Abstract
Oncogenic multidrug resistance is commonly intrinsic to renal cancer based on the physiological expression of detoxification transporters, particularly ABCB1, thus hampering chemotherapy. ABCB1 activity is directly dependent on its lipid microenvironment, localizing to cholesterol- and sphingomyelin (SM)-rich domains. As ceramides are the sole source for SMs, we hypothesized that ceramide synthase (CerS)-derived ceramides regulate ABCB1 activity. Using data from RNA-Seq databases, we found that patient kidney tumors exhibited increased CerS2 mRNA, which was inversely correlated with CerS6 mRNA in ABCB1+ clear cell carcinomas. Endogenous elevated CerS2 and lower CerS5/6 mRNA and protein resulted in disproportionately higher CerS2 to CerS5/6 activities (approximately twofold) in chemoresistant ABCB1high (A498, Caki-1) compared with chemosensitive ABCB1low (ACHN, normal human proximal convoluted tubule cell) cells. In addition, lipidomics analyses by HPLC–MS/MS showed bias toward CerS2-associated C20:0/C20:1-ceramides compared with CerS5/6-associated C14:0/C16:0-ceramides (2:1). SMs were similarly altered. We demonstrated that chemoresistance to doxorubicin in ABCB1high cells was partially reversed by inhibitors of de novo ceramide synthesis (l-cycloserine) and CerS (fumonisin B1) in cell viability assays. Downregulation of CerS2/6, but not CerS5, attenuated ABCB1 mRNA, protein, plasma membrane localization, rhodamine 123+ efflux transport activity, and doxorubicin resistance. Similar findings were observed with catalytically inactive CerS6-H212A. Furthermore, CerS6-targeting siRNA shifted ceramide and SM composition to ultra long-chain species (C22–C26). Inhibitors of endoplasmic reticulum–associated degradation (eeyarestatin I) and the proteasome (MG132, bortezomib) prevented ABCB1 loss induced by CerS2/6 downregulation. We conclude that a critical balance in ceramide/SM species is prerequisite to ABCB1 expression and functionalization, which could be targeted to reverse multidrug resistance in renal cancers.
Keywords: P-glycoprotein, cancer drug resistance, ABC transporter, unfolded protein response, sphingolipids
Abbreviations: ACACA, acetyl-CoA carboxylase alpha; ATP6α, activating transcription factor 6α; CerS, ceramide synthase; CHOP, C/EBP homologous protein; DOX, doxorubicin; ER, endoplasmic reticulum; ERAD, ER-associated degradation; FASN, fatty acid synthase; GRP78, glucose-regulated protein 78 kDa; HERPUD1, homocysteine-inducible ER protein with ubiquitin-like domain 1; HPCT, human proximal convoluted tubule; HMGCL, 3-hydroxymethyl-3-methylglutaryl-CoA lyase; LCC, long-chain ceramide; LD, lipid domain; MDR, multidrug resistance; Mff, mitochondrial fission factor; PDIA4, protein disulfide isomerase family A member 4; SM, sphingomyelin; SREBP, sterol-regulatory element binding protein; TG, thapsigargin; TM, transmembrane; TUN, tunicamycin; UCSC, University of California, Santa Cruz; UPR, unfolded protein response; VLC, very long chain; XBP1, X-box binding protein 1
The canonical multidrug resistance (MDR) protein ABCB1 belongs to the evolutionary-conserved ABC superfamily, a large group of transmembrane (TM) transporters that utilize energy to translocate various substrates across membranes (1). During tumor cell transformation, adaptive changes occur and confer the hallmarks of cancer, including evasion of apoptosis, to which ABCB1 contributes by extruding proapoptotic substrates and/or drugs for cancer treatment.
ABCB1 function is influenced not only by the lipid composition in its immediate vicinity but also by membrane fluidity, phospholipid headgroups, and fatty acyl chain lengths (reviewed in Ref. (2)). Small and specialized membrane lipid domains (LDs) (<100 nm) are highly enriched in cholesterol, glycosphingolipids, and phospholipids with saturated fatty acids, particularly sphingomyelin (SM), and provide optimal lipid composition for integral membrane protein function. The presence of SM is crucial to LD formation (3), permitting interactions between cholesterol and glycosphingolipids. ABCB1 localizes to LDs (4), likely because of its capacity to bind cholesterol (5) or, possibly, SM (6).
Cancer cell membranes differ in lipid composition. Indeed, SM is increased in multiple cancer tissues (7, 8), and in a rat hepatocarcinoma model, SM gradually increased and preceded ABCB1 appearance, which occurred in late tumor stages (9). Ceramide is the sole source for SM, the major sphingolipid in mammalian membranes, and would increase density and size of LDs and may lay the foundation for optimizing ABCB1 and thus MDR potential.
The major route of ceramide synthesis is the de novo pathway in the endoplasmic reticulum (ER), in which the family of ER-residing ceramide synthases (CerSs) plays a key role (10). Ceramides generated in the ER have a multitude of functions, such as regulating protein trafficking and signaling ER stress (reviewed in Ref. (11)). Each of the mammalian CerS1–6 isoforms has a unique tissue expression profile and uses fatty acyl CoAs with a specific fatty acid chain length varying between C16 and C24 (10, 12). For example, CerS2 has a preference for C22–C24 fatty acids, generating very long-chain (VLC) ceramides (VLCCs), whereas CerS5 and CerS6 prefer C16, producing LCCs. The significance of the acyl chain length in determining biological activity is becoming apparent, for example, in cell growth or apoptosis (13).
We hypothesized that ceramide and/or SM are crucial contributors to MDR development, ABCB1 functionalization and acquisition of the full MDR phenotype (6). In the current study, we report a switch to favor longer chain ceramides and SMs in the MDR phenotype, promoting ABCB1 functionalization and preventing its degradation. The data suggest that a correct balance in sphingolipid levels is necessary to confer MDR.
Results
CerS isoform expression in renal cancer
CerS2, CerS5, and CerS6 are predominantly expressed in the mouse kidney (10). Using a genomic analyses platform, the aforementioned CerS isoforms were significantly altered in patient renal tumors compared with normal adjacent tissue (Fig. 1A). From three renal cancer subtypes examined, two exhibited elevated CerS2 and CerS5 mRNAs (clear cell and papillary) or attenuated CerS6 mRNA (chromophobe and papillary). When CerS was correlated with ABCB1 gene expression, CerS2 (slope of 0.07; r2 = 0.06) and CerS6 (slope of 0.12; r2 = 0.07) showed slight positive correlation, whereas CerS5 (slope of −0.05; r2 = 0.08) did not (Fig. 1B). Moreover, in patient clear cell carcinomas with ABCB1 gene expression higher than the median (9.61), an inverse correlation between CerS2 and CerS6 mRNA was observed (slope, −0.46; r2 = 0.07) (Fig. 1C). These observations were confirmed in renal cancer cell lines: Drug-resistant ABCB1high cell lines A498 and Caki-1 harbored disproportionately more CerS2 mRNA (>50%) (Fig. 1D) and correlated with ABCB1 mRNA (Fig. S1A). Ratio of CerS2:CerS5 mRNA was elevated in A498 cells, whereas ratios for CerS2:CerS6 and CerS5:CerS6 were elevated in all renal cancer cells compared with a normal renal cell line derived from human proximal convoluted tubule (HPCT) (Fig. 1E). These observations were substantiated by studies in patient-derived metastatic renal cancer organoids wherein both CerS2 and CerS6 mRNAs were up to twofold higher compared with HPCT, CerS2:CerS5 mRNA ratio was similarly increased (∼2.4 in organoids; compared with ∼2.2 in A498 cells), and correlated with strong expression of ABCB1 mRNA (∼37-fold above HPCT) (R. Zarbock, M.L. Martin, W.K. Lee, unpublished observations).
Figure 1.
Ceramide synthase (CerS) expression, activity, and ceramide species are altered in MDR renal cancer cells.A, relative gene expression of CerS2 (LASS2), CerS5 (LASS5), and CerS6 (LASS6) in human normal and tumor tissues from the kidney from RNA-Seq studies available at UCSC Xena. Normal adjacent, n = 129; clear cell carcinoma, n = 530; chromophobe, n = 66; papillary carcinoma, n = 288; and normal noncancer, n = 27. B, scatter plots of CerS isoforms versus ABCB1 gene expression in human renal clear cell carcinomas. C, scatter plot of CerS2 and CerS6 gene expression in human renal clear cell carcinomas with high relative ABCB1 expression (above median 9.61). Databases (Genotype Tissue Expression and TCGA) were accessed through UCSC Xena. CerS isoform expression in normal human renal proximal tubule (HPCT), drug-sensitive ABCB1low renal cancer (ACHN), and MDR ABCB1high renal cancer (A498, Caki-1) cells was analyzed by quantitative PCR using isoform-specific primers. Numerical proportions (D) and ratios (E) of CerS2, CerS5, and CerS6 mRNA in each cell line are shown (n = 3–10). F, immunoblotting of ABCB1 and CerS isoforms in HPCT and renal cancer cells. F′, densitometry analyses were executed using Fiji/ImageJ (n = 4–10). G, resistance to anthracycline chemotherapeutic drug and ABCB1 substrate doxorubicin (DOX) after 48 h by MTT assay reflects expression of ABCB1 shown in (F) (n = 2–9). Mean IC50: ACHN, 0.67 μM; A498, 2.04 μM; Caki-1, 1.61 μM. H, contribution of de novo ceramide synthesis and CerS enzymes in conferring MDR phenotype was affirmed by application of DOX (1 μM, 24 h) in MTT assays following pharmacological serine palmitoyltransferase inhibitor, l-cycloserine (l-CS, 0.05 mM, n = 5) or CerS inhibitor fumonisin B1 (FB1, 3 μM, n = 5). I, trypan blue cell viability staining in combination with cell counting (n = 3) and (J) electrical impedance measurements of cell monolayers (where increased capacitance indicates cell detachment and loss of cell viability) (n = 3) confirmed findings with L-CS. K, in vitro CerS activity assay assessed CerS2 activity with acyl CoA 24:1 or CerS5/6 activities with acyl CoA 16:0 (K′). Blank (no cell extract) and FB1 (20 μM) were included as negative controls. Data were quantified by densitometry and standard curve analysis and normalized to HPCT cells. L, C16:0 to C24:1 activity ratio was plotted (n = 5–6). MDR, multidrug resistance; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; TCGA, The Cancer Genome Atlas; UCSC, University of California, Santa Cruz.
At the protein level, changes in CerS2 and CerS6 reflected the mRNA findings (Fig. 1, F–F′; a validated commercial antibody for endogenous CerS5 is unavailable). CerS2 was augmented in ABCB1high cells, whereas CerS6 was decreased in all renal cancer cell lines compared with HPCT. As expected, ABCB1 was elevated only in the MDR cell lines, A498 and Caki-1, at both the mRNA (Fig. S1A) and protein (Fig. 1, F–F′) levels. Moreover, increased resistance to the chemotherapeutic drug doxorubicin (DOX) (Fig. 1G) in ABCB1high cells was reversed by the de novo ceramide synthesis/serine palmitoyltransferase inhibitor l-cycloserine and the CerS inhibitor, fumonisin B1 (Fig. 1, H–J), suggesting ceramide contribution to the MDR phenotype.
MDR renal cancer cells switch ceramide and SM species composition
In all renal cancer cell lines, whether chemoresistant or not, CerS2 activity was elevated compared with HPCT, whereas CerS5/6 activities were increased in ACHN but diminished in Caki-1 (Fig. 1K), and resulting in significantly lower CerS5/6:CerS2 activity ratio by twofold and more in ABCB1high cells (Fig. 1L).
HPLC–MS/MS analyses confirmed that C14:0/C16:0-ceramides are lowered (by approximately 40%), whereas C20:0/C20:1-ceramides, which are also products of CerS2 (10), are elevated by ∼2.5-fold in ABCB1high versus ABCB1low cells (Table 1). Based on these data, the C14:0/C16:0 to C20-ceramide ratio was 4.3-fold lower in A498 compared with ACHN, corresponding to enzyme activity ratios (Fig. 1L). Disrupting the ceramide species balance by natural egg ceramides, whose main component is C16-ceramide (14), diminishes drug resistance (Fig. S1B). Since ceramide is the sole source of SM, elevated C20-SM, C24-SM, and C26-SM by 1.4-fold to twofold as well as lowered C14–C18 SMs by about 50% in A498 compared with ACHN (Table 2) reflected the changes in ceramide species (Table 1).
Table 1.
Lipidomics (HPLC–MS/MS) analyses of ceramide species in ACHN and A498 cells
| Ceramide species | ACHN | A498 | p < 0.05? |
|---|---|---|---|
| C14-Cer | 26.9 ± 3.4 | 16.0 ± 1.7 | Y |
| C16-Cer | 272.0 ± 13.3 | 165.9 ± 28.9 | Y |
| C18-Cer | 13.4 ± 4.1 | 21.9 ± 8.3 | N |
| C18:1-Cer | 4.7 ± 0.3 | 5.7 ± 1.1 | N |
| C20-Cer | 10.0 ± 0.9 | 26.3 ± 1.4 | Y |
| C20:1-Cer | 1.4 ± 0.1 | 3.5 ± 0.6 | Y |
| C20:4-Cer | 0.05 ± 0.02 | 0.02 ± 0.01 | N |
| C22-Cer | 135.2 ± 22.5 | 108.0 ± 11.9 | N |
| C22:1-Cer | 36.7 ± 11.5 | 36.8 ± 4.8 | N |
| C24-Cer | 757.5 ± 101.0 | 418.0 ± 35.1 | Y |
| C24:1-Cer | 733.6 ± 160.1 | 392.8 ± 32.0 | N |
| C26-Cer | 34.0 ± 4.3 | 30.3 ± 2.9 | N |
| C26:1-Cer | 58.3 ± 10.4 | 45.2 ± 5.4 | N |
Cells were grown for 2 to 3 days to reach 80% confluency prior to collection for lipidomics analyses as described in the Experimental procedures section (n = 2–4). Values are given in picomole/mg protein. Means ± SE are shown.
Abbreviations: N, no; Y, yes.
Table 2.
Lipidomics (HPLC–MS/MS) analyses of SM species in ACHN and A498 cells
| SM species | ACHN | A498 | p < 0.05? |
|---|---|---|---|
| C14-SM | 1441.9 ± 320.1 | 718.4 ± 81.3 | N |
| C16-SM | 31,373.1 ± 10,463.3 | 22,380.1 ± 5822.3 | N |
| C18-SM | 606.2 ± 71.9 | 258.1 ± 15.5 | Y |
| C18:1-SM | 370.5 ± 147.4 | 161.6 ± 50.9 | N |
| C20-SM | 116.7 ± 6.4 | 181.0 ± 10.7 | Y |
| C20:1-SM | 26.1 ± 3.0 | 39.0 ± 5.1 | N |
| C22-SM | 625.9 ± 70.8 | 711.3 ± 35.4 | N |
| C22:1-SM | 255.8 ± 31.7 | 303.6 ± 34.2 | N |
| C24-SM | 1274.1 ± 190.7 | 1829.4 ± 62.9 | Y |
| C24:1-SM | 4170.2 ± 754.8 | 3967.5 ± 341.0 | N |
| C26-SM | 18.3 ± 0.8 | 51.6 ± 8.5 | Y |
| C26:1-SM | 58.6 ± 3.2 | 173.0 ± 21.7 | Y |
| Lyso-SM | 1.5 ± 0.9 | 0.4 ± 0.3 | N |
Cells were grown for 2 to 3 days to reach 80% confluency prior to collection for lipidomics analyses as described in the Experimental procedures section (n = 4). Values are given in picomole/mg protein. Means ± SE are shown.
Abbreviations: N, no; Y, yes.
Taken together, ABCB1high cells exhibit a shift to predominantly longer chain ceramide and SM species, correlating with the MDR phenotype.
CerS2 and CerS6 govern plasma membrane ABCB1 localization, function, and chemoresistance
We next asked whether a direct relationship between CerS enzymes and ABCB1 existed. Alteration of CerS expression and activity by siRNA or heterologous expression was validated (Figs. 2, A and B and S2, A and B). We initially focused on CerS6 in MDR because of its reported involvement in apoptotic resistance of cancer cells (13, 15) by using silencing or overexpression of CerS6 (validated in Figs. S2–S4). CerS6 siRNA attenuated ABCB1 mRNA (Fig. S4A) and protein levels by >50% after 48 to 72 h in both A498 (Figs. 2C and S4B) and Caki-1 (Fig. S4C) cells, in an ABCB1high phenotype-specific manner (Fig. 2, D–D′). Although overexpression of CerS6 increased ceramide levels in A498 cells (Fig. S3), no significant effect on cell viability could be determined (Fig. S4F), which could be explained by saturation of the system wherein CerS6/ABCB1-mediated MDR is already maximal. Conversely, CerS6-WT expression in ABCB1low ACHN cells rendered them less susceptible to DOX toxicity (Fig. S4F). Similarly to CerS6, CerS2 downregulation resulted in loss of ABCB1 by ∼50% in ABCB1high cells (Fig. 2, D–D′). In contrast, ABCB1 was not affected by CerS5 siRNA (Figs. 2, D–D′ and S4C) with which no compensatory increase in CerS6 mRNA was observed (Fig. S4D). Heterologous overexpression of CerS enzymes in MDR cells confirmed that CerS can upregulate ABCB1 (Fig. 2E).
Figure 2.
ABCB1 expression and activity are dependent on CerS. CerS siRNA transfection efficiency. CerS2 or CerS6 siRNA (10 nM) was applied for up to 72 h to A498 cells, and efficiency was determined by (A) quantitative PCR (n = 3–5) or (B) immunoblotting (n = 6). C, CerS6 siRNA was transfected for 72 h, and CerS6 and ABCB1 were assessed by immunoblotting (n = 4). HPCT and renal cancer cell lines were transiently transfected with CerS-specific siRNA (10 nM, 72 h). ABCB1 and CerS immunoblots (D, D′) were performed (n = 2–9). E, heterologous expression of hemagglutinin (HA)-tagged CerS increased ABCB1. Immunoblot is representative of n = 3 to 4. F, F′, surface immunofluorescence staining and threshold analysis of ABCB1 in nonfixed nonpermeabilized A498 cells (siRNA 10 nM, 72 h). The scale bar represents 10 μm. Images (n = 8–10 per treatment) were analyzed from two independent experiments. G, rhodamine 123+ efflux assay in A498 cells transfected with CerS-specific siRNA (10 nM) for 72 h (n = 5–9). Statistical analyses compare CerS siRNA to control siRNA at each time point. H, MTT assay of A498 cells transfected with CerS6 siRNA (10 nM, 48 h) and exposed to 1 μM DOX for 24 h (n = 3). I, electrical impedance measurements of A498 cells transfected with CerS isoform-specific siRNA (10 nM, 48 h) and plated with 1 μM DOX into ECIS arrays. Capacitance at 40 kHz was monitored for 4 days. Slope analyses were performed from the steepest portion of each curve (n = 6). CerS, ceramide synthase; DOX, doxorubicin; ECIS, Electric cell–substrate impedance sensing; HPCT, human proximal convoluted tubule; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Plasma membrane ABCB1, detected by an antibody directed against an extracellular domain (Fig. 2, F–F′) and its transport activity (Fig. 2G), was strongly diminished by both CerS2 and CerS6 siRNA but not by CerS5 siRNA, confirming Figure 2, D and F, and resulted in chemosensitization to DOX (Fig. 2, H and I).
To discern ABCB1 requirement for CerS expression or activity, the CerS6 catalytically inactive mutant H212A was utilized, which attenuated ABCB1 by ∼50% at both mRNA (Fig. S4E) in ACHN cells as well as protein (Fig. 3, A–A′) levels and transport activity (Fig. 3B) in A498 cells. Hence, the product(s) of the CerS6 enzyme (either direct or indirect) is crucial to ABCB1 maintenance. Correspondingly, CerS6 siRNA-transfected A498 cells exhibited decreased C14–C20-ceramides (<30%) and strikingly increased C22–C26-ceramides (∼50%), determined by HPLC–MS/MS (Fig. 3C). Expectedly, corresponding SM species were similarly altered (Fig. 3D), suggesting wider impact of CerS6 activity on sphingolipid levels.
Figure 3.
CerS6 activity rather than expression impacts ABCB1.A, A′, expression of CerS6-WT and catalytically inactive mutant CerS6 H212A resulted in attenuation of ABCB1 with only CerS6 H212A after 72 h (n = 5). B, rhodamine 123+ efflux assay in A498 cells for 72 h (n = 5). Statistical analyses compare CerS6 H212A to CerS6 WT at each time point. Lipidomics (HPLC–MS/MS) analyses for ceramides (C) and sphingomyelins (D) in A498 cells transfected with CerS6 siRNA for 72 h (n = 2). C numbers indicate fatty acid chain lengths. CerS, ceramide synthase.
ER-resident CerSs prevent loss of ABCB1
CerS enzymes are normally localized to the ER where de novo ceramide synthesis takes place. Based on the multifactorial nature of MDR, we hypothesized that altered CerS localization, forming functional clusters to potentially remain in proximity to ABC transporters and maintain their lipid microenvironment, could also be a contributing factor to the MDR phenotype. In contrast to a report for CerS1 redistribution (16), colocalization of CerS2 (Fig. 4A) or CerS6 (Fig. 4B) with ER markers, calnexin or KDEL, respectively, in ABCB1high cells ruled out altered CerS redistribution to form functional clusters with ABC transporters and indicate they act from within the ER. Loss of CerS can engage the unfolded protein response (UPR) (13, 17). Downregulation of CerS increases glucose-regulated protein 78 kDa (GRP78), an ER stress marker in both A498 (Fig. 5, A and A′) and Caki-1 (data not shown) cells. Interestingly, the UPR target gene CHOP (C/EBP homologous protein, gene DDIT3) was not elevated by CerS6 siRNA despite intact ER stress/UPR, as indicated by the positive control thapsigargin (TG) (Fig. 5B).
Figure 4.
ER localization of CerS in renal cancer cells. Indirect immunofluorescence costaining of CerS2 (A) and CerS6 (B) and the ER markers calnexin (A) or KDEL (B). Colocalization analyses between CerS and ER marker (A′, B′) were performed with thresholded binary images using Fiji/ImageJ. Box plots with median lines and outliers are shown (analyzed from 8 to 24 images per treatment, n = 2–4). The scale bar represents 10 μm (A) or 20 μm (B). CerS, ceramide synthase; ER, endoplasmic reticulum.
Figure 5.
ER stress and unfolded protein response by CerS downregulationactivates ATF6α in MDR cells. Cells were transiently transfected with CerS-specific siRNA for 72 h or treated with SERCA inhibitor thapsigargin (TG) (3 μM for 6 h) as positive control. A, analysis of GRP78 by immunoblotting in A498 cells. A′, densitometry analysis of CerS-specific siRNA transfected A498 cells after 72 h (n = 5–6). B, quantitative PCR analysis of ABCB1 and CHOP mRNA in A498 cells (n = 6). Immunoblots and densitometry analyses for full length (FL, 90 kDa) and cleaved (50 kDa) ATF6α in CerS siRNA (C, C′) and CerS6 overexpressing (D) cells after 72 h (representative immunoblots of n = 7–9). Values indicate densitometry analysis with correction to GAPDH. Immunoblots in C for CerS2, CerS6, and GAPDH are reused from Figure 2D because target proteins were analyzed from the same set of samples. E, E′, immunofluorescence staining and quantification of nuclear ATF6α using threshold analysis in CerS siRNA cells after 72 h in fixed A498 cells. A minimum of 500 nuclei per treatment were analyzed from thresholded images using Fiji/ImageJ from two independent experiments. The scale bar represents 10 μm. ATF6α, activating transcription factor 6α; CerS, ceramide synthase; CHOP, C/EBP homologous protein; ER, endoplasmic reticulum; GRP78, glucose-regulated protein 78 kDa; MDR, multidrug resistance; SERCA, sarco-/endoplasmic reticulum calcium ATPase.
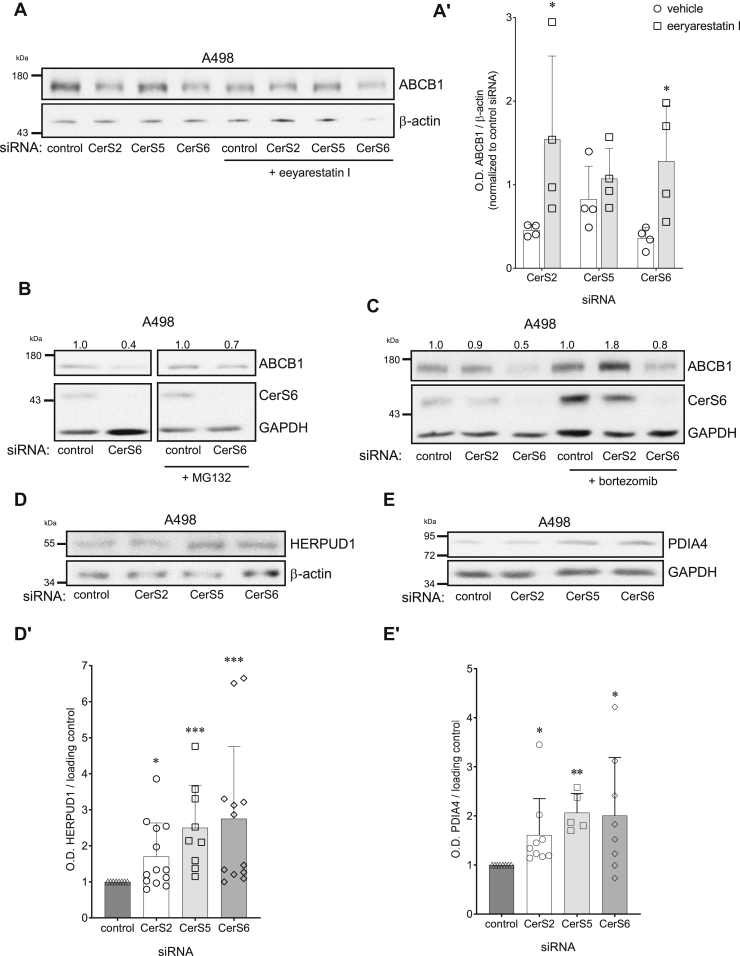
Cleavage of full-length activating transcription factor 6α (ATF6α) (90 kDa) in the Golgi apparatus and nuclear translocation as a 50 kDa fragment initiates the ER stress response via transcription (18) and can be independent of CHOP induction to promote the cellular recovery response to long-term ER stress through ER-associated degradation (ERAD) and ER chaperones (19). Loss of full-length ATF6α and/or increase in truncated ATF6α was evidenced in CerS siRNA or CerS6 H212A mutant-transfected A498 (Fig. 5, C and D) and Caki-1 (Fig. S5, A and B) cells, and increased nuclear ATF6α presence was confirmed by immunostaining (Fig. 5, E–E′). Activated ATF6α engages the ERAD pathway to target misfolded and/or damaged proteins for degradation by the proteasome. The ERAD inhibitor eeyarestatin I reversed loss of ABCB1 by CerS2 and CerS6 siRNA (Fig. 6, A–A′) or ER stressors TG and tunicamycin (TUN) (Fig. S5C). Moreover, proteasome inhibitors prevented ABCB1 loss (Fig. 6, B and C), and the ERAD proteins HERPUD1 (Fig. 6D) and PDIA4 were increased (Fig. 6E) after CerS downregulation, suggesting that a critical balance between ceramide species regulates ABCB1 expression.
Figure 6.
Loss of CerS induces ER-associated degradation of ABCB1. ABCB1 immunoblot and densitometry analyses in A498 cells transfected with 10 nM CerS-specific siRNA for 72 h. ERAD inhibitor eeyarestatin I (2.5 μM) (A, A′), or proteasome inhibitors MG132 (50 nM) (B) or bortezomib (2.5 nM) (C) were added 16 h post-transfection (n = 3–5). Assessment of ERAD proteins HERPUD1 (D, D′) or PDIA4/ERp72 (E, E′) in A498 cells after 72 h transfection with CerS isoform-specific siRNA (n = 6–10). CerS, ceramide synthase; ER, endoplasmic reticulum; ERAD, ER-associated degradation; HERPUD1, homocysteine-inducible ER protein with ubiquitin-like domain 1; PDIA4, protein disulfide isomerase family A member 4.
Discussion
Here, we have presented evidence that the oncogenic chemoresistant phenotype attributed to the MDR transporter ABCB1 is not only upregulated in MDR but also requires a defined lipid microenvironment for ER processing and functionalization in plasma or vesicular membranes, preventing chemotherapeutic drug targeting (20, 21).
TM proteins, such as drug transporters associated with MDR (e.g., ABCB1, ABCC1, ABCG2), require a stable and robust environment that depends on lipid composition (2). How could the shift in ceramide/SM species impact MDR? First, highly specific sphingolipid–protein interactions (22) could anchor and stabilize ABCB1 in the lipid bilayer (6). Second, VLC-SMs could contribute to LD formation for ABCB1 functionalization. Third, by potentially surrounding the substrate-binding pocket, which captures substrates through the hydrophobic portion of the lipid bilayer (23), they could govern substrate permissibility/accessibility, flexibility of the inward-facing conformation, or changes of the binding pocket during the transport cycle (24). Finally, they could act as protective chaperones against potential oxidative damage to help process and rectify misfolded ABCB1 (25).
ABCB1 overexpression alone is insufficient to confer MDR in some cell types, highlighting the requirement for additional factors. Perturbations in ceramide/SM are well evidenced in multiple types of cancer cells and preceded ABCB1 expression in a drug-induced liver carcinogenesis model (reviewed in Ref. (6)), suggesting that altered CerS expression could be an early event in oncogenic transformation. For completion of the full MDR phenotype, increased glycosphingolipids seem to be required to mislocalize ABCB1 to an intracellular vesicular pool, trapping further chemotherapeutic drug (reviewed in Ref. (6)).
CerS2 and CerS5/6 have important roles in membrane properties and cell survival, for example, loss of CerS2 results in perturbations in membrane biophysics (26), elevated C24-ceramides are correlated with more aggressive behavior of skull base chordoma cancers (27), and loss of CerS6 leads to cancer cell apoptosis (13). Both CerS5 and CerS6 can dimerize with CerS2 (28), and their complex regulation creates hurdles in discerning their individual contributions, their concerted activities as dimers, and/or feedback regulations (29).
UPR activation is resultant of proteotoxic or lipid bilayer stress (30). Intriguingly, ATF6α is specifically activated by an increase in sphingolipids (dihydrosphingosine and dihydroceramide) (31), whereas lipid bilayer stress activates other arms of the UPR (32), which were not significantly activated by CerS siRNA (data not shown). Downregulation of CerS would result in accumulation of dihydrosphingosine that may result in ATF6α activation and ERAD, which is in line with findings made for CerS6 (13, 33). Furthermore, C16-ceramide is involved in selective activation of ATF6α by Golgi membrane fragmentation (13). ATF6α appears to influence the protein kinase RNA-like endoplasmic reticulum kinase UPR arm as well as XBP1s to affect CHOP expression in addition to coordinating the cellular response to long-term ER stress. Exposure to ER stressors TG and TUN attenuated ABCB1 mRNA and protein, which was reversed by the ERAD inhibitor eeyarestatin I (Fig. S5C), suggesting that ERAD contributes to ABCB1 degradation during ceramide imbalance.
Given the established positive connection between ER stress and de novo lipogenesis, we investigated the role of the transcription factor sterol-regulatory element binding protein (SREBP), which has been shown to be inhibited by sphingolipid depletion (reviewed in Ref. (34)). SREBP1 precursor and mature forms as well as its target genes HMGCL, FASN, and ACACA were indeed downregulated by CerS siRNA after 72 h (Fig. S6). Hence, enhanced CerS activity in MDR cells could contribute to elevated lipid/cholesterol delivery through SREBP activation to support tumor cell growth and possibly, the incorporation of MDR-associated drug transporters, such as ABCB1, into the lipid membrane.
It is becoming increasingly apparent that absolute levels of single ceramide species are not the critical factor in governing cell behavior. Rather, the proportion between ceramide species is determinant in protein function and impacts cell fate. Disruption of this delicate balance can negatively affect cellular processes. For instance, overexpression of CerS2 with CerS4 or CerS6 could reverse proliferation inhibition and apoptosis induction by LCCs (35), or increased C24-ceramide prevented formation of a proapoptotic ceramide channel in mitochondria (36). In agreement, cells exhibiting an ABCB1high/MDR phenotype shift the ceramide/SM balance toward longer chain ceramides/SMs, implying a prosurvival characteristic. This phenomenon could potentially be relevant for other TM proteins, such as normally ER-resident GRP78, which is trafficked to the cell surface during carcinogenesis (37). Downregulation of CerS leads to increased GRP78 (Fig. 5A) and could be a result of ceramide-mediated disturbances in trafficking (38, 39), such that more GRP78 appears at the surface. Intriguingly, CerS5 did not affect ABCB1. Despite having the largest homology among CerS isoforms and same preference for generation of C16-ceramides, CerS5 and CerS6 appear to affect sphingolipid pools in distinct manners. This is highlighted in a recent study wherein C16:0-ceramides derived from CerS6, but not CerS5, interacted with and recruited mitochondrial fission protein Mff at the outer mitochondrial membrane, culminating in mitochondrial dysfunction (40). In addition to CerS6 enzymatic products, specific interaction with CerS6 could further contribute to differences between CerS5 and CerS6. In leukemic cells, CerS6 interaction with CD95/Fas prevents formation of the death signaling complex that contributes to their resistance to chemotherapy drug and pan-Bcl-2 inhibitor ABT-737 (41).
In conclusion, we posit that early phase oncogenic transformation modulates CerS expression, generating a distinct shift in C16/C20-ceramide balance, which mitigates ER stress/ERAD activation whilst increasing production of VLC-SMs, expansion of LDs, and paving the way for expression and functionalization of ABCB1. These alterations act in concert to confer MDR through ABCB1-mediated expulsion and sequestration of chemotherapeutic drugs.
Experimental procedures
Genomic data analysis
Analysis of gene expression patterns in normal and human kidney cancer tissues was performed using freely accessible databases through the Xena platform from the University of California, Santa Cruz (UCSC) (42). Data for normal tissues from noncancer patients were analyzed from the Genotype Tissue Expression project, and data for normal tumor-adjacent and tumor tissues were analyzed from The Cancer Genome Atlas using a filter for kidney. Subgroup comparisons were made using data from The Cancer Genome Atlas Kidney Clear Cell Carcinoma study. All studies have been reanalyzed by the UCSC RNA-Seq Compendium using the same RNA-Seq pipeline. Gene expression data are DESeq2 standardized and reported as log2(norm_count+1).
Cell culture and treatments
HPCTs were kindly provided by Dr Ulrich Hopfer (Case Western Reserve University) and propagated as previously described (43). The human primary renal cell carcinoma A498, human pleural effusion–derived kidney renal cell adenocarcinoma ACHN, human primary clear cell adenocarcinoma 786-O, and human skin metastasis-derived clear cell renal cell carcinoma Caki-1 cell lines were obtained from Cell Lines Service, authenticated by genotyping, and cultured as previously reported (43).
Treatments were performed in standard culture medium, unless otherwise stated. TG (Biozol), 1.5 μM for 3 to 6 h; TUN (Sigma–Aldrich), 6 μM for 3 to 6 h; eeyarestatin I (Santa Cruz Biotechnology, Inc), 1 to 2.5 μM for 56 h; MG132 (Enzo Life Sciences, Inc) and bortezomib (Sigma–Aldrich), 50 nM and 2.5 nM, respectively, for 56 h. DOX and daunorubicin (Sigma–Aldrich) were incubated for up to 48 h in serum-free medium.
Transient transfections
Cells were transfected with 10 nM control or CerS-specific siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's instructions. Predesigned targeting heterogeneous mixtures of siRNA sequences were obtained from Sigma–Aldrich (CerS2, EHU078471; CerS5, EHU001981; and CerS6, EHU156551) or nontargeting control siRNA SIC001 from Sigma–Aldrich or SR-CL000-005 from Eurogentec.
For overexpression studies, plasmids were transfected using Lipofectamine LTX Plus (Thermo Fisher Scientific) following manufacturer's instructions. C-terminal hemagglutinin-tagged human CerS (44), human CerS6-WT, and catalytically inactive mutant CerS6-H212A plasmids (13) were generated as described. For empty vector controls, the same amount of pcDNA3.1 was used.
PCR analyses
Total RNA extraction, complementary DNA synthesis, and quantitative PCRs were performed as previously described (43). Primers are listed in Table 3 and were obtained from Eurofins Genomics.
Table 3.
Primers for quantitative PCR
| Gene | Accession number | Forward primer (5′→3′) | Reverse primer (5′→3′) | Final primer concentration (nM) | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| ABCB1 | NM_000927.4 | AAATTGGCTTGACAAGTTGTATATGG | CACCAGCATCATGAGAGGAAGTC | 300 | 85 | RTPrimerDB ID: 3512 |
| ACACA | NM_198838 | CATGCGGTCTATCCGTAGGTG | GTGTGACCATGACAACGAATCT | 300 | 77 | PrimerBank MGH-PGA ID: 38679973c2 |
| ACTB | NM_001101.3 | ACTGGGACGACATGGAGAAA | ATAGCACAGCCTGGATAGCA | 300 | 189 | (51) |
| DDIT3 (CHOP) | NM_001195053.1 | TTCGGGACACTGTCCA | CAGCCAAGCCAGAGAAG | 300 | 162 | (52) |
| B2M | NM_004048.2 | CTCCGTGGCCTTAGCTGTG | TTTGGAGTACGCTGGATAGCCT | 300 | 69 | RTPrimerDB ID 3534 |
| FASN | NM_004104 | AAGGACCTGTCTAGGTTTGATGC | TGGCTTCATAGGTGACTTCCA | 300 | 106 | PrimerBank MGH-PGA ID 41872630c1 |
| GAPDH | NM_002046.3 | GCCAAAAGGGTCATCATCTC | GTAGAGGCAGGGATGATGTTC | 300 | 287 | (50) |
| HMGCL | NM_001166059 | GTGTCTCCTAAGTGGGTTCCC | TGGGTAGTTGATGCCAGGAAA | 300 | 84 | PrimerBank MGH-PGA ID 260654707c2 |
| RNA18SN1 | NR_145820.1 | CTTAGAGGGACAAGTGGCG | ACGCTGAGCCAGTCAGTGTA | 300 | 107 | (53) |
| TBP | NM_001172085.1 | CACGAACCACGGCACTGATT | TTTTCTTGCTGCCAGTCTGGAC | 300 | 89 | RTPrimerDB ID 8098 |
| TUBB2A | NM_001069.3 | ATCAGCAAGATCCGGGAAGAG | CCGTGTCTGACACCTTGGGT | 300 | 82 | RTPrimerDB ID 2758 |
Immunoblotting
Cell pellets were homogenized by sonication in an isosmotic sucrose buffer (0.25 M sucrose, 40 nM Mops, 0.1 mM EGTA, 0.1 mM MgSO4, and pH 7.4) with protease inhibitor cocktail (Sigma–Aldrich). Protein concentration was determined by Bradford protein assay.
SDS-PAGE and immunoblotting were performed according to standard procedures of wet transfer or electroblotted by rapid semidry transfer (Trans-Blot Turbo; Bio-Rad Laboratories) (43). Primary antibodies are listed in Table 4. Densitometry analyses were performed using Fiji/ImageJ software (GNU General Public License) (45).
Table 4.
Primary antibodies
| Target | Application | Dilution | Host species and clonality | Source | Catalog number |
|---|---|---|---|---|---|
| ABCB1 | IF | 1:200 | Mouse monoclonal (UIC2) | Santa Cruz Biotechnology, Inc | sc-73354 |
| ABCB1 | WB | 1:1000 | Mouse polyclonal (C219) | Enzo Life Sciences, Inc | ALX-801-002-C100 |
| β-Actin | WB | 1:80,000 | Mouse monoclonal | Sigma–Aldrich | A5316 |
| ATF6α | IF | 1:200 | Mouse monoclonal | Cosmo Bio Co Ltd | 73-500 |
| ATF6α | WB | 1:800 | Mouse monoclonal | Cosmo Bio Co Ltd | 73-500 |
| Calnexin | IF | 1:500 | Mouse monoclonal (AF18) | Thermo Fisher Scientific | MA3-027 |
| Ceramide | IF | 1:1000 | Mouse IgM (MID15B4) | Enzo Life Sciences, Inc | ALX-804-196-T050 |
| CERS2 | IF | 1:300 | Rabbit polyclonal | Sigma–Aldrich | HPA027262 |
| CERS2 | WB | 1:40,000 | Rabbit polyclonal | Sigma–Aldrich | HPA027262 |
| CERS6 | IF | 1:500 | Mouse monoclonal (5H7) | Abnova | H00253782 |
| CERS6 | WB | 1:5000 | Mouse monoclonal (5H7) | Abnova | H00253782 |
| GAPDH | WB | 1:80,000 | Rabbit monoclonal (14C10) | Cell Signaling Technology | 2118S |
| GRP78 | WB | 1:1000 | Rabbit polyclonal | Abcam | ab21685 |
| HERPUD1 | WB | 1:1000 | Rabbit polyclonal | Cell Signaling Technology | 26730 |
| KDEL | IF | 1:500 | Rabbit polyclonal | Thermo Fisher Scientific | PA1-013 |
| Lamin A/C | WB | 1:20,000 | Mouse monoclonal (4C11) | Cell Signaling Technology | 4777S |
| PDIA4/ERp72 | WB | 1:40,000 | Rabbit polyclonal | Thermo Fisher Scientific | PA-1007 |
| SREBP1 | WB | 1:2000 | Mouse monoclonal (2A4) | Santa Cruz Biotechnology, Inc | sc-13551 |
Source and dilutions of primary antibodies used in immunofluorescence (IF) staining and immunoblotting (Western blot).
CerS activity assay
CerS assays were performed as described elsewhere (46). Briefly, equal amounts of protein were combined with defatted bovine serum albumin (A6003; Sigma–Aldrich), N-[6-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)–sphinganine (810206P; Avanti Polar Lipids) and C16:0 (870716P) or C24:1 acyl CoA (870725P; Avanti Polar Lipids) and incubated at 37 °C. C16:0 was used to assess CerS5/6 activities, whereas C24:1 assessed CerS2 activity. The CerS inhibitor fumonisin B1 served as negative control. Lipids were extracted, dried, resuspended in chloroform:methanol (9:1, v/v), and separated by thin layer chromatography with a mobile phase consisting of chloroform:methanol:2 M ammonia (40:10:1, v/v/v). N-[6-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl) fluorescence was determined at 488 nm (Amersham Typhoon 5; GE Healthcare).
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde/PBS and permeabilized with 1% SDS/PBS (CerS6 and KDEL) or 0.5% saponin/PBS (CerS6 and calnexin). For CerS2 and calnexin staining, cells were fixed and permeabilized in acetone:methanol (1:1 [v/v]; −20 °C). After blocking with 1% bovine serum albumin/PBS, primary antibodies (Table 4) diluted in blocking buffer were incubated overnight at 4 °C. Species-specific secondary antibodies were diluted 1:500 in PBS. Nuclei were counterstained with 0.8 μg/ml Hoechst-33342 (Hoechst), and coverslips were embedded using fluorescent mounting medium (Agilent Dako). Images were acquired with widefield imaging systems consisting of either a Zeiss Axiovert 200M microscope equipped with Solar II light source and CoolSnap ES camera or a Nikon Eclipse Ci-L microscope equipped with C-LEDFI Epi-Fl LED Illuminator and DS-Ri2 camera. Image processing and analyses were performed using Fiji/ImageJ. For colocalization analyses, pixel overlap between thresholded binary images was assessed using the “AND” Boolean function. For nuclear ATF6α analysis, regions encompassing the nuclear perimeter in Hoechst images were transferred to corresponding thresholded ATF6α images. The percent area above the threshold within these regions was measured.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide viability assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide viability assay has been previously reported (47).
Electric cell–substrate impedance sensing
Cells (1.5 × 105) grown in 8W10E arrays (Ibidi) were monitored using an ECIS1600R (ECIS, electric cell–substrate impedance sensing) instrument (Applied BioPhysics, Inc) (48), and ECIS software, version 1.2.186.0 PC. An increase in capacitance at 64 kHz mirrors a loss of monolayer integrity translating as cell detachment and death.
Rhodamine 123+ efflux assay
Rhodamine 123+ is a cationic substrate of ABCB1. Efflux of rhodamine 123+ was determined as previously reported (47).
Lipidomic analyses of sphingolipids
Synthetic standards and samples underwent ethyl acetate/isopropanol liquid–liquid extraction followed by HPLC–MS/MS analysis using a Vanquish uHPLC system coupled to a Quantum Access Max triple quadrupole mass spectrometer equipped with an ESI probe operating in the multiple reaction monitoring positive ion mode (Thermo Fisher Scientific).
Chromatographic separations were obtained under a gradient elution on a C8 column using a mobile phase with ammonium formate, formic acid in water and methanol, as previously described (49). Peaks were recorded and processed using the instrument's software. Plotting the analyte/internal standard peak area ratios against analyte concentrations generates the sphingolipid-specific calibration curves.
Statistical analyses
Unless otherwise indicated, biological replicates were executed at least three times with independent cultures. Means ± SD are shown. For comparison of two groups, statistical analysis was carried out with two-tailed unpaired Student's t test with Excel (Microsoft). For more than two groups, one-way ANOVA with Tukey, Dunn, or Dunnett's post hoc test, assuming equality of variance, was applied (Sigma Plot 14.5; Systat Software, Inc). Results with p < 0.05 were considered statistically significant. Significance levels are labeled in the figures as follows: ∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001.
Data availability
All data are available in the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Tammar Joseph (Weizmann Institute of Science) for expert technical assistance.
Author contributions
W.-K. L. conceptualization; W.-K. L. methodology; W.-K. L., J. S. P., and B. O. formal analysis; W.-K. L., M. M., A. Q., H. P., E.-C. P., N. P., and J. L. K. investigation; W.-K. L., A. H. F., and F. T. resources; W.-K. L., A. H. F., and F. T. writing–original draft; W.-K. L., J. S. P., B. O., A. H. F., and F. T. writing–review & editing; W.-K. L. visualization; W.-K. L., A. H. F., and F. T. supervision; W.-K. L. project administration; W.-K. L., J. S. P., B. O., A. H. F., and F. T. funding acquisition.
Funding and additional information
This work was financially supported by the Westermann-Westdorp Foundation (to W.-K. L.), the Intramural Research Funding Program (IFF2018-52) (to W.-K. L.), the Centre for Biomedical Education and Research (ZBAF) at Witten/Herdecke University (to W.-K. L. and F. T.), and from the National Institutes of Health (CA173687, CA214641, DE016572, and P01 CA203628 to B. O.). Erasmus Plus traineeships were awarded to A. Q., H. P., and E.-C. P. Lipidomics analyses were performed at the MUSC Lipidomics Shared Resource, which is supported by NIH (C06 RR015455), Hollings Cancer Center support grant (P30 CA138313), or Center of Biomedical Research Excellence in Lipidomics and Pathobiology (P30 GM103339). A. H. F. is the Joseph Meyerhoff Professor of Biochemistry at the Weizmann Institute of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Dennis Voelker
Supporting information
References
- 1.Borst P., Schinkel A.H. P-glycoprotein ABCB1: A major player in drug handling by mammals. J. Clin. Invest. 2013;123:4131–4133. doi: 10.1172/JCI70430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharom F.J. Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: Its role in modulating protein function. Front. Oncol. 2014;4:41. doi: 10.3389/fonc.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinn P.J. Sphingolipid symmetry governs membrane lipid raft structure. Biochim. Biophys. Acta. 2014;1838:1922–1930. doi: 10.1016/j.bbamem.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Klappe K., Hummel I., Hoekstra D., Kok J.W. Lipid dependence of ABC transporter localization and function. Chem. Phys. Lipids. 2009;161:57–64. doi: 10.1016/j.chemphyslip.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Wang E., Casciano C.N., Clement R.P., Johnson W.W. Cholesterol interaction with the daunorubicin binding site of P-glycoprotein. Biochem. Biophys. Res. Commun. 2000;276:909–916. doi: 10.1006/bbrc.2000.3554. [DOI] [PubMed] [Google Scholar]
- 6.Lee W.K., Kolesnick R.N. Sphingolipid abnormalities in cancer multidrug resistance: Chicken or egg? Cell. Signal. 2017;38:134–145. doi: 10.1016/j.cellsig.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campanella R. Membrane lipids modifications in human gliomas of different degree of malignancy. J. Neurosurg. Sci. 1992;36:11–25. [PubMed] [Google Scholar]
- 8.D'Angelo G., Moorthi S., Luberto C. Role and function of sphingomyelin biosynthesis in the development of cancer. Adv. Cancer Res. 2018;140:61–96. doi: 10.1016/bs.acr.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Nakatsukasa H., Evarts R.P., Burt R.K., Nagy P., Thorgeirsson S.S. Cellular pattern of multidrug-resistance gene expression during chemical hepatocarcinogenesis in the rat. Mol. Carcinog. 1992;6:190–198. doi: 10.1002/mc.2940060304. [DOI] [PubMed] [Google Scholar]
- 10.Levy M., Futerman A.H. Mammalian ceramide synthases. IUBMB Life. 2010;62:347–356. doi: 10.1002/iub.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelnik I.D., Ventura A.E., Kim J.L., Silva L.C., Futerman A.H. The role of ceramide in regulating endoplasmic reticulum function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158489. doi: 10.1016/j.bbalip.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Tidhar R., Zelnik I.D., Volpert G., Ben-Dor S., Kelly S., Merrill A.H., Jr., Futerman A.H. Eleven residues determine the acyl chain specificity of ceramide synthases. J. Biol. Chem. 2018;293:9912–9921. doi: 10.1074/jbc.RA118.001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senkal C.E., Ponnusamy S., Bielawski J., Hannun Y.A., Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do U.H., Ramachandran S. Mild alkali-stable phospholipids in chicken egg yolks: Characterization of 1-alkenyl and 1-alkyl-sn-glycero-3-phosphoethanolamine, sphingomyelin, and 1-alkyl-sn-glycero-3-phosphocholine. J. Lipid Res. 1980;21:888–894. [PubMed] [Google Scholar]
- 15.Suzuki M., Cao K., Kato S., Komizu Y., Mizutani N., Tanaka K., Arima C., Tai M.C., Yanagisawa K., Togawa N., Shiraishi T., Usami N., Taniguchi T., Fukui T., Yokoi K., et al. Targeting ceramide synthase 6-dependent metastasis-prone phenotype in lung cancer cells. J. Clin. Invest. 2016;126:254–265. doi: 10.1172/JCI79775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Sridevi P., Alexander H., Laviad E.L., Min J., Mesika A., Hannink M., Futerman A.H., Alexander S. Stress-induced ER to Golgi translocation of ceramide synthase 1 is dependent on proteasomal processing. Exp. Cell Res. 2010;316:78–91. doi: 10.1016/j.yexcr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spassieva S.D., Mullen T.D., Townsend D.M., Obeid L.M. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 2009;424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 19.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesman M.M., Fojo T., Bates S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 21.Seebacher N., Lane D.J., Richardson D.R., Jansson P.J. Turning the gun on cancer: Utilizing lysosomal P-glycoprotein as a new strategy to overcome multi-drug resistance. Free Radic. Biol. Med. 2016;96:432–445. doi: 10.1016/j.freeradbiomed.2016.04.201. [DOI] [PubMed] [Google Scholar]
- 22.Ernst A.M., Brugger B. Sphingolipids as modulators of membrane proteins. Biochim. Biophys. Acta. 2014;1841:665–670. doi: 10.1016/j.bbalip.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Aller S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P.M., Trinh Y.T., Zhang Q., Urbatsch I.L., Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward A.B., Szewczyk P., Grimard V., Lee C.W., Martinez L., Doshi R., Caya A., Villaluz M., Pardon E., Cregger C., Swartz D.J., Falson P.G., Urbatsch I.L., Govaerts C., Steyaert J., et al. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13386–13391. doi: 10.1073/pnas.1309275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo T.W., Clarke D.M. Correction of defective protein kinesis of human P-glycoprotein mutants by substrates and modulators. J. Biol. Chem. 1997;272:709–712. doi: 10.1074/jbc.272.2.709. [DOI] [PubMed] [Google Scholar]
- 26.Silva L.C., Ben David O., Pewzner-Jung Y., Laviad E.L., Stiban J., Bandyopadhyay S., Merrill A.H., Jr., Prieto M., Futerman A.H. Ablation of ceramide synthase 2 strongly affects biophysical properties of membranes. J. Lipid Res. 2012;53:430–436. doi: 10.1194/jlr.M022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Corte E., Dei Cas M., Raggi A., Patane M., Broggi M., Schiavolin S., Calatozzolo C., Pollo B., Pipolo C., Bruzzone M.G., Campisi G., Paroni R., Ghidoni R., Ferroli P. Long and very-long-chain ceramides correlate with A more aggressive behavior in skull base chordoma patients. Int. J. Mol. Sci. 2019;20:4480. doi: 10.3390/ijms20184480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laviad E.L., Kelly S., Merrill A.H., Jr., Futerman A.H. Modulation of ceramide synthase activity via dimerization. J. Biol. Chem. 2012;287:21025–21033. doi: 10.1074/jbc.M112.363580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegner M.S., Schiffmann S., Parnham M.J., Geisslinger G., Grosch S. The enigma of ceramide synthase regulation in mammalian cells. Prog. Lipid Res. 2016;63:93–119. doi: 10.1016/j.plipres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Halbleib K., Pesek K., Covino R., Hofbauer H.F., Wunnicke D., Hanelt I., Hummer G., Ernst R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell. 2017;67:673–684.e8. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Tam A.B., Roberts L.S., Chandra V., Rivera I.G., Nomura D.K., Forbes D.J., Niwa M. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev. Cell. 2018;46:327–343.e7. doi: 10.1016/j.devcel.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fun X.H., Thibault G. Lipid bilayer stress and proteotoxic stress-induced unfolded protein response deploy divergent transcriptional and non-transcriptional programmes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158449. doi: 10.1016/j.bbalip.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Senkal C.E., Ponnusamy S., Manevich Y., Meyers-Needham M., Saddoughi S.A., Mukhopadyay A., Dent P., Bielawski J., Ogretmen B. Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. J. Biol. Chem. 2011;286:42446–42458. doi: 10.1074/jbc.M111.287383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worgall T.S. Sphingolipid synthetic pathways are major regulators of lipid homeostasis. Adv. Exp. Med. Biol. 2011;721:139–148. doi: 10.1007/978-1-4614-0650-1_9. [DOI] [PubMed] [Google Scholar]
- 35.Hartmann D., Wegner M.S., Wanger R.A., Ferreiros N., Schreiber Y., Lucks J., Schiffmann S., Geisslinger G., Grosch S. The equilibrium between long and very long chain ceramides is important for the fate of the cell and can be influenced by co-expression of CerS. Int. J. Biochem. Cell Biol. 2013;45:1195–1203. doi: 10.1016/j.biocel.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Stiban J., Perera M. Very long chain ceramides interfere with C16-ceramide-induced channel formation: A plausible mechanism for regulating the initiation of intrinsic apoptosis. Biochim. Biophys. Acta. 2015;1848:561–567. doi: 10.1016/j.bbamem.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Farshbaf M., Khosroushahi A.Y., Mojarad-Jabali S., Zarebkohan A., Valizadeh H., Walker P.R. Cell surface GRP78: An emerging imaging marker and therapeutic target for cancer. J. Control Release. 2020;328:932–941. doi: 10.1016/j.jconrel.2020.10.055. [DOI] [PubMed] [Google Scholar]
- 38.Utermohlen O., Herz J., Schramm M., Kronke M. Fusogenicity of membranes: The impact of acid sphingomyelinase on innate immune responses. Immunobiology. 2008;213:307–314. doi: 10.1016/j.imbio.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Bian F., Xiong B., Yang X., Jin S. Lipid rafts, ceramide and molecular transcytosis. Front. Biosci. (Landmark Ed) 2016;21:806–838. doi: 10.2741/4422. [DOI] [PubMed] [Google Scholar]
- 40.Hammerschmidt P., Ostkotte D., Nolte H., Gerl M.J., Jais A., Brunner H.L., Sprenger H.G., Awazawa M., Nicholls H.T., Turpin-Nolan S.M., Langer T., Kruger M., Brugger B., Bruning J.C. CerS6-derived sphingolipids interact with Mff and promote mitochondrial fragmentation in obesity. Cell. 2019;177:1536–1552.e23. doi: 10.1016/j.cell.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Verlekar D., Wei S.J., Cho H., Yang S., Kang M.H. Ceramide synthase-6 confers resistance to chemotherapy by binding to CD95/Fas in T-cell acute lymphoblastic leukemia. Cell Death Dis. 2018;9:925. doi: 10.1038/s41419-018-0964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., Zhu J., Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee W.K., Thévenod F. Oncogenic PITX2 facilitates tumor cell drug resistance by inverse regulation of hOCT3/SLC22A3 and ABC drug transporters in colon and kidney cancers. Cancer Lett. 2019;449:237–251. doi: 10.1016/j.canlet.2019.01.044. [DOI] [PubMed] [Google Scholar]
- 44.Laviad E.L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A.H., Jr., Futerman A.H. Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 2008;283:5677–5684. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R.N., Futerman A.H. Kinetic characterization of mammalian ceramide synthases: Determination of K(m) values towards sphinganine. FEBS Lett. 2007;581:5289–5294. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 47.Lee W.K., Torchalski B., Kohistani N., Thévenod F. ABCB1 protects kidney proximal tubule cells against cadmium-induced apoptosis: Roles of cadmium and ceramide transport. Toxicol. Sci. 2011;121:343–356. doi: 10.1093/toxsci/kfr071. [DOI] [PubMed] [Google Scholar]
- 48.Thévenod F., Wolff N.A., Bork U., Lee W.K., Abouhamed M. Cadmium induces nuclear translocation of beta-catenin and increases expression of c-myc and Abcb1a in kidney proximal tubule cells. Biometals. 2007;20:807–820. doi: 10.1007/s10534-006-9044-9. [DOI] [PubMed] [Google Scholar]
- 49.Bielawski J., Pierce J.S., Snider J., Rembiesa B., Szulc Z.M., Bielawska A. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 2009;579:443–467. doi: 10.1007/978-1-60761-322-0_22. [DOI] [PubMed] [Google Scholar]
- 50.Wu H., Hait W.N., Yang J.M. Small interfering RNA-induced suppression of MDR1 (P-glycoprotein) restores sensitivity to multidrug-resistant cancer cells. Cancer Res. 2003;63:1515–1519. [PubMed] [Google Scholar]
- 51.Thanasai J., Limpaiboon T., Jearanaikoon P., Sripa B., Pairojkul C., Tantimavanich S., Miwa M. Effects of thymidine phosphorylase on tumor aggressiveness and 5-fluorouracil sensitivity in cholangiocarcinoma. World J. Gastroenterol. 2010;16:1631–1638. doi: 10.3748/wjg.v16.i13.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu P., Struijs M.C., Mei J., Witte-Bouma J., Korteland-van Male A.M., de Bruijn A.C., van Goudoever J.B., Renes I.B. Endoplasmic reticulum stress, unfolded protein response and altered T cell differentiation in necrotizing enterocolitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Day C.J., Kim M.S., Stephens S.R., Simcock W.E., Aitken C.J., Nicholson G.C., Morrison N.A. Gene array identification of osteoclast genes: Differential inhibition of osteoclastogenesis by cyclosporin A and granulocyte macrophage colony stimulating factor. J. Cell. Biochem. 2004;91:303–315. doi: 10.1002/jcb.10780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article.