: capillary, ABI PRISM 2 (61 cm by 50 μm; part no. 402840; Perkin-Elmer Applied Biosystems 310 Genetic Analyzer Capillaries); temperature, 50°C; and electric field, 15 kV. Results were analyzed by GeneScan Analysis Software 2.1 (Perkin-Elmer).
Detection-limit determination of capillary PCR with pure culture.
Initially, we determined the CFU of Salmonella serovars Enteritidis, Typhimurium, and Gallinarum by plating 10 μl from 10-fold dilutions of their 5-ml TTB stock cultures to XLT4 agar. The dilutions were performed up to 10−12 for each culture. We determined the stock concentrations of the above-mentioned strains as 3 × 108, 3 × 109, and 9 × 108 CFU ml−1, respectively. At the same time, we sampled 1 ml from each dilution to use for DNA extractions and as templates in PCR.
Sensitivity and detection-limit determination of capillary PCR with experimental samples.
In order to determine the detection sensitivity of PCR, we used PE cultures (TTB) of artificially spiked samples with Salmonella serovars Enteritidis, Typhimurium, and Gallinarum. Briefly, 0.2 and 1 g of Salmonella-free chicken iliocecal samples (determined by a bacteriological method and PCR) were minced and added into 10 ml of TTB. Tenfold dilutions from 3 × 103 CFU ml−1 to 3 CFU ml−1 of serovars Enteritidis and Typhimurium and 7 × 103 CFU ml−1 to 7 CFU ml−1 of serovar Gallinarum were inoculated into TTB with intestinal homogenate and were incubated at 37°C for 18 h. PCR was performed by using 1 ml from each culture as described above. Salmonella growth was confirmed by plating from each dilution.
Specificity determination of PCR.
Citrobacter sp., E. coli, Klebsiella sp., P. aeruginosa, and Streptococcus sp. were cultured aerobically at 37°C for 18 h in brain heart infusion (BHI) broth. The DNA extraction procedure and PCR method used were the same as described for Salmonella.
Capillary PCR with natural samples.
A total of 0.2 g from each of 53 chicken iliocecal samples from five different flocks harboring Salmonella and 53 Salmonella-free samples, determined by the bacteriological method described in the National Poultry Improvement Plan of the U.S. Department of Agriculture, Animal Health Inspection Service (27), were subjected to our capillary PCR. Also, a rapid slide agglutination test was performed with blood samples from these chickens to diagnose Salmonella serovar Gallinarum and Salmonella serovar Gallinarum biotype Pullorum infections. Salmonella serovar Enteritidis TTB cultures and uninoculated TTB were used as positive and negative controls in PCR, respectively.
Bacteriological examination of natural samples.
PCR results were compared with those of the bacteriological examination described in the National Poultry Improvement Plan, U.S. Department of Agriculture (27). Natural samples were inoculated into both TTB and TTBH and were incubated at 41°C for 24 h. Twenty microliters from these cultures were plated onto XLT4 agar and were incubated at 37°C for 24 h. For delayed secondary enrichment (DSE), broth cultures were additionally incubated at 22°C for 5 days, and 1 ml from 5-day incubated broth cultures was transferred into 10 ml of freshly prepared TTB or TTBH. After 24 h of incubation at 37°C, these cultures were streaked onto XLT4 agar. The plates were examined for Salmonella colonies after 18 h of incubation at 37°C. Suspected Salmonella colonies were subjected to biotyping (27) and serogrouping (Difco Manual, Difco Laboratories, Detroit, Mich.).
RESULTS
Evaluation of capillary gel electrophoresis.
Electrophoresis for one sample was completed in 35 min. For each sample, the fluorescence intensity was plotted on the y axis and the size in base pairs (bp) on the x axis. The values on the x axis are to denote GeneScan 500 size-standard fragments and to compare the specific PCR products where the peaks were detected. In the electropherograms, we could detect precisely the 281-bp specific PCR product analyzed by the GeneScan 672 software.
Comparison of different primary enrichment broths.
Using the 18-h pure cultures of Salmonella serovar Enteritidis in BPW, NB, TTB, and TTBH, the bacterium was isolated by the bacteriological method and was detected by capillary PCR. By contrast, when intestinal homogenate (artificially spiked with Salmonella) was added into the PE broths, bacteriological reisolation and specific PCR amplification was observed only in TTB and TTBH cultures. We did not observe any detection superiority of TTBH over TTB, and we pursued the tests with natural samples by using only TTB.
Detection limit of capillary PCR with pure Salmonella cultures.
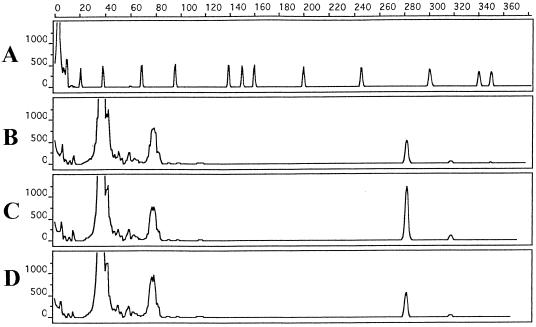
PCR could detect 3 CFU ml−1 of serovar Enteritidis, 3 CFU ml−1 of serovar Typhimurium, and 9 CFU ml−1 of serovar Gallinarum (Fig. 1). These numbers correspond to the dilutions of 10−8 for serovar Enteritidis, 10−9 for serovar Typhimurium, and 10−8 for serovar Gallinarum stock cultures. PCR was able to detect Salmonella DNA from these high dilutions because we used 1 ml from the dilutions for DNA preparation. However, because of the high dilution, when 10 μl was plated from these dilutions, we did not observe any colonies in the corresponding plates.
FIG. 1.
Sensitivity determination of TTB enrichment-capillary PCR-capillary gel electrophoresis with pure Salmonella enterica serovars Enteritidis, Typhimurium, and Gallinarum. x axis, size in base pairs; y axis, relative fluorescence intensity. Electropherograms: (A) peak patterns of GeneScan 500 size-standard fragments run under denaturing conditions; (B, C, and D) 281-bp PCR product detected from 3, 7, and 9 CFU of pure serovar Enteritidis, serovar Typhimurium, and serovar Gallinarum cultures/ml, respectively. The GeneScan 500 molecular weight marker dye was scaled up five times.
Sensitivity and detection limit of capillary PCR with experimental samples.
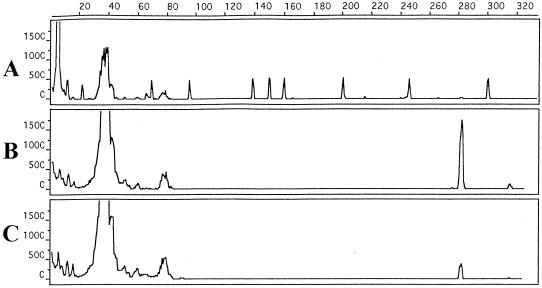
The detection limit of TTB enrichment containing intestinal homogenate with capillary PCR was as follows: 3 CFU ml−1 for serovars Enteritidis (Fig. 2) and Typhimurium (Fig. 3), and 7 CFU ml−1 for serovar Gallinarum (Fig. 4). Also, PCR was not inhibited from the intestinal homogenate amount. Salmonella was recovered by plating from each dilution. This indicated and confirmed that Salmonella was present as a template in each PCR.
FIG. 2.
Sensitivity determination of TTB enrichment-capillary PCR-capillary gel electrophoresis with chicken intestinal homogenate artificially contaminated with Salmonella serovar Enteritidis. x axis, size in base pairs; y axis, relative fluorescence intensity. Electropherograms: (A) peak patterns of GeneScan 500 size-standard fragments run under denaturing conditions; (B and C) 281-bp PCR product detected from 3 × 103 and 3 × 102 CFU of serovar Enteritidis cultures with chicken intestinal homogenate/ml. The GeneScan 500 molecular weight marker dye was scaled up five times.
FIG. 3.
Sensitivity determination of TTB enrichment-capillary PCR-capillary gel electrophoresis with chicken intestinal homogenate artificially contaminated with Salmonella serovar Typhimurium. x axis, size in base pairs; y axis, relative fluorescence intensity. Electropherograms: (A) peak patterns of GeneScan 500 size-standard fragments run under denaturing conditions; (B, C, and D) 281-bp PCR product detected from 3 × 103, 3 × 102, and 3 CFU of serovar Typhimurium cultures with chicken intestinal homogenate/ml, respectively. The GeneScan 500 molecular weight marker dye was scaled up four times.
FIG. 4.
Sensitivity determination of TTB enrichment-capillary PCR-capillary gel electrophoresis with chicken intestinal homogenate artificially contaminated with Salmonella serovar Gallinarum. x axis, size in base pairs; y axis, relative fluorescence intensity. Electropherograms: (A) peak patterns of GeneScan 500 size-standard fragments run under denaturing conditions; (B and C) 281-bp PCR product detected from 7 × 102 and 7 CFU of serovar Gallinarum cultures with chicken intestinal homogenate/ml, respectively. The GeneScan 500 molecular weight marker dye was scaled up two times.
Specificity of PCR.
None of the non-Salmonella strains yielded a PCR product. A 281-bp PCR product, which is close to the expected product size of 284 bp (18), was detected only in Salmonella strains, indicating that this PCR was specific for the Salmonella genus.
PCR with natural samples.
Capillary PCR could detect Salmonella on the flock basis. When evaluated individually for each flock, a specific PCR product was detected in 35 of 53 samples (Table 1). In 9 of these 35 PCR-positive samples (A10, A12, D43, D44, D45, D46, D48, D49, and E51) Salmonella could not be detected bacteriologically either after PE (A10, D44, and E51) or after PE and DSE (A12, D43, D45, D46, D48, and D49).
TABLE 1.
Comparison of the primary enrichment, delayed secondary enrichment, and PCR results
| Flock/sample no. | Result (+ or −)a
|
Serotype isolated | ||
|---|---|---|---|---|
| PE | DSE | PCR | ||
| A/1 | + | + | + | Enteritidisb |
| A/2 | + | + | − | Enteritidis |
| A/3 | + | + | + | Enteritidis |
| A/4 | + | + | − | Enteritidis |
| A/5 | + | + | + | Enteritidis |
| A/6 | + | + | + | Enteritidis |
| A/7 | + | + | + | Enteritidis |
| A/8 | + | + | − | Enteritidis |
| A/9 | − | + | − | Enteritidis |
| A/10 | − | + | + | Enteritidis |
| A/11 | + | + | − | Enteritidis |
| A/12 | − | − | + | NDc |
| A/13 | + | − | + | Enteritidis |
| A/14 | − | + | − | Enteritidis |
| A/15 | − | + | − | Enteritidis |
| A/16 | + | + | + | Enteritidis |
| A/17 | − | + | − | Enteritidis |
| A/18 | + | + | + | Enteritidis |
| A/19 | − | + | − | Enteritidis |
| A/20 | + | + | + | Enteritidis |
| A/21 | + | + | + | Enteritidis |
| B/22 | + | + | + | Enteritidis |
| B/23 | + | + | + | Enteritidis |
| B/24 | + | + | + | Enteritidis |
| B/25 | + | + | + | Enteritidis |
| B/26 | + | + | + | Enteritidis |
| B/27 | + | + | + | Enteritidis |
| B/28 | + | + | + | Enteritidis |
| B/29 | + | + | + | Enteritidis |
| C/30 | + | + | + | Serogroup C1 |
| C/31 | + | + | + | Serogroup C1 |
| C/32 | + | + | + | Serogroup C1 |
| C/33 | + | + | + | Serogroup C1 |
| C/34 | − | + | − | Serogroup C1 |
| C/35 | − | + | − | Serogroup C1 |
| C/36 | − | + | − | Serogroup C1 |
| C/37 | − | + | − | Serogroup C1 |
| C/38 | − | + | − | Serogroup C1 |
| C/39 | − | + | − | Serogroup C1 |
| C/40 | − | + | − | Serogroup C1 |
| C/41 | − | + | − | Serogroup C1 |
| C/42 | − | + | − | Serogroup C1 |
| D/43 | − | − | + | ND |
| D/44 | − | + | + | Enteritidis |
| D/45 | − | − | + | ND |
| D/46 | − | − | + | ND |
| D/47 | + | + | + | Enteritidis |
| D/48 | − | − | + | ND |
| D/49 | − | − | + | ND |
| E/50 | + | + | + | Enteritidis |
| E/51 | − | + | + | Enteritidis |
| E/52 | + | + | + | Enteritidis |
| E/53 | + | + | + | Enteritidis |
| Total no. of positive samples | 30 | 46 | 35 | |
For PE and DSE results, + indicates that Salmonella colonies were observed after plating; − indicates that no Salmonella growth was observed after plating. For PCR results, + indicates that a specific PCR product was detected; − indicates that a specific PCR product was not detected.
Salmonella enterica serovar Enteritidis.
ND, not determined.
In the remaining 18 PCR-negative samples, 4 samples (A2, A4, A8, and A11) were found to harbor Salmonella after both PE and DSE (Table 1) and 14 samples (A9, A14, A15, A17, A19, C34, C35, C36, C37, C38, C39, C40, C41, C42) were positive after DSE (Table 1).
Fifty-three additional chicken intestinal homogenate samples, which were negative in their PE and DSE in bacteriological examination, were found to be also negative in their PCRs. All the chickens examined for agglutinins to serovar Gallinarum and serovar Gallinarum biotype Pullorum by a slide agglutination test were negative.
DISCUSSION
The primary goal of this work was the rapid detection of Salmonella from chicken intestinal homogenates by capillary PCR. We chose to use two Salmonella genus-specific primers (18) in capillary PCR (29–31) and detected the specific products by capillary gel electrophoresis. We decided to use PE of intestinal homogenates in TTB and performed the bacterial lysis method (21) to obtain Salmonella DNA. PCRs performed with Salmonella templates by this procedure from artificially prepared samples consistently yielded positive results with high sensitivity. We also applied this procedure to clinical samples, and 35 of 53 samples yielded a specific amplicon. DSE results indicated that 46 of these samples harbored Salmonella. There may be several reasons for the false-negative PCR results, such as the presence of inhibitory substances in the feces and enrichment broth contents (5, 12, 23). We investigated the possible roles of these factors in our study by artificially contaminating the samples, and we observed that TTB enrichment and sample amount did not diminish the sensitivity of the PCR. Individual false-negative results in naturally contaminated samples are not important for flock-based Salmonella screening because infected flocks can be easily detected using a PE and PCR combination (Table 1). We can suggest that DSE be pursued if only the real infection rate (individual-based screening) in a flock needs to be determined. Regardless, we decided that the PE and capillary PCR combination was useful for rapid and primary screening of Salmonella in chicken flocks.
Incubation in an enrichment broth (5, 12, 23) increases the number of viable organisms in the sample to allow detection by PCR. It is also inexpensive and requires little manipulation compared to other selective enrichment systems (20, 28). By using pure Salmonella serovar Typhimurium cultures, Stone et al. (23) found that Rappaport-Vassiliadis and TTB were inhibitory to PCR, whereas BHI and selenite-cystine broths were not. In our study, we used TTB, NB, and BHI broths with pure Salmonella cultures and found that none of these enrichment broths were inhibitory to PCR. However, from BHI and NB we could not detect any specific PCR product in natural and artificially spiked samples, but from TTB, we did. In addition, we reisolated serovar Enteritidis from TTB with artificially spiked samples but not from BPW and NB. Therefore, we decided that TTB was probably the most suitable environment for the growth of Salmonella as a pure culture both in artificially contaminated and in natural samples. The superior selectivity of TTB for avian fecal samples over NB and BHI, which are nonselective PE broths, may have permitted the growth of Salmonella and inhibited the competitive flora, thus resulting in a positive PCR. Also, selection of a PE medium for Salmonella PCR may depend on the sample type (5, 23), because composition differences in samples may greatly alter the enrichment environment and may even inhibit the growth of Salmonella. For example, Stone et al. (23) found BHI to be a favorable PE medium to detect Salmonella from mammalian (bovine and equine) feces and tissue samples, while Lin and Tsen (13) reported the advantage of using combined lactose-tetrathionate broth for human fecal and food samples.
There may possibly be differences in the initial numbers of Salmonella in the feces of clinically diseased and subclinically infected animals. This study aimed to detect birds that were subclinically infected with Salmonella. Therefore, a PCR procedure with high sensitivity was required. Sensitivity and detection-limit results revealed that our optimized capillary PCR had detection limits of 3, 3, and 9 CFU for cultures of pure Salmonella serovars Enteritidis, Typhimurium, and Gallinarum, respectively. For artificially prepared samples, the detection limits were 3, 3, and 7 CFU, respectively, with the same Salmonella strains.
With poultry, control of infection depends largely on the identification of the infection in the early stages. Standard bacteriological methods may require 5 to 11 days to isolate and identify Salmonella (27) from avian fecal samples. These time-consuming methods should be complemented with a rapid and primary screening procedure such as PCR. In the PCR studies with avian samples for Salmonella, preparation of the sample, PCR, and detection of the amplicon without and with (required for subclinically infected birds) PE may well take up to 6 and 24 h per sample, respectively (26, 33). In this study, we combined capillary PCR and capillary gel electrophoresis to more quickly determine if an avian intestinal sample harbors Salmonella. Capillary PCR has the advantage of reducing amplification time and mispriming and of significantly increasing the specific amplicon yield because temperature changes during the denaturation, annealing, and extension steps occur within seconds in the air-thermal cycler (29–31). As a result, with the capillary PCR method we used, approximately 20 h was required (per sample: primary enrichment, 18 h; DNA extraction, 25 min; capillary PCR, 35 min; and capillary gel electrophoresis, 45 min). If samples are from clinically diseased birds, the total time for PCR and detection is reduced to 2 h, since the 18-h PE is not required.
Capillary gel electrophoresis is also highly sensitive. We reanalyzed with conventional gel electrophoresis several selected PCR samples in which we detected low peak heights from the electropherograms. We could not observe any specific PCR product band from these samples. This demonstrates that capillary gel electrophoresis, probably due to the low amount of amplicon with its high detection capacity, aided in visualizing these low amplicons, which could not otherwise be detected with conventional gel electrophoresis (data not shown).
We suggest that use of the bacterial lysis method (18, 21) is cheap and rapid when compared to immunomagnetic separation (19, 28) and other DNA extraction protocols (21, 22). The combination of TTB enrichment with the bacterial lysis method and capillary PCR with capillary gel electrophoresis is suitable for the rapid detection of salmonellae in chicken feces and, thus, to diagnose Salmonella-infected flocks. Future plans are to develop standardized clinical PCR assays for other avian pathogens by continously monitoring DNA amplification (32), which would allow us to rapidly optimize the amplification conditions.
ACKNOWLEDGMENTS
This work was supported by the Scientific and Technical Research Council of Turkey (TUBITAK) grant TOGTAG/TARP-2203.
We thank Medge Owen Unal for critical comments and Sevket Gunay for laboratory assistance.
REFERENCES
- 1.Aabo S, Andersen J K, Olsen J E. Research note: detection of Salmonella in minced meat by the polymerase chain reaction method. Lett Appl Microbiol. 1995;21:180–182. doi: 10.1111/j.1472-765x.1995.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 2.Bennett A R, Greenwood D, Tennant C, Banks J G, Betts R P. Rapid and definitive detection of Salmonella in foods by PCR. Lett Appl Microbiol. 1998;26:437–441. doi: 10.1046/j.1472-765x.1998.00368.x. [DOI] [PubMed] [Google Scholar]
- 3.Carli K T, Kahraman M M, Sen A, Sonmez G. Septicemia and blindness by Salmonella typhimurium, Salmonella enteritidis and Salmonella essen in adult chicken. Veterinarium. 1996;7:22–26. [Google Scholar]
- 4.Chen S, Yee A, Griffiths M, Wu K Y, Wang C-N, Rahn K, De Grandis S A. A rapid, sensitive and automated method for detection of Salmonella species in foods using AG-9600 AmpliSensor Analyzer. J Appl Microbiol. 1997;83:314–321. doi: 10.1046/j.1365-2672.1997.00226.x. [DOI] [PubMed] [Google Scholar]
- 5.Chiu C H, Ou J T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen N D, Martin L J, Simpson B, Wallis D E, Neibergs H E. Comparison of polymerase chain reaction and microbiological culture for detection of Salmonellae in equine feces and environmental samples. Am J Vet Res. 1996;57:780–786. [PubMed] [Google Scholar]
- 7.Fricker C R. The isolation of salmonellas and campylobacters. J Appl Bacteriol. 1987;63:99–116. doi: 10.1111/j.1365-2672.1987.tb02692.x. [DOI] [PubMed] [Google Scholar]
- 8.Gast R K. Paratyphoid infections. In: Calnek B W, Barnes H J, Beard C W, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 97–121. [Google Scholar]
- 9.Gouws P A, Visser M, Brözel V S. A polymerase chain reaction procedure for the detection of Salmonella spp. within 24 hours. J Food Prot. 1998;61:1039–1042. doi: 10.4315/0362-028x-61.8.1039. [DOI] [PubMed] [Google Scholar]
- 10.Humbert F, Carraminana J J, Lalande F, Salvat G. Bacteriological monitoring of Salmonella enteritidis carrier birds after decontamination using enrofloxacin, competitive exclusion and movement of birds. Vet Rec. 1997;141:297–299. doi: 10.1136/vr.141.12.297. [DOI] [PubMed] [Google Scholar]
- 11.Kimura B, Kawasaki S, Fujii T, Kusunoki J, Itoh T, Flood S J A. Evaluation of TaqMan PCR assay for detecting Salmonella in raw meat and shrimp. J Food Prot. 1999;62:329–335. doi: 10.4315/0362-028x-62.4.329. [DOI] [PubMed] [Google Scholar]
- 12.Kongmuang U, Luk J M C, Lindberg A A. Comparison of three stool-processing methods for detection of Salmonella serogroups B, C2, and D by PCR. J Clin Microbiol. 1994;32:3072–3074. doi: 10.1128/jcm.32.12.3072-3074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J S, Tsen H Y. Development and use of polymerase chain reaction for the specific detection of Salmonella typhimurium in stool and food samples. J Food Prot. 1999;62:1103–1110. doi: 10.4315/0362-028x-62.10.1103. [DOI] [PubMed] [Google Scholar]
- 14.Luk J M, Kongmuang U, Tsang R S W, Lindberg A A. An enzyme-linked immunosorbent assay to detect PCR products of the rfbS gene from serogroup D salmonellae: a rapid screening prototype. J Clin Microbiol. 1997;35:714–718. doi: 10.1128/jcm.35.3.714-718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahon J, Lax A J. A quantitative polymerase chain reaction method for the detection in avian feces of salmonellas carrying the spv gene. Epidemiol Infect. 1993;111:455–464. doi: 10.1017/s0950268800057186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandrell R E, Wachtel M R. Novel detection techniques for human pathogens that contaminate poultry. Curr Opin Biotech. 1999;10:273–278. doi: 10.1016/s0958-1669(99)80048-2. [DOI] [PubMed] [Google Scholar]
- 17.Notermans S, van de Giessen A, Henken A M. Future requirements for diagnosing and monitoring of pathogenic micro-organisms in poultry and eggs. In: Thorns C J, Jones P, editors. COST Action 97 pathogenic micro-organisms in poultry and eggs. 2. Monitoring procedures, rapid detection methods and techniques. Luxembourg, Belgium: Office for Official Publications of the European Communities; 1997. pp. 9–14. [Google Scholar]
- 18.Rahn K, De Grandis S A, Clarke R C, McEwen S A, Galan J E, Ginocchio C, Curtis III R, Gyles C L. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 19.Rijpens N, Herman L, Vereecken F, Jannes G, Smedt J D, Zutter L D. Rapid detection of stressed Salmonella spp. in dairy and egg products using immunomagnetic separation and PCR. Int J Food Microbiol. 1999;46:37–44. doi: 10.1016/s0168-1605(98)00171-8. [DOI] [PubMed] [Google Scholar]
- 20.Ripabelli G, Sammarco M L, Grasso G M. Evaluation of immunomagnetic separation and plating media for recovery of Salmonella from meat. J Food Prot. 1999;62:198–201. doi: 10.4315/0362-028x-62.2.198. [DOI] [PubMed] [Google Scholar]
- 21.Soumet C, Ermel G, Fach P, Colin P. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett Appl Microbiol. 1994;19:294–298. doi: 10.1111/j.1472-765x.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 22.Soumet C, Ermel G, Rose V, Rose N, Droin P, Salvat G, Colin P. Identification by a multiplex PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol. 1999;29:1–6. doi: 10.1046/j.1365-2672.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- 23.Stone G G, Oberst R D, Hays M P, McVey S, Chengappa M M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J Clin Microbiol. 1994;32:1742–1749. doi: 10.1128/jcm.32.7.1742-1749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone G G, Oberst R D, Hays M P, McVey S, Galland J C, Curtiss III R, Kelly S M, Chengappa M M. Detection of Salmonella typhimurium from rectal swabs of experimentally infected beagles by short cultivation and PCR-hybridization. J Clin Microbiol. 1995;33:1292–1295. doi: 10.1128/jcm.33.5.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuchili L M, Kodama H, Izumoto Y, Mukamoto M, Fukata T, Baba T. Detection of Salmonella gallinarum and S. typhimurium DNA in experimentally infected chicks by polymerase chain reaction. J Vet Med Sci. 1995;57:59–63. doi: 10.1292/jvms.57.59. [DOI] [PubMed] [Google Scholar]
- 26.Tuchili L M, Kodama H, Sharma R N, Takatori I, Pandey G S, Kabilika S, Mukamoto M, Tsuji S, Baba T. Detection of Salmonella DNA in chicken embryos and environmental samples by polymerase chain reaction. J Vet Med Sci. 1996;58:881–884. doi: 10.1292/jvms.58.881. [DOI] [PubMed] [Google Scholar]
- 27.United States Department of Agriculture Animal and Plant Health Inspection Service. The National Poultry Improvement Plan. CFR Part 147. Auxiliary provisions on National Poultry Improvement Plan. Subpart B. Bacteriological examination procedure. § 147-11. Laboratory procedure recommended for the bacteriological examination of salmonella. Washington, D.C.: United States Department of Agriculture; 1996. pp. 14–19. [Google Scholar]
- 28.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of Salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittwer C T, Fillmore G C, Hillyard D R. Automated polymerase chain reaction in capillary tubes with hot air. Nucleic Acids Res. 1989;17:4353–4357. doi: 10.1093/nar/17.11.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittwer C T, Fillmore G C, Garling D J. Minimizing the time required for DNA amplification by efficient heat transfer to small samples. Anal Biochem. 1990;186:328–331. doi: 10.1016/0003-2697(90)90090-v. [DOI] [PubMed] [Google Scholar]
- 31.Wittwer C T, Garling D J. Rapid cycle DNA amplification: time and temperature optimization. Biotechniques. 1991;10:76–83. [PubMed] [Google Scholar]
- 32.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 33.Woodward M J, Kirwan S E S. Detection of Salmonella enteritidis in eggs by the polymerase chain reaction. Vet Rec. 1996;138:411–413. doi: 10.1136/vr.138.17.411. [DOI] [PubMed] [Google Scholar]