Graphical abstract
Abbreviations: A, Adenine; BiLSTM, Bi-directional Long-Short Term Memory; C, Cytosine; CDF, Cutting frequency determination; CNN, Convolutional Neural Networks; CRISPR/Cas9, Clustered Regularly Interspaced Short Palindromic Repeats / CRISPR associated protein 9; CRISPR-IP, CRISPR model based on Identity and Position; DNN, Dense Neural Networks; G, Guanine; gRNA, Guide RNA; GRU, Gate Recurrent Unit; LOGOCV, Leave-one-gRNA-out cross-validation; LSTM, Long-Short Term Memory; PAM, Protospacer adjacent motif; PR-AUC, Area Under the Precision-Recall Curve; RNN, Recurrent Neural Networks; ROC-AUC, Area Under the Receiver Operating Characteristic Curve; T, Thymine; U, Uracil
Keywords: CRISPR-Cas9, Off-target prediction, Deep learning, Encoding scheme
Abstract
The CRISPR/Cas9 gene-editing system is the third-generation gene-editing technology that has been widely used in biomedical applications. However, off-target effects occurring CRISPR/Cas9 system has been a challenging problem it faces in practical applications. Although many predictive models have been developed to predict off-target activities, current models do not effectively use sequence pair information. There is still room for improved accuracy. This study aims to effectively use sequence pair information to improve the model's performance for predicting off-target activities. We propose a new coding scheme for coding sequence pairs and design a new model called CRISPR-IP for predicting off-target activity. Our coding scheme distinguishes regions with different functions in the sequence pairs through the function channel. Moreover, it distinguishes between bases and base pairs using type channels, effectively representing the sequence pair information. The CRISPR-IP model is based on CNN, BiLSTM, and the attention layer to learn features of sequence pairs. We performed performance verification on two data sets and found that our coding scheme can represent sequence pair information effectively, and the CRISPR-IP model performance is better than others. Data and source codes are available at https://github.com/BioinfoVirgo/CRISPR-IP.
1. Introduction
CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) system is a powerful genome editing technology for editing various species and cells [1], [2], [3]. The CRISPR/Cas9 system has two key components: Guide RNA (gRNA) and Cas9 endonuclease. Guide RNA is an RNA chimera composed of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). The crRNA contains a guide sequence that can accurately guide the Cas9 protein to the corresponding target of the genome. The 20-nucleotide guide sequence in the guide RNA is complementary to the DNA target sequence, and the Cas9 nuclease cuts the DNA upstream of the 3-nucleotide protospacer adjacent motif (PAM) to form a blunt-ended DNA double-strand break [4], [5], [6]. Although the CRISPR/Cas9 system has many advantages, its possible off-target risk has affected the in-depth study of gene-editing technology [7]. Therefore, improving off-target prediction methods' performance is critical to help isolate the exact location of DNA cleavage.
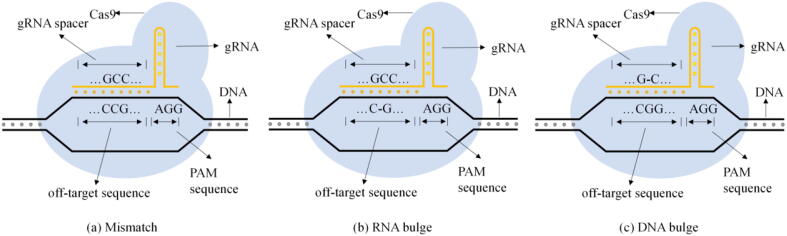
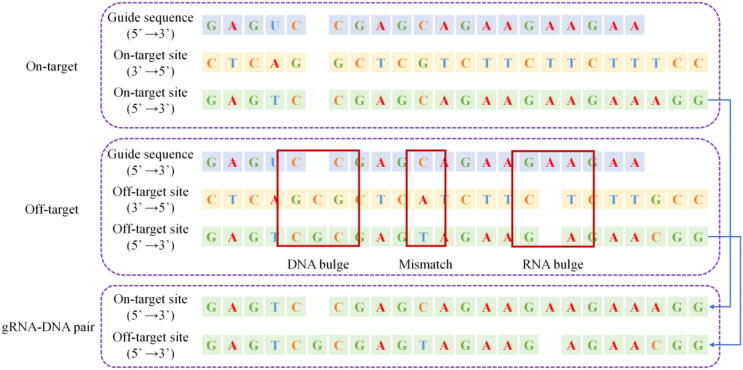
The off-target effect is that the Cas9 protein binds to an unexpected genomic site for cutting [8]. Off-target sites can be divided into three categories, as shown in Fig. 1: (1) Mismatches, (2) RNA bulges (insertion), and (3) DNA bulges (deletion) [9].
Fig. 1.
Three cases of off-target types.
In recent years, researchers have studied the off-target prediction of the CRISPR/Cas9 system and proposed various methods. These methods can mainly be divided into alignment-based methods and scoring-based methods [10]. (1) Alignment-based methods determine the integrity of the search for potential off-target sites through the maximum number of mismatches, allowed PAM, and other conditions, such as Cas-OFFinder [11] and CRISPRitz [12]. Therefore, they are often used to find possible off-target sites from the entire genome. (2) Score-based methods are used to predict the off-target activity of gRNA-DNA pairs, which can be divided into three categories. Early models used hypothesis-driven methods (evaluating off-target activities based on formulas), such as MIT [13] and CDF [14]. These methods calculate the probability of off-target activity at potential off-target sites based on hand-made rules. Further, some researchers attempted to use machine learning methods to construct models, such as CRISTA [15] and Elevation [16]. These methods used the number of GCs, mismatch positions, and other artificially constructed features to predict the probability of off-target activity. Recently, researchers have proposed some prediction models based on deep learning methods, such as AttnToMismatch_CNN [17] and CRISPR-Net [18]. These methods can learn features automatically from the sequence pairs and utilize these features for prediction.
Although existing studies have attempted complex off-target prediction models, they do not effectively use the sequence pair information. How to utilize sequence pair information effectively is still a challenging problem. We can divide the off-target prediction problem based on deep learning into two tasks. (1) Convert the gRNA-DNA sequence pair into a vector or matrix representation. (2) Use deep learning models to learn high-order features from vector or matrix representations and make predictions for sequence pairs.
For the first task, a series of coding schemes were proposed. Lin et al. coded gRNA and DNA into a 4-dimensional one-hot vector and obtained the corresponding base pair code through the 'OR' operation [19]. Charlier et al. pointed out that the 'OR' operation of the coding scheme would lead to information loss and proposed using the concatenating operation instead of the 'OR' operation [20]. Neither of these two coding schemes considers the off-target situation with bulges. Lin et al. proposed a new coding scheme to encode sequence pairs containing bulges and mismatches [18]. However, they all ignore that the gRNA-DNA sequence pair contains two sequence regions with different functions. In addition, the coding scheme proposed by Lin et al. uses 'OR' operates in the type channel part, which will also cause a part of the information to be lost.
For the second task, a series of network models are proposed. As shown in Table 1, these models use two or three of the four types of network layers. The convolutional layer learns local features through the convolution kernel [21], the recurrent layer learns the context features of the sequence by saving the sequence state [22], and the attention layer learns global features by calculating the attention score [23], [24]. Finally, the dense layer maps the features to the sample label space. Different types of network layers have a different focus on learning features. However, no model simultaneously uses four types of network layers for off-target prediction.
Table 1.
The model's association with the network layer.
| Model | Convolution | Recurrent | Attention | Dense |
|---|---|---|---|---|
| CRISPR-Net [18] | Yes | Yes | No | Yes |
| CRISPR-OFFT [25] | Yes | No | Yes | Yes |
| AttnToMismatch_CNN [17] | Yes | No | Yes | Yes |
| CNN_std [19] | Yes | No | No | Yes |
| DeepCRISPR [26] | Yes | No | No | Yes |
Notes: 'Yes' means that the model uses this kind of network layer, and 'NO' means it does not.
In this study, we propose a new coding scheme for gRNA-DNA sequence pairs. That distinguishes between bases and base pairs in sequence pairs through the type channels, distinguishes regions with different functions in the sequence pairs through the function channel, and solves the information loss problem in Lin’s coding scheme. In addition, we develop a new model CRISPR-IP (CRISPR model based on Identity and Position), which learns the identity features of base pairs through CNN, learns the position features of base pairs through BiLSTM, and learns the sequence pair features through the attention layer. Finally, the model predicts the possibility of off-target activity for each potentially off-target gRNA-DNA pair. The experiments have proved that our coding scheme effectively represents sequence pair information, which helps improve the model’s prediction performance. Moreover, the performance of our proposed model is better than several advanced off-target prediction models. Therefore, it is expected to become a potential tool to guide the CRISPR/Cas9 system experiments.
2. Results
The experiments in the study are designed as follows: First, we evaluate the two coding schemes through the CRISPR-IP model and three types of neural networks. Second, we compared the CRISPR-IP with other advanced off-target models and analyzed how the four parts of the CRISPR-IP affect the model’s performance through ablation experiments. Then, we studied the impact of samples with bulges on models and augmented epigenetics factor features to evaluate models. Finally, we studied the impact of over-sampling and under-sampling methods on the prediction performance of the CRISPR-IP.
2.1. Comparison of coding schemes
We use leave-one-gRNA-out cross-validation (LOGOCV) on the CIRCLE-Seq data set and SITE-Seq data set to evaluate the prediction performance of the two coding schemes in the same model. For evaluation details, please refer to the section “Performance evaluation”. The average values of the cross-validation results in the CIRCLE-Seq data set and the SITE-Seq data set are shown in Table 2 and Table 3.
Table 2.
Performance for each predictive model on the CIRCLE-seq data set.
| Metric | CRISPR-IP | FNN3 | FNN5 | FNN10 | CNN3 | CNN5 | LSTM | GRU | Encoding |
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | 0.989 | 0.962 | 0.955 | 0.988 | 0.988 | 0.988 | 0.984 | 0.983 | encoding scheme 1 |
| Accuracy | 0.989 | 0.983 | 0.976 | 0.981 | 0.988 | 0.988 | 0.986 | 0.976 | encoding scheme 2 |
| F1 score | 0.375 | 0.148 | 0.110 | 0.169 | 0.004 | 0.005 | 0.250 | 0.284 | encoding scheme 1 |
| F1 score | 0.383 | 0.138 | 0.096 | 0.090 | 0.019 | 0.037 | 0.171 | 0.240 | encoding scheme 2 |
| PR-AUC | 0.483 | 0.150 | 0.102 | 0.161 | 0.244 | 0.239 | 0.230 | 0.265 | encoding scheme 1 |
| PR-AUC | 0.494 | 0.103 | 0.065 | 0.069 | 0.242 | 0.226 | 0.331 | 0.330 | encoding scheme 2 |
| Precision | 0.666 | 0.291 | 0.229 | 0.477 | 0.057 | 0.240 | 0.396 | 0.401 | encoding scheme 1 |
| Precision | 0.675 | 0.240 | 0.146 | 0.190 | 0.334 | 0.418 | 0.427 | 0.424 | encoding scheme 2 |
| ROC-AUC | 0.961 | 0.897 | 0.840 | 0.770 | 0.940 | 0.935 | 0.873 | 0.907 | encoding scheme 1 |
| ROC-AUC | 0.968 | 0.856 | 0.812 | 0.782 | 0.937 | 0.929 | 0.936 | 0.901 | encoding scheme 2 |
| Recall | 0.295 | 0.330 | 0.278 | 0.128 | 0.002 | 0.002 | 0.244 | 0.320 | encoding scheme 1 |
| Recall | 0.320 | 0.123 | 0.113 | 0.099 | 0.010 | 0.020 | 0.161 | 0.308 | encoding scheme 2 |
Notes: Better results are indicated in bold. Encoding scheme 1 was proposed by Lin et al., and coding scheme 2 was proposed by us.
Table 3.
Performance for each predictive model on the CIRCLE-seq data set.
| Metric | CRISPR-IP | FNN3 | FNN5 | FNN10 | CNN3 | CNN5 | LSTM | GRU | Encoding |
|---|---|---|---|---|---|---|---|---|---|
| Accuracy | 0.990 | 0.670 | 0.796 | 0.955 | 0.982 | 0.982 | 0.988 | 0.987 | encoding scheme 1 |
| Accuracy | 0.990 | 0.971 | 0.966 | 0.984 | 0.990 | 0.989 | 0.988 | 0.988 | encoding scheme 2 |
| F1 score | 0.621 | 0.144 | 0.133 | 0.364 | 0.222 | 0.255 | 0.560 | 0.518 | encoding scheme 1 |
| F1 score | 0.644 | 0.364 | 0.403 | 0.472 | 0.533 | 0.501 | 0.569 | 0.531 | encoding scheme 2 |
| PR-AUC | 0.695 | 0.209 | 0.082 | 0.302 | 0.316 | 0.319 | 0.665 | 0.616 | encoding scheme 1 |
| PR-AUC | 0.751 | 0.350 | 0.339 | 0.391 | 0.641 | 0.587 | 0.682 | 0.676 | encoding scheme 2 |
| Precision | 0.808 | 0.117 | 0.087 | 0.325 | 0.691 | 0.672 | 0.669 | 0.688 | encoding scheme 1 |
| Precision | 0.791 | 0.529 | 0.439 | 0.538 | 0.882 | 0.887 | 0.725 | 0.796 | encoding scheme 2 |
| ROC-AUC | 0.973 | 0.767 | 0.775 | 0.855 | 0.891 | 0.885 | 0.970 | 0.965 | encoding scheme 1 |
| ROC-AUC | 0.982 | 0.892 | 0.900 | 0.904 | 0.968 | 0.961 | 0.971 | 0.973 | encoding scheme 2 |
| Recall | 0.526 | 0.711 | 0.614 | 0.566 | 0.194 | 0.211 | 0.569 | 0.504 | encoding scheme 1 |
| Recall | 0.593 | 0.473 | 0.583 | 0.555 | 0.396 | 0.364 | 0.570 | 0.506 | encoding scheme 2 |
Notes: Better results are indicated in bold. Encoding scheme 1 was proposed by Lin et al., and coding scheme 2 was proposed by us.
For the CRISPR-IP model, the evaluation results show that the off-target prediction performance of the model using the new coding scheme is improved. On the CIRCLE-Seq data set, the CRISPR-IP model using our coding scheme achieved better results on PR-AUC, ROC-AUC, F1-score, Precision, and Recall, with an increase of 1.1%, 0.7%, 0.8%, 0.9%, 2.5%, respectively. Accuracy is the same as the result of the model using Lin’s coding scheme, and both are 0.989. That also reflects that the imbalance of the samples will cause some evaluation metrics to be unable to evaluate the prediction results of the model objectively. On the SITE-Seq data set, the CRISPR-IP model using our coding scheme achieved better results on PR-AUC, ROC-AUC, F1-score, and Recall, with an increase of 5.6%, 0.9%, 2.3%, and 6.7%. Although Precision has declined by 1.7%, the improvement of the F1-score shows that the CRISPR-IP model using our coding scheme has improved the overall predictive performance.
To compare the two coding schemes more objectively, we evaluated them through three types of network models. For details of the network, see the section “Neural networks for comparing coding schemes”. On the SITE-Seq data set, the CNN and RNN networks using our coding scheme have achieved better results in six performance metrics, which shows that the use of our coding scheme can improve the performance of CNN and RNN networks. Although the DNN network using our coding scheme has declined in Recall, the improvement of the F1-score shows that the performance of the DNN network’s off-target prediction is overall improved. However, on the CIRCLE-Seq data set, the prediction results of the three types of networks using our coding scheme and Lin’s coding scheme have advantages and disadvantages in six performance metrics. Because the sample imbalance problem of the CIRCLE-Seq data set is more serious, making models challenging to learn useful features from the coding, resulting in poor prediction results. In the performance of the two data sets, we found that the prediction results on the SITE-seq data set are better than the results on the CIRCLE-Seq data set under the same coding scheme and the same model. The problem of sample imbalance affects the model’s ability to learn information from the code. In short, the results of Table 2 and Table 3 show that our coding scheme can more effectively express the information of sequence pairs, which helps the model to achieve better results.
2.2. Comparison of different models
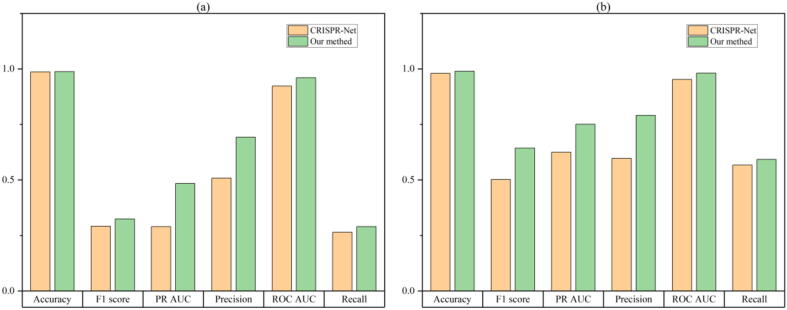
This section compares the CRISPR-IP model with other advanced off-target prediction models. The experimental results of the CRISPR-IP and the CRISPR-Net on the two data sets are shown in Fig. 2.
Fig. 2.
Performance evaluation of CRISPR-IP and CRISPR-Net. The result of CIRCLE-Seq dataset is (a), the result of SITE-Seq dataset is (b).
It can be observed from Fig. 2 that compared with the CRISPR-Net model, the performance of our model has greatly improved. On the CIRCLE-Seq data set, CRISPR-IP has increased by 0.1%, 19.4%, 3.7%, 3.4%, 18.5%, 2.5% in Accuracy, PR-AUC, ROC-AUC, F1-score, Precision, Recall, respectively. On the SITE-seq data set, CRISPR-IP has increased by 1%, 12.6%, 2.9%, 14.2%, 19.4%, 2.6% in Accuracy, PR-AUC, ROC-AUC, F1-score, Precision, Recall, respectively.
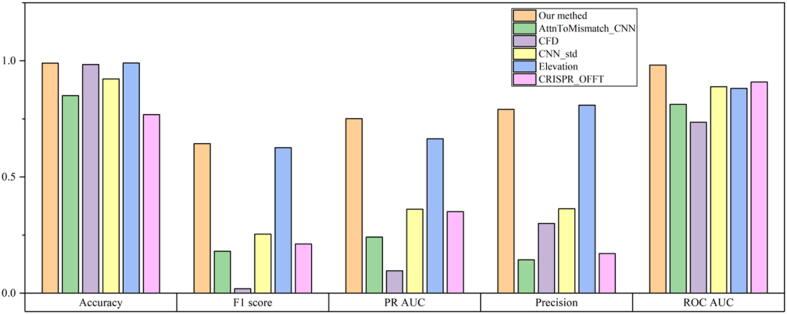
The results of the CFD, AttnToMismatch_CNN, CNN_std, CRISPR-OFFT, and Elevation methods on the SITE-seq data set are shown in Fig. 3. Compared with CFD, AttnToMismatch_CNN, CRISPR-OFFT, and CNN_std, our model significantly improves the results of the other five evaluation metrics, except for Accuracy, which is seriously affected by sample imbalance problems. Compared with the Elevation method, the CRISPR-IP model has improved PR-AUC, ROC-AUC, F1-score, and Recall by 8.7%, 10.1%, 1.8%, and 6.5%, respectively. Accuracy, Precision has decreased by 0.1% and 1.8%. The Elevation method has better results on Precision. Because the Elevation method is more inclined to predict samples as negative samples, resulting in lower Recall and F1-score indicators. We conclude that CRISPR-IP has better predictive capabilities than other advanced models in Fig. 2 and Fig. 3.
Fig. 3.
Evaluation of performance for CRISPR-IP and other models on SITE-seq data set.
2.3. Ablation study
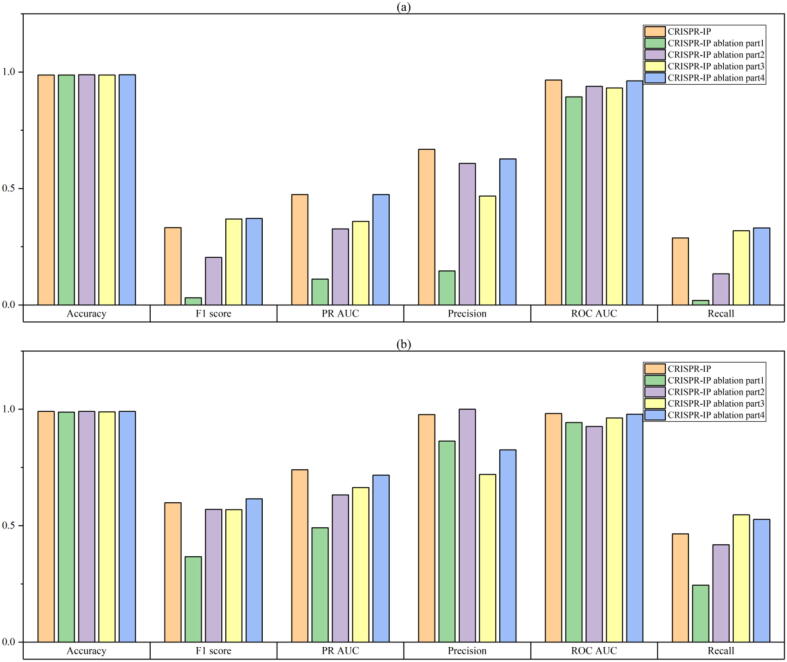
In this section, we design ablation experiments to verify the influence of different parts of CRISPR-IP on the prediction performance. The details of CRISPR-IP architecture are in the “CRISPR-IP architecture” section. By deleting the four parts of the CRISPR-IP model, four ablation models were constructed: “ablation part1″, ”ablation part2″, “ablation part3″, and ”ablation part4″. Among them, “ablation part4″ does not delete all of the dense layers and retains a dense layer as the output layer. The purpose of the ablation experiment is mainly to show whether the four parts of the model improve performance. The experimental results are shown in Fig. 4.
Fig. 4.
Results of ablation experiments. The result on CIRCLE-Seq dataset is (a), the result on SITE-Seq dataset is (b).
Due to the imbalance of the sample, models can easily obtain a high value of Accuracy. The model’s Accuracy has a small gap and is not distinguishable. Therefore, we gave up the comparison of Accuracy values. In addition, CRISPR-IP and the four ablation models have advantages and disadvantages in F1-score, Precision, and Recall. These metrics are affected by the threshold and have limitations that are difficult to reflect the overall performance. According to PR-AUC and ROC-AUC that can reflect the overall situation, we found that CRISPR-IP has better results than the four ablation models. In PR-AUC and ROC-AUC, we focus more on the comparison results of PR-AUC. PR-AUC can better reflect the global performance of classification when samples are imbalanced. We observe from PR-AUC results that removing the convolution layer and pooling layer (ablation part1) has the most significant impact on the model’s performance, which is reduced by 36.4% on the CIRCLE-Seq data set, 24.9% on the SITE-Seq data. It shows that learning the identity features of nucleotide pairs through the convolutional layer plays an essential role in the model’s off-target predicting. The significant impacts are to delete the BiLSTM part of the model (ablation part2) and delete the model’s attention and global pooling layers (ablation part3). The PR-AUC of the model with the BiLSTM part removed decreases 14.7% on the CIRCLE-Seq data set and 10.8% on the SITE-Seq data. The PR-AUC of the model that removes the attention and global pooling layers reduces 11.5% on the CIRCLE-Seq data set and 7.6% on the SITE-Seq data. It shows that learning the sequence features of nucleotide pairs containing position information through the BiLSTM layer and the focused learning of nucleotide pairs features through the attention layer can improve the model’s performance. The most negligible impact on model performance is the dense layer (ablation part4). The PR-AUC of the model that deletes the dense layer part is reduced by 0.03% on the CIRCLE-Seq data set and 2.3% on the SITE-Seq data, slightly lower than the CRISPR-IP model. That shows that the dense layer improves performance, although the degree of improvement is not precise.
2.4. Impact of sequence pairs with bulges
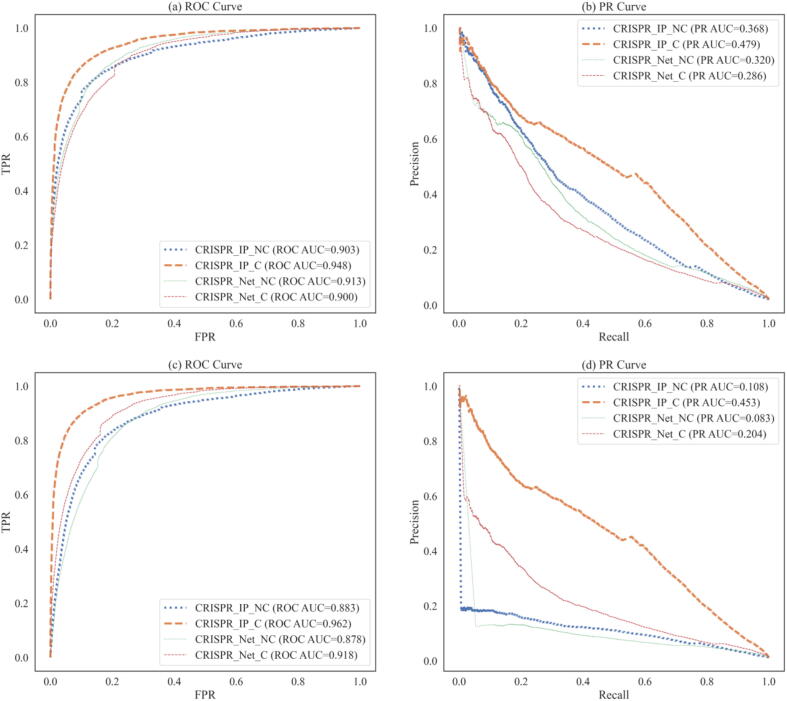
In this part, we study the impact of samples (sequences) with bulges on the model's prediction performance on the CIRCLE-Seq dataset. We named the original CIRCLE dataset as Dataset_C, which contains samples with bulges. We delete the samples containing bulges in the CIRCLE-Seq dataset and construct the dataset named Dataset_NC. We used LOGOCV to evaluate CRISPR-IP and CRISPR-Net on two datasets. The evaluation results are shown in Fig. 5.
Fig. 5.
Results of models on the Dataset_C and Dataset_NC. The results of the Dataset_NC are (a) and (b), and the results of the Dataset_C are (c) and (d). Model_Name_C are models trained on the Dataset_C, and so are Model_Name_NC.
From Fig. 5(a) and (b), the prediction performance of the models trained on the Dataset_C has increased or decreased in the Dataset_NC. CRISPR_IP_C compared with CRISPR_IP_NC ROC-AUC and PR-AUC increased by 4.5% and 11.1%. In contrast, CRISPR_Net_C was reduced by 1.3% and 3.4%, respectively, compared with CRISPR_Net_NC. That shows it is possible to learn effective features from samples (sequence pairs) with bulges and help classify and predict samples that do not contain bulges by designing coding schemes and models. Fig. 5(c) and (d) show that the models trained on the Dataset_NC are difficult to extend to the Dataset_C. Compared with CRISPR_IP_C, results of CRISPR_IP_NC were lower. ROC-AUC and PR-AUC were reduced by 7.9% and 34.5%, respectively. CRISPR_Net_NC was reduced by 4.0% and 12.1%, respectively, compared with CRISPR_Net_C. In addition, in Fig. 5, we found that under the same conditions, our model's performance is better than CRISPR-Net, which also proves the superiority of our model.
To analyze the reasons why it is difficult to expand, we analyzed the top N sequence pairs (TopN) predicted to maybe off-target on the Dataset_C of CRISPR_IP_NC and CRISPR_IP_C, and the results are shown in Table 4. Taking Top7000 as an example, the CRISPR_IP_NC gives higher prediction scores for unknown samples (sequence pairs with bulges). Therefore, the number of sequence pairs with bulges (NB) contained in Top7000 is as high as 5633, ten times more than CRISPR_IP_C's NB. However, the accuracy of CRSIPR_IP_NC predicting these samples is very low, affecting the overall performance of the model. As a result, the number of off-target sequence pairs (NOT) predicted by CRSIPR_IP_NC is 2204 fewer than predicted by CRISPR_IP_C. In addition, we found that both Dataset_C and Dataset_DC have sample imbalance problems. The positive–negative sample ratio of Dataset_C is about 1:79, and the positive–negative sample ratio of Dataset_NC is about 1:47. The sample imbalance problem of Dataset_C is more serious, which causes CRISPR_IP_C to give samples a lower prediction score. As shown in Table 4, the mean of predicted scores (MPS) of CRISPR_IP_C is lower than that of CRISPR_IP_NC.
Table 4.
The results of TopN.
| Metric | Model | 1000 | 2000 | 3000 | 4000 | 5000 | 6000 | 7000 |
|---|---|---|---|---|---|---|---|---|
| NOT | CRISPR_IP_NC | 189 | 369 | 545 | 733 | 885 | 1036 | 1204 |
| NOT | CRISPR_IP_C | 768 | 1329 | 1882 | 2351 | 2759 | 3109 | 3408 |
| NB | CRISPR_IP_NC | 880 | 1708 | 2536 | 3323 | 4118 | 4893 | 5633 |
| NB | CRISPR_IP_C | 40 | 101 | 136 | 206 | 274 | 350 | 441 |
| MPS | CRISPR_IP_NC | 1.000 | 0.999 | 0.998 | 0.995 | 0.990 | 0.983 | 0.974 |
| MPS | CRISPR_IP_C | 0.968 | 0.891 | 0.799 | 0.721 | 0.658 | 0.606 | 0.563 |
Note: NOF: Number of off-target sequence pairs. NB: Number of sequence pairs with bulges. MPS: Mean of the predicted scores.
2.5. Evaluation of CRISPR-IP with epigenetic information
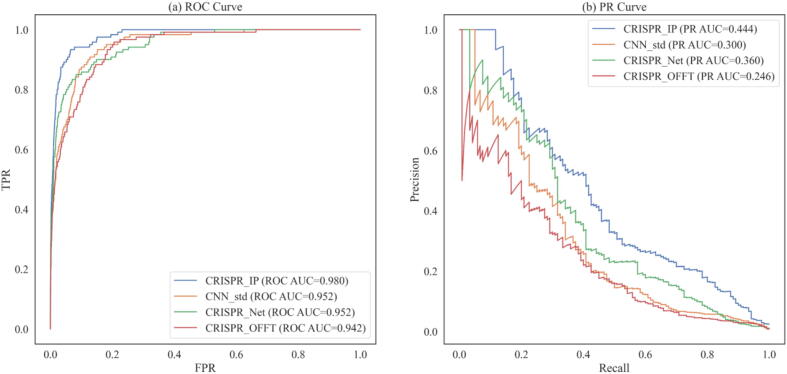
Epigenetic factors are factors that affect gRNA off-target prediction. We study the off-target prediction performance of CRISPR-IP with four epigenetic features (CTCF, DNase, H3K4me3, and RRBS) compared the modified CNN_std, CRISPR-Net, and CRISPR-OFFT on two off-target datasets from HEK293T and K562 cell types. Since the K562 data set has only 20,319 samples, it is difficult to use for deep model learning, so we trained models on the HEK293T dataset and verified them on the K562 dataset. The results are shown in Fig. 6. CRISPR-IP achieved the highest ROC-AUC (0.980) and PR-AUC (0.444). All in all, compared with advanced deep learning models, CRISPR-IP with sequence information and epigenetic factors has outstanding performance in ROC-AUC and PR-AUC.
Fig. 6.
Results of CRISPR-IP, CRISPR-Net, CRISPR-OFFT and CNN_std on K562 Dataset.
2.6. Impact of sampling method
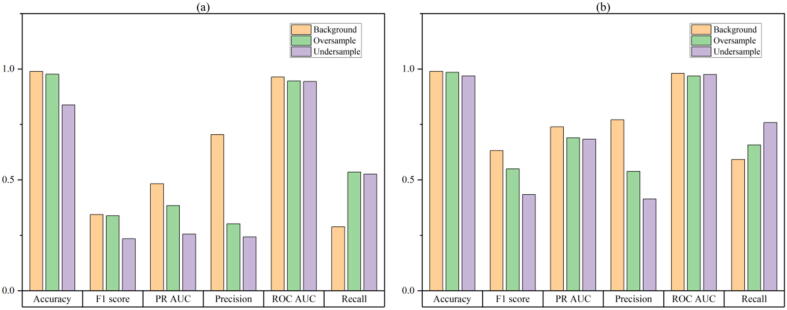
From previous experiments, we found that the sample imbalance problem affected the prediction performance of our model. Therefore, we designed this experiment to test the influence of two commonly used resampling methods to deal with sample imbalance on the model. For details of the resampling method, please refer to the “Sampling Method” section. The experimental results are shown in Fig. 7. First of all, on the two data sets, training models without resampling have better results on Accuracy, PR-AUC, ROC-AUC, F1-score, and Precision, and only a decrease in Recall. That means that using oversampling methods causes excessive noise of positive samples. Using undersampling methods causes discarding many samples, resulting in the inability to learn all the features of negative samples. These two problems are more severe than the sample imbalance problem, seriously affecting the model's prediction performance. The sample imbalance problem with the CIRCLE-Seq data set is more severe than the SITE-Seq data set. The positive–negative sample ratio is 1:78, larger than the 1:57 of the SITE-Seq data set. We found that oversampling has improved the model's accuracy, PR-AUC, F1-score, and Precision compared to undersampling on the SITE-Seq data set, while ROC-AUC and Recall have decreased. On the CIRCLE-Seq dataset, oversampling has improved all of the performance metrics compare to undersampling. Therefore, using the undersampling method, the CIRCLE-Seq dataset will lose more negative samples, resulting in the model learning fewer negative sample features and worse model prediction performance.
Fig. 7.
Performance evaluation of no processing and two resampling methods on CRISPR-IP model. The result of CIRCLE-Seq dataset is (a), the result of SITE-Seq dataset is (b).
3. Discussion and conclusion
The guide sequence and potential off-target sequence pairs play an essential role in off-target prediction. Effective use of sequence pair information can improve the model performance of off-target prediction. The process can be divided into two parts: (1) Use coding schemes to encode sequence pairs and effectively express information. (2) Use the off-target prediction model to learn features from the coding to perform off-target prediction automatically.
The main contribution of our study is to propose a new coding scheme and network model. Our coding scheme is the first to consider distinguishing different regions of function in sequence pairs. It improves the problem of information loss in the coding scheme proposed by Lin et al. Our experiments prove that it can express more sequence information for network model learning and prediction. In our experiments, we tested three types of deep learning networks, i.e., CNN, FNN, and RNN, to show the effectiveness and robustness of our coding scheme. Our experiments have proved the practicality of integrating three types of network layers for feature extraction, which can better use sequence information and obtain better predictions. We proved the necessity of three network layers to extract features through ablation experiments, and the absence of any one of them will affect the model's prediction performance.
In addition, we studied whether using the data with bulges for training affects the prediction of the data with bulges. We found that using data that does not contain bulges for training, the model cannot learn the features of bulges and cannot be used to predict samples that contain bulges. We evaluate the model's performance with epigenetic information and the impact of the sampling method on the model through experiments. Compared with advanced deep learning models, CRISPR-IP with sequence information and epigenetic factors has outstanding performance. Undersampling and oversampling methods will reduce the model's prediction performance.
Deep learning models trained on imbalanced data tend to achieve high accuracies for the majority class. However, the learning models generally perform worse for the minority class, which is noteworthy in this case [25]. Data imbalance is a common problem for off-target prediction, and efficient computational techniques can help address the issue [27]. The two resampling methods will reduce the CRISPR-IP's prediction performance. However, the improvement in Recall shows that the model is still affected by sample imbalance. How to alleviate the impact of sample imbalance is our research direction in the future.
4. Materials and methods
4.1. Dataset
The experimental methods of whole-genome detection of off-target sites are currently divided into in vitro and in vivo methods [28]. This paper uses the experimental data sets obtained by two in vitro detection methods for model training and verification to compare the sequence pairs' influence on off-target activities and exclude the influence of the complex environment in the cell. The two data sets are the experimental results based on the SITE-seq method [29] and the CIRCLE-seq method [30]. The CIRCLE-Seq dataset contains gRNA-DNA pairs from 10 guide sequences, of which 7371 are active off-target (430 with bulges). The SITE-Seq dataset contains gRNA-DNA pairs from 9 guide sequences, of which 3767 are active off-target (no bulges). Given the guide sequence, Cas-offinder [11], a versatile tool for searching for potential off-target sites, obtains inactive off-target sites in the genome. The CIRCLE-Seq dataset obtained 577,578 inactive off-target sites. The SITE-Seq data set obtained 213,966 inactive off-target sites. For the classification model, the active off-target sites are labeled as “1″, and the inactive off-target sites are labeled as ”0″. In addition, we use the HEK293T and K562 datasets collected by Chuai et al. to evaluate CRISPR-IP with epigenetic information [26].
4.2. Coding scheme
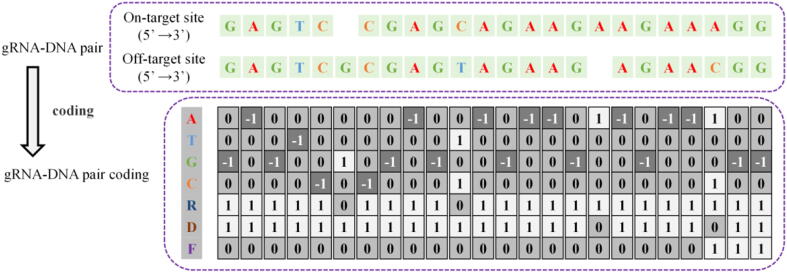
The potential off-target sequence of the guide sequence is a sequence that is similar but not the same as the target sequence in other positions of the genome. Although the guide sequence and the potential off-target sequence have mismatches, insertions, or deletions, they can guide and activate the Cas9 nuclease to cut. Therefore, we can predict whether off-target activities occur at the potential off-target site based on the guide sequence and the potential off-target sequence. This study represents gRNA-DNA pairs by corresponding target sequence and potential off-target sequence pairs, as shown in Fig. 8. The targeting sequence in the DNA represents the guide sequence. Thymine (T) replaces uracil (U), which retains the original information and avoids redundant coding.
Fig. 8.
Representation for the gRNA-DNA pair.
To our knowledge, the coding scheme proposed by Lin et al. [18] is the first and only coding scheme proposed to encode sequence pairs with bulges. Lin’s coding scheme includes five type channels (A, T, C, G, _) and two direction channels. The type channels are adenine (A), guanine (G), cytosine (C), thymine (T), and base deletion (-). Encode bases through one-hot vectors, and encode base pairs through OR operations. For example, adenine is encoding as (1,0,0,0,0), adenine and guanine pair is encoding as (1,0,0,1,0), adenine and adenine pair is encoding as (1,0,0,0,0). The encoding of the adenine pair is the same as adenine’s encoding in the adenine channel. Therefore, the label ‘1′ of the adenine channel cannot distinguish that it is an adenine or an adenine pair. That will inevitably lead to the loss of some information. In addition, there are two sequence regions with different functions in the gRNA-DNA pair. The guide sequence region is responsible for accurately guiding the Cas9 protein to the corresponding target of the genome. The PAM sequence region has no guiding function but has an important influence on the role of the Cas9 protein. The coding does not reflect that the base pairs in different regions have different functions. That will also lead to some information loss.
We propose a new coding scheme to solve information loss, as shown in Fig. 9. We constructed four type channels (A, T, G, C), using one-hot vectors (1,0,0,0), (0,1,0,0), (0,0,1,0), (0,0,0,1) respectively represent adenine, thymine, cytosine, and guanine. When there is a bulge on DNA or RNA (corresponding base deletion on RNA or DNA), use (0,0,0,0) to indicate the base deletion at that position. When two bases in a base pair are different, or one base in a base pair is deletion, our coding scheme performs an OR operation on the two vectors representing the base pair. When two bases in a base pair are the same, our coding scheme performs an OR operation and reverses it. For example, the base pair “AC” is represented as (1,0,0,1), “AA” is represented as (−1,0,0,0).
Fig. 9.
An example of gRNA-DNA pair coding.
In this article, the first base of base-pair is on gRNA and the second base is on DNA. For example, “AC” means A is on gRNA, C is on DNA. However, it is impossible to distinguish the bases on different sequences after encoding the base pair. For example, “AC” and “CA” are the same code (1,0,0,1). To distinguish the base in different sequences, we have added two-position channels (R, N). The first distinguishes whether it is on gRNA, and the second distinguishes whether it is on DNA—the high priority base position in the type channels. The priority order is A, T, G, C. Therefore, adenine in the “AC” has a high priority, and on gRNA, the direction vector is (1,0). In the “CA,” adenine on DNA is highly prioritized, and the direction vector is (0,1). In the “AA” base pair, adenine has a high priority, and on DNA and gRNA, the direction vector is (1,1). In addition, to distinguish the different regions of the gRNA-DNA sequence pair, we added a function channel. The guide sequence area is defined by '0′, and the PAM sequence area is defined by '1′.
4.3. CRISPR-IP architecture
Many models have been proposed to predict off-target activities, but they have not effectively used sequence pair information. The unfocused learning features from sequence pairs will be disturbed by irrelevant information. This paper proposes a neural network model CRISPR-IP to predict off-target activities. The idea of CRISPR-IP is to extract sequence features based on the identity and position of base pairs to predict each potential off-target gRNA-DNA pair. The research of Doench et al. [14] and Listgarten et al. [16] inspire this idea. Doench et al. found that the identity and position of mismatched nucleotide pairs played an important role in determining off-target activity and proposed cutting frequency determination (CFD) score to calculate potential off-target activity scores. Listgarten et al. built a machine learning model Elevation-score to predict off-target activity based on the position and identity of mismatched nucleotide pairs. They found that a single feature that merges the identity and position of mismatched nucleotide pairs is more important. However, the internal mechanism of the CRISPR/cas9 system is not presently clear and explicit. Manually designed features may negatively affect the prediction results. Therefore, we proposed the CRISPR-IP model, which automatically learns the features of sequence pairs based on the identity and position of base pairs and predicts the off-target activity of gRNA-DNA pairs.
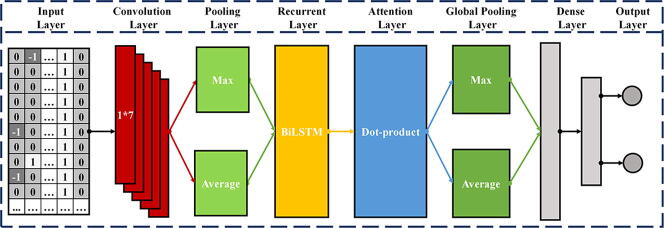
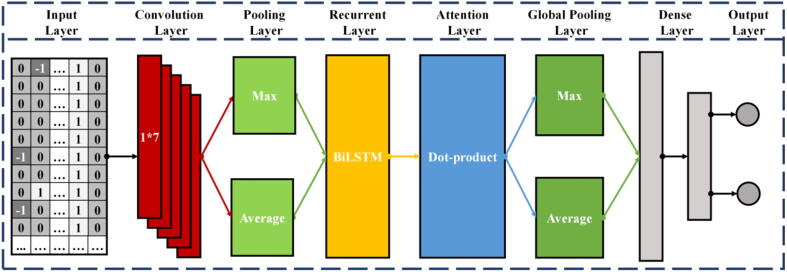
Fig. 10 describes the network architecture of CRISPR-IP. CRISPR-IP can be divided into four parts, (1) Convolutional layer and pooling layers, (2) BiLSTM layer, (3) Attention layer and global pooling layers, (4) Dense layers. The input of CRISPR-IP is the coding matrix after gRNA-DNA pair coding. The dimension of the coding matrix is (T, 7), where T is the sequence length and 7 is the coding dimension of nucleotide pairs.
Fig. 10.
Architecture of the CRISPR-IP.
First, the input of the model goes through the convolutional layer. Convolution is another representation of the input and convolution of the input data to obtain new features. The convolution kernel is also called a filter, and it is usually a two-dimensional matrix used to extract data features. Superimpose multiple convolution kernels to form the convolutional layer. Through the convolution layer, the model performs convolution operation with the coding of each nucleotide pair in turn to learn the identity features of each nucleotide pair. The pooling layer is also called the down-sampling layer, usually after the convolutional layer. Pooling first divides the features obtained by convolution into several regions and then calculates the average or maximum value in the regions, respectively. The small transformation of the input through the pooling operation becomes approximately unchanged, which improves the model’s generalization ability.
Second, the features extracted by the convolutional and pooling layers will be input to the BiLSTM layer. BiLSTM is a variant of LSTM [31] and consists of two parallel LSTMs: an input forward sequence and an input reverse sequence, which can obtain the features representation of the sequence forward and reverse information. LSTM is a type of recurrent neural network (RNN), which adds gate control (input gate, forget gate, and output gate) based on a recurrent neural network to determine the storage and discarding of information. LSTM solves the problem of gradient explosion of the RNN model and can better describe sequence data. The basic formula of the LSTM model is as follows:
Assuming that the input sequence is , the state of the hidden layer is . In the formula, is the forget gate at position to prevent the introduction of too much information; is the input gate at position , used for information selection; is the state of the cell unit at the current position; is the output of the cell unit at the current position; is the output gate at position ; is the predicted value at position in the sequence. Through the BiLSTM layer, the corresponding feature of each nucleotide pair in the sequence will learn the sequence information before and after it, which contains the position information of the nucleotide pair.
Subsequently, the features extracted by the BiLSTM layer are input into the attention layer. The attention layer is based on Luong-style attention [32], and the calculation formula is as follows:
, , and are query, key, and value vectors, respectively. The attention mechanism applies human attention patterns to neural networks, emphasizing critical information and eliminating the interference of unimportant details. We use a variant of the attention mechanism, the self-attention mechanism [23], to learn sequence features. The self-attention mechanisms’ , , and are the same, reducing the dependence on external information and better capturing the internal correlation of data or features. Through the attention layer, the features of each base pair are based on the “attention” to learn the remaining base pair information in the selected sequence. Then the global maximum pooling and global average pooling are used to reduce information redundancy and prevent excessive Fitting.
Finally, the sequence features extracted by the attention layer and the global pooling layers are input to the dense layers to predict the possibility of off-target activities.
4.4. Performance evaluation
All experiments are evaluated using leave-one-gRNA-out cross-validation (LOGOCV). LOGOCV divides the entire data set into two non-overlapping subsets, uses data from one gRNA for testing and from the remaining gRNA to train the model. For the objectivity of the experiment, we use data from different gRNAs in turn as the test set in the verification.
We used Accuracy, Precision, Recall, F1 Score, PR-AUC (Area Under the Precision-Recall Curve), ROC-AUC (Area Under the Receiver Operating Characteristic Curve) as the model’s performance metrics. The accuracy rate, precision rate, recall rate, and F1 Score use 0.5 as the threshold to divide the active off-target and the inactive off-target. Since the off-target prediction data set has the problem of sample imbalance, we pay more attention to the performance metrics of low impact of sample imbalance: PR-AUC [33]. The higher the value, the proves that the model has better performance on class imbalance.
4.5. Model parameters
Through LOGOCV, we determine the locally optimal parameters of the CRISPR_IP model. The essential parameters of the four parts of the CRISPR-IP model are as follows: (1) Convolutional layer and pooling layers. The number of convolutional layer filters is 60, the convolution kernel size is 1*7, and the step size is 1. The pooling window size of the pooling layer is 2, and the step size is 2. (2) BiLSTM layer. BiLSTM consists of two unidirectional LSTM layers, forward and reverse. Each unidirectional LSTM returns the entire sequence with neural units are 30 and dropout is 0.25. (3) Attention layer and global pooling layers. The attention layer uses dot product self-attention. The global pooling layer performs an average and maximum pooling operation on the sequence dimensions in the sequence data. (4) Dense layers. The dense layers have three layers. The first layer of neural units is 100, using the ‘relu’ activation function and batch regularization. The number of units in the second layer is 200, using the ‘relu’ activation function, and the dropout is 0.9. The third layer is the output layer, the neural unit is 2, using the ‘softmax’ activation function.
4.6. Neural networks for comparing coding schemes
We used the CRISPR_IP model to evaluate the effects of our coding scheme and Lin’s coding scheme on the CIRCLE-Seq dataset and the SITE-Seq dataset. To compare coding schemes more objectively, we use Dense Neural Networks (DNN), Convolutional Neural Networks (CNNs), and Recurrent Neural Networks (RNNs), three basic neural networks for evaluation. These three types of networks have been widely used in off-target prediction [20]. We built several different DNN, CNN, and RNN networks and evaluated them on two data sets to measure our coding scheme and Lin’s coding scheme. The brief structure description of the neural network used in our research is shown in Table 5, and the detailed description is shown in Supplementary Table S1.
Table 5.
Taxonomy of the models used in coding schemes experiments and their respective architectures.
| Name | Type | Architecture |
|---|---|---|
| DNN3 | DNN | 3 dense layers |
| DNN5 | DNN | 5 dense layers |
| DNN10 | DNN | 10 dense layers |
| CNN2 | CNN | 1 convolutional layer, 1 dense layer |
| CNN3 | CNN | 2 convolutional layer, 1 dense layer |
| LSTM | RNN | 1 LSTM layer, 2 dense layers |
| GRU | RNN | 1 GRU layer, 2 dense layers |
4.7. Comparison of other models
We compare CRISPR-IP with six advanced off-target prediction models, CRISPR-Net [18], CFD(cutting frequently determination) [14], CNN_std [19], Elevation [16], CRISPR-OFFT [25], and AttnToMismatch_CNN [17].
To our knowledge, CRISPR-Net is the first and only model that predicts potential off-target sequence pairs with bulges. Therefore, we evaluate CRISPR-IP and CRISPR-Net on the CIRCLE-Seq dataset and the SITE-Seq dataset. The CRISPR-Net model uses the hyperparameters proposed by the author and uses LOGOCV to retrain and evaluate.
For the previous off-target prediction model that only predicted potential gRNA-DNA sequence pairs that contained mismatches, we evaluated them and CRISPR-IP on the SITE-Seq dataset. These models include CFD, AttnToMismatch_CNN, CNN_std, Elevation, CRISPR-OFFT. Among them, the CFD uses the coefficient matrix provided by the author for score calculation. The AttnToMismatch_CNN, CRISPR-OFFT, and CNN_std models are retrained and evaluated based on LOGOCV on the SITE-Seq data set. The first layer model of Elevation reuses the author’s model and parameters, and the second layer of Elevation uses AdaBoostRegressor [34] to retrain and test on the SITE-Seq dataset based on LOGOCV. In addition, some researchers have proposed DeepCRISPR [26] and Chen’s models [7] based on pre-training. Since authors do not provide pre-trained models, we cannot fine-tune them for off-target prediction tasks based on the SITE-Seq data set, so they did not participate in the comparison.
4.8. Sampling methods
The two data sets used in the study have sample imbalance problems. The positive and negative sample ratios of the CIRCLE-Seq data set and the SITE-Seq data set are about 1:78 and 1:57, respectively. Imbalanced samples may affect the training results of the model. Due to different models deal with sample imbalance in different ways. For example, the CRISPR-Net model does not use resampling methods because resampling will cause performance degradation. AttnToMismatch_CNN uses an oversampling method to balance the samples to get higher prediction performance.
Therefore, we studied the effect of using over-sampling and under-sampling methods to balance samples on model performance. Assume that the number of positive samples is M, negative samples is N, and M ≪ N. The oversampling method repeatedly selects positive sample data until positive samples equal negative samples (the number is N). The undersampling method randomly selects M samples from all negative samples and discards the remaining negative samples.
5. Availability of data and materials
Additional data and source codes are available at https://github.com/BioinfoVirgo/CRISPR-IP.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was partially supported by grants from the National Key R&D Program of China (2019YFA0110802 and 2019YFA0802800), the Fundamental Research Funds for the Central Universities. The funding bodies did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.01.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., et al. CRISPR RNA maturation by trans -encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ran F.A., Hsu P., Lin C.Y., Gootenberg J., Konermann S., Trevino A.E., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 7.Chen D., Shu W., Peng S. 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; 2020. Predicting CRISPR-Cas9 Off-target with Self-supervised Neural Networks; pp. 245–250. [Google Scholar]
- 8.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.-S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42(11):7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Zhang X., Cheng L., Luo Y. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 2020;17(1):13–22. doi: 10.1080/15476286.2019.1669406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancellieri S., Canver M.C., Bombieri N., Giugno R., Pinello L., Hancock J. CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing. Bioinformatics. 2020;36(7):2001–2008. doi: 10.1093/bioinformatics/btz867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abadi S., Yan W.X., Amar D., Mayrose I., Xu H. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol. 2017;13(10):e1005807. doi: 10.1371/journal.pcbi.1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Listgarten J., Weinstein M., Kleinstiver B.P., Sousa A.A., Joung J.K., Crawford J., et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018;2(1):38–47. doi: 10.1038/s41551-017-0178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., He D.i., Xie L., Segata N. Prediction of off-target specificity and cells-pecific fitness of CRISPR-Cas system using attention boosted deep learning and network-based gene feature. PLoS Comput Biol. 2019;15(10):e1007480. doi: 10.1371/journal.pcbi.1007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J., Zhang Z., Zhang S., Chen J., Wong K.C. CRISPR-Net: a recurrent convolutional network quantifies CRISPR off-target activities with mismatches and indels. Adv Sci. 2020;7(13):1–17. [Google Scholar]
- 19.Lin J., Wong K.C. Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics. 2018;34(17):i656–i663. doi: 10.1093/bioinformatics/bty554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlier J., Nadon R., Makarenkov V., Kelso J. Accurate deep learning off-target prediction with novel sgRNA-DNA sequence encoding in CRISPR-Cas9 gene editing. Bioinformatics. 2021;37(16):2299–2307. doi: 10.1093/bioinformatics/btab112. [DOI] [PubMed] [Google Scholar]
- 21.Albawi S., Mohammed T.A., Al-Zawi S. 2017 International Conference on Engineering and Technology (ICET) 2017. Understanding of a convolutional neural network. [Google Scholar]
- 22.Schuster M., Paliwal K.K. Bidirectional recurrent neural networks. IEEE Trans Signal Process. 1997;45(11):2673–2681. [Google Scholar]
- 23.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., et al. Proceedings of the 31st International Conference on Neural Information Processing Systems. 2017. Attention is All you Need; pp. 5998–6008. [Google Scholar]
- 24.Brody S, Alon U, Yahav E. How Attentive are Graph Attention Networks? arXiv Prepr arXiv210514491. 2021;1–24.
- 25.Zhang G., Zeng T., Dai Z., Dai X. Prediction of CRISPR/Cas9 single guide RNA cleavage efficiency and specificity by attention-based convolutional neural networks. Comput Struct Biotechnol J. 2021;19:1445–1457. doi: 10.1016/j.csbj.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., et al. DeepCRISPR: Optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19(1):80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y., Chuai G., Yu W., Qu S., Liu Q. Data imbalance in CRISPR off-target prediction. Brief Bioinform. 2020;21(4):1448–1454. doi: 10.1093/bib/bbz069. [DOI] [PubMed] [Google Scholar]
- 28.Naeem M., Majeed S., Hoque M.Z., Ahmad I. Latest developed strategies to minimize the off-target effects in CRISPR-cas-mediated genome editing. Cells. 2020;9:1–23. doi: 10.3390/cells9071608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M., et al. Mapping the genomic landscape of CRISPR–Cas9 cleavage. Nat Methods. 2017;14(6):600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 30.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 32.Luong M.T., Pham H., Manning C.D. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. 2015. Effective Approaches to Attention-based Neural Machine Translation; pp. 1412–1421. [Google Scholar]
- 33.Liu Q., Cheng X., Liu G., Li B., Liu X. Deep learning improves the ability of sgRNA off-target propensity prediction. BMC Bioinf. 2020;21:51. doi: 10.1186/s12859-020-3395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drucker H. 14th International Conference on Machine Learning. 1997. Improving regressors using boosting techniques; pp. 107–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional data and source codes are available at https://github.com/BioinfoVirgo/CRISPR-IP.