Abstract
Ferroptosis and necroptosis are two pro-inflammatory cell death programs contributing to major pathologies and their inhibition has gained attention to treat a wide range of disease states. Necroptosis relies on activation of RIP1 and RIP3 kinases. Ferroptosis is triggered by oxidation of polyunsaturated phosphatidylethanolamines (PUFA-PE) by complexes of 15-Lipoxygenase (15LOX) with phosphatidylethanolamine-binding protein 1 (PEBP1). The latter, also known as RAF kinase inhibitory protein, displays promiscuity towards multiple proteins. In this study we show that RIP3 K51A kinase inactive mice have increased ferroptotic burden and worse outcome after irradiation and brain trauma rescued by anti-ferroptotic compounds Liproxstatin-1 and Ferrostatin 16-86. Given structural homology between RAF and RIP3, we hypothesized that PEBP1 acts as a necroptosis-to-ferroptosis switch interacting with either RIP3 or 15LOX. Using genetic, biochemical, redox lipidomics and computational approaches, we uncovered that PEBP1 complexes with RIP3 and inhibits necroptosis. Elevated expression combined with higher affinity enables 15LOX to pilfer PEBP1 from RIP3, thereby promoting PUFA-PE oxidation and ferroptosis which sensitizes Rip3K51A/K51A kinase-deficient mice to total body irradiation and brain trauma. This newly unearthed PEBP1/15LOX-driven mechanism, along with previously established switch between necroptosis and apoptosis, can serve multiple and diverse cell death regulatory functions across various human disease states.
Keywords: Lipid signaling, Whole body irradiation, Traumatic brain injury, Regulated necrosis, Necroptotic death, Ferroptotic death
Graphical abstract
Highlights
-
•
PEBP1 acts as an endogenous RIP3 kinase inhibitor
-
•
PEBP1 suppresses necroptosis and sensitizes cells to ferroptosis
-
•
RIP3 kinase inactive K51A mutant enhances pro-ferroptotic HpETE-PE generation
-
•
As a necroptosis/ferroptosis switch, PEBP1 worsens outcomes of γ-IR and TBI
1. Introduction
Irreparably damaged cells are destined to die via one of several genetically pre-determined and evolutionarily conserved programs [1]. Failure of one of these processes inevitably triggers another death pathway, thus altering the spread of perilous materials from damaged cells to their local communities. This can be beneficial in facilitating functional restoration or detrimentally disseminate pro-death signals [2]. Key switching mechanisms between regulated cell death pathways are themselves genetically predetermined [3]. One important program of cell demise is ferroptosis [4]. We previously identified several enzymatic players essential for ferroptosis, but its overall regulatory schema and interactions with other cell death programs are only beginning to emerge. Ferroptosis is inexorably linked to functional deficiency of the selenoprotein, glutathione peroxidase 4 (GPX4) – the only GPX isoform capable of reducing phospholipid hydroperoxides (PL-OOH) [5]. Execution of ferroptosis is also dependent on Acyl-Coenzyme A (CoA) Synthetase Long Chain 4 (ACSL4), which yields arachidonoyl (C20:4, AA)- or adrenoyl (C22:4, AdA)-CoA substrates necessary for AA-phosphatidylethanolamine (PE; AA-PE) and AdA-PE synthesis. Under pro-ferroptotic conditions, AA- and AdA-PE are enzymatically oxidized by 15-lipoxygenase (15LOX) complexed with a scaffold protein, phosphatidylethanolamine-binding protein 1 (PEBP1) [6,7]. The resultant hydroperoxy-AA-PE (e.g., hydroperoxy-eicosatetraenoyl-PE, HpETE-PE) and hydroperoxy-AdA-PE serve as specific ferroptotic signaling effectors [8].
Ferroptosis is implicated in several common and severe disease states (e.g., traumatic brain injury (TBI), ischemia-reperfusion, asthma, acute kidney injury) [6]. Necroptosis is similarly well represented in acute and chronic pathologies [9] and often found commingling with other cell death pathways. Mechanistically divergent from ferroptosis, necroptosis relies upon the (de)ubiquitination and (auto)phosphorylation-driven assembly of Receptor Interacting Kinase 1 (RIP1) and RIP3 complexes, which culminates in (1) Mixed Lineage Kinase domain-like protein (MLKL) phosphorylation, (2) rapid membrane permeabilization, and (3) release of nuclear factor kappa-light chain enhancer of activated B cells (NF-кB)-associated cytokines and pro-inflammatory damage-associated molecular patterns (DAMPs) [10,11]. Owing to its central role in necroptosis, inhibitors of RIP3 have gained attention to treat wide range of acute and chronic disease states [12]. The essentiality of elimination of fatally injured cells implies that failure to execute the necroptotic program must trigger an alternative type of regulated cell death. Indeed, the switch from necroptosis to apoptosis has been identified [13]. It has been shown that high concentrations of several RIP3 kinase inhibitors (e.g., GSK872) and RIP3 kinase inactivating mutation D161 N induce apoptosis while RIP3 K51A kinase-dead knock-in mice are viable and immunocompetent [14]. Whether inhibition of necroptosis sensitizes cells to ferroptosis and the potential mechanisms of the switch between these two regulated cell death pathways remain to be elucidated.
PEBP1, also known as RAF kinase inhibitory protein, is an endogenous serine-/threonine-kinase inhibitor and displays regulatory promiscuity towards multiple protein partners [[15], [16], [17]]. When complexed with 15LOX, PEBP1 enables 15LOX to utilize AA-PE and AdA-PE as the peroxidation substrates instead of polyunsaturated free fatty acids (PUFA) [18]. In this study we show that RIP3 K51A kinase inactive mice have increased ferroptotic burden and worse outcome after irradiation (IR) and TBI that are rescued by anti-ferroptotic compounds Liproxstatin-1 and Ferrostatin 16-86. Given a distinct structural homology between RAF kinase and RIP3 kinase, we hypothesized that PEBP1 may act as a switch in a ferroptosis-necroptosis connection. Using genetic, biochemical, redox lipidomics and computational approaches, we established that PEBP1 can alternatively bind and (i) inhibit RIP3 activity thus turning-off necroptosis or (ii) activate 15LOX-mediated AA-PE/AdA-PE oxidation and ferroptosis.
2. Materials and methods
2.1. Experimental model and subject details
2.1.1. Cell cultures
All cell lines used in this study were cultured in an atmosphere of 37°C, 5% CO2, 95% humidity. The cell lines used are outlined within individual legends and the STAR methods table (where information regarding the sex of cells is also found, if relevant). All cell lines were authenticated by Short Tandem Repeat (STR) profiling and confirmed negative for mycoplasma prior to experiments commencing. HT22 cells were a generous gift from Dr. David Schubert (The Salk Institute, La Jolla, CA). HT22, L929 (NTCT clone 929, ATCC, Manassas, VA), IEC18 (CRL-1589, ATCC), and HEK 293T (CRL-3216, ATCC) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, ATCC 30-2002) supplemented with 10% fetal calf serum (A3160401, US Origin, ThermoFisher, Waltham, MA) and 1× penicillin/streptomycin (15140122, ThermoFisher). Cells were passaged using 0.25% Trypsin-EDTA (25200056, ThermoFisher) at 80-90% confluence every 2-4 days. The methods for culturing and maintenance of primary long-term bone marrow (BM) cells and enteroid monolayers are described in further detail below.
2.1.2. Animal studies
All procedures were performed according to the protocols established by the Institutional Animal Care and Use Committee of the University of Pittsburgh. The Rip3K51A/K51A knock-in strategy was designed and performed by genOway as described previously [19]. Briefly, the RIP3 gene-targeting vector was constructed from the C57BL/6 mouse genome. The K51A point mutation was inserted into RIP3 exon 2, while a neomycin resistance gene cassette was inserted in intron 3, flanked by FRT sites for further Flp-mediated excision. Exon 2 including the K51A point mutation was flanked by loxP sites enabling access to constitutive or conditional deletion using Cre-mediated recombination. Similarly, Rip1K45A/K45A mice were created by using a RIP1 gene-targeting vector constructed from the C57BL/6 mouse genome. The K45A point mutation was inserted into RIP1 exon 3. A neomycin cassette was placed in intron 3 and flanked by FRT sites for FLP excision. LoxP sites were placed flanking exon 3 to allow for Cre mediated deletion.
Liproxstatin-1 (Lip-1, 50 mg/kg), Ferrostatin16-86 (Fer16-86, SRS16-86, 15 mg/kg), or vehicle (1:3, [50:50 Cremophor:Ethanol] to normal saline) was administered intraperitoneally (i.p.) at 24h before and 24h after whole-body gamma irradiation (γ-IR). Eight to twelve week-old wild-type C57BL/6, Rip1K45A/K45A and Rip3K51A/K51A mice were irradiated to 9.25Gy using a Shepherd Mark 1 Model 68 cesium irradiator at a dose rate of 310 cGy/minute, as described [20]. Mice were monitored for the development of severe hematopoietic or gastrointestinal syndrome and were sacrificed after losing >20% original body weight for >24 hours or >15% weight loss while demonstrating signs of a moribund state. At the time of sacrifice, animals were transcardially perfused with 5 ml calcium-/magnesium-free (CCF) phosphate buffered saline (PBS) and ileum was snap frozen. Tissue was mechanically homogenized in cold 1x PBS buffer containing 1x Protease & Phosphatase Inhibitor cocktail (ThermoFisher). Proteolyic enzyme activity (i.e. caspase-3/7) was measured in homogenate prepared without protease/ phosphatase inhibitor using a luminescence Caspase–GloTM 3/7 assay kit (Promega) according to the manufacturer's instructions. For histological analysis, mice were perfused with heparinized saline followed with 2% paraformaldehyde. Ileum was excised and immersion-fixed for 2 hours, and cryoprotected in 30% sucrose, then frozen in dry ice-cooled methalbutane. Transverse ileum sections were cut on a freezing sliding microtome with a thickness of 5 μm.
A CCI model was used as previously described [21]. Briefly, eight to twelve week-old wild-type C57BL/6 and Rip3K51A/K51A were anesthetized with 4.5% isoflurane (Anaquest) in 70% nitrous oxide and 30% oxygen using a Fluotec 3 vaporizer (Colonial Medical). The mice were placed in a stereotaxic frame and a 5-mm craniotomy was made over the parieto-temporal cortex using a drill and a trephine. The bone flap was removed and discarded, and a pneumatic cylinder with a 3-mm flat tip impounder with velocity 6 m/s, depth 1.2 mm, and duration 100 ms was used to induce CCI. The scalp was sutured closed and the mice were returned to their cages to recover.
2.1.3. CCI Lesion volume measurement
Coronal brain sections were stained with hematoxylin were collected as described [22]. Lesion area within each hemisphere was determined using morphometric image analysis in ImageJ (NIH, Bethesda, MD). The area of the injured hemisphere (left) was subtracted from the area of the uninjured hemisphere, and the difference was multiplied by 0.5 to obtain the volume of brain tissue loss (mm3), then normalized to mean sham brain tissue volume, and expressed as percent tissue loss
2.1.4. Behavioral testing
Behavioral testing was conducted during the light phase of the circadian cycle by experimenters blinded to the treatment conditions. Prior to each test, mice were acclimatized to the room for at least 30 min. Mice were tested in a battery of assays, according to the schedule as previously described [22]. Vestibulo-motor ability was evaluated by wire grip and rotarod assays. Wire grip was performed on days 1, 3, 7, 10, 14, and 21 following CCI by placing mice on a 45-cm-long metal wire suspended 45 cm above the ground and were allowed to traverse the wire for 60 sec. The latency to fall within the 60-sec interval was measured, and a wire grip score was quantitated using a 5-point scale. Testing was performed in triplicate and an average value calculated for each mouse on each test day. Rotarod testing was performed by training three times per day for 3 days with 5-min rest intervals to establish baseline performance. The average daily scores of each subject were used for pairwise statistical analyses. Post-CCI performance was tested for each mouse three times each day on days 21, 22, and 23post-CCI. Briefly,mice were placed on an automated rotarod apparatus (Harvard Apparatus, Holliston, MA), which accelerated from 4 to 40 r/min over 60 sec. Maximum trial duration was 300 sec or until the mouse fell off the rotarod. The average latency to drop and the average r/min speed attained over the three trials was recorded for each day of testing.
The Morris water maze (MWM) testing was performed as previously described [22] starting with pretraining prior to injury and retesting on day 21 post-CCI. Spatial learning was assessed at approximately the same time each day. Each mouse was subjected to seven hidden platform trials per day using a random set of starting positions at any one of the four quadrants. A given trial consisted of the average latency from each of the four starting positions. Mean swim speed (m/sec) and time in target quadrant was recorded. If a mouse failed to find the platform within 90 sec, then it was placed on the platform for approximately 10 sec. Probe trials were performed 24 hours after the last hidden platform trial by allowing mice to swim in the tank for 30 seconds with the platform removed. Finally, two visible platform trials with the platform raised 0.5 cm above the water were performed.
2.1.5. Total lipid extraction and quantitation phosphatidylethanolamine (PE) by liquid chromatography mass spectrometry
Pieces of ileum (2-10 mg of protein) or brain (2-10 mg of protein) tissues were homogenized and lipids were extracted by using the Folch method (41). To avoid ex-vivo oxidation chloroform-methanol mixture containing 0.01% butylated hydroxytoluene was used. Total phospholipid content was quantified using the micro method for phosphorus measurement [23] Phospholipids (1nmol/1 μL) were analyzed by LC/MS using a Dionex Ultimate 3000 HPLC system coupled on-line to a Q-Exactive hybrid Quadrupole-Orbitrap and Orbitrap Fusion and Orbitrap Fusion Lumos mass spectrometer (ThermoFisher) using a normal phase column (Luna 3 μm Silica (2) 100 Å, 150 × 2.0 mm, (Phenomenex)). The injection volume was 5 μL. The analysis was performed using gradient solvents (A and B) containing 10 mM ammonium formate at a flow rate of 0.2 ml/min. Solvent A contained isopropanol/hexane/water (285:215:5, v/v/v), and solvent B contained isopropanol/hexane/water (285:215:40, v/v/v). All solvents were LC/MS-grade. The column was eluted for 0-23 min with a linear gradient from 10% to 32% B; 23-32 min with a linear gradient of 32%–65% B; 32-35 min with a linear gradient of 65%–100% B; 35-62 min held at 100% B; 62-64 min with a linear gradient from 100% to 10% B; followed by an equilibration from 64-80 min at 10% B. Analysis was performed in negative ion mode at a resolution of 140,000 for the full MS scan in a data-dependent mode. Analysis of raw LC/MS data was performed using software package Compound DiscovererTM 2.1 (ThermoFisher) with an in-house generated analysis workflow and oxidized PL database. Peaks with S/N ratio of more than 3 were identified and searched against oxidized PL database. Lipids were further filtered by retention time and confirmed by a fragmentation mass spectrum. Values for m/z were matched within 5.5 ppm to identify the lipid species. Peak areas were used for quantification of oxygenated PL species. Data were presented as pmol normalized to μmol of total PLs. To quantify PE species deuterated PE-16:0D31/18:1 (1-hexadecanoyl(d31)-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine) (Avanti Polar Lipids) was used as internal standard. Internal standard was added directly to the MS sample to a final concentration of 1 μM. To quantitatively assess oxygenated PE species, calibration curves were obtained using 1-stearoyl-2-15(S)-HpETE-sn-glycero-3-phosphoethanolamine and 1-stearoyl-2-15(S)-HETE-sn-glycero-3-phospho-ethanolamine as reference standards (Cayman Chemicals). 1-stearoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine and 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine (Avanti Polar Lipids) were used as a reference standard to quantitatively estimate the amount of non-oxidized PE species.
2.1.6. Identification of phosphatidylethanolamine (PE) oxygenated species
Targeted single ion monitoring (tSIM) and MS/MS analysis was used to identify oxygenated phospholipids. Phospholipids were separated by solid phase extraction (SPE) using a sequential combination of silica gel/aminopropyl-silica gel SPE cartridges as described previously [24]. Fraction of PE was collected, solvent was evaporated under N2 and 20 μL of 100% of mobile phase B were added (see below). PE fraction was mixed on a vortex to completely dissolve the analytes and then analyzed by LC/MS/MS. The injection volume was 5 μL. PE species were separated on a C30 reverse phase column (Accucore, 2.1 mm x 25 cm, 2.6 μm particle size, Thermo Scientific). Solvent A: acetonitrile/water (50/50); Solvent B: 2-propanol/acetonitrile/water (85/10/5). Both A and B solvents contained 5 mM ammonium formate and 0.1% formic acid as modifiers. Gradient method was as follows: 0-40 min, 15%-50% B (linear, 5); 40-130 min, 50-100% B (linear, 5); 130-135 min, hold at 100% B; 135-140 min, 15% B (linear, 5); 140-150 min, 15% B for equilibration. The flow was maintained at 100 ul/min. The LC system was a Thermo Ultimate 3000 complete with a WPS-3000 autosampler. Column temperature was set at 35C. Targeted (tSIM) and MS/MS analysis of specific oxidized masses was performed on a Fusion Lumos tribrid mass spectrometer (Thermo). A tSIM analysis (isolation mode quadrupole) for ions of m/z 796.5134, 798.5193 and 826.5603 in negative ion mode (profile) was performed at a resolution of 120,000. The maximum injection time of 128 ms with 1 microscan and a normalized AGC target of 400% using the orbitrap as the detector. MS2 analysis was performed using high energy collisional dissociation (HCD) with collision energy set to 24 with an isolation window of 1.2 m/z and a resolution of 30,000, also using the orbitrap as the detector. Capillary spray voltage was set at 3900V, and ion transfer tube temperature was 300C. Sheath, auxiliary and sweep gasses were set to 30, 23 and 1 (arbitrary units), respectively.
2.1.7. LC-MS/MS analysis of PUFA
PUFA were analyzed by LC/MS using a Dionex Ultimate 3000 HPLC system coupled on-line to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher) using C30 reverse phase column (Accucore, 2.1 mm x 25 cm, 2.6 μm particle size, Thermo Scientific). The analysis was performed using gradient solvents (A and B) containing 5 mM ammonium formate at a flow rate of 100 μL/min. All solvents were LC-MS-grade. Solvent A: acetonitrile/water (50/50); Solvent B: 2-propanol/acetonitrile/water (85/10/5). Both A and B solvents contained 5 mM ammonium formate and 0.1% formic acid as modifiers. Gradient method was as follows: 0-40 min, 15%-50% B (linear, 5); 40-130 min, 50-100% B (linear, 5); 130-135 min, hold at 100% B; 135-140 min, 15% B (linear, 5); 140-150 min, 15% B for equilibration. Spectra were acquired in negative ion mode at a resolution of 140,000 for the full MS scan in a data-dependent mode. The scan range for MS analysis was m/z 150-600 with a maximum injection time of 100 ms using 1 microscan. An isolation window of 1.0 Da was set for the MS and MS2 scans. Capillary spray voltage was set at 2.6 kV, and capillary temperature was 250 °C. The S-lens RF level was set to 60. Analytical data were acquired and analyzed using Xcalibur 4.2 Quan Browser (Thermo Fisher Scientific). C17:0 (Sigma-Aldrich) was used as internal standard.
2.1.8. Far-western blotting
The interaction between PEBP1 and RIPK3 in vitro was examined by far-western blotting. Recombinant human RIPK3 (Abcam, Cat. Ab125566) and bovine serum albumin (Sigma, Cat. A2153) were separated on 8∼16% Tris-Glycine SDS-PAGE and electrically transferred to a nitrocellulose membrane. Proteins were renatured by incubation of the membrane in denaturing and renaturing buffer (100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% skim milk powder and 1 mM DTT) containing decreasing concentration of guanidine HCl (6 M, 3 M, 1 M, 0.1 M, 0 M) and the membrane was blocked with 5% skim milk. Then the membrane was incubated in 0.5 mL of protein-binding buffer (100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% skim milk powder and 1 mM DTT) containing 1 μg/mL of recombinant PEBP1 (see supplemental methods PEBP1 expression/purification) at 4 °C overnight. Excess PEBP1 was washed away and bound PEBP1 was immunodetected with PEBP1-specific antibodies (Santa Cruz, #SC-28837, Rabbit, 1:1000) after 1.5 hr incubation at room temperature. HRP-conjugated goat anti-rabbit/mouse IgG H&L (Sigma, #A0545, 1:1000) was applied as the secondary antibody.
2.1.9. Coimmunoprecipitation
Adherent cells were collected with trypsin-EDTA, collected (200 xg, 5 min.), and washed with PBS. Cells were then snap-frozen. Thawed pellet was resuspended in IP lysis buffer containing 150 mM NaCl, 50 mM Tris pH 7.4, 0.1% NP40, 0.5 mM EDTA along with protease and phosphatase inhibitor cocktail (ThermoFisher, 78440) and PMSF (1 mM) on ice for 30 min. The cell lysate was centrifuged at 14000 rpm, 4°C for 12 min and the supernatant was collected and used for IP (0.5-1 mg per sample, kept constant for a given experiment) after estimation of protein concentration by BCA protein determination kit. Cell lysate was incubated with no antibody, control, PEBP1, RIP3, RIP1, or MAPK14 antibody overnight at 4°C. Alternatively, antibody was first crosslinked to magnetic beads (ThermoFisher, 88805) according to manufactures direction (7 μg antibody/reaction), and then incubated overnight with cell lysate. Beads (ThermoFisher, 88802) were washed with 1% BSA in IP lysis buffer for an additional 4 hours, followed by several quick washes (4x, 1 min). Immunocomplexes were dissociated protein loading buffer or using a low pH elution approach (followed by neutralization and addition of 4X loading buffer). All samples were heated to 95°C for 10 min before loading on tris-glycine SDS-Page gel.
2.1.10. Cell culture γ-Irradiation
Mouse intestinal enteroid monolayers and bone marrow cells were prepared in fresh medium with Liproxstatin-1 (effective concentration: 1-10 μM), Ferrostatin-1 (0.5-10 μM), Nec-1s (20-40 μM), or z-VAD-fmk (10-25 μM) for 30-60 min. before irradiation. Samples were γ-irradiated with a Shepherd model 143-45A irradiator (J.L. Shepherd & Associates, CA) at the indicated dose (5, 10, 20 Gy). Irradiated cells were returned to 37°C in 5% CO2 incubator (timepoint: 0 hours post-IR) for the indicated duration.
2.1.11. Cell death assays
Cell death was assessed either by LDH release using CytoTox-ONE kit (Promega) according to manufacturer’s instructions or by flow cytometry-based Annexin V-FITC + PI (BioVision, Milpitas, CA) labeling. For LDH assay, the activity of enzyme present in culture medium was used as an index of cell death. To determine the upper limit of LDH activity, adherent cells were lysed in 0.9% Triton X-100 (100% death index). Cell death was calculated as the ratio between treatment-induced LDH release and the range between Triton X-100 and untreated culture medium-associated LDH signal. For flow cytometric cell death determination, harvested cells were washed in cold PBS and were stained with Annexin V-fluorescein isothiocyanate (FITC) and PI for 15 min in the dark before counting. Dead and dying cells stained for either/both AnnexinV or PI. Cell debris, represented by low forward and side scatter, were gated out of analysis. A minimum of ten thousand events were collected for each analyzed group using a FACScan Flow Cytometer (BD Biosciences, Rutherford, NJ) and associated FACSDiva software.
2.1.12. Isolation of crypts and small intestinal culture
Small intestinal organoids were established following the Stem Cell Technologies protocol. In brief, small intestinal organoids were established by isolating 10 cm of the most proximal portion of the small bowel from an adult C57BL/6 (n=3) or Rip3K51A/K51A (n=3) animals and placing it in a petri dish containing 10 mL of ice-cold DPBS. The small intestine was flushed three times with 1 mL DPBS to removal luminal contents before opening the tissue longitudinally and cutting into 2 mm2 sized pieces. The intestinal pieces were washed 15 times with ice-cold DPBS or until the supernatant was clear. Tissue fragments were transferred to a new tube containing 25mL of Gentle Dissociation Buffer and placed on a rotating plating platform (20rpm) for 15 minutes. Tissues were resuspended in 10ml of DPBS supplemented with 0.1% BSA and pipetted vigorously to dislodge intact crypts before filtering through 70-um strainer to remove tissue debris. Isolated crypts were centrifuged at 290g for 5 minutes and resuspended in 10mL ice-cold DMEM/Advanced F12 supplemented with 2mM Glutmax, 10mM HEPES, and 1x Pen/Strep (Mouse Wash Media). Crypts were resuspended in growth factor reduced matrigel at a density of 5-10 crypts/μL and 45ul droplets were plated on a pre-warmed 24-well plate. Once the matrigel solidifed, small intestinal organoid growth media (Mouse Wash Media supplemented with 1x R-spondin conditioned media, 1mM N-acetyl cysteine, 50ng/mL EGF, 100ng/mL Noggin, 1x N2 supplement, 1x B27 supplement, and 10uM Y27632 was added to each well and changed every other day after. Small intestinal organoids were passaged twice before using in any downstream experimentation.
2.1.13. 2-D monolayers from established small intestinal organoids
Culturing of 2-D monolayers from murine small intestinal tissue has been previously described in a detailed protocol established by Kozuka et al. Briefly, intestinal organoids were extracted from the Matrigel by placing 1mL of Gentle Disassociation Buffer in each well for 1 minute before gently pipetting 20 times and transferring suspension into a 15mL conical tube. Each well was washed with an additional 1mL of Gentle Disassociation buffer, which was added to the previous 15mL conical tube. The organoid suspension was placed on a rotating platform (20rpm) for 10 minutes prior to centrifugation at 290g for 5 minutes, 4C. The supernatant was decanted and the organoids were resuspended in 10mL ice-cold Mouse Wash Buffer before centrifugation at 200g for 5 minutes, 4C. The intact crypts were resuspended in small intestinal monolayer growth media (Mouse Wash Media supplemented with 1x R-spondin conditioned media, 50ng/mL EGF, 100ng/mL Noggin, 1x N2 supplement, 1x B27 supplement, 10uM Y27632, 2mM Valproic Acid, and 2.5uM CHIR99021) at 1 crypt unit/ul concentration and 150ul was placed on the apical side of a 24-well transwell (0.4um transparent PET membrane) coated with a 1:30 dilution of matrigel in Mouse Wash Media. 500ul of monolayer growth media was added to the basal side of the transwell. Media was changed 18-24 hours later and 150ul(apical) and 500ul(basal) of Differentiation Media (Mouse Wash Media supplemented 50ng/mL EGF, 300ng/mL BMP-4, 1x N2 supplement, 1x B27 supplement, and 10uM Y27632) was placed in each well. Monolayers were cultured an additional 48 hours before using in downstream assays.
2.1.14. RNA Isolation and cDNA synthesis
Total RNA was isolated from murine small intestinal monolayers using the RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The concentration and purity of each RNA sample were measured via spectrophotometry (ND-2000 spectrophotometer; NanoDrop Technologies, Inc., Wilmington, DE, USA). First-strand cDNA (0.5 μg of RNA) was prepared by using QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s instructions. cDNA was diluted to 0.5 ng/μl and stored at -20°C until further use.
2.1.15. Quantitative Real-Time PCR (qPCR) and analysis
Gene expression was measured relative to the housekeeping gene 50s ribosomal subunit protein L15 (RPLO) as previously described [25, 26]. In brief, cDNA was amplified using a Applied Biosystems (ABI) QuantStudio 3 real-time PCR detection system (ABI, Model # A28131), with a final reaction volume of 10 μl containing 2.5 ng of cDNA, primers (500 nM final concentration), and 1x PowerUp SYBR Mastermix, ROX (ABI, Cat # A25741). The amplification conditions for the qPCR reactions were as follows: 1 cycle of initial denaturation at 95°C for 4 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 56°C for 30 seconds, and elongation at 72°C for 30 seconds. All reactions were carried out in triplicate. Primer efficiencies were validated to be similar, which allowed the qPCR data to be analyzed using the comparative Ct method (2-ΔΔCt method).
2.1.16. Primary IL-3 Dependent Long-Term Bone Marrow (BM) Culture
Non-adherent IL-3 dependent cells were segregated from the mixed population long-term culture of bone marrow cells derived from wild type (Rip3+/+) and Rip3K51A/K51A mice. These were cultured in six-well tissue culture plates with 4.0 mL of Iscoves modified Eagles medium supplemented with 20% fetal calf serum (US Origin, ThermoFisher), 1× penicillin/streptomycin, and 1.0 ng/mL of recombinant IL-3 (Peprotech, Rocky Hill, NJ, USA). The cells were passaged at high density weekly for 10 weeks by cytocentrifugation at 300 xg and replenished with 4.0 mL medium. At week 10, the culture was split in two and frozen at -80°C for one week. Cells were then thawed, cultured, and passaged in the same medium as described above. BM were passaged every 3-4 days and maintained at high density. For RSL3 experiments, cells were plated in 35mm dishes containing RSL3 (0-500 nM), Fer-1 (500-1000 nM) and/or other compounds in normal growth media for 18-24h before cell death assay. For irradiation experiments, 8·104 cells in 3-4 mL media were plated in 60mm dishes with 1-5% FBS for 48h, followed by media change into normal 20% FBS media prior to IR. Drug dose ranges are listed with γ-irradiation exposure methods.
2.1.17. PEBP1 knockdown and necroptosis induction
PEBP1 knockdown in L929 cells was accomplished by transfected using RNAiMax (Invitrogen) according to manufacturer’s instruction 48 hours before secondary treatment. Necroptosis was induced in PEBP1 siRNA-treated cells via combination of z-VAD-fmk (10 μM), SM164 (100 nM), and TNFα (1, 5, 10 ng/mL) in the presence or absence of Nec-1s (20-40 μM). CRISPR-mediated PEBP1 knockdown was performed using a CRISPR double nickase (sc-42393-NIC, SCBT) or non-targeted control (sc-418922, SCBT) system according to the manufacturer's protocols. Two clones were created and screened using puromycin. Necroptosis induction in CRISPR clones was accomplished by combination of z-VAD-fmk (10 μM), SM164 (100 nM), and TNFα (1ng/mL) ± Nec-1s. LDH cytotoxicity was measured at 18-24 h following necroptosis induction.
2.1.18. Ferroptosis synergy in HT22, IEC18, and enteroid monolayers
For GSK’872 ferroptosis experiments, HT22 or IEC18 cells were plated in 24-well plates (∼2·104 cells/well). After 18h, cells were treated with vehicle (0.1% DMSO), RSL3 (50-75 nM) ± GSK’872 (25-50 μM) ± Lip-1 or Fer-1 (1 or 10 μM). For enteroid monolayers, 50 enteroids were seeded in 24-well transwells and treated on day 3 post-differentiation, using the above drug dose ranges and those specific in legends. For RAF inhibitor-related ferroptosis experiments, HT22 cells were plated in 24-well plate (1.7 – 2·104 cells/well). After 8-12 h, cells were treated with RSL3 (75 nM) for 18-20h ± Raf-1 inhibitors (5 μM: Sorafenib, PLX4720, or SB590885) ± Fer-1 (400 nM). Collectively, cell death was determined by measuring released lactate dehydrogenase (LDH) activity at 18-24h post-exposure.
2.1.19. PEBP1 expression and purification
Full-length human PEBP1 and related mutants were cloned into a pET21-derived (EMD Millipore, Billerica, MA) bacterial expression plasmid modified to express PEBP1 with N-terminal His10- and mRuby2 tags as described previously [6]. All PEBP1 constructs were cloned into the modified pET21-mRuby2 vector by Gibson Assembly (New England Biolabs, Billerica, MA) using primers with homology at the upstream (sense) NdeI site (5′-GGTCTGAGGGGATACACTCATATG-3′) and downstream (antisense) EcoRI site (5′-GCTTGTCGACGGAGCTCGAATTC-3′) of the vector. All primers were ordered from Life Technologies (Carlsbad, CA). Clones were evaluated by diagnostic restriction digest and sequencing (Genewiz, South Plainfield, NJ). Protein expression was performed in the Escherichia coli strain BL21 (DE3) Codon+ (Agilent, Santa Clara, CA). Following initial growth to OD600 of 0.6, cells were induced using 0.2 mM isopropylthio-beta-galactosidase (IPTG) and cultured overnight at room temperature. Cells were harvested by centrifugation and lysed in buffer containing 20 mM Tris (pH 8.0), 500 mM NaCl, 5 % glycerol, 5 mM imidazole, 1 mM β-mercaptoethanol, and protease inhibitors. Lysates were cleared by centrifugation at 29,000 xg. Protein was initially purified by nickel affinity chromatography (Qiagen, Hilden, Germany), followed by overnight digestion with TEV protease to remove the N-terminal His10-mRuby2 tag. After digestion, PEBP1 was isolated via a second round of nickel affinity chromatography. The protein was then dialyzed overnight at 4 °C into a buffer containing 20mM sodium acetate (pH 5.5) before cation exchange chromatography was performed in sodium acetate (pH 5.5) using a HiTrap SP column (GE Healthcare, Little Chalfont, UK). The resulting fractions were further refined by size exclusion chromatography in 20 mM HEPES (pH 8.0), 500 mM NaCl buffer using a Sephacryl S-200 column (GE Healthcare). Before performing experiments, PEBP1 was desalted into 5 mM Bis-Tris (pH 6.5), 25 mM NaCl as described previously [6].
2.1.20. Kinase activity assay
Full-length human RIP3 (Abcam, Cambridge, UK) activity, alone (200-400 ng/reaction) or preequilibrated (30 min, RT) in the presence of equimolar concentrations of recombinant PEBP1 or a specific RIP3 inhibitor, GSK’872 (100 nM), was determined using the Promega Kinase Glo-Max assay (Promega, Madison, WI) per manufacturer’s instructions. Activity of recombinant human RIP1 (SignalChem, R07-10G-10) and MAPK14 (ThermoFisher, PV3304) in the presence or absence of PEBP1 was also assessed. Reactions containing a final concentration of 200 μg/mL myelin basic protein (Abcam) and 12.5 μM ATP as the kinase substrates proceeded for 1 hour at room temperature in an opaque white 96-well plate. Relative inhibition is presented as percent loss of maximum observed kinase ATP consumption in the presence of equimolar PEBP1 or GSK’872.
2.1.21. Measurement of glutathione
Glutathione (GSH) concentration was assessed with Thiol Fluorescent Probe IV (Millipore-Sigma, 595504) according to manufacturer’s instruction. Briefly, to calculate amount of GSH the difference in Thiol Fluorescent Probe IV was measured in untreated homogenates or those pre-treated with bovine GSH peroxidase (1 U / 50 μL) (Sigma, G6137) in the presence of cumene hydroperoxides (1 mM) (Sigma) for 30 min at 25 °C. GSH concentration was calculated using a standard curve and normalized to homogenate total protein level (μg / 50 μL homogenate).
2.1.22. Fluorescent microscopy
Immunostaining and Confocal Microscopy. Cells were fixed in 2 % paraformaldehyde at 4 °C for 15 min and then permeabilized with 0.1 % Triton X-100 in PBS+ 0.5 % BSA (PBB) for 15 min. Tissue sections and cells were blocked with 5 % donkey serum for 45 min and incubated for 2 h at room temperature with the primary antibodies:15LOX2 (Santa Cruz sc67143), PEBP1 (Abcam ab196832), RIP3 (ProSci 2283). Alexa 488, cy3 and cy5 conjugated secondary antibodies were from Thermo-Fisher Scientific and Jackson Immuno. Cells were counterstained with Hoescht (Sigma B2883) 1mg/100ml dH20 and mounted using Gelvatol. Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labelling (TUNEL) was accomplished using the TMR Red kit (Cat. 12156792910) according to the manufacturer’s instructions (Roche, Basel, Switzerland). Images were collected in 200 nm z-steps using a Nikon A1R equipped with GAsP detectors and a 60X (1.4NA) objective.
Object-Based Co-localization Analyses. Object-based methods take into account the spatial and intensity distribution of the fluorescent signal and is superior to pixel (intensity) based approaches at colocalization – such as Pearson’s correlations [27]. 3D confocal (Nikon A1, 60X, 1.4NA) stacks (200 nm optical sections) were processed using blind deconvolution (10 iterations, NIS Elements, Nikon Inc., Melville NY) in order to maximize the information in each image and improve the accuracy of the subsequent object-based analysis [28]. Puncta (objects) were then segmented based on size and intensity using the 3D spot detection tool in NIS Elements General Analysis 3 (GA3), which generated a binary layer containing the spatial positioning of each object in the x-, y- and z- axis for each individual protein label (i.e. 15LOX, PEBP1 and RIP3). Co-localization was tested using a Boolean “having” operation. This logical argument tests the intersection of selected binary layers and will identify objects that have both protein A “and” protein B (e.g. 15LOX/PEBP1 or RIP3/PEBP1).
Fluorescence Resonance Energy Transfer. FRET based analyses can resolve the proximity of interacting proteins within 1–10 nm. The FRET phenomenon is the non-radiative (dipole-dipole) energy transfer from a fluorescent donor to an acceptor fluorophore. Because the efficiency of FRET decreases as the sixth power of the distance between the donor and acceptor, when the proteins are separated by >10 nm, no FRET occurs. PEBP1 and RIP3 were labelled with cy3 (donor) and cy5 (acceptor) and imaged at 10X zoom using a Leica SP8 spectral confocal microscope equipped with a 60X (1.4NA) optic. FRET was confirmed by acceptor (cy5) photo-bleaching. With this method, if true FRET is present, the donor will show an increased intensity in the bleached region.
2.1.23. Molecular docking and molecular dynamics simulations
Protein-protein docking of RIP3, PEBP1, and RAF1. Simulations of RIP3/PEBP1 complex predictions have been performed using the CLUSPRO [29] respectively with PDB structures: RIP3(4M66) [30], PEBP1(1BEH) [31], RAF1(3Q4C) [32]. Molecular docking has been performed with the SMINA package, which is a fork of AutoDock Vina [33]; customized to better support scoring function development and high-performance energy minimization. For each structure, 10 independent runs were performed.
All-atom Molecular Dynamics Simulations. All-atom MD simulations were performed for human PEBP1 (1BEH) in complex with RIP3 (4M66), RIP3-K51A, or RIP1 (4ITJ) [34] using the GROMACS MD simulation software [35] with CHARMM27 force fields [35]. The PEBP1 to RIP3/RIP3K51A/RIP1 protein interaction has been identified using the molecular docking approach described above. In preparatory simulations with explicit water models (TIP3P), we adopted the following protocol: 0.2 ns of water equilibration, 10,000 steps of minimization, 0.35 ns of heating from 0 to 300 K, and 0.15 ns equilibration of the whole system before initiating production MD run. Three independent MD trajectories were generated, each 200 ns duration with 2 fs time step. A cutoff of 12 Å for non-bonded interactions was applied. Langevin dynamics and the Langevin piston algorithm were used to maintain the temperature at 300 K and the pressure at 1 atm. PYMOL were used for visualization.
Coarse-Grained Molecular Dynamics Simulations. The MARTINI force field version 2.2 [36] was used to simulate protein-protein interactions at the Coarse-grained level. Here, atoms are grouped together to form a single group, reducing the size of the system to be simulated, and reducing computational time The crystal structures of 15LOX (4NRE) [37], RIP3 (4M66) and PEBP1 (1BEH) were used to build the coarse-grained models. Three simulation systems were set up: All the three systems included 15LOX and PEBP1, system 1 had an additional one RIP3 molecule, system 2 had two RIP3 molecules and system 3 had three RIP3 molecules. The purpose being to observe if the PEBP1’s preferred binding partners. For each system, three different orientations of the proteins was created, to generate three trajectories (MD1, MD2, MD3).
The CHARMM-GUI interface [38] was used to set up the simulation models. GROMACS version 5.1.4 was used to perform the molecular dynamics simulations with the standard parameter settings for the MARTINI force field. The system was simulated using the isothermal-isobaric (NPT) ensemble Prior to the production run of 150 ns, the simulation system was energy minimized and a series of equilibration simulations were performed to relax the protein and the solvent system. During the equilibration phase the force-constant of 4000 kJ mol−1 nm−2 is used and a standard value of 0.9 nm is the cut-off. The simulation temperature was 303.15 K. The v-rescale thermostat was used to control the temperature with a coupling constant τp = 1.0 ps. The pressure was isotropically coupled to an external bath of τp = 5.0 ps and compressibility of 4.5 x 10−5 bar−1 using the Berendsen barostat. The electrostatic interactions were calculated using a shifted potential with a cut-off of 1.2 nm, while for the van der Waals interactions, a shifted potential was used with a cutoff of 1.2 nm and switch at 0.9 nm. The interaction between the proteins is calculated as a minimum distance between any atom pair on the respective proteins.
2.1.24. Statistical analysis
Data are expressed as Mean ± S.D. or Mean ± S.E.M. as indicated in Fig. legends. Differences between treatment groups were detected using Student’s t-test (two groups) or ANOVA (more than two groups, corrected for multiple comparisons) – or their non-parametric counterparts for non-normally distributed data. Differences are considered statistically significant when p<0.05. The log-rank test (Mantel-Cox) was used for analysis of overall animal survival that is defined as the time from the date of radiation to the 30 days post-radiation between drug vs. vehicle groups.
3. Results
3.1. Increased ferroptotic burden in necroptosis-deficient Rip3K51A/K51A mice following irradiation and traumatic brain injury
To investigate the interplay between necroptosis and ferroptosis, we utilized animal models of whole body γ-irradiation (γ-IR) and a controlled cortical impact (CCI) model of -TBI- that induce a time-dependent mix of regulated necrotic cell death [[6], [21], [39]]. Our previous work established that necroptosis inhibitor necrostatin-1 (Nec-1) is a potent radiomitigator and radioprotectant, able to improve survival of wild-type C57bl/6 mice exposed to γ-IR [20]. Since Nec-1 is also known to have anti-ferroptotic effects [40], we explored the possible contribution of ferroptosis in γ-IR-induced lethality. To this end we utilized kinase dead RIP3 (Rip3K51A/K51A) and RIP1 (Rip1K45A/K45A) mice. We found that Rip3K51A/K51A kinase-inactive knock-in animals (Fig. 1A) experienced greater γ-IR mortality versus Rip3+/+. No survival advantage was observed in Rip1K45A/K45A (Fig. S1A) compared to wild-type. Similarly, Castle et al. reported that Rip3−/− mice were not protected against IR-induced death vs wild type animals [41]. RIP3 is the master regulator of necroptosis, and its kinase activity is indispensable for necroptosis execution. With this in mind, we aimed to understand the paradoxical mechanism of RIP3 kinase inactivation on IR survival. The fast dividing epithelial cells of the small intestinal mucosa are highly γ-IR sensitive and their death affects the overall survival of animals [42]. Ileal villi from Rip3K51A/K51A contained more nuclear terminal deoxynucleotidyl transferase dUTP nick end labeled (TUNEL) cells (Fig. 1B–D) vs. wild-type tissue within 3 days of γ-IR. Notably, caspase activity was not increased at this timepoint (Fig. 1E). Given that cells dying via regulated necrosis can also display TUNEL positivity [43], we asked whether an alternative non-apoptotic death pathway mediated radio-sensitization in necroptosis-deficient mice. Therefore, we used high resolution liquid chromatography tandem mass spectrometry (LC-MS/MS) to quantify AA-PE and AdA-PE and their hydroperoxy-metabolites (PEox) – the most sensitive and specific markers of ferroptosis in vivo. Prior to γ-IR, levels of GPX4, 15LOX, PEBP1 (Fig. S1B), AA-PE, AdA-PE (Fig. S1C), or PUFA (linoleic acid [C18:2], linolenic acid [C18:3], arachidonic acid [C20:4], eicosapentaenoic acid [C20:5] adrenic acid [C22:4], docosapentaenoic acid [C22:5] and docosahexaenoic acid [C22:6]) (Fig. S1D) were not different in the ileum of WT and Rip3K51A/K51A animals. We observed a significant decrease in GPX4 expression following γ-IR (Fig. S1B) in both Rip3+/+ and Rip3K51A/K51A ileum, but there was no difference between the groups. Levels of 15LOX did not change significantly following post-γ-IR, but it tended to be higher in Rip3K51A/K51A vs Rip3+/+ on day 3 post-γ-IR. PEBP1 levels were higher in Rip3K51A/K51A vs Rip3+/+ on day 3 post-γ-IR. Levels of oxidized PE (PEox) including proferroptotic 15-HpETE-PE were significantly increased in Rip3K51A/K51A ileum (Fig. 1F–K, S1E-F) in comparison to γ-IR wild-type tissue on day 3 post-γ-IR.
Fig. 1.
Rip3K51A/K51Amice display worse survival and increased ileal PEox levels after ionizing whole body irradiation.Rip3K51A/K51A kinase inactivated mice died at a significantly higher rate (A) compared to Rip3+/+ control animals following γ-IR. Log-rank (Mantel-Cox) test, p < 0.05, n = 11–12 animals/group. Ileal mucosal injury was greater in (B) Rip3K51A/K51A vs. (C) Rip3+/+ on post-γ-IR (day 3), demonstrated by (D) an increased proportion of TUNEL+ nuclei. TUNEL (red), hoechst (blue). Mean ± SEM, *p < 0.05, n = 6–8 animals/per group, scale: 100 μm. (E) Caspase-3/7 activity in ileum was similar in Rip3K51A/K51A vs. Rip3+/+ on day 3 post-γ-IR. Relative luminescence normalized to total protein concentration, Mean ± SEM. (F) Representative full mass spectrum demonstrating the ion identities and relative abundances of phosphatidylethanolamine (PE) and plasmalogen-PE (PEp) species obtained from naïve Rip3K51A/K51A ileum. Peaks are labeled as PE(X:Y) and PEp(X:Y) where “X” indicates the number of acyl carbons (sn-1 & sn-2 positions), and “Y” represents the total number of unsaturated acyl chain bonds. Precursors of oxygenated PE species (ferroptotic cell death signals) are shown in red. (G) Extracted base-peak chromatograms of pro-ferroptotic hydroperoxy-eicosatetraenoyl-PE (m/z 798.5293, 15-HpETE-PE) obtained from ileum of Rip3+/+ (upper panel) and Rip3K51A/K51A at various times post-γ-IR. Extracted base-peak chromatogram of the internal standard (m/z 747.7143) for each sample is shown by dashed lines. (H–K) Levels of pro-ferroptotic PEox were greater in Rip3K51A/K51A vs. Rip3+/+ ileum on day 1, 3, and day 5 after γ-IR. Volcano plots: log2 (fold-change: Rip3K51A/K51A versus Rip3+/+) vs. -log10 (statistical p-value); red = increased levels, blue = decreased levels. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next utilized two anti-ferroptotic agents, Liproxstatin-1 (Lip-1) and a novel pharmacokinetically advanced homolog of ferrostatin-1 (Fer-1), Ferrostatin16-86 (Fer16-86), in vivo to determine the contribution of ferroptotic death in radiosensitization of Rip3K51A/K51A mice. Both Lip-1 (Fig. 2A) and Fer16-86 (Fig. S2B) protected Rip3K51A/K51A mice against lethal γ-IR. Lip-1 significantly attenuated γ-IR induced increases in pro-ferroptotic cell death signals PE(38:4)-OOH (Fig. 2B) and PE(40:4)-OOH (Fig. 2C) in Rip3K51A/K51A ileum (Fig. S2C). Collectively, these data demonstrate that (i) RIP3K51A expression enhances γ-IR mortality and (ii) increased synthesis of pro-ferroptotic PEox, mitigatable by Lip-1-.
Fig. 2.
Liproxstatin-1 rescues Rip3K51A/K51Abone marrow cells, enteroids and mice from ionizing radiation and attenuates pro-ferroptotic PEox generation. (A) Liproxstatin-1 (Lip-1) was radioprotective and reduced γ-IR mortality of Rip3K51A/K51A. Log-rank (Mantel-Cox) test, p < 0.05, n = 10–12 animals/group/experiment, data from 2 to 3 independent experiments/group. Lip-1 attenuated γ-IR induced increases in pro-ferroptotic cell death signals PE(38:4)-OOH (B), and PE(40:4)-OOH (C), n = 5 animals/group, Mean ± SEM, *p < 0.05 vs. 0 Gy, #p < 0.05 vs. γ-IR + vehicle. Shown are MS2 spectra of PE(38:4)-OOH (B, middle panel), and PE(40:4)-OOH (C, right panel). Fragments formed during MS2 fragmentation that are attributed -by the polar head of PE with m/z 140.066 and 196.066 along with others are shown and associated with the identification of hydroperoxy-PE species. MS3 spectra of molecular ion with m/z 317.21 corresponding to hydroperoxy-arachidonic acid with loss of water is shown in figure B, right panel. (D) Rip3K51A/K51A primary BM cells are more sensitive to γ-IR injury compared to Rip3+/+ wild-type cells. Rip3K51A/K51A BM cells benefited from anti-ferroptotic agents, Fer-1 or Lip-1. Nec-1s was only protective in wild type cells but not Rip3K51A/K51A, confirming the functional necroptosis knockout. Cell death measured by Annexin V-FITC + Propidium iodide (PI) flow cytometry, Mean ± SD, *p < 0.05 vs. same genotype γ-IR + DMSO group, †p < 0.05 vs. respective DMSO-only (no γ-IR), #p < 0.05 vs. wild-type γ-IR + DMSO group, n = 4–6 independent experiments. (E) Primary differentiated enteroid monolayers derived from Rip3K51A/K51A mice are significantly more sensitive to γ-IR-induced (5 Gy) death compared to Rip3+/+ wild-type cells. Rip3K51A/K51A enteroid mortality was reduced with anti-ferroptotic Lip-1. Cell death measured by LDH release at 22–24 h, Mean ± SEM, *p < 0.05 vs. 0 Gy group, #p < 0.05 vs. matched γ-IR + vehicle group, n = 3–5 independent experiments.
This enhancement of γ-IR-induced ferroptosis was also documented in in vitro using radiosensitive cells from bone marrow (BM) and ileal enteroids of Rip3+/+ and Rip3K51A/K51A mice. Like animals, Rip3K51A/K51A primary BM cells (Fig. 2-D) and enteroids (Fig. 2-E, S2D-F) were more sensitive to γ-IR injury compared to Rip3+/+ cells. Anti-ferroptotic Fer-1 and 1 Lip-1 provided radioprotection in Rip3K51A/K51A BM and enteroids. As expected, necrostatin-1s (Nec-1s), a specific RIP1 inhibitor, did not protect mutant necroptosis-deficient Rip3K51A/K51A BM cells (Fig. 2D).
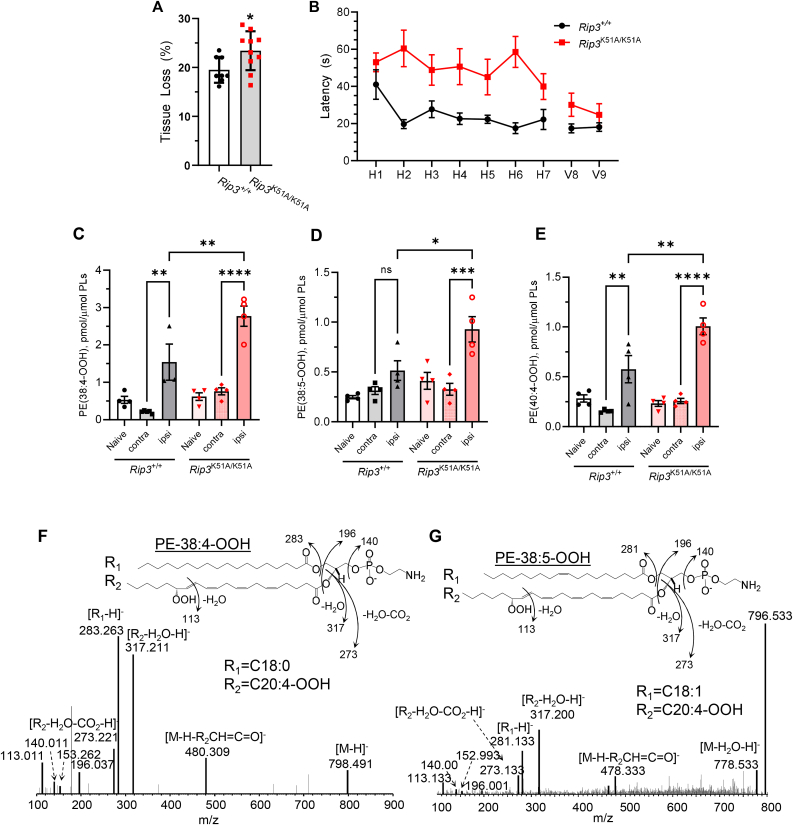
We further employed another in vivo model, controlled cortical impact (CCI), in which the role of ferroptosis in brain injury has been established by prior studies [44]. We found that Rip3K51A/K51A animals were more susceptible to CCI-induced ipsilateral cortical tissue loss (Fig. 3A) and demonstrated worse cognitive outcomes (Fig. 3B) despite the lack of differences in vestibulo-motor function (Figs. S3A–C) compared to Rip3+/+ mice. Notably, pro-ferroptotic PEox species PE(38:4)-OOH, PE(38:5)-OOH and PE(40:4)-OOH that contain hydroperoxy-arachidonic (15-HpETE) and hydroperoxy-adrenic (17-HpDTE) acids were significantly elevated following CCI in the ipsilateral cortex vs contralateral cortex or non-injured counterparts in both Rip3+/+ and Rip3K51A/K51A mice, their content in the injured Rip3K51A/K51A mice was significantly higher vs injured Rip3+/+ mice. (Fig. 3C–G, S3D).
Fig. 3.
Rip3K51A/K51Amice exhibit more tissue injury, worse functional deficits and increased PEox following CCI. (A) More ipsilateral cortical tissue loss occurs in Rip3K51A/K51A animals vs. Rip3+/+ following CCI. Data are Mean ± SEM, n = 8–10/group, *p < 0.05. (B) Neurological function was assessed after CCI in Rip3+/+ and Rip3K51A/K51A mice. Line graph showing the MWM swim latency to reach the hidden (H) platform on days 1–7 and visible (V) platform on day 8 and 9 following CCI. Data are Mean ± SEM, n = 10/group, *p < 0.05. LC/MS-based quantitative assessment of ferroptotic cell death signals PE(38:4)-OOH (C), PE(38:5)-OOH (D) and PE(40:4)-OOH (E) in cerebral cortical tissue of naïve and CCI exposed Rip3+/+ and Rip3K51A/K51A mice. Contra and ipsi denote the opposite side and the same side of the injured cerebral hemisphere, respectively, in the CCI mice. Shown are the MS2 spectra of PE(38:4)-OOH (F), and PE(38:5)-OOH (G). Fragments formed during MS2 fragmentation that are attributed to polar head of PE with m/z 140.0 and 196.0 along with others are shown and associated with identification of hydroperoxy-PE species. Data are Mean ± SEM, n = 5/group, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.2. PEBP1 forms complexes with RIP3
As we established that Rip3K51A/K51A mutant cells and animals had higher sensitivity to ferroptotic death, we explored the molecular mechanism(s), including the role of the K51A mutation. RIP kinase family members are structurally homologous to RAF kinases. However, RIP3 demonstrated the greatest degree of structural catalytic site homology with RAF (Fig. 4A). While RAF and RIP3 have expectedly similar Ser/Thr kinase domains, we noted that their similarities also included a conserved (i) ASP_GLY_PHE (DGF, Mg2+ binding site) motif, (ii) HIS_ARG_ASP_LEU motif (HRDL, ATP binding site), and (iii) a key arginine at their respective dimerization interfaces (RIP3K69, RAFR401) (Figs. S4A–C) [45]. Normally RAF kinase is strongly suppressed by a specialized inhibitory protein, RKIP1 or PEBP1 [46]. Therefore, we reasoned that PEBP1 can inhibit RIP3 kinase. Indeed, our molecular docking simulations showed the interaction of PEBP1 with RIP3 was energetically favorable (Fig. 4B). The top 10 lowest energy binding poses all oriented such that (i) PEBP1 docks proximal to RIP3’s catalytic site with key interfacial interactions involving RIP3’s R69 at the necroptosis-obligate homodimerization interface [47], and (ii) PEBP1 bound RIP3 via its heterodimerization loop region that includes residues 127–150 [31]. Full atomistic molecular dynamics (FAMD) simulations showed the formation of PEBP1 complexes within tens of nanoseconds (ns) with wild-type RAF (Fig. 4C), RIP3 (Fig. 4D) – but not with RIP1 (Fig. 4E) and RIP3K51A mutant (Fig. 4F). The specific RIP3 kinase-inactivating K51A mutation disrupts salt-bridges between E61 on αHC helix and D161 on the conserved DFG motif (at the catalytic site). This lack of complex formation in FAMD between PEBP1 and RIP3K51A suggests that these salt-bridges underlie the RIP3→PEBP1 interaction interface. Notably, a specific RIP3 inhibitor, GSK’872, had a similar salt-bridge disrupting effect (Fig. S4D).
Fig. 4.
PEBP1 associates with RIP3. (A) Structures of RAF (pdbid: 3c4c) and RIP3 (pdbid: 4m66) are highly conserved in sequence and tertiary structure. Catalytic regions shown enclosed in dashed region. (B) Lowest energy binding pose of PEBP1 (cyan) on RIP3 (grey) from docking simulations. Key interfacial interactions between PEBP1’s heterodimerization loop region (residues 127–150, including D144-H145) and RIP3’s α-helix C (αC), specifically R69 (spacefilling model) are highlighted. (C–F) Full Atomistic Molecular Dynamics simulations between PEBP1 with RAF, RIP3, RIP1, or RIP3K51A. Grey dotted line indicates the threshold maximum distance positive protein-protein interaction (≤0.5 nm). Favorable interaction is predicted between (C) PEBP1 and RAF or (D) RIP3WT, but not (E) RIP1 or (F) RIP3K51A mutant. N = 3–4 independent simulations. (G–H) Colocalization between RIP3 and PEBP1 is reduced in Rip3K51A/K51A vs. Rip3+/+ primary bone marrow cells. (G) PEBP1_RIP3 colocalization normalized to cell number, Mean ± SD, *p < 0.05. (H): “Merge” [left panels]: PEBP1 (red), RIP3 (green), and hoechst (blue). “Colocalized objects” [right panels]: PEBP1_RIP3 colocalized objects (yellow), n = 3 independent experiments, scale: 20 μm. (I-J) FRET analysis showing close physical proximity (≤10 nm) of RIP3 with PEBP1 in HT22 cells. (I) FRET effect was confirmed through acceptor (cy5) photobleaching (white arrows) and reciprocal Cy3 donor fluorophore unquenching. FRET ratio (donor/acceptor relative fluorescent intensity (RFU)) is pseudo-colored (range 0–10, violet-red); (J) Representative RFU vs. single excitation (Ex) wavelength. Cy3 vs. Cy5 emission (Em) wavelengths are indicated. N = 3 independent experiments. (K) Far western blotting demonstrating specific interaction of recombinant human PEBP1 with RIP3. Representative non-denaturing immunoblots showing PEBP1 (0.5 μg protein per incubation) binds to membrane-immobilized RIP3 (left) but not GST or BSA control proteins. No PEBP1 signal is detected if the blot is not incubated with recombinant PEBP1 (middle). Protein loading was verified by Ponceau S prior to incubation with PEBP1 (right). N = 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To experimentally test these computational predictions, we used three biochemical and cell-based methods to evince PEBP1’s direct interaction with RIP3: (1) object-based colocalization analysis, (2) fluorescent resonance energy transfer (FRET), (3) far western blotting. We used a 3D (voxel) object based colocalization method, which preserves Z-plane spatial and fluorescent intensity information, unlike traditional 2-dimensional Pearson correlation strategies [6]. We found that RIP3 and PEBP1 colocalized in Rip3+/+ primary BM cells (Fig. 4G and H), but RIP3K51A_PEBP1 colocalization in Rip3K51A/K51A cells was significantly reduced vs. wild type. We further applied FRET techniques to assess whether RIP3_PEBP1 colocalization signified close proximity. Because the efficiency of FRET decreases as the sixth power of the distance between the donor and acceptor fluorophores, labeled proteins separated by > 10 nm will not produce a FRET response. We detected a FRET response between RIP3 and PEBP1 (Fig. 4I and J), corroborating our colocalization finding. While FRET can define an upper distance limit (10 nm), we wished to determine whether PEBP1 and RIP3 are in immediate physical proximity with each other. To address this, we used a far western blotting method – in which an antibody-detectable “bait” protein (PEBP1) is used to detect the membrane-immobilized target “prey” protein (RIP3). We noted that PEBP1 was bound to and detected with RIP3 (Fig. 4K) but not with GST or BSA (as potential non-specific partners).
3.3. PEBP1 negatively regulates necroptosis
To define the functional role of the PEBP1_RIP3 interaction, we modulated PEBP1 content and measured resultant RIP3 activity. An equimolar amount of PEBP1 substantially reduced RIP3’s kinase activity but not that of RIP1 or MAPK14 (p38α) (Fig. 5A). Compared to non-targeted (NT) siRNA-treated control L929 cells, PEBP1 knockdown by siRNA enhanced [TNFα + z-VAD-fmk + SM164]-induced activation and phosphorylation of RIP3 (pRIP3, murine S231/232) and MLKL (pMLKL, murine S345) at 4 h (1.8- and 1.5-fold, respectively, Figs. S5A) and 12 h (4.7- and 1.8-fold, respectively, Fig. 5B). Levels of pRIP1 (S166) were unchanged by PEBP1 siRNA. Matching this, necroptotic cell death was enhanced in PEBP1KD cells, obtained by either siRNA (Fig. 5C) or CRISPR methods (Fig. 5D, Fig. S5B). We further tested PEBP1’s necroptotic role in TNFα gradient experiments. Increasing TNFα dose produced marginally greater death in control NT siRNA L929 cells. In contrast, cell death in PEBP1KD was increased with increasing TNFα concentrations. In both cases, Nec-1s was completely protective.
Fig. 5.
PEBP1 regulates necroptotic death. (A) Human recombinant RIP3 kinase activity is specifically and potently inhibited by equimolar PEBP1 or the small molecule inhibitor, GSK’872. Human recombinant RIP1 kinase and another canonical serine/threonine kinase, MAPK14 (p38α), are not inhibited by equimolar PEBP1. PEBP1 alone has no effect on ATP concentration. Mean ± SD, *p < 0.05 vs. uninhibited enzyme, n = 3 independent experiments. (B) Representative immunoblots showing reduced PEBP1 expression is associated with greater pRIP3 (pS231/pT232) and pMLKL (pS345) levels 12 h following TNFα, z-VAD-fmk, and SM-164 (T/Z/S) necroptosis induction. L929 cells were transfected with PEBP1 or non-targeted (NT) siRNA for 48 h prior to T/Z/S treatment. Representative of 3 independent experiments. (C) PEBP1 siRNA knockdown L929 cells experienced a greater increase in necroptosis at higher TNFα (1, 5, 10 ng/mL) doses compared to NT siRNA cells. Cell death was specifically rescued by Nec-1s (see Fig. S5A). Z/S: z-VAD-fmk + SM164; cell death measured by LDH release at 16–20 h, Mean ± SD, *p < 0.05 vs. NT siRNA, n = 3 independent experiments. (D) PEBP1 knockdown CRISPR sensitizes L929 cells to necroptotic death. T/Z: TNFα (10 ng/mL) + z-VAD-fmk. Cell death measured by LDH release at 18 h, Mean ± SD, *p < 0.05 vs. non-targeted (NT) CRISPR, n = 3 independent experiments.
3.4. Increased PEBP1 availability sensitizes to ferroptosis whereas formation of PEBP1-RIP3 kinase complex suppresses necroptosis
Given the duality of PEBP1 interactions with RIP3 and 15LOX, we studied the role of PEBP1 in necroptosis-ferroptosis crosstalk. We assumed that conditions influencing PEBP1 to alternatively bind/inhibit RIP3 or bind/activate 15LOX may serve as a rheostat between necroptosis and ferroptosis. Weakening of PEBP1’s interaction with alternative partners and increased expression of 15LOX would enhance the 15LOX’s capacity to bind PEBP1 resulting in pro-ferroptotic response. Modeling and experimental data indicated that PEBP1 does not associate with K51A mutant RIP3 to the same extent as with wild type. Primary BM expressing the weak PEBP1 partner, RIP3K51A mutant, were highly sensitive to RSL3-induced death while wild type BM were resistant (Fig. 6A). Increased PEBP1_15LOX co-localization preceded death (Fig. S6A). This cell death was rescued by anti-ferroptotic Lip-1 or Fer-1, whereas other programmed death inhibitors – z-VAD-fmk, Nec-1s, or Bafilomycin-A1 (BafA1, anti-autophagic death) – did not prevent Rip3K51A/K51A-associated sensitivity to RSL3 (Fig. 6B). Furthermore, γ-IR promoted formation of the pro-ferroptotic PEBP1-15LOX interactions in Rip3K51A/K51A ileum demonstrated through immunocytochemistry and three-dimensional object-based co-localization analysis (Figs. S6C–D).
Fig. 6.
RIP3K51Amutant and inhibitor GSK’872 promote IR and RSL3 induced ferroptosis. (A) Rip3K51A/K51A (red bars) primary BM cells were dose-dependently sensitive to the GPX4 inhibitor and ferroptosis-inducer, RSL3 (dose range: 0–500 nM). Mean ± SD, *p < 0.05 vs. respective genotype DMSO control (0 nM RSL3) group, n = 4 independent experiments. (B) Ferrostatin-1 (Fer-1, 1 μM), but not z-VAD-fmk, Nec-1s, or BafA1 reverse Rip3K51A/K51A BM cell death upon exposure to RSL3. Cell death measured by Annexin V-FITC + Propidium iodide (PI) flow cytometry, Mean ± SD, *p < 0.05 vs. respective genotype RSL3 group, n = 4 independent experiments. (C) GSK’872 synergistically enhances RSL3-induced ferroptosis in HT22 cells. Toxicity was only observed with the GSK’872 + RSL3 cotreatment, not individually. Synergistic injury was rescued completely by Fer-1. Cell death measured by LDH release, Mean ± SD, *p < 0.05 vs. vehicle, n = 3 independent experiments. (D) RIP3 inhibitor, GSK’872, synergistically enhances RSL3-induced ferroptotic cell death in wild type primary mouse small intestinal enteroid monolayers. Compared to RSL3 or GSK’872 monotreatment, cotreatment significantly increased enteroid death. The synergistic injury was rescued by anti-ferroptotic Lip-1. Cell death measured by LDH release at 22–24 h, Mean ± SD, *p < 0.05 vs. RSL3 only group, n = 3 independent experiments. (E) GSK’872 sensitized immortalized wild-type rat intestinal epithelial cells (IEC18) to ferroptosis. Lip-1 reversed the GSK’872 + RSL3 cotreatment effect. Cell death measured by LDH release at 18–20 h, Mean ± SD, *p < 0.05 vs. RSL3 only group, n = 3 independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To verify that ferroptosis sensitivity is modulated by 15LOX’s access to PEBP1, we used pharmacologic means to liberate PEBP1 from RIP3 or RAF. We hypothesized that, similar to Rip3K51A/K51A mutant cells, increased PEBP1 availability for 15LOX binding would prime cells for ferroptotic signaling. Previous modeling work indicated that GSK’872 disrupts RIP3’s catalytically important salt bridges, giving rise to its inhibitory activity. However, binding of GSK’872 causes an out-rotation of wild type RIP3’s R69-proximate αHelix-C (αHC) (Fig. S4D), which is also seen with the RIP3K51A mutant. Indeed, non-toxic GSK’872 concentrations rapidly reduced RIP3-PEBP1 colocalization (Figs. S7A–B). This effect, combined with low dose individually non-toxic RSL3 cotreatment, lead to increased 15LOX_PEBP1 complex formation (Figs. S7C–D) and a synergistic increase in ferroptosis (Fig. 6C). This GSK’872 ferroptosis-sensitizing effect was observed in several models, including wild type primary enteroid monolayer cells (Fig. 6D) and rat small IEC18 intestinal epithelial cells (Fig. 6E).
Further, we used FAMD simulations to model the effects of several RAF inhibitors on the PEBP1_RAF interaction. Our MD simulations revealed that sorafenib and, to a lesser extent, PLX4720 disrupted the RAF_PEBP1 interaction (Fig. 7A–C). These predicted dissociating effects of sorafenib were in line with the experimentally observed reduced PEBP1_RAF colocalization (Figs. S7E–F). Cotreatment with sorafenib and RSL3 prompted increased PEBP1_15LOX colocalization (Figs. S7G–H), lipid peroxidation (BODIPY-C11, Fig. 7D), and synergistically enhanced ferroptotic cell death (Fig. 7E). Given the reported ability of sorafenib to suppress the cystine/glutamate system Xc− [48], we determined GSH concentrations and found that low concentrations of sorafenib used did not inhibit GSH synthesis (Fig. 7F). PLX4720 also synergistically enhanced RSL3-triggered death, but to a lesser degree, in line with our FAMD predictions. The computationally predicted ineffectiveness of another RAF inhibitor, SB590885, as a disruptor of the RAF_PEBP1 interaction, corresponded with its experimentally documented inability to enhance ferroptosis (Fig. 7E).
Fig. 7.
Disruption of RAF_PEBP1 complexes interaction promotes 15LOX_PEBP1 mediated ferroptosis. (A-C) FAMD simulations between PEBP1 and [inhibitor]-bound RAF. Minimum distance between any atom on PEBP1 and RAF. The dotted line indicates the threshold maximum distance for positive protein-protein interaction (≤0.5 nm). In the presence of RAF inhibitors sorafenib (C) and to a lesser extent PLX4720 (A), PEBP1 does not associate with RAF. This frees PEBP1 for alternative protein interactions (e.g., 15LOX). (B) With SB590885, PEBP1 remains bound to RAF, suggesting the inhibitor does not dissociate the PEBP1_RAF complex. Representative of 2–3 independent simulations. (D) Individually, sorafenib or low dose RSL3 had no effect on the redox state of the reporter lipid (BODIPY-C11). Cotreatment of RSL3 and sorafenib increased lipid peroxidation in HEK 293T cells. Mean ± SD, *p < 0.05, n = 3 independent experiments. (E) Sorafenib and, to a lesser extent, PLX4720 synergistically enhance RSL3-induced death in HT22 cells. Sorafenib’s effects were independent of system Xc− inhibition (see Fig. S7H). Synergistic death was rescued completely by Fer-1. Cell death measured by LDH release, Mean ± SD, *p < 0.05 vs. vehicle, n = 3 independent experiments. (F) Sorafenib does not inhibit system Xc− (SLC7A11) mediated GSH synthesis at the dose used to sensitize cells to ferroptosis. Monotreatment with erastin (10 μM), a specific system Xc− inhibitor, reduced GSH levels. Mean ± SD, *p < 0.05, n = 3 independent experiments.
4. Discussion
Fatally injured cells die by one of several tightly controlled pathways. The mechanisms of programmed cell death touch closely on one-another; up- or downregulation of one death pathway affects the execution of others. We demonstrated that RIP3 kinase can interact with the promiscuous endogenous serine-/threonine-kinase inhibitor, PEBP1. The nature of the PEBP1_RIP3 interaction is likely analogous to that of PEBP1_RAF kinase. PEBP1 binding inhibits RIP3’s essential kinase function, thus regulating the necroptotic program. Our previous work established that 15LOX is yet another partner of PEBP1 [6,18]. Binding of PEBP1 with 15LOX alters the dioxygenase catalytic selectivity, leading to the generation of pro-ferroptotic 15-HpETE-PE signals [7]. Computational assessment of the relative affinities of PEBP1 for RIP3 versus 15LOX (Figs. S8A–C), indicated that 15LOX is the preferred PEBP1 partner. A 3:1 ratio of RIP3:15LOX was required for RIP3 to outcompete 15LOX for PEBP1 in our simulations. This suggests the PEBP1_RIP3 interaction may be transient and subordinate to the formation of the ferroptotic 15LOX_PEBP1 complex. Enhancing 15LOX’s access to PEBP1 by genetically or pharmacologically disrupting alternative interactions of PEBP1 (i.e., with RAF, RIP3) enhances this pro-ferroptotic effect. Indeed, we observed that pharmacologically disrupted RIP3_PEBP1 or RAF1_PEBP1 interactions with GSK’872 or sorafenib/PLX4720, respectively, sensitized cells to secondary RSL3-induced ferroptosis.
Cells closely regulate the initiation and propagation of cell death programs. Our work shows that binding of PEBP1 with RIP3 results in kinase inhibition and may represent yet another high-fidelity regulatory mechanism to prevent unintentional necroptosis [49]. However, under disease conditions leading to GPX4/GSH deficiency, irreparably damaged cells destined to die can activate ferroptosis by complex formation between PEBP1 and 15LOX that results in the formation of PEox. The switch to ferroptosis does not come without cost. Ferroptotic signaling results in a paracrine-like spreading death effect [50,51]. These effects offer an explanation of why the switch to ferroptosis results in enhanced γ-IR mortality and worse neurological function after CCI. Necroptosis and ferroptosis act in parallel under several common and complex disease pathologies, for example in various forms of acute kidney injury (AKI). Folic acid (FA)-induced AKI is notably pro-ferroptotic [52]. Likewise, the weakened interaction between RIP3K51A and PEBP1 is permissive to PEBP1_15LOX complex formation.
Mandal et al. demonstrated that several mutations in RIP3’s kinase domain combined with high-concentrations of kinase inhibitors (e.g., GSK’872) caused RIP1-dependent apoptosis rescued by caspase inhibitor z-VAD [[53], [54], [55]] The RIP3 inhibitor activated caspase 8(Casp8) via RIP homotypic interaction motif-driven recruitment of RIPK1 to yield a Casp8-FADD-cFLIP complex and was independent of the pro-necrotic kinase activities. Among several RIP3 mutants, a kinase-dead D161N mutant induced spontaneous apoptosis independently of the inhibitors, whereas D161G, D143N, and K51A mutants, like wild-type, induced apoptosis only in the presence of the inhibitors [54,55]. Furthermore, Rip3K51A/K51A mutants were viable and fertile, in contrast to the Casp8-dependent perinatal lethality of Rip3D161N/D161N mice. Our work shows worse outcomes in Rip3K51A/K51A mice following pathological insults of γ-IR and CCI. We also found that enhanced mortality after γ-IR in K51A animals was attenuated by antiferroptotic compounds Lip-1 and Fer 16–86 (Figs. 2A and S2A) but not by z-VAD-fmk (Fig. S8D), suggesting that ferroptosis was largely responsible for the mortality. Combined, these data show that therapeutic targeting of RIP3 inhibition for radioprotection or neuroprotection is challenging. Although we did not explore in the current work future studies could reveal a potential therapeutic role for RIP3 kinase inhibitors in radiosensitization for cancer treatment.
There may also be alternative ferroptosis-necroptosis connections outside the direct PEBP1_RIP3 mechanism. A number of proteins have been shown to associate with the activated RIP3-immunocomplex, including the glutamine transporter, SLC1A5 [56]. Activation of SLC1A5 and glutamine (Gln) transporters promotes ferroptosis by upregulation of glutaminolysis [57]. Altering RIP3’s interaction with these Gln transporters by pharmacologic or genetic means may similarly affect ferroptosis. Overall, simultaneous inhibition of both necroptosis and ferroptosis may be necessary to maximize protection and likelihood of clinical benefit in disease states where either programmed necrotic pathway is suspected to operate.
In summary, we demonstrated that RIP3 K51A kinase inactivation enhances ferroptotic death resulting in worse outcome after γ-IR and TBI. Consequently, anti-ferroptotic compounds Liproxstatin-1 and Ferrostatin 16-86 afford protection. PEBP1 serves as a regulator of cell death by inhibiting pro-necroptotic RIP3 activity while activating 15LOX to generate pro-ferroptotic HpETE-PE signals. These newly established regulatory functions of PEBP1 may serve multiple and diverse roles across various human disease states.
Author contributions
HB, MJW and VEK designed/planned the study and wrote the paper; AML, ZH, GW, HD, GS, and QY performed experiments with different cell types and enteroids, analyzed data; MWE, LW, JSG, and MJW performed whole body irradiation experiments and analyzed data; ZH and MJW performed controlled cortical impact experiments and analyzed data; YYT, TSA, AML, AAA, and ZEH performed different MS measurements and analyzed data; AML, CSC and SW performed imaging analysis; AML, ZH, JCR, AAK, HLM, QY, GVS, and APV performed in vitro biochemical experiments; IHS performed computational modeling; IB supervised computational studies and participated in paper writing; KPM, PJG and JB participated in the discussion of experiments and paper writing.
Declaration of competing interest
The authors have declared that no conflict of interest exists.
Acknowledgements
This work was supported by NIH funding (U19AI068021, U01AI156924, U01AI156923, R01NS061817, R01NS076511, F30HL142130, R01GM139297, P41GM103712). The authors would like to thank Mark Ross and Morgan Jessup for their microscopy technical assistance, Renee Fisher for technical assistance in irradiation experiments, Karolina Mikulska-Ruminska for simulation modelling assistance, - Valentyna Kapralova for her technical assistance in the tissue total lipid extraction, and Carla Johnson for assistance with immnoblot.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102232.
Contributor Information
Valerian E. Kagan, Email: kagan@pitt.edu.
Michael J. Whalen, Email: mwhalen@mgh.harvard.edu.
Hülya Bayır, Email: bayihx@upmc.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Conrad M., Angeli J.P.F., Vandenabeele P., Stockwell B.R. Regulated necrosis: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2016;15:348. doi: 10.1038/nrd.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez J. Prix fixe: efferocytosis as a four-course meal. Curr. Top. Microbiol. Immunol. 2017;403:1–36. doi: 10.1007/82_2015_467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berghe T.V., Linkermann A., Jouan-Lanhouet S., Walczak H., Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 4.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., Stockwell B.R. Ferroptosis: an iron-dependent form of non-apoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angeli J.P.F., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Rådmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Förster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O’Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel S.E., Tyurina Y.Y., Zhao J., St Croix C.M., Dar H.H., Mao G., Tyurin V.A., Anthonymuthu T.S., Kapralov A.A., Amoscato A.A., Mikulska-Ruminska K., Shrivastava I.H., Kenny E.M., Yang Q., Rosenbaum J.C., Sparvero L.J., Emlet D.R., Wen X., Minami Y., Qu F., Watkins S.C., Holman T.R., VanDemark A.P., Kellum J.A., Bahar I., Bayir H., Kagan V.E. PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals. Cell. 2017;171(3):628–641. doi: 10.1016/j.cell.2017.09.044. e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S., Dar H.H., Liu B., Tyurin V.A., Ritov V.B., Kapralov A.A., Amoscato A.A., Jiang J., Anthonymuthu T., Mohammadyani D., Yang Q., Proneth B., Klein-Seetharaman J., Watkins S., Bahar I., Greenberger J., Mallampalli R.K., Stockwell B.R., Tyurina Y.Y., Conrad M., Bayir H. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13(1):81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A., Prokisch H., Trumbach D., Mao G., Qu F., Bayir H., Fullekrug J., Scheel C.H., Wurst W., Schick J.A., Kagan V.E., Angeli J.P., Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13(1):91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou W., Yuan J. Necroptosis in health and diseases. Semin. Cell Dev. Biol. 2014;35:14–23. doi: 10.1016/j.semcdb.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 10.Grootjans S., Vanden Berghe T., Vandenabeele P. Initiation and execution mechanisms of necroptosis: an overview. Cell Death Differ. 2017;24(7):1184–1195. doi: 10.1038/cdd.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasparakis M., Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517(7534):311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Liu T., Lei T., Zhang D., Du S., Girani L., Qi D., Lin C., Tong R., Wang Y. RIP1/RIP3-regulated necroptosis as a target for multifaceted disease therapy (Review) Int. J. Mol. Med. 2019;44(3):771–786. doi: 10.3892/ijmm.2019.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsch M., Günther S.D., Schwarzer R., Albert M.C., Schorn F., Werthenbach J.P., Schiffmann L.M., Stair N., Stocks H., Seeger J.M., Lamkanfi M., Krönke M., Pasparakis M., Kashkar H. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575(7784):683–687. doi: 10.1038/s41586-019-1770-6. [DOI] [PubMed] [Google Scholar]
- 14.Mandal P., Berger S.B., Pillay S., Moriwaki K., Huang C., Guo H., Lich J.D., Finger J., Kasparcova V., Votta B., Ouellette M., King B.W., Wisnoski D., Lakdawala A.S., DeMartino M.P., Casillas L.N., Haile P.A., Sehon C.A., Marquis R.W., Upton J., Daley-Bauer L.P., Roback L., Ramia N., Dovey C.M., Carette J.E., Chan F.K., Bertin J., Gough P.J., Mocarski E.S., Kaiser W.J. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell. 2014;56(4):481–495. doi: 10.1016/j.molcel.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiss K., Kisker C., Lohse M.J., Lorenz K. Raf kinase inhibitor protein (RKIP) dimer formation controls its target switch from Raf1 to G protein-coupled receptor kinase (GRK) 2. J. Biol. Chem. 2012;287(28):23407–23417. doi: 10.1074/jbc.M112.363812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung K.C., Rose D.W., Dhillon A.S., Yaros D., Gustafsson M., Chatterjee D., McFerran B., Wyche J., Kolch W., Sedivy J.M. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol. cell. Biol. 2001;21(21):7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbit K.C., Trakul N., Eves E.M., Diaz B., Marshall M., Rosner M.R. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J. Biol. Chem. 2003;278(15):13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 18.Anthonymuthu T.S., Kenny E.M., Shrivastava I., Tyurina Y.Y., Hier Z.E., Ting H.C., Dar H.H., Tyurin V.A., Nesterova A., Amoscato A.A., Mikulska-Ruminska K., Rosenbaum J.C., Mao G., Zhao J., Conrad M., Kellum J.A., Wenzel S.E., VanDemark A.P., Bahar I., Kagan V.E., Bayir H. Empowerment of 15-lipoxygenase catalytic competence in selective oxidation of membrane ETE-PE to ferroptotic death signals, HpETE-PE. J. Am. Chem. Soc. 2018;140(51):17835–17839. doi: 10.1021/jacs.8b09913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayaan E., Moser A., MacKerell A.D., Jr., York D.M. CHARMM force field parameters for simulation of reactive intermediates in native and thio-substituted ribozymes. J. Comput. Chem. 2007;28(2):495–507. doi: 10.1002/jcc.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z., Epperly M., Watkins S.C., Greenberger J.S., Kagan V.E., Bayır H. Necrostatin-1 rescues mice from lethal irradiation. Biochim. Biophys. Acta. 2016;1862(4):850–856. doi: 10.1016/j.bbadis.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bermpohl D., You Z., Lo E.H., Kim H.-H., Whalen M.J. TNF alpha and fas mediate tissue damage and functional outcome after traumatic brain injury in mice. J. Cerebr. Blood Flow Metabol. 2007;27(11):1806–1818. doi: 10.1038/sj.jcbfm.9600487. [DOI] [PubMed] [Google Scholar]
- 22.Sîrbulescu R.F., Chung J.Y., Edmiston W.J., Poznansky S.A., Poznansky M.C., Whalen M.J. Intraparenchymal application of mature B lymphocytes improves structural and functional outcome after contusion traumatic brain injury. J. Neurotrauma. 2019;36(17):2579–2589. doi: 10.1089/neu.2018.6368. [DOI] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., Sloane Stanley G. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 24.Fauland A., Trötzmüller M., Eberl A., Afiuni-Zadeh S., Köfeler H., Guo X., Lankmayr E. An improved SPE method for fractionation and identification of phospholipids. J. Separ. Sci. 2013;36(4):744–751. doi: 10.1002/jssc.201200708. [DOI] [PubMed] [Google Scholar]
- 25.Kozuka K., He Y., Koo-McCoy S., Kumaraswamy P., Nie B., Shaw K., Chan P., Leadbetter M., He L., Lewis J.G., Zhong Z., Charmot D., Balaa M., King A.J., Caldwell J.S., Siegel M. Development and characterization of a human and mouse intestinal epithelial cell monolayer platform. Stem Cell Rep. 2017;9(6):1976–1990. doi: 10.1016/j.stemcr.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham K.E., Novak E.A., Vincent G., Siow V.S., Griffith B.D., Ranganathan S., Rosengart M.R., Piganelli J.D., Mollen K.P. Calcium/calmodulin-dependent protein kinase IV (CaMKIV) activation contributes to the pathogenesis of experimental colitis via inhibition of intestinal epithelial cell proliferation. Faseb. J. : Off. Publ. Fed. Am. Soc. Exp. Biol. 2019;33(1):1330–1346. doi: 10.1096/fj.201800535R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy, American journal of physiology. Cell physiol. 2011;300(4):C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biggs D.S. 2010. 3D Deconvolution Microscopy, Current Protocols in Cytometry; pp. 1–20. Chapter 12. Unit 12.19. [DOI] [PubMed] [Google Scholar]
- 29.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie T., Peng W., Yan C., Wu J., Gong X., Shi Y. Structural insights into RIP3-mediated necroptotic signaling. Cell Rep. 2013;5(1):70–78. doi: 10.1016/j.celrep.2013.08.044. [DOI] [PubMed] [Google Scholar]
- 31.Banfield M.J., Barker J.J., Perry A.C., Brady R.L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure (London, England. 1998;6(10):1245–1254. doi: 10.1016/s0969-2126(98)00125-7. 1993. [DOI] [PubMed] [Google Scholar]
- 32.Xie P., Streu C., Qin J., Bregman H., Pagano N., Meggers E., Marmorstein R. The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase. Biochemistry. 2009;48(23):5187–5198. doi: 10.1021/bi802067u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie T., Peng W., Liu Y., Yan C., Maki J., Degterev A., Yuan J., Shi Y. Structural basis of RIP1 inhibition by necrostatins. Structure (London, England : 1993) 2013;21(3):493–499. doi: 10.1016/j.str.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 37.Kobe M.J., Neau D.B., Mitchell C.E., Bartlett S.G., Newcomer M.E. The structure of human 15-lipoxygenase-2 with a substrate mimic. J. Biol. Chem. 2014;289(12):8562–8569. doi: 10.1074/jbc.M113.543777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 39.Tyurina Y.Y., Tyurin V.A., Epperly M.W., Greenberger J.S., Kagan V.E. Oxidative lipidomics of gamma-irradiation-induced intestinal injury. Free Radic. Biol. Med. 2008;44(3):299–314. doi: 10.1016/j.freeradbiomed.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Radmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Forster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O'Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castle K.D., Daniel A.R., Moding E.J., Luo L., Lee C.L., Kirsch D.G. Mice lacking RIP3 kinase are not protected from acute radiation syndrome. Radiat. Res. 2018;189(6):627–633. doi: 10.1667/RR15001.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somosy Z., Horvath G., Telbisz A., Rez G., Palfia Z. Morphological aspects of ionizing radiation response of small intestine. Micron. 2002;33(2):167–178. doi: 10.1016/s0968-4328(01)00013-0. Oxford, England. 1993. [DOI] [PubMed] [Google Scholar]
- 43.Linkermann A., Chen G., Dong G., Kunzendorf U., Krautwald S., Dong Z. Regulated cell death in AKI. J. Am. Soc. Nephrol. 2014;25(12):2689–2701. doi: 10.1681/ASN.2014030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenny E.M., Fidan E., Yang Q., Anthonymuthu T.S., New L.A., Meyer E.A., Wang H., Kochanek P.M., Dixon C.E., Kagan V.E., Bayir H. Ferroptosis contributes to neuronal death and functional outcome after traumatic brain injury. Crit. Care Med. 2019;47(3):410–418. doi: 10.1097/CCM.0000000000003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavoie H., Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 2015;16:281. doi: 10.1038/nrm3979. [DOI] [PubMed] [Google Scholar]
- 46.Hagan S., Garcia R., Dhillon A., Kolch W. Raf kinase inhibitor protein regulation of raf and MAPK signaling. Methods Enzymol. 2006;407:248–259. doi: 10.1016/S0076-6879(05)07021-7. [DOI] [PubMed] [Google Scholar]
- 47.Raju S., Whalen D.M., Mengistu M., Swanson C., Quinn J.G., Taylor S.S., Webster J.D., Newton K., Shaw A.S. Kinase domain dimerization drives RIPK3-dependent necroptosis. Sci. Signal. 2018;11(544):eaar2188. doi: 10.1126/scisignal.aar2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon S.J., Patel D.N., Welsch M., Skouta R., Lee E.D., Hayano M., Thomas A.G., Gleason C.E., Tatonetti N.P., Slusher B.S., Stockwell B.R. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3 doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kearney C.J., Cullen S.P., Clancy D., Martin S.J. RIPK1 can function as an inhibitor rather than an initiator of RIPK3-dependent necroptosis. FEBS J. 2014;281(21):4921–4934. doi: 10.1111/febs.13034. [DOI] [PubMed] [Google Scholar]
- 50.Kim S.E., Zhang L., Ma K., Riegman M., Chen F., Ingold I., Conrad M., Turker M.Z., Gao M., Jiang X., Monette S., Pauliah M., Gonen M., Zanzonico P., Quinn T., Wiesner U., Bradbury M.S., Overholtzer M. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 2016;11(11):977–985. doi: 10.1038/nnano.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linkermann A., Skouta R., Himmerkus N., Mulay S.R., Dewitz C., De Zen F., Prokai A., Zuchtriegel G., Krombach F., Welz P.S., Weinlich R., Vanden Berghe T., Vandenabeele P., Pasparakis M., Bleich M., Weinberg J.M., Reichel C.A., Brasen J.H., Kunzendorf U., Anders H.J., Stockwell B.R., Green D.R., Krautwald S. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2014;111(47):16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller T., Dewitz C., Schmitz J., Schröder A.S., Bräsen J.H., Stockwell B.R., Murphy J.M., Kunzendorf U., Krautwald S. Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell. Mol. Life Sci. 2017;74(19):3631–3645. doi: 10.1007/s00018-017-2547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandal P., Berger S.B., Pillay S., Moriwaki K., Huang C., Guo H., Lich J.D., Finger J., Kasparcova V., Votta B., Ouellette M., King B.W., Wisnoski D., Lakdawala A.S., DeMartino M.P., Casillas L.N., Haile P.A., Sehon C.A., Marquis R.W., Upton J., Daley-Bauer L.P., Roback L., Ramia N., Dovey C.M., Carette J.E., Chan F.K.-M., Bertin J., Gough P.J., Mocarski E.S., Kaiser W.J. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell. 2014;56(4):481–495. doi: 10.1016/j.molcel.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pazdernik N.J., Donner D.B., Goebl M.G., Harrington M.A. Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-κB. Mol. cell. Biol. 1999;19(10):6500–6508. doi: 10.1128/mcb.19.10.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton K., Dugger D.L., Wickliffe K.E., Kapoor N., de Almagro M.C., Vucic D., Komuves L., Ferrando R.E., French D.M., Webster J., Roose-Girma M., Warming S., Dixit V.M. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science (New York, N.Y.) 2014;343(6177):1357. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 56.Fettweis G., Di Valentin E., L'Homme L., Lassence C., Dequiedt F., Fillet M., Coupienne I., Piette J. RIP3 antagonizes a TSC2-mediated pro-survival pathway in glioblastoma cell death. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864(1):113–124. doi: 10.1016/j.bbamcr.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 57.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59(2):298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.