Abstract
Panicle blast is the most severe type of rice blast disease. Screening of rice genotypes for panicle blast resistance at the field level requires an efficient and robust method of inoculation. Here, we standardized a method that can be utilized for both small- and large-scale screening and assessment of panicle blast infection and disease reaction. The method involves inoculation of Magnaporthe oryzae spore culture in the neck of the rice panicle using a syringe and covering the inoculation site with wet cotton wrapped with aluminum foil to provide the required humidity for spore germination. The method was standardized using panicle blast-resistant cv. Tetep and susceptible cv. HP2216 inoculated with Mo-ni-025 isolate of M. oryzae. The method was evaluated at phenotypic as well as molecular level by expression analysis of disease responsive pathogenesis-related (PR) genes. We found this method simple, robust, reliable, and highly efficient for screening of large germplasm sets of rice for panicle blast. This was validated by screening the wild rice germplasm for panicle blast response in the field using three M. oryzae strains and subsequently with the most virulent strain in 45 EMS-induced mutants of Nagina 22 shortlisted based on field screening in a blast hotspot region. We identified five novel blast disease-resistant wild rice genotypes and 15 Nagina 22 mutants that can be used in breeding programmes.
Keywords: Rice, Panicle blast, Magnaporthe, Syringe inoculation, Disease resistance, PR genes, Wild rice, Mutants
Introduction
Rice (Oryza sativa L.) adds more than 20% of total calories in a regimen and is a foundation of food energy for more than 3.5 billion people in the world (Shikari et al. 2014). Cultivation of rice under various ecological conditions experiences various biotic and abiotic stresses. Among the several biotic stresses in rice such as sheath blight, bacterial blight, false smut, kernel bunt, etc., the most important and devastating disease around the world is the rice blast caused by Magnaporthe oryzae. It accounts for 10–30% yield losses every year, even higher in certain blast-prone regions (Shikari et al. 2014). Infection of M. oryzae spores on leaf surfaces forms necrotic lesions on leaf leading to leaf blast whereas, the panicle blast is developed when spores infect panicles during the flowering stage. Panicle blast of rice is comparatively more devastating than leaf blast in terms of yield losses. During the panicle blast, pathogen damages the tissues responsible for water and nutrient supply, resulting in poor grain filling and yield loss (Shim et al. 2005). Due to the evolution of pathogen, changing pattern of pathogenicity, and changing climatic conditions, resistance to rice blast is notoriously short-lived and there is a need to identify and use improved management strategies for a durable disease resistance (Zhu et al. 2005). The natural infection process of M. oryzae spores on rice tissues involves attachment of conidia to the host surface, its germination, appressorium formation, and penetration into the host surface by a penetration peg leading to invasive growth in the host tissue (Kim et al. 2001). The infection thus, results in the formation of typical lesions. During the milky stage of grains, the infection continues for 25 days until the maturity of grains, which results in reduced grain filling, causing 35–40% of yield loss (Filippi and Prabhu 1998; Kobayashi et al. 2001).
M. oryzae needs specific temperature, humidity, and other environmental conditions for its growth and causing disease in the host plant. Because of the specificity of its desirable conditions, it only spreads in specific geographical locations around the world. However, due to the fluctuation of climatic conditions and pathogenicity every year, there is no uniform pattern of disease incidence and it is difficult to evaluate the same with accuracy. Thus, the failure to carry out a uniform evaluation of panicle blast disease limits the phenotypic and genetic studies and screening of germplasm to identify resistant sources (Fujii et al. 2000). Therefore, to date only one gene (Pb1) and few robust QTLs (qPb6–1, qPbh-7–1 and Pb-bd1) have been identified for panicle blast resistance (Fang et al. 2016, 2019; Hayashi et al. 2010). Recently, four genes OsGF14b, OsOXO2, OsOXO3 and OsOXO4 were found to be responsible for positive regulation of defence-related genes against the rice panicle blast disease and one novel locus qPBR10-1 was identified that controls the rice panicle blast resistance (Yan et al. 2021; Dong et al. 2021; Wu et al. 2021). Alternatively, the screening of germplasm can be conducted under controlled conditions with defined temperature, humidity, soil moisture and other factors. However, it is an expensive affair to develop such huge structures to screen a large collection of germplasm. Moreover, screening for panicle blast is to be conducted at the milky stage of seeds and therefore plants need to be grown until the reproductive stage for disease phenotyping. To deal with such limitations, it was necessary to develop a simple technique for blast inoculation for screening of large germplasm sets in natural field conditions. We have been working on this important area to identify and characterize resistant resources for panicle blast and leaf blast over the last few years (Kumar et al. 2021; Rawal et al. 2018; Sureshkumar et al. 2019). Here, we have standardized a syringe based panicle blast inoculation method for large scale screening of germplasm resources. A similar method earlier demonstrated by Puri et al. uses the method of spore inoculation in sheath base of photosynthetic leaf. In this method chances of other infections, i.e. pod borer, bacterial infection to panicle, retains, as the panicle flower is still inside the leaf cover (Puri et al. 2009). The other method where researchers used the cotton-wrapping method of inoculation, they have to spray water on that cotton for 3–4 min after every 3 h to maintain humidity (Liu et al. 2016; Dong et al. 2021). This method was both labourious, time consuming and the percent of humidity can not be maintained steadily. The success of an inoculation technique depends on the reproducible occurrence of disease as assessed by both phenotypic observations as well as analysis of molecular response to the disease. The molecular response to disease can be characterized by enhanced expression of disease-responsive genes. Among such genes, pathogenesis-related (PR) protein-coding genes are specifically expressed as a protection mechanism against different biotic stresses (Agrios 1997). PR-proteins have direct and indirect effects on plant resistance mechanism, against the fungal infection, which includes hydrolyzing fungal cell walls (chitinases and β-1,3-glucanases) and formation of oligosaccharide elicitors to produce phytoalexins (Ebrahim et al. 2011). These PR proteins were first reported in tobacco plants infected with Tobacco Mosaic Virus (van Loon and van Kammen 1970). In the case of rice, expression analysis of PR protein-coding genes has been done in M. oryzae-infected rice samples (Kitajima and Sato 1999). The PR protein genes are induced after the infection of M. oryzae. Hence, these genes can be utilized for the analysis of the molecular response of plants to confirm the infection of M. oryzae.
The present study aimed at standardizing the syringe inoculation technique for panicle blast infection wherein we demonstrate the occurrence of disease at both phenotypic level in the form of symptoms and also at the molecular level by expression of PR genes. We screened the pathogenicity of different M. oryzae strains for panicle blast infection and confirmed the pathogenicity spectrum of the selected strains on known blast-resistant and susceptible rice cultivars. We also screened 12 wild rice genotypes and 45 EMS induced mutants of Nagina 22 with this method to identify novel resistance sources to utilize in the rice breeding programs for blast disease resistance.
Materials and methods
Plant growth and conditions
Seeds of Tetep, a well-known blast-resistant genotype, HR12 and HP2216 which are blast susceptible genotypes were sown in autoclaved soil-rite. Another genotype NKSWR-2, a wild rice accession (Tripathy et al. 2018) of unknown disease reaction was also used. Fifteen-day-old seedlings were then transplanted in pots filled with soil and grown in the glasshouse with controlled conditions at temperature 25 ± 2 °C and 80–90% relative humidity. All the plants were grown till the reproductive stage of panicle formation. To further test the inoculation method in the field conditions as well as to screen the wild rice genotype collection for panicle blast disease reaction, 12 wild rice accessions including NKSWR-2 were selected from the wild rice germplasm collected from different agro-climatic zones of India (Singh et al. 2018; Tripathy et al. 2018). Besides the wild rice resources, we also used the EMS-induced mutant resources of Nagina 22 available in 2017 (Sevanthi et al. 2018). We identified 60 blast-resistant lines using 1.5 kg M2 generation mutant seeds (approximately 100 thousand seeds) screened in the blast nursery at ICAR-NRRI-Central Rainfed Upland Rice Research Station (CRURRS), Hazaribagh, Jharkhand, India. In 2018 and 2019, these 60 mutants were again screened for leaf and panicle blast resistance at ICAR-NIPB, New Delhi and leaf blast under natural epiphytotic conditions at CRURRS and ICAR-Research Complex for NEH Region, Umiam, Meghalaya, the two major hotspots for this disease in India. In 2020, we screened three progeny lines of the most promising 45 stable mutant lines at ICAR-NIPB with the artificial inoculation procedure described in this study.
Preparation of Magnaporthe oryzae inoculum
Three different isolates of M. oryzae viz, Dehradun (Mo-ni-025), Sikkim-6 (Mo-nwi-053), and N22 (Mo-nwi-022) were used in the present study. The fungi cultures were revived using 4–6% of sucrose solution. The mycelia from these samples were then cultured on Potato Dextrose Agar (PDA) media (HiMedia, India) slants for vegetative growth and incubated at 25 °C in light-deprived conditions for 15 days. The vegetative growth of mycelia was then macerated in 5 ml of autoclaved double distilled water and plated on Mathur’s media (HiMedia, India) for reproductive growth and incubated at 25 °C under blue and white fluorescence light for 8–10 days. After the incubation period, a thick layer of pathogen hyphae and spores was observed. The spores were collected by scraping the growth in double distilled water to make spore suspension. The suspension was then passed through two layers of muslin cloth. The spore count was carried out using 10 µl spore suspension on a glass slide and observation under a light microscope Olympus IX81 (Olympus Corporation, Tokyo, Japan) at 40× magnification. A concentration of 0.02% Tween-20 was added to the suspension with a spore concentration of about 105 spores/ml. This suspension culture was used for inoculation.
Magnaporthe oryzae inoculation in the panicle neck
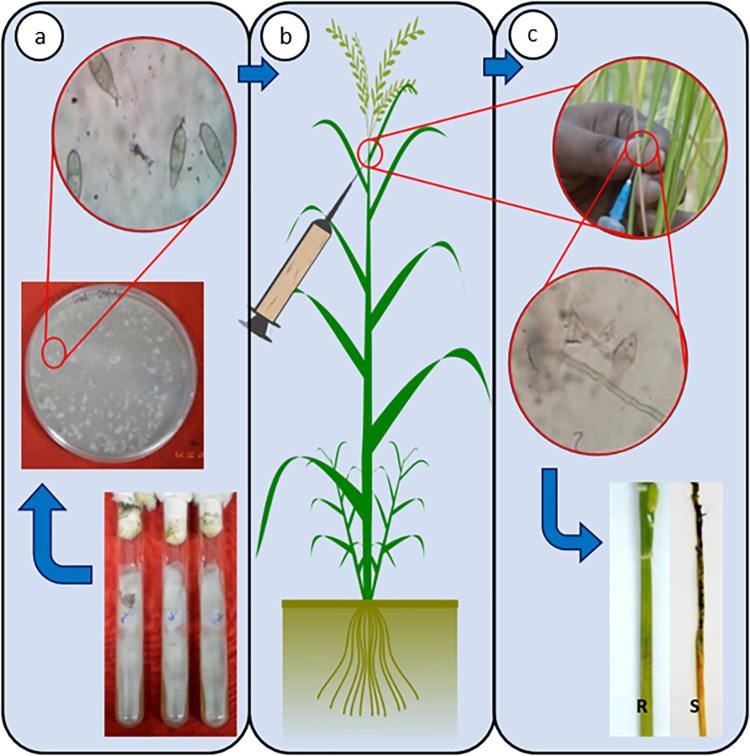
Three panicles each from five plants, thus in total 15 panicles representing each genotype under study, were inoculated with three strains of M. oryzae. Approximately 20 µl of the spore suspension per panicle was used for injecting into the neck of the panicle at the booting stage using a 1 ml syringe. Inoculation was done in the peduncle at approximately 2 inch above the node of the flowering panicle in such a way that the spore suspension moves upward into the whole panicle. The injected site of the peduncle was then covered with wet cotton soaked in autoclaved double distilled water to maintain the required humidity and darkness (Fig. 1). This ensured a favorable microenvironment required for the growth and development of fungal spores. The cotton swab was further covered with aluminum foil to avoid drying out. All the inoculated plants were continued to grow in controlled conditions at 25 ± 2 °C and 80–90% relative humidity with 16/8 h light and dark conditions in the blast phenotyping facility of the institute.
Fig. 1.
a Culturing of M. oryzae strain (Dehradun) on Potato Dextrose Agar for 10–15 days, followed by sporulation of the M. oryzae strain on Mathur's media for 10–15 days at 25 °C incubation after which plates were washed with distilled water to make spore suspension which was checked under microscope; b The spore suspension is then injected into the neck of rice panicle for infection at 60–70% humidity and temperature of 24–25 °C; c Resistance genotype of rice shows only marks of infection while the susceptible genotypes shows the effect of severe damage cause by M. oryzae infection
Disease phenotyping and evaluation of the infected panicles
Disease pathogenicity of the fungal strains was categorized into three groups, viz. resistant, moderate, and severe based on observed symptoms. The severity of the disease was evaluated visually for each panicle from the average size of the lesion in three biological replicates. Panicles with no disease lesions were considered as resistant genotypes while those with lesions as susceptible genotypes. For molecular analysis, samples were taken at 24, 48, 72, and 96 h post-inoculation (hpi) with a mock negative control inoculated with autoclaved distilled water. Further, the seed set in the infected panicles was observed at maturity to confirm the disease reaction phenotype recorded.
RNA isolation, cDNA synthesis, and qRT-PCR
Samples of the panicle tissues of Tetep and HP2216 inoculated with Mo-ni-025 strain were collected at four time points after inoculation, snap-frozen in liquid nitrogen, and stored at − 80 °C to be used later for RNA isolation. Extraction of total RNA from the panicle tissues was carried out using Spectrum Plant Total RNA Kit (Sigma-Aldrich, USA) according to the manufacturer’s protocol. The quality and quantity of isolated RNA were checked by gel electrophoresis and UV–Vis Spectrophotometer (Nano-Drop 2000, Thermo-Scientific, USA), respectively. cDNA synthesis was carried out using Applied Biosystems™ High-Capacity cDNA reverse transcription kit. qRT-PCR primers were designed for selected inducible PR protein genes using the PrimerQuest tool on IDT-DNA and synthesized from Sigma, India (Table 1). The rice actin gene was used as an internal control. qRT-PCR was carried out in LightCycler 480 II Real-Time PCR Instrument (Roche) using a reaction volume of 10 µl containing a required concentration of diluted cDNA, 5 µl SYBR green (Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix by Agilent Technologies), 0.4 µl of ROX dye, and 0.3 µl (10 µmol) each primer. The PCR program used was denaturation at 95 °C for 30 s, annealing at 58 °C for 15 s, and extension at 72 °C for 20 s, for 40 cycles. Differential expressions of genes were analyzed using the fold change method (Livak and Schmittgen 2001). Three biological replicates and three technical replicates for each sample were used for the analysis.
Table 1.
List of pathogenesis-related genes used in the expression profiling of rice genotypes against rice blast disease caused by M. oryzae strain Mo-ni-025
| PR genes | Accession no. | Gene locus Id | Gene product name |
|---|---|---|---|
| PR2 | AP014957 | LOC_Os01g71340 | Glycosyl hydrolases family 17, putative, expressed |
| PR3 | AP014960 | LOC_Os04g41620 | CHIT2—Chitinase family protein precursor, expressed |
| PR7 | AP014958 | LOC_Os02g53860 | OsSub22—Putative subtilisin homologue, expressed |
| PR8 | AP014957 | LOC_Os01g64110 | Glycosyl hydrolase, putative, expressed |
| PR10 | D82066 | LOC_Os12g36880 | Pathogenesis-related Bet v I family protein, putative, expressed |
| PR14 | AP014966 | LOC_Os10g32030 | Retrotransposon protein, putative, unclassified, expressed |
| PR-pha | AP014960 | LOC_Os04g43800 | Phenylalanine ammonia-lyase, putative, expressed |
Results
Syringe inoculation method for blast infection
For standardization of the syringe inoculation method, four different rice genotypes were inoculated with Mo-ni-025 strain. The blast-resistant genotype Tetep did not show any symptoms of the disease even after 72 hpi. Only the symptoms of hypersensitive response were observed. A similar phenotype was noticed in the case of wild rice genotype NKSWR-2 and hence it was considered as blast-resistant. For successful inoculation by this method, symptoms should appear on susceptible genotypes used as a positive control. Thus, the success of the method could be assessed by observing the phenotypes of well-known blast susceptible genotypes, HP2216 and HR12. Here, both the genotypes showed the formation of long lesions after infection with M. oryzae. The lesion symptoms started to appear on panicles at 48 hpi and increased up to 96 hpi showing the success of the inoculation method (Fig. 2). The lesions formed on the neck of panicle at the site of inoculation in three replicates were measured in centimetres (cm) and average of the lesions was taken. After infection in Tetep cv. known for blast resistance, no lesions can be seen after inoculation of M. oryzae spores in neck of the panicle at all three 48, 72, and 96 hpi. While NKSWR-2 genotype was found to be resistant against the blast disease with minimal lesions at the site of inoculation, i.e. of 0.1 cm at 48 hpi, and of 0.3 cm at 72 and 96 hpi. Whereas in case of blast susceptible rice cultivars HR12, lesions with average size of 5.3, 12.1 and 18.2 cm were observed at 48, 72 and 96 hpi, respectively. Also, lesions size increased in second blast susceptible cultivar HP2216, with average size increases from 8.5, 23.2 and 29.6 cm at 48, 72 and 96 hpi, respectively.
Fig. 2.
Disease reaction phenotypes of Tetep, NKSWR-2, HR-12 and HP2216 at different time intervals following inoculation with M. oryzae. (hpi- hours post inoculation)
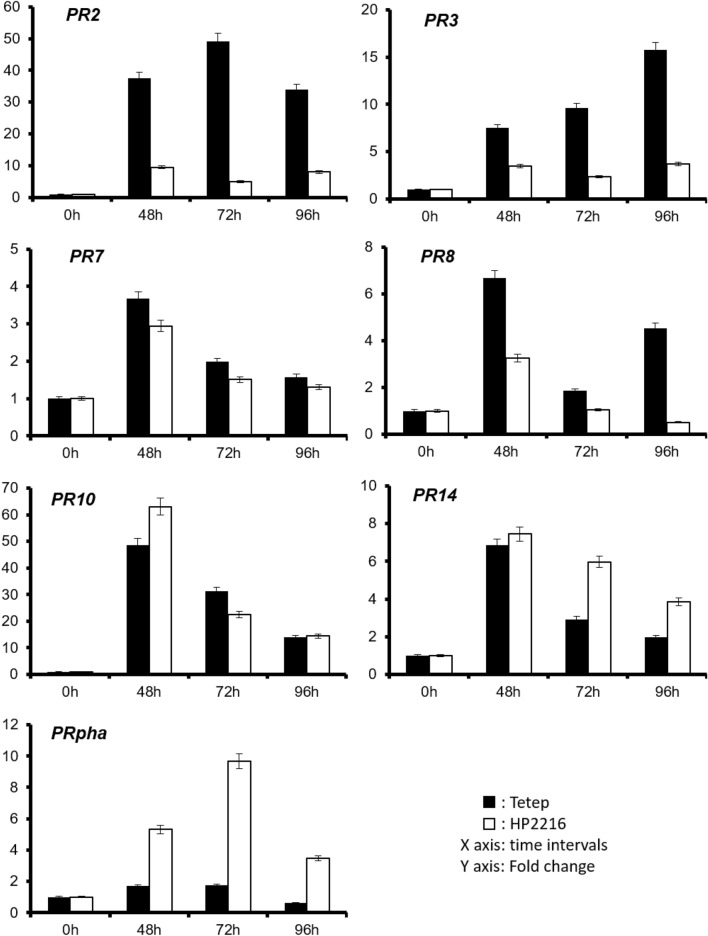
Disease confirmation at molecular level by expression analysis of PR protein genes
To confirm whether the phenotypic observations of disease reactions were also reflected at the molecular level, expression analysis of seven disease responsive PR protein genes was carried out by qRT-PCR in Tetep and HP2216 genotypes inoculated with Mo-ni-025 strain. All the genes showed induced expression in panicle tissues after M. oryzae infection in comparison with mock-inoculated control samples (Fig. 3). Among the analyzed genes, PR2 (Glycosyl Hydrolases family 17 protein), PR3 (CHIT2—Chitinase family protein), PR7 (OsSub22—Putative Subtilisin homolog protein), and PR8 (Glycosyl Hydrolase protein) showed a higher level of up-regulation in the blast-resistant genotype Tetep as compared to the blast susceptible genotype HP2216. On the other hand, expression of the remaining 3 genes PR10 (Pathogenesis-Related Bet v I family protein), PR14 (Retrotransposon protein), and PR18 (Phenylalanine Ammonia-lyase) was higher in HP2216 than in Tetep.
Fig. 3.
Expression analysis of pathogenesis related (PR) genes in blast disease resistant Tetep (T) and susceptible HP2216 (H) rice genotypes at different time intervals 0, 48, 72 and 96h after inoculation with fungus M. oryzae strain Mo-ni-025
Screening of wild rice germplasm for identification of novel resources for blast resistance
After successfully demonstrating the syringe inoculation method for panicle blast infection in controlled conditions in the blast phenotyping facility, the method was implemented at the field level for screening of rice germplasm for blast resistance. The method was found equally efficient for disease inoculation at the field level and several plants showed disease symptoms depending on the genotype. Based on the level of symptoms on the infected panicles, plants were categorized into blast-resistant and blast-susceptible genotypes (Fig. 4, Table 2). The wild rice genotypes NKSWR-2, NKSWR-9, NKSWR-104, NKSWR-398 and NKSWR-401 were having lesion size less than 0.1 cm or having only syringe inoculated wound of blast infection and thus were considered as blast-resistant. In contrast, NKSWR-13, NKSWR-39, NKSWR-396, and NKSWR-403 genotypes were found to have lesion size of more than eight cm and considered to be blast susceptible. The comparison of grain production in these genotypes further confirmed the resistant nature of the genotypes. However, the grain formation was largely reduced in susceptible genotypes. The remaining three genotypes, namely NKSWR-446, NKSWR-273 and NKSWR-279 showed differential reactions to different strains.
Fig. 4.
Phenotypic screening of rice blast disease at different wild rice (NKSWR) genotypes inoculated with M. oryzae strain Mo-ni-025. This figure shows the level of infection on different wild rice genotypes after 10 days of inoculation. NKSWR-2 shows minimal effect of blast infection while NKSWR-430 showing the severe effect of blast disease on rice panicle
Table 2.
Disease reactions as observed in the wild rice genotypes inoculated with three different strains of M. oryzae
| Rice genotypes | M. oryzae reaction on rice panicle | ||
|---|---|---|---|
| Sikkim-6 (Mo-nwi-53) | N22 (Mo-nwi-022) | Dehradun (Mo-ni-025) | |
| NKSWR-2 | R | R | R |
| NKSWR-9 | R | R | R |
| NKSWR-104 | R | R | R |
| NKSWR-398 | R | R | R |
| NKSWR-401 | R | R | R |
| NKSWR-446 | R | S | S |
| NKSWR-273 | R | S | S |
| NKSWR-279 | R | R | S |
| NKSWR-013 | S | S | S |
| NKSWR-039 | S | S | S |
| NKSWR-396 | S | S | S |
| NKSWR-403 | S | S | S |
Screening of Nagina 22 mutants for identification of novel resources for blast resistance
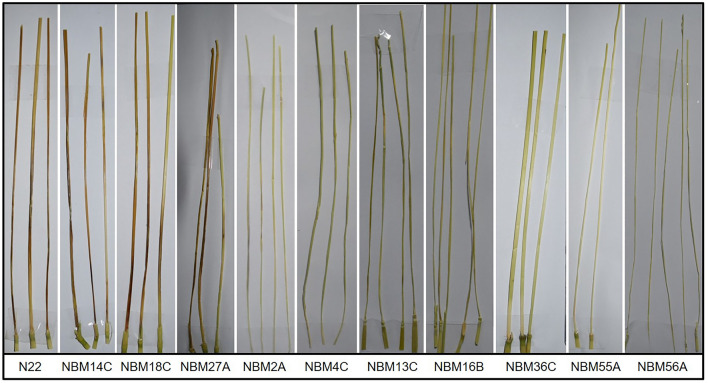
Screening of 45 putative resistant Nagina 22 (N22) mutants against panicle blast using newly developed method revealed that, 16 of the N22 mutants (NBM) were highly resistant against panicle blast inoculation with no lesion or of size less than 0.2 cm, 15 were moderately resistant having lesion size upto 0.5 cm and 14 were highly susceptible to panicle blast disease with lesion size greater than 2 cm. (Fig. 5; Table 3). When screened for leaf blast at seedlings, 10 mutants were found to be resistant, whereas NBM6C, 7B, 13B, 16C, 22C, 35A and 53C showed susceptible reaction (Table 3; Supplementary Fig. 1). Out of the entire set of putative blast-resistant mutants, initially identified in hot spot field screening of the M2 mutant population, NBM13C, 13D, 16A, 55C, 56A, 56B and 56C were found to be resistant to both leaf and panicle blast (Table 3, Fig. 5; Supplementary Fig. 1).
Fig. 5.
Phenotypic screening of rice blast disease in susceptible Nagina 22 and different mutants after 21 days of inoculation with M. oryzae strain Mo-ni-025. NBM14C, NBM18C and NBM27A showed susceptible reaction whereas rest of the mutants showed resistance reaction
Table 3.
Disease reactions observed in Nagina 22 mutants inoculated with M. oryzae strain Mo-ni-025 using new syring inoculation technique
| Mutant IDs | Number of mutants | Disease reaction |
|---|---|---|
| NBM2A, 4C, 7A, 13C, 16A, 16B, 16C, 50C, 51C, 52A, 52C, 55A, 55C, 56A, 56B, 56C | 16 | Highly resistant to panicle blast |
| NBM4A, 4B, 6C, 7B, 8C, 9A, 13D, 15A, 22C, 23A, 23B, 36B, 36C, 32A, 53C | 15 | Moderately resistant to panicle blast |
| NBM5C, 14C, 18C, 21A, 21B, 25C, 27A, 27B, 37A, 41C, 43B, 45C, 47A, 60C | 14 | Susceptible to panicle blast |
| NBM13C, 13D, 16A, 22B, 53A, 53B, 55C, 56A, 56B, 56C | 10 | Resistant to leaf blast |
| NBM6C, 7B, 13B, 16C, 22C, 35A and 53C | 7 | Susceptible to leaf blast |
Discussion
Owing to the magnitude of losses caused by rice panicle blast disease, it is necessary to identify resistant genotypes for inclusion in the breeding programs. The foremost requirement for this is to screen the available rice germplasm for panicle blast resistance. The large-scale screening of the blast disease at the field level is limited by non-uniformity and fluctuation in disease occurrence and, therefore, it is difficult to evaluate blast disease with accuracy (Fujii et al. 2000). Response of panicle blast depends on the developmental stages and time intervals of infection. The infection of neck blast on resistance and susceptible rice cultivars at preliminary heading and full heading stages of rice at the field-level did not show proper symptoms (Hao et al. 2012a, b). The pathogen needs specific temperature and humidity levels which are not controlled in the field and hence do not show reproducible results. Previously, different leaf blast inoculation methods such as spot inoculation, filter paper inoculation, and inoculation by the spraying of spores on leaves have been used for blast fungus in rice (Challagulla et al. 2015). However, all of these methods have their limitations. The method of panicle blast inoculation using syringe developed by Puri et al. (2009) requires controlled environmental conditions in green house and, therefore, it cannot be implemented at field level. Liu et al. (2016) and more recently Dong et al. (2021) have devised the cotton-wrapping inoculation method where a cotton having fungus is wrapped onto the panicle neck and plants were grown in controlled environmental conditions for growth of the fungus. However, this method needs to provide humidity at every 3 h of interval. Moreover, the possibility of infection is low as it depends on the ability of the fungus to penetrate the panicle tissue. Thus, all these methods either lacks maintenance of humidity or effective penetration by fungus for disease infection. Moreover, these earlier methods are restricted to screening in the controlled conditions and thus, there was no standard method developed for panicle inoculation with blast fungus for large-scale screening at field level. Considering the necessity of a standard, simple and efficient panicle blast inoculation method for screening of large germplasm sets in both controlled conditions as well as at field level, we developed the syringe inoculation method described here. In contrast to earlier methods of Liu and Dong, in our method the spores of M. oryzae were directly injected inside the neck of the panicle at the peduncle base with the help of a syringe which eliminates the need for spores to penetrate the physical barrier. This enhances the chances of occurrence of disease to many folds. In addition, the wet cotton wrap helps in maintaining the required humidity for M. oryzae spores to germinate and infect the panicle. Thus, this method can also be applied in areas with low humidity and unfavorable conditions for the growth of fungus. The added advantage of the method is avoidance of the possible spread of fungal spores to other plants in the field and admixture of different strains when evaluating more than one strain that normally occur in the spray method (Sørensen et al. 2016). Besides this, we have also earlier tested this method under controlled conditions and used them for transcriptome studies (Kumar et al. 2021) before standardizing it for field conditions. The success of this method depends on some of the factors namely temperature, humidity, pathogenic spore count, spore pathogenicity, stage of host and host susceptibility. The growth stage and concentration of fungal spores is an important factor that decides the disease severity. In addition, the moisture content of the cotton wrapped around the injected site should be sufficient enough to provide the required humidity but should not result in rotting of the panicle. Another most important factor is the stage of the panicle to be infected. Therefore, it is important to look for these factors for successful inoculation.
Identification of the most virulent strain of the pathogen is key to the success of any screening program. Use of the most virulent strain for screening will result in the selection of the best blast-resistant germplasm. Therefore, three different strains were used to understand any strain-specific results of infection. In the present study, Mo-ni-025 which was isolated from the Dehradun region in the northern part of India was found to be the most virulent. The occurrence of disease symptoms on infected panicles of HP2216 and HR12 genotypes confirmed the success of the method of inoculation using M. oryzae spores. The phenotypic observations of disease occurrence reflected at the molecular level is expected to confirm the disease response of the plant. Disease inducible PR protein genes are one of the best reporter genes for assessing the disease response in plants and have been reported earlier (Meng et al. 2019). Our analysis showed the blast responsive expression of the PR genes, thus confirming the blast infection at the molecular level.
Once the syringe inoculation method was established in controlled conditions, it was also replicated in the field level for screening of the rice germplasm. Out of the 12 wild rice genotypes inoculated with the blast fungus, five genotypes were identified as blast resistant. Similarly, in the case of 45 N22 mutants, shortlisted as rice blast resistant in hot spot screening, 15 were found to be resistant to panicle blast on the syringe method. In the resistant genotypes, plants did not show any symptoms of fungal infection even after 21 days of inoculation. The moderately resistant plants do show lesions; however, their effect on plant growth and yield was less as compared to the susceptible plants. The expression of blast-responsive PR protein genes in these plants manages to restrict the spread of infection and also helps in adaptation to biotic stress conditions (Jain and Khurana 2018). In contrast, in susceptible plants, lesions were developed on the whole neck of the panicle which disrupts the supply of nutrients and water to the developing flowers and grains. This results in total yield loss of the affected plants.
NKSWR-446, NKSWR-273, and NKSWR-279 were resistant to the Sikkim-6 (Mo-nwi-53) strain but became susceptible to Dehradun (Mo-ni-025) strain. Thus, the resistance of the rice genotypes is also strain specific indicating the presence of R genes for corresponding effectors of M. oryzae strains in resistant lines and vice versa. Five genotypes NKSWR-2, NKSWR-9, NKSWR-104, NKSWR-398, and NKSWR-401 showed strong resistance to all the three strains confirming their broad-spectrum nature. In the case of the mutant resources, since we used the leaf blast resistant lines identified in M2 generation and tested them in M5 generation with the new method, we could identify a number of highly resistant and moderately resistant genotypes. Thus, the 5 wild rice and 15 mutant genotypes identified as highly resistant to panicle blast infection are elite candidates for utilization in rice blast resistance breeding programs.
Conclusion
The present study has standardized a simple and effective syringe inoculation method for panicle blast infection in rice that can be used for large-scale panicle blast screening. The method applies to both controlled as well as field conditions as the wet cotton and covered aluminum foil provide the required humidity for spores to germinate and infect. The method is demonstrated in field conditions for screening of wild rice germplasm and N22 mutant population for panicle blast disease reactions leading to the identification of rice genotypes with broad-spectrum blast resistance. Thus, the method can be effectively utilized for identification of blast resistant genotypes from landraces, varieties and wild germplasm as well as for screening mapping populations for panicle blast resistant gene discovery. In addition, the method can be replicated in phenotyping of other similar diseases with disease-specific standardizations.
Acknowledgements
The Rajiv Gandhi National Fellowship from the University Grant Commission, The government of India, for VK, is acknowledged.
Author contributions
Conceptualization: AS, AM and TS; methodology: VK, SB and PS; validation: VK and MT; investigation: AS, AM and MA; resources: NS, AS and AM; writing—original draft preparation: VK, SK, and AS; writing—review and editing: AS, AM, MA, NS and TS; supervision: AS; project administration: AS and AM; funding acquisition, AS, AM, AR and TS.
Funding
The work was supported by the Indian Council of Agricultural Research (ICAR)-Centre for Agricultural Bioinformatics, IASRI (CABin Scheme), ICAR-National Institute for Plant Biotechnology, New Delhi.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Vishesh Kumar, Email: visheshkumar08@gmail.com, https://www.researchgate.net/profile/Vishesh-Kumar-6.
Pankaj K. Singh, Email: pkmolbio@gmail.com, https://www.researchgate.net/profile/Pankaj-Singh-49
Suhas Gorakh Karkute, Email: S.Karkute@icar.gov.in, https://www.researchgate.net/profile/Suhas-Karkute.
Mohd. Tasleem, Email: mohdtasleem99@gmail.com, https://www.researchgate.net/profile/Mohd-Tasleem-2
Someshwar Bhagat, Email: bhagatsomeshwar@gmail.com, https://www.researchgate.net/profile/Someshwar-Bhagat.
M. Z. Abdin, Email: mzabdin@jamiahamdard.ac.in, https://www.researchgate.net/profile/M-Abdin
Amitha Mithra Sevanthi, Email: amithamithra.nrcpb@gmail.com, https://www.researchgate.net/profile/Amitha-Mithra-Sevanthi.
Anil Rai, Email: anil.rai@icar.gov.in, https://www.researchgate.net/profile/Anil-Rai-6.
Tilak Raj Sharma, Email: trsharma1965@gmail.com, https://www.researchgate.net/profile/Tilak-Sharma-2.
Nagendra K. Singh, Email: nksingh4@gmail.com, https://www.researchgate.net/profile/Nagendra-Singh-6
Amolkumar U. Solanke, Email: amol.solanke@icar.gov.in, Email: amolsgene@gmail.com, https://www.researchgate.net/profile/Amolkumar-Solanke
References
- Agrios GN. Plant pathology. 4. San Diego: Academic Press; 1997. pp. 93–114. [Google Scholar]
- Challagulla V, Bhattarai S, Midmore DJ. In-vitro vs in-vivo inoculation: screening for resistance of Australian rice genotypes against blast fungus. Rice Sci. 2015;3(22):132–137. doi: 10.1016/j.rsci.2015.05.017. [DOI] [Google Scholar]
- Dong J, Zhou L, Feng A, Zhang S, Fu H, Chen L, Zhao J, Yang T, Yang W, Ma Y, Wang J. The OsOXO2, OsOXO3 and OsOXO4 positively regulate panicle blast resistance in rice. Rice. 2021;14(1):1–6. doi: 10.1186/s12284-021-00494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim S, Usha K, Singh B. Pathogenesis related (PR) proteins in plant defense mechanism. Sci against Microb Pathog. 2011;2:1043–1054. doi: 10.1007/978-981-10-7371-7_12. [DOI] [Google Scholar]
- Fang N, Wang R, He W, Yin C, Guan C, Chen H, Huang J, Wang J, Bao Y, Zhang H. QTL mapping of panicle blast resistance in japonica landrace Heikezijing and its application in rice breeding. Mol Breed. 2016;36(12):1–8. doi: 10.1007/s11032-016-0603-7. [DOI] [Google Scholar]
- Fang N, Wei X, Shen L, Yu Y, Li M, Yin C, He W, Guan C, Chen H, Zhang H, Bao Y. Fine mapping of a panicle blast resistance gene Pb-bd1 in Japonica landrace Bodao and its application in rice breeding. Rice (n y) 2019;12(1):18. doi: 10.1186/s12284-019-0275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi MC, Prabhu AS. Relationship between panicle blast severity and mineral nutrient content of plant tissue in upland rice. J Plant Nutr. 1998;21(8):1577–1587. doi: 10.1080/01904169809365505. [DOI] [Google Scholar]
- Fujii K, Hayano-Saito Y, Saito K, Sugiura N, Hayashi N, Tsuji T, Izawa T, Iwasaki M. Identification of a RFLP marker tightly linked to the panicle blast resistance gene, Pb1, in rice. Breed Sci. 2000;50(3):183–188. doi: 10.1270/jsbbs.50.183. [DOI] [Google Scholar]
- Hao Z, Wang L, Huang F, Tao R. Expression of defense genes and antioxidant defense responses in rice resistance to neck blast at the preliminary heading stage and full heading stage. Plant Physiol Biochem. 2012;57:222–230. doi: 10.1016/j.plaphy.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Hao Z, Wang L, Huang F, Tao R. Expression patterns of defense genes in resistance of the panicles exerted from the caulis and from the tillers to neck blast in rice. Plant Physiol Biochem. 2012;60:150–156. doi: 10.1016/j.plaphy.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, Yano M. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64(3):498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- Jain D, Khurana JP. Molecular aspects of plant–pathogen interaction. Singapore: Springer; 2018. Role of pathogenesis-related (PR) proteins in plant defense mechanism; pp. 265–281. [Google Scholar]
- Kim S, Ahn IP, Lee YH. Analysis of genes expressed during rice-Magnaporthe grisea interactions. Mol Plant Microbe Interact. 2001;14(11):1340–1346. doi: 10.1094/MPMI.2001.14.11.1340. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Sato F. Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J Biochem. 1999;125(1):1–8. doi: 10.1093/oxfordjournals.jbchem.a022244. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kanda E, Kitada K, Ishiguro K, Torigoe Y. Detection of rice panicle blast with multispectral radiometer and the potential of using airborne multispectral scanners. Phytopathology. 2001;91(3):316–323. doi: 10.1094/PHYTO.2001.91.3.316. [DOI] [PubMed] [Google Scholar]
- Kumar V, Jain P, Venkadesan S, Karkute SG, Bhati J, Abdin MZ, Sevanthi AM, Mishra DC, Chaturvedi KK, Rai A, Sharma TR. Understanding rice-Magnaporthe oryzae interaction in resistant and susceptible cultivars of rice under panicle blast infection using a time-course transcriptome analysis. Genes. 2021;12(2):301. doi: 10.3390/genes12020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yang J, Zhang S, Zhao J, Feng A, Yang T, Wang X, Mao X, Dong J, Zhu X, Leung H. OsGF14b positively regulates panicle blast resistance but negatively regulates leaf blast resistance in rice. Mol Plant Microbe Interact. 2016;29(1):46–56. doi: 10.1094/MPMI-03-15-0047-R. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meng Q, Gupta R, Min CW, Kwon SW, Wang Y, Je BI, Kim YJ, Jeon JS, Agrawal GK, Rakwal R, Kim ST. Proteomics of Rice—Magnaporthe oryzae interaction: what have we learned so far? Front Plant Sci. 2019;10:1383. doi: 10.3389/fpls.2019.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, An JM, Shin YC, Kim KJ, Lee BJ, Paek KH. Molecular characterization of pepper germin-like protein as the novel PR-16 family of pathogenesis-related proteins isolated during the resistance response to viral and bacterial infection. Planta. 2004;219(5):797–806. doi: 10.1007/s00425-004-1290-x. [DOI] [PubMed] [Google Scholar]
- Puri KD, Shrestha SM, Chhetri GB, Joshi KD. Leaf and neck blast resistance reaction in tropical rice lines under green house condition. Euphytica. 2009;165(3):523–532. doi: 10.1007/s10681-008-9771-9. [DOI] [Google Scholar]
- Rawal HC, Mithra SA, Arora K, Kumar V, Goel N, Mishra DC, Chaturvedi KK, Rai A, Devi SV, Sharma TR, Solanke AU. Genome-wide analysis in wild and cultivated Oryza species reveals abundance of NBS genes in progenitors of cultivated rice. Plant Mol Biol Report. 2018;36(3):373–386. doi: 10.1007/s11105-018-1086-y. [DOI] [Google Scholar]
- SES I . Standard evaluation system. Manila: International Rice Research Institute; 2002. pp. 11–30. [Google Scholar]
- Sevanthi A, Kandwal P, Kale PB, Prakash C, Ramkumar MK, Yadav N, Mahato AK, Sureshkumar V, Behera M, Deshmukh RK, Jeyaparakash P. Whole genome characterization of a few EMS-induced mutants of upland rice variety Nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front Plant Sci. 2018;9:1179. doi: 10.3389/fpls.2018.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikari AB, Rajashekara H, Khanna A, Krishnan SG, Rathour R, Singh UD, Sharma TR, Prabhu KV, Singh AK. Identification and validation of rice blast resistance genes in Indian rice germplasm. Indian J Genet Plant Breed. 2014;74(3):286–299. doi: 10.5958/0975-6906.2014.00846.3. [DOI] [Google Scholar]
- Shim HS, Hong SJ, Yeh WH, Han SS, Sung JM. Damage analysis of rice panicle blast on disease occurrence time and severity. Plant Pathol J. 2005;21(2):87–92. doi: 10.5423/PPJ.2005.21.2.087. [DOI] [Google Scholar]
- Singh B, Singh N, Mishra S, Tripathi K, Singh BP, Rai V, Singh AK, Singh NK. Morphological and molecular data reveal three distinct populations of Indian wild rice Oryza rufipogon Griff species complex. Front Plant Sci. 2018;9:123. doi: 10.3389/fpls.2018.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CK, Thach T, Hovmøller MS. Evaluation of spray and point inoculation methods for the phenotyping of Puccinia striiformis on wheat. Plant Dis. 2016;100(6):1064–1070. doi: 10.1094/PDIS-12-15-1477-RE. [DOI] [PubMed] [Google Scholar]
- Sureshkumar V, Dutta B, Kumar V, Prakash G, Mishra DC, Chaturvedi KK, Rai A, Sevanthi AM, Solanke AU. RiceMetaSysB: a database of blast and bacterial blight responsive genes in rice and its utilization in identifying key blast-resistant WRKY genes. Database. 2019 doi: 10.1093/database/baz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy K, Singh B, Singh N, Rai V, Misra G, Singh NK. A database of wild rice germplasm of Oryza rufipogon species complex from different agro-climatic zones of India. Database. 2018 doi: 10.1093/database/bay058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Van Kammen A. Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacumvar “Samsun” and “Samsun NN”—I. Phytochemistry. 1970;7(10):1727–1735. doi: 10.1016/S0031-9422(00)86642-X. [DOI] [PubMed] [Google Scholar]
- Wu Y, Xiao N, Li Y, Gao Q, Ning Y, Yu L, Cai Y, Pan C, Zhang X, Huang N, Zhou C. Identification and fine mapping of qPBR10-1, a novel locus controlling panicle blast resistance in Pigm-containing P/TGMS line. Mol Breed. 2021;41(12):1–4. doi: 10.1007/s11032-021-01268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Liu Q, Naake T, Huang W, Chen M, Kong Q, Zhang S, Li W, Li X, Liu Q, Yang J. OsGF14b modulates defense signaling pathways in rice panicle blast response. Crop J. 2021;9(4):725–738. doi: 10.1016/j.cj.2020.10.007. [DOI] [Google Scholar]
- Zhu YY, Fang H, Wang YY, Fan JX, Yang SS, Mew TW, Mundt CC. Panicle blast and canopy moisture in rice cultivar mixtures. Phytopathology. 2005;95(4):433–438. doi: 10.1094/PHYTO-95-0433. [DOI] [PubMed] [Google Scholar]