Summary
In the pathogenesis of autoimmune disorders, the modulation of leukocytes′ trafficking plays a central role, still poorly understood. Here, we focused on the effect of TLR2 ligands in trafficking of T helper cells through reshuffling of CD44 isoforms repertoire. Concurrently, strain background and TLR2 haplotype affected Wnt/β-catenin signaling pathway and expression of splicing factors. During EAE, mCD44v9-v10 was specifically enriched in the forebrain and showed an increased ability to bind stably to osteopontin. Similarly, we observed that hCD44v7 was highly enriched in cells of cerebrospinal fluid from MS patients with active lesions. Moreover, TLRs engagement modulated the composition of CD44 variants also in human T helper cells, supporting the hypothesis that pathogens or commensals, through TLRs, in turn modulate the repertoire of CD44 isoforms, thereby controlling the distribution of lesions in the CNS. The interference with this mechanism(s) represents a potential tool for prevention and treatment of autoimmune relapses and exacerbations.
Subject areas: Molecular biology, Immunology, Cell biology
Graphical abstract
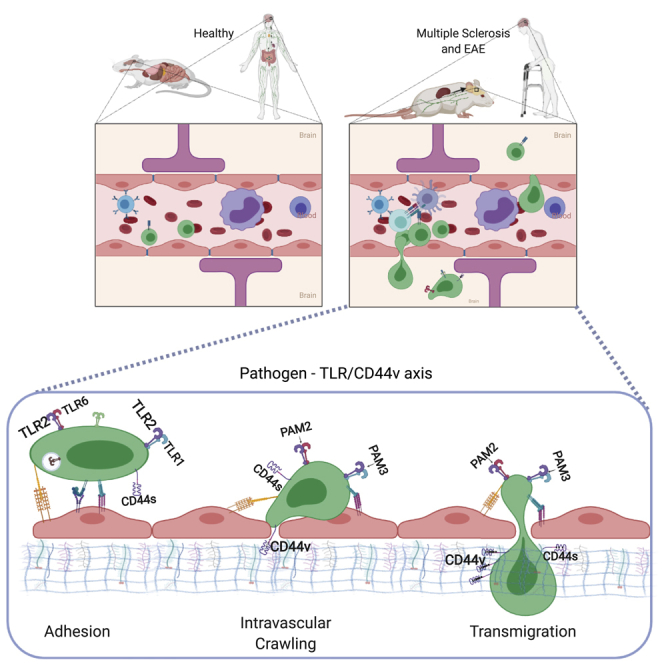
Host-Pathogen interaction modulating T cell trafficking and tissue infiltration through CD44 variants (CD44v). Circulating T cells through lymphoid and non-lymphoid organs, regulate homeostasis of immune system. The environment could lead an imbalance, also acting on the ability of T cells to move. The expression of certain CD44 isoforms can explain the ability of T cells to infiltrate into the CNS of patients affected by multiple sclerosis, crossing the blood brain barrier. This effect seems to be associated with microbial products, able to modulate CD44 splicing on T cells, that regulate their ability to move (Vestweber, 2015). Created with Biorender.com.
Highlights
-
•
Environment and genetic are both involved in the regulation of T cell motility
-
•
Pathogens and commensals impact on T cell trafficking through a TLRs/CD44v axis
-
•
Regulation of CD44 isoforms by TLRs is a new pathogenetic mechanism of autoimmunity
-
•
Modulation of CD44 isoform can be a new target for therapy of multiple sclerosis
Molecular biology; Immunology; Cell biology
Introduction
T cell trafficking is a central and highly regulated process during the immune response. Chemokines, chemokine receptors, integrins, selectins, matrix-associated proteins, and related receptors, all play a specific role in the coordinated process of guiding specific subsets of T cells to the most appropriate site for action (Oukka and Bettelli, 2018; Sandor et al., 2019; Strazza et al., 2015). Interfering with this process currently represents an attractive and efficient tool for therapeutic intervention in autoimmune diseases by preventing relapses and exacerbations (Calvier et al., 2020; Mousavi, 2020). Among autoimmune disorders, understanding disease pathogenesis in multiple sclerosis (MS) is challenging, because the distribution of lesions within the CNS is difficult to predict. Several factors have been called to play a role including antigen (Ag) density, T cell phenotype, and local milieu at the site of infiltration (Gross et al., 2017; Lindner et al., 2018).
We previously reported that pathogen recognition receptors (PRR) have a role in the control of lymphocytes′ trafficking. Mobilization of activated Ag-specific T cells critically depends on the amount of the stimuli from the environment in combination with the genetic background. Indeed, we demonstrated that the amount of Mycobacterium tuberculosis (Mtb) in the adjuvant (Nicolò et al., 2013) impacts immune cells trafficking in different mouse strains, based on a polymorphic residue at position 82 of mouse TLR2 (82Ile in SJL/J versus 82Met in C57Bl/6 mice): high concentrations of Mtb in the adjuvant are needed for early mobilization of activated T cells in the SJL/J strain, while challenge of C57Bl/6 leads to early mobilization independently of the amount of Mtb (Nicolò et al., 2013; Piermattei et al., 2016). In addition, we observed that pathogens were able to directly regulate T cell trafficking by binding TLR2 expressed by T cells following Ag-driven activation (Piermattei et al., 2016), whereas their ability to modulate the Th phenotype of activated T cells relies on indirect activation through TLR2 expressed by APCs (Fallarino et al., 2016; Luz et al., 2015).
Interestingly, we also observed that TLR2 polymorphic residue at position 82 also associated with differences in the distribution of CNS lesions during experimental autoimmune encephalomyelitis (EAE) (Piermattei et al., 2016), suggesting a more complex role for TLR2 in regulating T cell trafficking.
CD44 represents one of the best markers for bone marrow-derived cells infiltrating the CNS (Brennan et al., 1997; Fagone et al., 2018). It is expressed in several alternatively spliced isoforms, the most common of which, herewith CD44s (standard), is the shortest one, lacking all variant exons and accounting for most of the repertoire of mRNAs specific for CD44. All CD44 isoforms share trans-membrane, intracytoplasmic, and ligand-binding (NH-terminal) domains. The closest region to the membrane of the extracellular domain is the most variable and is involved in the regulation of cell–cell and cell–matrix interactions, homing, lymphopoiesis, and cell activation (Di Sante et al., 2013). Some of these functions are linked to the ability of CD44 to bind to the hyaluronic acid (HA) (Galluzzo et al., 1995). Moreover, it has been previously described a specific role of CD44 isoforms in the pathogenesis of immunomodulated diseases (Di Sante et al., 2013), whereas involvement of CD44v5 was shown in the pathogenesis of asthma (Yang et al., 2012). In addition, CD44v3/v6 was proposed as biomarker of disease activity in systemic lupus erythematosus (SLE) (Latini et al., 2021; Novelli et al., 2019). Decrease of CD44 expression leads to a reduction of adhesive interactions with lymph node matrix, thereby facilitating the exit of T cells from lymph nodes (Brennan et al., 1997; McDonald and Kubes, 2015). However, it is not clear the role that the different isoforms play in the regulation of cell trafficking, nor which stimuli modify the composition of the CD44 repertoire in T cells. The role of CD44 isoforms on activated T cells was explored during chronic-relapsing EAE, and the results showed that anti-CD44 antibodies inhibit the migratory capacity of T cells (Brennan et al., 1999), whereas the ablation of exons v7 and v10 significantly reduced EAE severity (Ghazi-Visser et al., 2013).
In this work, we examined the role of TLR2 in the regulation of trafficking of T cells, focusing on its ability to reshuffle the role of CD44 and the alternative splicing of its pre-mRNA. In addition, we also showed that stimulation of T cells by LPS or CpG (activating other TLRs also expressed by T cells) modified the alternative splicing of CD44, leading to a repertoire of CD44 isoforms-specific mRNAs that varied specifically depending on the type of stimulus. Notably, T cells infiltrating distinct areas of the CNS during EAE displayed different levels of mRNAs coding for specific CD44 isoforms. Finally, we report that overexpression of an isoform of CD44 in cells of the cerebrospinal fluid (CSF) was associated with presence of active inflammatory lesions in human multiple sclerosis (MS).
Results
Mtb- and TLR2-dependent, early mobilization of PLP139-151-specific, activated T cells associated with lower surface expression of CD44
We first examined the molecular mechanisms translating TLR2 stimulation into regulation of T cell trafficking from the lymph nodes (LN), and performed microarray analysis, comparing mRNA from T cells obtained from the LN of SJL/J mice challenged with adjuvant containing 50 versus 200 μg/20g of heat-inactivated Mtb (Figure 1A). We also compared the results with those obtained from challenging F1 mice of SJL/JwtxC57Bl/6wt and SJL/JwtxC57Bl/6tlr2− that were previously described (Figure 1F), thus reproducing the conditions of the experiments reported in (Piermattei et al., 2016). However, the analysis of the results did not show a parallel regulation of mobility for any of the proteins most obviously linked to T cell mobility (e.g., CD49d, CD11a, S1P-R and CD62L, chemokines, or chemokine receptors). Indeed, we therefore proceeded to examine the expression of these markers known to play a relevant role in the interaction between leukocytes and endothelia on antigen-specific activated T cells (Figures 1B–1D and 1G–1I).
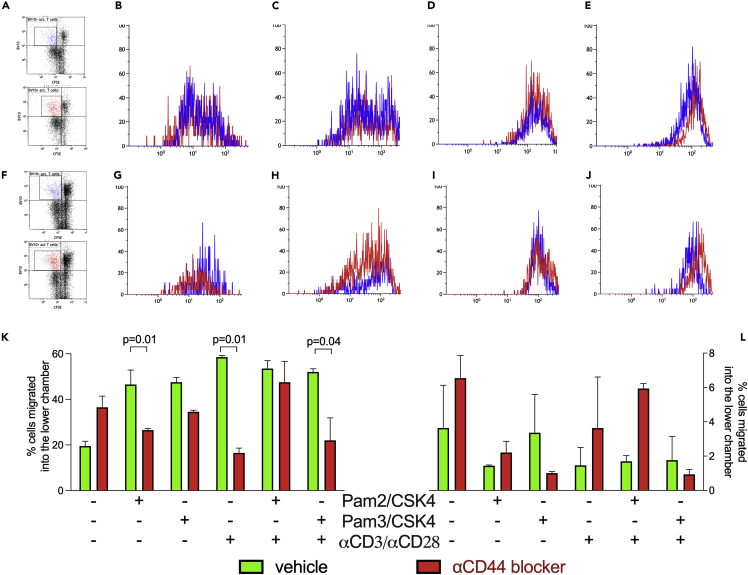
Figure 1.
The amount of Mtb and TLR2 haplotype regulates surface expression of CD44 on antigen-specific proliferating T cells
(A–J) The expression levels of CD49d (B and G), CD11a (LFA-1, C and H), CD62L (D and I), and CD44 (E and J), all APC conjugated on PLP139-151-specific (TCR-Vβ10+-PE) proliferating (CFSE-FITClow) T cells were evaluated by flow cytometry. (A and F) Gating strategies. (B–E) Two groups of four SJL/Jvβ10+ mice were immunized with an emulsion of PLP139-151 (PLP) in CFA containing two different concentrations of heat-killed Mtb: 200 (fast mobilizer, blue) or 50 μg/mouse (slow mobilizer, red). (G–J) Two groups of four F1 mice, SJL/JVβ10+xC57Bl/6wt (fast mobilizer, blue) and SJL/JVβ10+xC57Bl/6tlr2− (slow mobilizer, red) were challenged with PLP in CFA containing 50 μg/mouse of Mtb. T cells from draining LN were labeled with CFSE(-FITC) and stimulated in vitro with PLP. Three days later, cells were recovered stained for vβ10-PE and the expression of markers upon described was compared on Vβ10+ CFSElow cells, i.e., T cells that had proliferated in response to PLP.
(K and L) CD4+ CD62LlowT cells were isolated from SJL/J (n = 15 mice, in three different experiments, K) and C57Bl/6 (n = 4, L) mice′ spleens. T cells were plated on Matrigel® and stimulated with different conditions and in the presence (red bars) or absence (green bars) of and α-pan-CD44 mAb. After 24 h, cells that passed the Matrigel® were harvested and counted by flow cytometry. Data are expressed as percentage of cells recovered in the lower chamber. Statistical analysis was performed by two-way ANOVA, Tukey′s multiple comparisons. Data are shown as mean ± SD. Only significant p values are displayed.
To study the level of expression of these adhesion molecules, we took advantage of a mouse SJL/J strain transgenic for the β-chain of a PLP139-151-specific TCR (vβ10, SJL/Jvβ10+) (Nicolò et al., 2006, 2013), that is not spontaneously activated in vivo, but upon immunization (Penitente et al., 2008). The staining of lymph nodes-derived vβ10+CFSElowT cells with CD49d, CD11a (LFA-1), and CD62L, revealed no differences of expression in conditions leading to fast or low mobilization (Figures 1B–1D), while a decrease (approximately, 5-fold) of the expression of CD44 was observed when cells from fast mobilizer condition were examined (Figure 1E).
The levels of these cell surface markers were more variable between F1 (SJL/Jvβ10+xC57Bl/6wt) and F1 (SJL/Jvβ10+xC57Bl/6tlr2−/−), probably due to interferences between the different genetic backgrounds (Figures 1G–1I). However, again we found that F1 (SJL/Jvβ10+xC57Bl/6wt), the fast mobilizer genetic background, associated with a lower expression of CD44 on the surface of activated T cells (Figure 1J), suggesting that conditions leading to fast or slow mobilization from lymph nodes of activated T cells associated with the expression of low or high levels of CD44 on T cells, respectively.
Stimulation of activated CD4+ T cells by TLR2 ligands reshuffled the role of CD44 in T cell trafficking
To better understand the effect of TLR2 ligation on trafficking properties of T cells due to CD44 regulation, we measured the ability of CD4+CD62Llow T cells to cross Matrigel in a standard assay, upon cognate stimulation by biotinylated microparticles loaded with mouse αCD3 and αCD28 antibodies (Ab), in the presence or absence of synthetic TLR2 ligands (a cocktail of Pam2/CSK4 and Pam3/CSK4) and of α-panCD44 mAb, to assess its contribution to modification of trafficking.
As expected, α-panCD44 mAb increased the number of unstimulated T cells crossing the Matrigel in both SJL/J and C57Bl/6 mice (Figures 1K and 1L). These observations confirmed the data reported in the literature showing that interaction of CD44 with extracellular matrix results in an inhibition of T cell mobility (Baaten et al., 2010; Govindaraju et al., 2019).
A simultaneous stimulation of TLR2/TLR6 (with Pam2/CSK4) and of TLR2/TLR1 (with Pam3/CSK4) heterodimers of SJL/J-derived T cells led to an increase of the number of cells crossing the Matrigel, confirming that stimulation through this pathway was efficient in promoting cell mobility. Unexpectedly, however, in these conditions, blocking CD44 resulted in a significant decrease in the number of T cells crossing the Matrigel, in the SJL/J strain (One-way ANOVA test, p = 0.01 with PAM2/CSK4 and αCD3/αCD28, and p = 0.04 with combined PAM3/CSK4 and αCD3/αCD28). The same result was observed when SJL/J T cells were stimulated by microparticles (αCD3/αCD28). Thus, upon stimulation by TLR2 ligands, CD44 reshuffled its functional properties and T cell trafficking through the Matrigel became CD44-dependent (Figure 1K). However, such a reshuffling of CD44 function did not occur in T cells from the C57Bl/6 strain (Figure 1L).
Cognate stimulus and engagement of TLR2 modified the expression and function of CD44 and modulated alternative splicing of CD44 pre-mRNA
As mentioned above, CD44 comprises several isoforms, differing in their sequences due to alternative splicing of exons. mRNA coding for the shortest isoform represented the most easily detected product of alternative splicing of CD44 pre-mRNA. As expected, mRNA specific for the shortest isoform (CD44s) was the most represented in unstimulated T cells (Figure S1).
The activation of T cells via CD3/CD28 led to an increase of CD44s mRNA (Figures 2A and 2B). Likewise, triggering of TLR2 (but also of TLR9 and TLR4) led to an increase of mRNA specific for CD44s. However, we consistently observed that simultaneous triggering of both CD3/CD28 and TLR2/TLR6 dimer (Figure 2C) reduced the mRNA specific for this isoform. This observation suggests that cognate interaction leads to a decrease of CD44s to license activated T cells to exit LN, only when a concomitant signal through TLR-dependent pathway is also triggered.
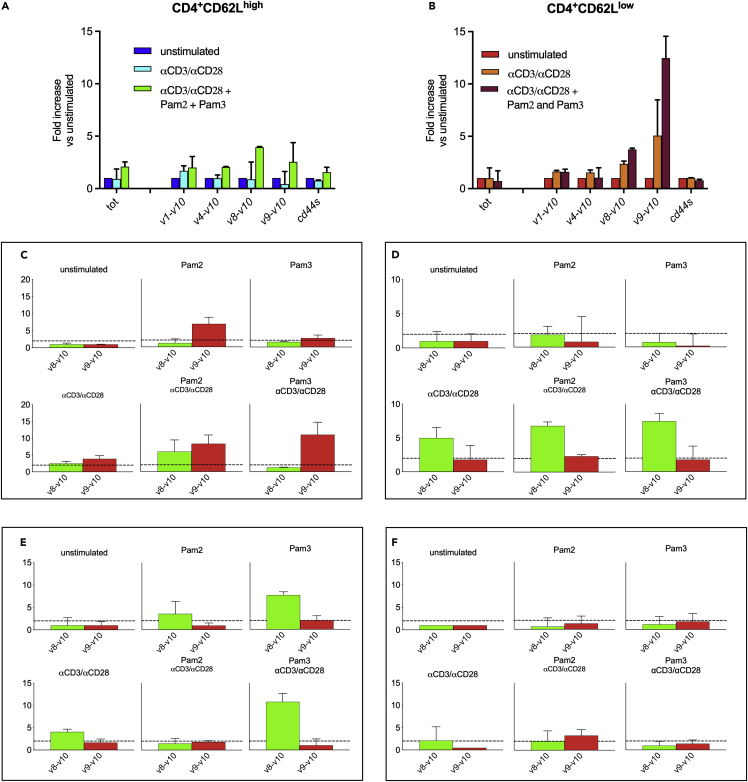
Figure 2.
TLR2 activation modulated CD44 alternative splicing, promoting upregulation of isoforms CD44v8-v10 and CD44v9-v10 in SJL/J strain and only isoform CD44v8-v10 in C57Bl/6 strain
(A and B) CD4+CD62Llow and CD4+CD62Lhigh cells were isolated from the spleens of naÏve female SJL/J (n = 5) and stimulated with αCD3/αCD28, PAM2/CSK4, and PAM3/CSK4. The repertoire of CD44 mRNA isoforms was analyzed by RT-qPCR. A consistent upregulation of CD44v9-v10 and a slighter upregulation of CD44v8-v10 occurred upon stimulation of CD4+CD62Llow T cells with αCD3/αCD28 and PAM2/CSK4 or PAM3/CSK4.
(C and D) TLR2/6 and TLR2/1 display non-overlapping abilities to favor the production of CD44v8-v10 and CD44v9-v10 isoforms in SJL/J versus C57Bl/6 mice. CD4+CD62Llow cells were isolated from the spleens of SJL/J (C) or C57Bl/6 (D) mice and stimulated as described. (C) PAM2/CSK4 or PAM3/CSK4 alone are both able to induce an upregulation of CD44v9-v10 (Red Bars), albeit modest, but cognate stimulation plus TLR2 promotes a much stronger increase of both, CD44v8-v10 (Green Bars) and CD44v9-v10, in SJL/J strain (n = 4). (D) Only isoform CD44v8-v10 was upregulated in C57Bl/6 mice (n = 4) and required cognate stimulation.
(E and F) CD44 repertoire regulation on T cells in part depends on TLR2 polymorphism. CD4+CD62Llow cells were isolated from the spleen of F1 SJL/JwtxC57Bl/6wt (n = 3, E) or F1 SJL/JwtxC57Bl/6tlr2− (n = 3, F) mice, and stimulated as described in the figure. Data show that TLR2 of SJL/J is able to restore the expression of isoform CD44v9-v10 when C57Bl/6 TLR2 is not present (F). All data are shown as mean ± SD.
The results reported in Figure 1K implied that a major reshuffling of CD44 function occurred in SJL/J T cells upon TLR2 stimulation. We therefore tested the hypothesis that such a stimulus was also modifying the composition of the CD44 repertoire. In a first type of experiments, we examined the effect of TLR2 engagement on CD44 in the presence of cognate stimulation (αCD3/αCD28), mRNA specific for two isoforms, CD44v8-v10 and, to a much higher extent, CD44v9-v10 were selectively promoted, especially in CD4+CD62Llow T cells (Figures 2A and 2B). Next, we examined the role of each stimulus, individually (cognate, TLR2/TLR6 dimer, and TLR2/TLR1 dimer), only in CD4+CD62Llow T cells (Figure 2C panel). Cognate activation alone led to a modest up-regulation of mRNA specific for isoform CD44v9-v10. Additional stimulation of TLR2 led to a much stronger upregulation of mRNAs specific for isoforms CD44v8-v10 and CD44v9-v10.
However, the type of TLR2 dimer triggered alternatively spliced CD44 pre-mRNA. In fact, mRNA specific for isoform CD44v9-v10 was upregulated via activation of TLR2/TLR6 (PAM2/CSK4) and of TLR2/TLR1 (PAM3/CSK4), whereas mRNA specific for isoform CD44v8-v10 was upregulated only by TLR2/TLR1 dimer. Together, our results show that signals coming from TCR and from TLR2 reshuffled the alternative splicing of pre-mRNA specific for CD44, with a pattern depending on the type of dimer involvement.
T cells from C57Bl/6 mice selectively upregulated mRNA specific for isoform CD44v8-v10, in a TLR2(I82M)-dependent manner
We have previously shown that the ability of Mtb to modulate trafficking properties of CD4+ T cells was regulated in a strain-specific manner and depends on a single non-synonymous polymorphism at position 82 of TLR2 (Piermattei et al., 2016). The levels of CD44 on T cell surface was modified according to TLR2 polymorphism (Figure 1E). We therefore next examined if mouse strain and TLR2 polymorphism have a role on the alternative splicing of pre-mRNA specific for CD44.
The regulation of alternative splicing of pre-mRNA of CD44 differed in C57Bl/6 mice compared to SJL/J mice. In fact, both TLR2/TLR1 and TLR2/TLR6 dimers upregulated effectively and to the same extent the production of mRNA specific for isoform CD44v8-v10, while production of mRNA specific for isoform CD44v9-v10 was not upregulated above the 2-fold threshold by any of the two dimers (Figure 2D panel).
To evaluate the contribution of TLR2 polymorphism at position 82 in the differences observed in the regulation of CD44 pre-mRNA splicing, we next examined CD44 regulation on T cells from F1 (SJL/JwtxC57Bl/6wt, which have TLR2 from both SJL/Jwt(82Ile) and C57Bl/6wt (82Met)), versus F1 (SJL/JwtxC57Bl/6tlr2−) which t have only TLR2 of SJL/J, as previously described by our group (Nicolò et al., 2013; Piermattei et al., 2016). The results (Figures 2E and 2F panels) indicated that TLR2 polymorphism together with other genetic traits play a role in determining the differences between the two mouse strains. In fact, the haplotype of SJL/J origin predominantly inhibited the upregulation of CD44v8-v10–specific mRNA by TLR2/TLR6 occurring when TLR2 of C57Bl/6wt origin is also present (Figure 2E), but was not sufficient to restore the ability of TLR2 to upregulate the production of mRNA specific for isoform CD44v9-v10 (Figure 2C). However, when TLR2 of C57Bl/6wt origin was absent, the ability of TLR2/TLR6 dimers to upregulate the production of the mRNA specific for isoform CD44v9-v10 was restored, although to a lower extent (Figure 2F).
In conclusion, these results show that while TLR2 regulates alternative splicing of CD44-specific pre-mRNA in both SJL/Jwt and C57Bl/6wt mouse strains, the impact of TLR2 engagement on the relative composition of CD44 repertoire depends on the TLR2 isoform present and on other strain-specific characteristics.
Wnt/β-catenin pathway was regulated by strain background and TLR2
Several previous observations have shown that the Wnt/β-catenin system regulates CD44 expression and its alternative splicing in human cells (Goncalves et al., 2008; Idris et al., 2019; Vallée et al., 2018). Particularly, these studies showed that TLR3 engagement inhibits the phosphorylation of β-catenin by favoring ubiquitination of GSK-3β, in a TRAF6-dependent manner. We therefore examined if engagement of TLR2 on mice-derived CD4+CD62Llow T cells is also able to inhibit phosphorylation of β-catenin. The Western Blot analysis showed that the amount of total β-catenin was increased by 1-h stimulations via TLR2/6, TLR2/1, and αCD3/αCD28 (Figure 3A), while both TLR2 stimulations downregulated the levels of the phosphorylated form of β-catenin (Figure 3B). After 6 h of stimulation, total β-catenin showed to be still upregulated only by stimulus through TLR2/6 (Figure 3C), while phosphorylated β-catenin resulted downregulated by all stimuli (Figure 3D). Thus, TLR2 engagement immediately blocked the phosphorylation of β-catenin, thereby favoring total β-catenin accumulation and, consequently, its ability to translocate into the nucleus to act as a transcription factor.
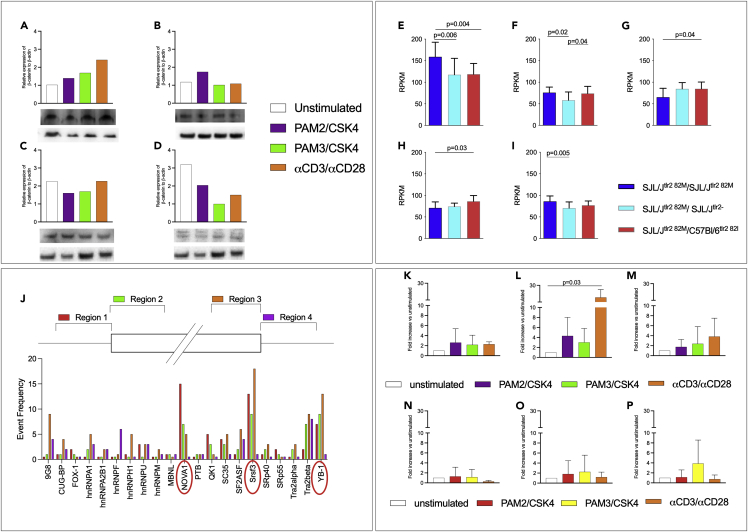
Figure 3.
TLR2 interacts with the Wnt/β-catenin pathway and promotes the expression of splicing factors involved in the CD44 alternative splicing
(A–D) Stimulation of TLR2/1 and TLR2/6 dimers blocks phosphorylation of β-catenin. CD4+CD62low T cells were enriched from the spleens of naive SJL/J mice (n = 3) and stimulated as described with PAM2/CSK4, PAM3/CSK4, or αCD3/αCD28 microparticles, for 1 (A and B) or 6 (C and D) hours. Proteins were purified and examined by Western blot for total (A and C) and phosphorylated (B and D) β-catenin (displayed data are the result of a single Western Blot assay).
(E–I) The wnt/β-catenin pathway is influenced by strain, TLR2 gene dose, and TLR2 haplotype. Five SJL/Jtlr2 82M/M (deep blue bars), six SJL/Jtlr2 82M/- (light blue bars), and six SJL/Jtlr2 82M/I (red bars) were challenged s.c. with CFA containing 50 μg/mouse of heat-inactivated Mtb. Microarray analysis was performed on CD4+ cells enriched from draining LN 4 days after immunization. Data are normalized versus the highest value obtained for each mRNA. Significance of the differences was evaluated by one-way ANOVA and Tukey HSD test. Values (mean + SD) are shown for Wnt10a (A), Wnt16 (B), Ctnnal1 (C), Wnt10b (D), and β-catenin (E).
(J) Splicing factors prediction through SFMAP software. Following the methodology of Paz et al. (Paz et al., 2010) and using the software SFMAP at http://sfmap.technion.ac.il, it has been possible to predict the splicing factors most probably involved in the alternative splicing of CD44v9-v10 and CD44v8-v10 (red circles indicate the most relevant ones).
(K–P) TLR2 stimulation changes mRNAs of the splicing factors NOVA-1, SRSF3, and YB-1 in SJL/J-derived but not in C57Bl/6-derived T cells. T cells isolated from SJL/J (K–M, n = 3) or C57Bl/6 (N–P, n = 4) were stimulated with PAM2/CSK4, PAM3/CSK4, or αCD3/αCD28 microparticles (data are shown as fold increase ± SD and normalized on unstimulated cells). As potentially involved in the alternative splicing of CD44 favoring the production of CD44v9-10 mRNA specific of three splicing factors, NOVA-1 (K andN), SRSF3 (L andO), and YB-1 (M andP) has been tested in RT-qPCR. Although differently depending on the stimulus, the three splicing factors resulted consistently upregulated in the SJL/J strain (one-way ANOVA statistical analysis showed a significant p = 0.03 for SRSF-3, unstimulated vs αCD3/αCD28 stimulated cells), while no relevant upregulation was observed in the C57Bl/6 strain, in which in most conditions is shown a downregulation of the same splicing factors.
To examine if the expression of all Wnt, and of α- and β-catenins mRNAs is modulated in a strain and TLR2 haplotype-dependent manner, we retrospectively evaluated the RNA microarray analysis of lymph node CD4+ T cells from SJL/J, F1 (SJL/JwtxC57Bl/6tlr2−), and F1 (SJL/JwtxC57Bl/6wt) mice, reported here and in our previous work (Piermattei et al., 2016). Results are reported in Figures 3E–3I.
mRNAs levels specific for Wnt-10a and Wnt-16 (Figures 3E and 3F) did not differ between the two F1 mice, but both significantly differed from the parental SJL/J mice (One-way ANOVA test, p = 0.004 for SJLTlr2− vs parental SJL/J and p = 0.006 for SJLxC57Bl/6 vs parental SJL/J in Wnt-10; one-way ANOVA test, p = 0.02 for SJLTlr2− vs parental SJL/J and p = 0.04 for SJLxC57Bl/6 vs parental SJL/J in Wnt-16) indicating that regulation of these factors depended on the mouse strain (SJL/J vs SJL/JxC57Bl/6). Levels of mRNA specific for Wnt-10b (Figure 3G) showed no differences between the SJL/J and F1 hybrids (SJL/JwtxC57Bl/6wt, one-way ANOVA test, p = 0.11) mice, but both differed from the F1 (SJL/JwtxC57Bl/6tlr2−, one-way ANOVA test, p = 0.04) mice, indicating that regulation of that factor depended on gene dosage of TLR2.
Finally, we found no difference in mRNA levels for α-catenin1 (Figure 3H) between the SJL/J and F1 hybrids (SJL/JwtxC57Bl/6tlr2−, one-way ANOVA test, p = 0.8) mice, while they showed to differ between SJL/J and the F1 hybrids (SJL/JwtxC57Bl/6wt) mice (one-way ANOVA test, p = 0.03), thus indicating that regulation of this factor depended on the reported polymorphism of TLR2 between SJL/J and C57Bl/6 strains. Finally, the level of mRNA specific for β-catenin (Figure 3I) appeared to be significantly different between the parental SJL/J and the hybrid SJLtlr2− (one-way ANOVA test, p = 0.005), while no difference was found between SJL/J and the hybrid F1 (SJL/JxC57Bl/6, one-way ANOVA test, p = 0.11).
Engagement of TLR2 modulated the levels of mRNAs specific for CD44 splicing factors
Based on the DNA sequence of CD44 at the splicing site leading to the generation of CD44v9-v10 isoform, we next used a computational approach to determine the most likely splicing factors involved in the production of CD44v9-v10 and CD44v8-v10 (Figure 3J). We found that three splicing factors (Nova-1, Srsf3, and Yb-1) were potentially involved in the splicing of mRNA to produce CD44v9-v10 (Paz et al., 2010). As a support of the validity of this approach, one of these factors, Srsf3, has previously been shown to be regulated by β-catenin, in cancer cells (Bordonaro, 2013; Goncalves et al., 2008; Idris et al., 2019; Thorsen et al., 2011). Therefore, we compared modulation of their expression in CD4+CD62low T cells of SJL/J and C57Bl/6 mice. Each stimulus was able to produce a distinct modulation—mostly an upregulation—of the mRNAs specific of the three splicing factors, in the absence or presence of the cognate stimulus, in the SJL/J mouse (Figures 3K–3M). Confirming our observation that C57Bl/6 mice failed to upregulate isoform CD44v9-v10, none of the condition tested led to upregulation of NOVA-1, SRSF3, or YB-1 in T cells derived from this latter strain. Rather, in most conditions, TLR2 stimulation resulted in a downregulation of the levels of mRNA specific for the splicing factors tested (Figures 3N–3P). Thus, we cannot exclude that all tested splicing factors alone or in combination regulate the selective production of CD44v9-v10 isoform.
Taken together, the observations reported in Figure 3 corroborated the hypothesis of the existence of a pathway connecting TLRs via β-catenin to alternatively spliced CD44, as previously shown in cancer cells (Todaro et al., 2014).
Engagement of TLR4 and TLR9 led to modification of alternative splicing of CD44 pre-mRNA
In addition to TLR2, activated T cells also express TLR4 and TLR9. As all TLRs share the common signaling pathway that relies on adaptor protein MyD88, we assessed if also triggering of these other two TLRs could lead to reshuffling of the repertoire of the CD44-specific mRNA. We therefore stimulated isolated, SJL/J-derived T cells with CpG (selective ligand for TLR9), LPS (for TLR4), and PPD (as a mainly TLR2-restricted ligating agent). Results are reported in Figure 4A. As expected, stimulation with PPD resulted in the upregulation of isoforms CD44v9-v10 and CD44v8-v10, that was also dependent on the simultaneous cognate interaction. Stimulation with LPS and CpG led to modifications of the landscape of the CD44 isoform-specific mRNAs, which were distinct from those generated by engagement of TLR2. These results led to three main observations: first, induction of alternative splicing by LPS and CpG did not depend on αCD3/αCD28 microparticles stimulation; second, stimulation by both LPS and CpG led to upregulation also of mRNA specific for isoform CD44v1-v10, which was never promoted by TLR2; third, stimulation by LPS failed to promote the production of mRNA specific for isoform CD44v8-v10 also when T cells were simultaneously stimulated via CD3/CD28.
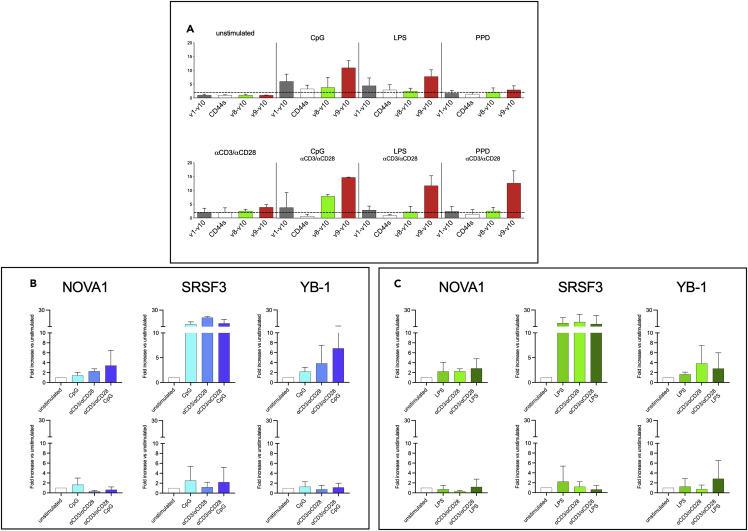
Figure 4.
Stimulation of TLR4, TLR9 modulated alternative splicing of CD44 in the SJL/J strain
(A–C) Because activated T cells also express TLR4 and TLR9, CD4+CD62low T cells from SJL/J (n = 3) were stimulated with LPS (TLR4), CpG (TLR9), and PPD (as a mainly TLR2-restricted ligating agent). mRNA of CD44 isoforms was analyzed by RT-qPCR. Compared to unstimulated condition, isoforms CD44v1-v10 and CD44v9-v10 resulted upregulated by LPS and CpG alone or in combination with αCD3/αCD28 microparticles (A). RT-qPCR analysis also showed that CpG and LPS modulate in distinct ways the splicing factors NOVA1, SRSF3, and YB-1 (B and C, upper panels, respectively). mRNA of CD4+CD62low T cells from C57Bl/6 mice (B and C, lower panels), stimulated in the same ways, were also analyzed and the stimulation resulted in down-regulation of the three splicing factors (data are normalized on unstimulated cells and displayed as fold increase ± SD).
We also tested in RT-qPCR the mRNA levels of the splicing factors NOVA-1, SRSF-3, and YB-1 in SJL/J (upper panel) andC57Bl/6 (lower panel) strains upon stimulation with CpG (Figure 4B) or LPS (Figure 4C), in presence or absence of cognate stimulation with αCD3/αCD28 beads. Data showed that CpG, alone and together with αCD3/αCD28 particles, was able to upregulate all the three splicing factors in the SJL/J strain, while any relevant difference was observed in the C57Bl/6 one. The same observations were reported in the mRNA level analysis of these splicing factors upon stimulation with LPS.
Thus, albeit signals from all TLRs converge on the MyD88 pathway, each pathogen receptor promoted a specific pattern of mRNAs specific for the various isoforms of CD44.
Cells infiltrating the forebrain at the onset of EAE overexpressed CD44v9-v10
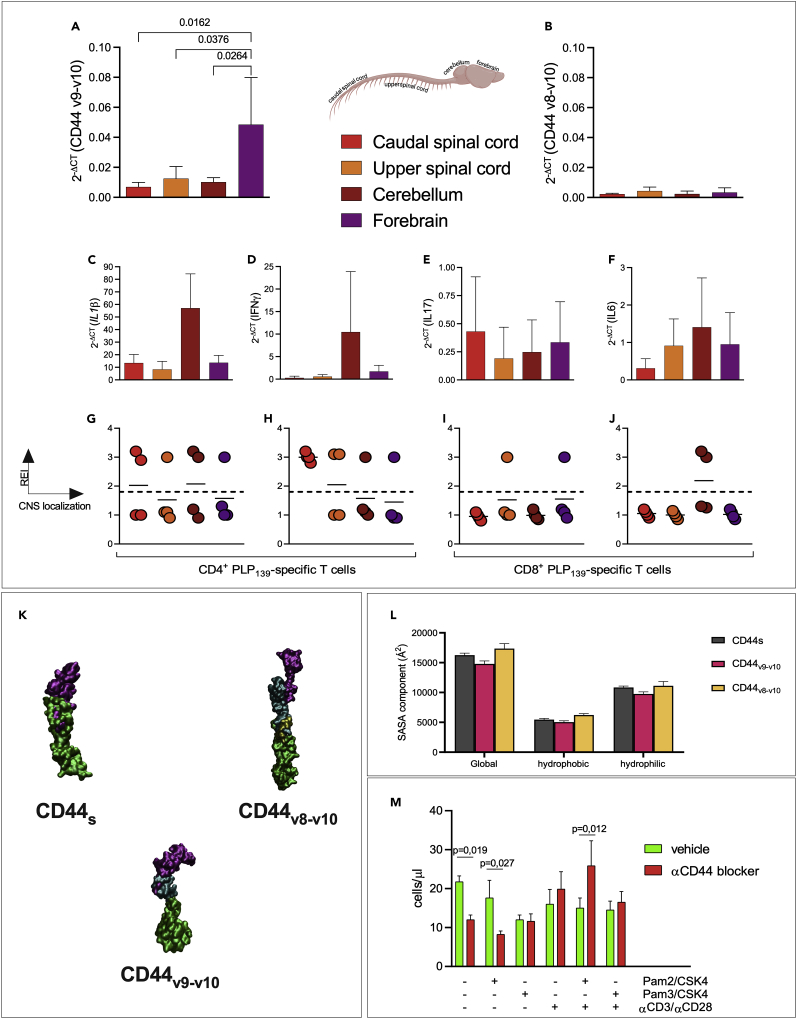
We previously reported that the polymorphism TLR2I82M influences the distribution of immune infiltrates in the CNS, with TLR282I promoting the presence of lesions in the forebrain and TLR-282M dominantly preventing it (Piermattei et al., 2016). Because TLR2 polymorphism led to differences in the processing of CD44 pre-mRNA as shown above, we next examined the hypothesis that isoforms CD44v9-v10 and CD44v8-v10 distributed differently in the various areas of the CNS during EAE. We therefore examined the amount of mRNA specific for the CD44 isoforms in four distinct sections of the CNS: caudal spinal cord, upper spinal cord, cerebellum, and forebrain at EAE onset. The amount of mRNA specific for CD44s was used as a measure of the number of infiltrating cells in each section. Therefore, to assess if any preferential expression of these isoforms existed among cells infiltrating each section, we compared the distribution of mRNA specific for isoforms CD44v8-v10 or CD44v9-v10, normalized for CD44s (results are shown in Figures 5A and 5B).
Figure 5.
EAE SJL/J mice showed CD44v9-v10 overexpression in the forebrain at the onset of the disease, and TLR stimulation modifies the binding of CD44 to osteopontin
(A–J) Central nervous systems from SJL/J mice (n = 4) at the onset of EAE were harvested and divided into four areas: caudal spinal cord (red bar), upper spinal cord (orange bar), cerebellum (dark red bar), and forebrain (purple bar). CD44 isoforms were analyzed by RT-qPCR and normalized to the mRNA specific for CD44s. A significant upregulation of isoform CD44v9-v10 was observed in the forebrain (A) compared to the caudal spinal cord (two-way ANOVA, Tukey′s multiple comparisons test p = 0.02), the upper spinal cord (two-way ANOVA, Tukey′s multiple comparisons test p = 0.04), and to the cerebellum (two-way ANOVA, Tukey′s multiple comparisons test p = 0.03). No significant changes in the brain areas were observed for isoform CD44v8-v10 (B). Amount of mRNA specific for IL-1β (C), INF-γ (D), IL-17 (E), and IL-6 (F) has been evaluated and normalized to the mRNA specific for CD44s. (G–J) Presence of PLP139-151-specific clonotypic CD4+ and CD8+ cells in the CNS areas at the onset of EAE in the same samples. cDNA from the same samples were analyzed by CDR3BV-BJ spectratyping as described in (Nicolò et al., 2006). Enrichment of the indicated TCR-β rearrangement is reported as rate enrichment index (REI) for each brain area with the same code color used above.
(K) In silico modeling of CD44s, CD44v8-v10, and CD44v9-v10. Region corresponding to CD44v8-v10 and CD44v9-v10 are shown in yellow and violet, respectively.
(L) Calculated solvent accessible surface area (SASA) of isoforms CD44s (gray), CD44v9-v10 (pink), and CD44v8-v10 (yellow). All SASA values of CD44v9-v10 resulted significantly higher compared to the ones of CD44s and CD44v8-v10 (one-way ANOVA test, p< 0.0001).
(M)TLR2 stimulation modifies interaction of T cells with osteopontin in a CD44-dependent manner. CD4+CD62Llow T cells were isolated from SJL/J spleen (n = 4 mice) and stimulated as described in the figure. T cells were plated on Matrigel® pre-coated with OPN and stimulated with PAM2/CSK4, PAM3/CSK4, and/or by αCD3/αCD28-coated microparticles, in the presence (red bars) or absence (green bars) of α-pan-CD44 mAb. After 6 h, cells that passed the Matrigel® were counted by flow cytometry. Data are expressed as number of cells recovered in the lower chamber. Statistical analysis was performed by two-way ANOVA, Tukey′s multiple comparisons. All data are shown as mean ± SD. Only significant p values are displayed.
We observed that the expression of CD44s-specific mRNA was similar in caudal CNS, while it was 10-fold lower in forebrain (Figure S2A). Likewise, the expression level of mRNAs specific for IL1β and IL6 (as indicators of the ongoing inflammatory response) showed the same distribution of CD44s (Figures 5C and 5F).
Supporting our hypothesis, the ratio between CD44v9-v10 and CD44s-specific mRNAs was significantly higher in the forebrain with respect to the other CNS areas (Figure 5A, one-way ANOVA test, forebrain vs caudal SC p = 0.02, forebrain vs upper SC p = 0.04 and forebrain vs cerebellum p = 0.03). On the contrary, CD44v1-v10, CD44v4-v10, and CD44v8-v10 were expressed in all examined areas at the same relative concentration (one-way ANOVA test, all p values resulted p≥ 0.7), excluding any specific association of other isoforms with cells homing preferentially to any CNS area (Figures S2B and S2C and 5B, respectively).
The observations reported in Figures 5C–5F could be explained by differences in clonal composition or in the T-helper phenotype of the T cell repertoire infiltrating the PFC. The analysis of CD4+ (Nicolò et al., 2006) and CD8+ (Penitente et al., 2008) clonotypes in the CNS (Figure 5G) shows that T cells infiltrating the forebrain did not belong to a single clonotype. Likewise, cells infiltrating the forebrain do not have a distinct cytokine phenotype and did not show any selective enrichment for Th1 or Th17 cells (Figures 5D and 5E).
Taken together, these data show that CD44v9-v10 promotes T cell homing in the forebrain. However, we did not find any evidence for a clonal or functional limitation of its expression and the available data do not suggest a mechanism for which isoform CD44v9-v10 is upregulated in vivo selectively only on particular T cell subsets.
All CD44 isoforms share the same ligand-binding domain and juxta- and intracellular domains. Yet, we observed that expression of isoform CD44v9-v10 was enriched on some cells that are prone to infiltrate the forebrain. Such observation implies that upregulation of this isoform modifies the binding of CD44 to some ligand(s) differentially expressed in the extracellular matrix of distinct CNS areas.
To address this, the three-dimensional structural models of the three isoforms of CD44, CD44s, CD44v9-v10, and CD44v8-v10 (Figure 5K) were generated and investigated by in silico approaches (Figures S3A and S3B). The best scoring models displayed a C-score value of −1.07, −0.67, and −1.75 for CD44s, CD44v8-v10, and CD44v9-v10, respectively. Figure 5L shows that, upon insertion of the 98 residues, the global SASA of the CD44v9-v10N-terminal domain decreased, if compared to CD44s (16,263 ± 334 Å2 and 14,793 ± 502 Å2, respectively). A comparable reduction is observed for the hydrophobic and polar SASA components.
Interestingly, the CD44v8-v10N-terminal domain displayed SASA values significantly higher (one-way ANOVA test, p< 0.0001) than the two other isoforms (17,361 ± 833 Å2, 6,231 ± 234 Å2, and 11,130 ± 705 Å2 for global, hydrophobic, and polar components, respectively). Considering that the SASA quantifies the isoform exposure to solvent molecules, the observed differences support the hypothesis that the various CD44 isoform are characterized by a different interaction pattern with ligands.
CNS extracellular matrix contains several ligands of CD44 under non-inflammatory conditions, and can be further modified by inflammation, e.g., through the action of metalloproteinases. Yet, few reports show differences in individual matrix proteins along the various areas. As shown in Figure 5M, we observed that TLR stimulation reshuffles the role of CD44 in the binding to low MW hyaluronic acid, the main constituent of Matrigel. Osteopontin (OPN) was observed to display distinct immunoreactivity in frontal versus caudal areas of human CNS, hinting distinct isoforms as its possible ligands (under normal conditions) in driving the specific homing of T cells. On the other hand, type I collagen (Coll I) has been shown to be present in the perivascular spaces of CNS and thus could be potentially involved in T cell migration.
The results of migration assay on Matrigel enriched with OPN (Figure 5M) demonstrated that TLR2/6 and αCD3/αCD28 stimuli increase the adhesion of CD44 to OPN. In fact, anti-CD44 antibody blocked migration of T cells through OPN-coated Matrigel, in unstimulated cells. Thus, in the absence of any further stimulation, OPN favors CD62lowT cell crossing in a CD44-dependent manner. Stimulation by αCD3/αCD28 reduces the crossing which is only modestly (or not at all) restored by αCD44. Thus, upon stimulation with αCD3/αCD28, there is an apparent loss of the role of OPN/CD44 binding in promoting T cell trafficking across Matrigel. When we further add PAM2/CSK4, we invert the effect of binding between OPN and CD44, because now αCD44 promotes T cell migration. Thus, simultaneous stimulation with αCD3/αCD28 and PAM2/CSK4 modifies the role of OPN/CD44 binding blocking T cells in the upper chamber or within the gel itself. As a control of specificity, binding to type I collagen was increased by the same stimuli, but in this case blocking CD44 did not reverse this effect (Figure S4). Thus, our data suggest that reshuffling of CD44 role in T cell trafficking possibly affected its ability to bind to OPN.
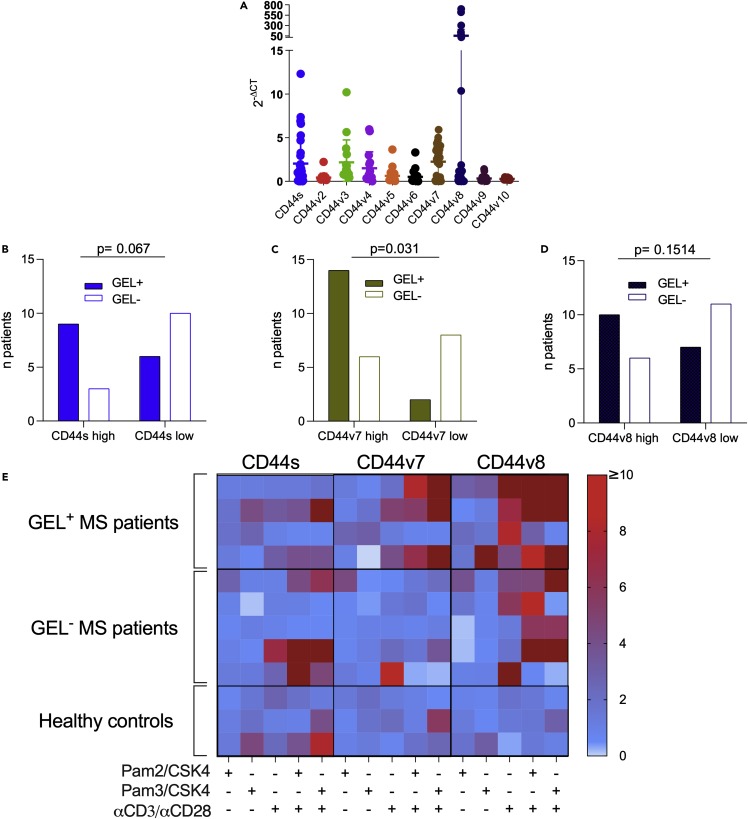
Human CD44v7-specific mRNA expressed in cells from CSF of patients with MS associated with presence of gadolinium-enhancing lesions
The data reported above in the mouse model suggested that CD44 isoforms might play a role in addressing T cells selectively to (distinct) CNS compartments. We therefore examined if the repertoire of mRNAs specific for CD44 isoform variants in all the type of cells obtained from human CSF of patients with MS (Table 1) showed any sign of skewing. In Figure 6A, we report that the expression of CD44v7 and CD44v8-specific mRNAs appear to have a bimodal distribution of values. Samples were divided into two roughly equivalent groups for CD44v7high and CD44v7low values, while high values for CD44v8 mRNA were expressed only by a small subgroup of samples. We did not find a correlation of mRNA expression levels among CD44v7 or CD44v8 and nuclear factors or cytokines characterizing Th cell phenotype, further supporting our findings in the mouse model. Although high expression of CD44v8 and s (and of any other variant CD44) was not found to be related to any clinical feature (Figures 6B and 6D), CD44v7high patients with MS show active inflammatory lesions, detected as gadolinium-enhancing lesions (GELs) at the CT scan (Figure 6C), significantly more frequently than CD44v7low ones (Fisher′s exact test, p = 0.031). Together, these observations suggest that regulation of CD44 alternative splicing is involved in disease development not only in the EAE model as shown above, but also in the pathogenesis of neuroinflammation in MS.
Table 1.
Demographic, clinical, and laboratoristic data of patients with MS for CSF analysis
| RR-MS patients N=34 | |
|---|---|
| Sex, n of female (%) | 24 (70.5) |
| Age, years (mean±SD) | 37.8±10.4 |
| Disease Duration, mo. (mean±SD) | 27,2±49.4 |
| EDSS (mean±SD) | 1,51±0.97 |
| Previous attacks, n of patients (%) | 27 (79.4) |
| Relapse within 3 months, n of patients (%) | 12 (35,3) |
| CSF - OB, n of patients (%) | 25 (73,4) |
| ESR, mm/h (mean±SD) | 9.6±9.0 |
| CRP, mg/dl (mean±SD) | 2.6±7.1 |
| CSF protein, mg/dl (mean±SD) | 29.1±10.3 |
| CSF glucose, mg/dl (mean±SD) | 61.2±9.9 |
| IgG index (mean±SD) | 0.95±0.59 |
| MRI+, n of patients (%) | 5 (14.7) |
| Total GDP+, n of patients (%) | 20 (58.8) |
| Encephalic GDP+, n. of patients (%) | 19 (55.9) |
EDSS: expanded disability status scale; Previous attacks are intended as a number of two or more attacks preceding the study entry (day of diagnosis). Relapse within three months is intended within 3 months from the study entry (day of enrollment). CSF: Cerebrospinal Fluid; OB: Oligoclonal Bands; ESR: ErythroSedimentation Rate; CRP: C Reactive Protein; MRI+: MRI positive lesions in the spinal cord at study entry (day of diagnosis); Total and encephalic GDP+: Gadolinium phosphide-enhancing positive lesions in both spinal cord and encephalon, or only encephalon at study entry (day of diagnosis).
Figure 6.
Alternative splicing of CD44 in human CD4+ cells from patients affected by multiple sclerosis
(A) Repertoire of alternatively spliced CD44vv in cells from CSF of patients with RR-MS (n = 34). Data are shown as mean ± SD.
(B–D) Correlation of CD44s (B), -v7 (C), and -v8 (D) expression (>median, full columns; <median, empty columns) in patients showing gadolinium-enhancing lesions (GEL+) in the CNS. Only CD44v7 resulted to be significantly correlated (Fisher′s exact tests, p = 0.031) to the presence of GEL+, while none of the other CD44 isoform showed to be related to any clinical features.
(E) mRNA expression levels of CD44s, v7, and v8 in CD4+ human T cells stimulated with PAM2/CSK4, PAM3/CSK4, and/or αCD3α/CD28 and isolated from peripheral blood of patients with MS (N = 9) and healthy donors (N = 3). The colored bar indicates high or low expression of the three CD44 isoforms. No relevant differences were observed between GEL+ (N = 4), GEL− (N = 5) patients with MS, and the healthy subjects in relation to CD44s and CD44v7, whereas CD44v8 resulted mostly upregulated in patients with MS, independently of the presence of active lesions.
We next evaluated if TLRs were able to modulate the repertoire of CD44 isoforms in human T cells also, with a special reference to -v7 and -v8. We therefore isolated CD4+CD62low cells from peripheral blood of 9 patients with MS (4 GELs+ and 5 GELs−) and three healthy donors (Table 2) and stimulated them with TLRs ligands in the presence or absence of biotinylated microparticles coated with human αCD3/αCD28/αCD2 antibodies to mimic antigen-presenting cells and simulate the cognate interaction, as described above for mouse model. The results for -s,-v7, and -v8 isoforms are reported in Figure 6E and show that TLR ligands poorly modulated the repertoire of these CD44 variants in the absence of cognate stimulus. Instead, cognate stimulus alone drove a selective upregulation of mRNA specific for -v7, -v8 (and -v9) variants, and of the standard isoform (CD44s). In the presence of the cognate stimulus, TLRs ligands were able to further modulate the alternative splicing of CD44 pre-mRNA in a TLR-specific fashion in a similar manner as in mouse T cells. However, the pattern of upregulated isoforms and the extent of stimulation was both variable among patients and controls. In general, cells from patients with MS appeared more prone to upregulating CD44 isoforms following TLR and cognate stimulus than controls.
Table 2.
Demographic, clinical, and laboratoristic data of patients with MS for PBMC analysis
| RR-MS patients N=9 | Healthy donors N=3 | |
|---|---|---|
| Sex, n of female (%) | 6 (66.6) | 1 (33.3) |
| Age, years (mean±SD) | 31.8±7.74 | 33.3±7.09 |
| Disease Duration, mo. (mean±SD) | 377.8±552.5 | NA |
| EDSS (mean±SD) | 1.11±0.91 | NA |
| Previous attacks, n of patients (%) | 3 (33.3) | NA |
| Relapse within 3 months, n of patients (%) | 5 (55.5) | NA |
| CSF - OB, n of patients (%) | 8 (88.9) | NA |
| ESR, mm/h (mean±SD) | 5.2±4.3 | NA |
| CRP, mg/dl (mean±SD) | 0.51±0.07 | NA |
| CSF protein, mg/dl (mean±SD) | 31.5±13.4 | NA |
| CSF glucose, mg/dl (mean±SD) | 64.3±8.5 | NA |
| IgG index (mean±SD) | 0.86±0.5 | NA |
| MRI+, n of patients (%) | 2 (22.2) | NA |
| Total GDP+, n of patients (%) | 4 (44.4) | NA |
| Encephalic GDP+, n. of patients (%) | 2 (22.2) | NA |
EDSS: expanded disability status scale; Previous attacks are intended as a number of two or more attacks preceding the study entry (day of diagnosis). Relapse within three months is intended within 3 months from the study entry (day of enrollment). CSF: Cerebrospinal Fluid; OB: Oligoclonal Bands; ESR: ErythroSedimentation Rate; CRP: C Reactive Protein; MRI+: MRI positive lesions in the spinal cord at study entry (day of diagnosis); Total and encephalic GDP+: Gadolinium phosphide-enhancing positive lesions in both spinal cord and encephalon, or only encephalon at study entry (day of diagnosis).
Discussion
In the present work, we defined a pathway through which infectious agents can directly modify trafficking properties of activated T cells.
CD44 is a family of closely related molecules obtained by alternative splicing of a single pre-mRNA and by post-transcriptional modifications of its product. All CD44 isoforms share the same ligand-binding site and trans-membrane and intracellular domains. The ligand-binding domain has a large repertoire of ligands, mostly consisting of the extracellular matrix molecules (Ponta et al., 2003). In general, it is believed that CD44 increases the adherence of cells to the extracellular matrix, leading to variation in homing properties. Our observation that conditions leading to early mobilization of T lymphocytes from the LN associate with a decrease in the level of CD44 expressed on the T cell surface is coherent with this hypothesis.
The complex modulation of alternatively spliced CD44 pre-mRNA has been the object of several studies, mostly in tumor cells (Zhang et al., 2021). The largest array of information regards its “proximal” regulation, highlighting the role of factors of the spliceosome in the production of variant isoforms of CD44 (Filippov et al., 2007; Goncalves et al., 2008; Loh et al., 2015; Midgley et al., 2017; Tripathi and Zhang, 2017). Although beyond the aim of current research, we highlighted a limited number of intermediate signaling pathways that led to modifications of CD44 alternative splicing. Work from several groups converge on a prominent role for the β-catenin pathway in regulating CD44 alternative splicing (Goncalves et al., 2008; Jiang et al., 2013; Todaro et al., 2014). It has been reported that TLR3 through TRAF-6 blocks the phosphorylation of β-catenin by ubiquitination of GSK-3β (Ko et al., 2015). Furthermore, β-catenin acts as a transcription factors for several splicing factors such as, among others, SRSF3 that has already been demonstrated to be involved in CD44 alternative splicing in cancer (Bordonaro, 2013; Goncalves et al., 2008; Thorsen et al., 2011). Our data indicate that this pathway is also operating in activated CD4+ T cells, where TLRs activation (frequently in tandem with cognate interaction) was able to block β-catenin phosphorylation and upregulated (at least in the SJL/J strain) the expression of three splicing factors, Nova-1, Yb-1, and Srsf3.
In addition, here we reported also that strain background and TLR2 haplotype (the genetic factors that modulate CD44 splicing) modulated the expression of mRNAs specific for proteins involved in the β-catenin pathway.
To the best of our knowledge, ours is the first report showing a role for TLRs in the regulation of CD44 alternative splicing, and on its regulation in T lymphocytes. We showed that several conditions concurred to determine the outcome in terms of CD44 repertoire, both in mouse and human: activation status, co-occurrence of the cognate interaction, type of TLR involved. In preliminary experiments, with both TLR2 and MyD88 KO mice, we observed absence of upregulation of isoform CD44v8-10, that occurs upon stimulation with PAM2/CSK4 of T cells from the C57Bl/6 mouse (Figure S5). Taken together, our results suggest that the pathway of signaling leading to reshuffling of the CD44 repertoire may proceed from TLR(s), through MyD88 to β-catenin and splicing factors (Martino et al., 2016).
CD44 variants are expressed by several types of cells, and in a cell-specific pattern (Weidle et al., 2011). In humans, lymphocytes express variant v6, i.e., the CD44 isoform that uses only exon 11. Variant v6 and other variants also encompassing exon 11 are also expressed by cancer cells with high metastatic potential, suggesting that the region encoded by exon 11 is involved in lymphocytes homing to LNs. Accordingly, here we report that a decreased expression of CD44 associates with early exit of T cells from LN and that upon antigen stimulation, blockade of CD44 increases the number of T cells able to cross Matrigel in vitro. In all experiments reported here in the mouse model, we observed a reduction of the expression levels of CD44s when cognate and TLR-mediated stimuli are simultaneously provided to T cells. However, we also show that a shift of the CD44 repertoire, promoting the expression of CD44v9-v10, occurs in these T cells. Moreover, this modification seemed to contribute to the mechanism(s) of crossing of Matrigel in a strain-specific manner. Overall, these data indicate that trafficking of T cells during immune responses is modulated in two distinct ways, by decreasing the expression of CD44 (presumably, mainly CD44s) and by reshuffling its isoform repertoire. Individual genetic variations of CD44 or in the regulation of splicing factors favor one mechanism over the other, as apparently verified in our studies in mice with SJL/J versus C57Bl/6 genetic background.
CD44 plays a dominant role in the ability of immune cells to enter the CNS. A detailed characterization of the distribution of 44-cell surface markers on cells from brain immune compartments indicated that expression of CD44 was the hallmark of all immune cells derived from the blood and able to enter the CNS in non-inflammatory conditions (Korin et al., 2017). CD44 collaborates with CD49 for the infiltration of CNS by leukocytes during EAE, and treatment with anti-CD44 blocks the development of the disease (Brennan et al., 1999). We observed that the presence of inflammatory infiltrates in the forebrain during EAE depends on TLR2 polymorphism at position 82 (Nicolò et al., 2013); we show here that the same polymorphism regulates specifically the production of CD44v9-v10 isoform, that in turn associated specifically to cell licensed to infiltrate the forebrain at the onset of EAE. Our results also indicate that TLR2 stimulation modified the binding of CD44 to OPN. OPN has a complex role, acting as an extracellular matrix protein, as well as a chemokine and cytokine. Interestingly, OPN is present in different isoforms along the CNS. Thus, we can reasonably propose that a CD44 isoform (most likely, v9-v10) could drive the selective homing of T cells into specific areas of the CNS by interacting with one specific isoform of OPN.
It is known that infectious episodes often drive flares of MS (Perry et al., 2007). The distribution of infiltrates and lesions during MS is particularly unpredictable within each acute bout of disease and over time, along the course of MS (Lindner et al., 2018). Our observations illustrate this mechanism for infectious agents in the determination of autoimmune diseases, adding to those of molecular mimicry and T cell phenotype modulation that were reported by Miyauchi and collaborators (Miyauchi et al., 2020), and that we suggested a few years ago in our previous work in which infection with a non-pathogenic mycobacterium cross-reactive with PLP139-151 was able to induce EAE (Nicolò et al., 2010). Here, we report that TLRs represent a path through which commensals as well as pathogenic infectious agents can modulate trafficking properties of T cells, ultimately leading to infiltration of the CNS. Our observation suggests that infectious agents can play a role in determining the distribution of lesions, acting along the pathway described in this work. In real life situation, however, the relation infectious agent-CNS infiltration will possibly be less linear, given the influence of individual genetic background and of the concomitant presence of a large commensal infectious flora.
The use of anti-CD44 antibodies in human therapy leads to development of severe side effects (Tijink et al., 2006), while the effectiveness of treatment with anti-CD49 of MS shows that interfering with the specific pathways involved in T cell trafficking into the CNS is a promising approach. Treatment aimed at the fine-tuning of the repertoire of CD44 or to prevent infiltration of leukocytes at specific sites may provide new tools for the treatment of MS and other autoimmune diseases.
In conclusion, our data show that pathogens and commensals can modify trafficking properties of activated T lymphocytes, by engaging TLRs and modifying the repertoire of CD44 isoforms. This pathway can play a relevant role in the determination of flares and/or remission of autoimmune diseases, even in the absence of direct cross-recognition by previously activated, self-specific T cells, potentially representing a new target for the therapy of brain inflammatory diseases. Future perspectives of this work and its possible evolutions will aim to explain the complex topic of the causative relations of each single alternatively spliced CD44vv in regulating T cell trafficking, exploring, for example, the influences of simultaneous expression of the standard isoform or of other isoforms on the same cell, or the interaction of other adhesion molecules such as (e.g.) CD49, or dissecting the impact of still poorly studied splicing factors specifically involved in the modulation of alternative splicing of CD44.
Limitations of the study
As stated above, ours is the first report describing a TLRs alternatively spliced CD44 axis in T lymphocytes. Although we suggest that the pathway of signaling leading to reshuffling of the CD44 repertoire possibly proceeds from TLR(s), through MyD88 to β-catenin and splicing factors, the specific pathway(s) downstream TLR has still to be fully described and detailed because engagement of different TLRs appears to lead to different CD44 isoforms repertoires. Moreover, we focused on T cells and on a specific disease model, but CD44v are also expressed by other cell types, also involved in autoimmune disorders. Thus, we expect that the mechanism here hypothesized is more complex and deserves to be deepened. Although we identified the potential ligand(s) of CD44 isoform(s) in the CNS, this aspect should be expanded, testing other possible components of the ECM that could have different affinity for each CD44v or that could interact with them in a complex manner dependent on the target organ. Finally, our data on CSF and PBMC from a cohort of RR-MS patients, although confirming our observations on EAE model, showed differences in terms of splice variants, suggesting, despite a similar mechanism, substantial differences between human and experimental disease that deserves to be deepened.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD49d (α4-integrin/VLA4) | BD Biosciences | Cat# 564397; RRID:AB_2738789 |
| CD11a (LFA-1) | BD Biosciences | Cat# 746310; RRID:AB_2743634 |
| CD62L (L-Selectin) | BD Biosciences | Cat# 746726; RRID:AB_2743990 |
| CD44 antibody | BD Biosciences | Cat# 746710, RRID:AB_2743977 |
| CD4 MicroBeads, human | Miltenyi Biotec | Cat#130-045-101,RRID:AB_2889919 |
| TCR vβ10 PE-conjugated | BD Biosciences | Cat# 553285, RRID:AB_394757 |
| APC-conjugated IgG | BD Biosciences | Cat# 554014, RRID:AB_395209 |
| β-Catenin Antibody | Cell Signaling Technology | Cat# 9562, RRID:AB_331149 |
| Phospho- β-Catenin (Ser675) | Cell Signaling Technology | Cat# 4176, RRID:AB_1903923 |
| β-Actin | Cell Signaling Technology | Cat# 12620, RRID:AB_2797972 |
| Anti-Rabbit secondary antibody | ThermoFisher Scientific | Cat# 31466, RRID:AB_10960844 |
| Biological samples | ||
| Cerebrospinal fluid (CSF) from MS patients | Università Cattolica del Sacro Cuore | N/A |
| Whole blood samples from MS patients and healthy donors | Università Cattolica del Sacro Cuore | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Pam2CSK4 | InvivoGen | tlrl-pm2s-1 |
| Pam3CSK4 | InvivoGen | tlrl-pms |
| LPS-B5 (Ultrapure) | InvivoGen | tlrl-pb5lps |
| ODN 1585 (CpG) | InvivoGen | tlrl-1585 |
| Critical commercial assays | ||
| Cell Trace™ CFSE Cell Proliferation Kit | Invitrogen | C34554 |
| CD4+ CD62L+ T Cell Isolation Kit mouse | Miltenyi Biotec | 130-106-643 |
| T Cell Activation/Expansion Kit mouse | Miltenyi Biotec | 130-093-627 |
| T Cell Activation/Expansion Kit human | Miltenyi Biotec | 130-091-441 |
| Bordetella Pertussis toxin | Sigma-Aldrich | P7208 |
| Freund′s Adjuvant, Complete | Sigma-Aldrich | F5881 |
| PLP (139-151) | Bio-techne | 2567/1 |
| Matrigel® Basement Membrane Matrix | Corning | 356234 |
| Collagen, Type I | Sigma-Aldrich | C3867 |
| Osteopontin | Sigma-Aldrich | O2260 |
| Cell Counting Kit (CCK) | Sigma-Aldrich | 03285 |
| WT Expression Kit | Thermo Fisher Scientific | 4411974 |
| GeneChip® WT Terminal Labeling kit | Thermo Fisher Scientific | 901524 |
| SYBR® Green Supermix | Biorad | 170-8882: |
| Experimental models: Cell lines | ||
| Primary Human CD4+ T cells cultures | This study | N/A |
| Primary Murine splenocyte cultures | This study | N/A |
| Experimental models: Organisms/strains | ||
| SJL/J | Charles River Laboratories | RRID:IMSR JAX:000686 |
| C57BL/6NCrl | Charles River Laboratories | RRID:IMSR_CRL:027 |
| C57BL/6TLR2−/− | Jackson Laboratories | RRID:IMSR_JAX:004650 |
| C57BL/6MYD88−/− | Jackson Laboratories | RRID:IMSR_JAX:009088 |
| SJL/Jvb10+ | Polygene Transgenetics | N/A |
| Oligonucleotides | ||
| All human primers used are listened in Table S1 | This paper | N/A |
| All mouse primers used are listened in Table S2 | This paper | N/A |
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com RRID:SCR_002798 |
| Kaluza Software | Beckman Coulter | https://www.beckman.it/flow-cytometry/software/kaluza RRID:SCR_016182 |
| TAC Software | Thermo Fisher Scientific | https://www.thermofisher.com/it/en/home/global/forms/life-science/download-tac-software.html RRID:SCR_016519 |
| I-TASSER | University of Michigan | www.zhanggroup.org/I-TASSER/RRID:SCR_014627 |
| Desmond | D. E. Shaw Research | https://www.deshawresearch.com/resources_desmond.html RRID:SCR_014575 |
| Alliance Q9 Advanced | UVITEC Cambridge | https://www.uvitec.co.uk/alliance-q9-advanced/N/A |
| SFmap | Paz et al. (2010) | http://sfmap.technion.ac.il/N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gabriele Di Sante (gabriele.disante@unipg.it)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human CSF and blood collection
Thirty-four RR-MS patients were recruited at MS Centers at the “Fondazione Policlinico Universitario A. Gemelli” - Catholic University of the Sacred Heart of Rome (demographic and clinical characteristics are summarized in Table 2). All patients were affected by Relapsing-Remitting (RR)MS defined according to revised McDonald′s criteria (Thompson et al., 2018). At the time of enrollment patients were free from immunomodulatory (never treated with disease-modifying drugs or therapy stopped by more than 6 months earlier) or immunosuppressive treatment (no immunosuppressive treatment for at least 2 years). From the whole cohort, we collected cerebrospinal fluid (CSF) samples that we analyzed for CD44 variants as described above.
Mice
For all experiments, only 6–8 weeks old female mice were used. The breeding, the backcrossing and the maintenance of mice were performed at the animal facility of Università Cattolica del Sacro Cuore, CenRis. All experimental work has been conducted in accordance with relevant national legislation on the use of animals for research, referring to the Code of Practice for the Housing and Care of Animals Used in Scientific Procedures and the protocol was approved by the Ethics Committee of animal welfare organization (OPBA) of the “Università Cattolica del Sacro Cuore” of Rome and by the Italian Ministry of Health (authorization number 321/2017-PR, protocol number 1F295.34/04-11-2016, date of approval April, 12th 2017).
Methods details
Animal procedures
The RR-EAE have been induced by immunizing SJL/J mice subcutaneously (s.c.) at the base of the tail with an emulsion of myelin antigen PLP139-151 (75 μg/20g, purchased from Primmbiotech, Italy) and 4X-concentrated complete Freund′s adjuvant (CFA, 200μg of Mycobacterium Tuberculosis wall for each mouse of about 20g, purchased from Sigma-Aldrich, Merck, Germany). To help breaking the BBB, a double dosage of Bordetella Pertussis toxin (BDT, total 300 ng/20g mouse, purchased from Sigma-Aldrich, Merck, Germany) have been administered at day 0 and 3. After treatment, mice of all groups have been daily monitored for body weight and the development of clinical signs and symptoms (CSS) following the score scale described in (Baker and Amor, 2012; Camponeschi et al., 2021; Miller et al., 2010; Nicolò et al., 2006). According to this score scale, the EAE scale comprises a range of value between 0 (no clinical symptoms observed) and 3 (mouse paralysis). After perfusion with PBS, from each EAE affected mouse different samples were collected: spleen, blood and CNS of which one hemisphere and the spinal cord have been employed for morphological analyses and the other for molecular analyses. Control mice were sacrificed accordingly. The cellular studies were performed after the collection of splenocytes of different mouse strains: SJL/Jwt, C57BL/6wt, C57BL/6TLR2−/−, F1(SJL/Jwt× C57BL/6TLR2−/−), F1(SJL/Jwt× C57BL/6wt), C57BL/6MYD88−/−, SJL/Jvb10+. The TLR2-KO mice was purchased from Charles River (Nicolò et al., 2013) while the insertion of the vβ10-transgene was generated by Polygene Transgenetics (Rümlang, Switzerland) in C57BL/6 mice and then transferred (after more than 10 generation) to SJL/J strain (Piermattei et al., 2016). For flow cytometric analysis the strains of mice listed above were immunized s.c. with PLP139-151 (50 μg/20g) emulsified alternatively with 1X or 4X CFA (50 or 200 μg/20g). Draining popliteal lymph nodes (LN) were collected 8 days after the immunizations.
Cell culture
We performed an in vitro experiment, on naÏve splenocytes collected from 8-10 weeks old female SJL/Jwt, C57Bl/6wt and F1 SJL/J hybridized C57Bl/6wt or C57Bl/6tlr2−. After the enrichment of T helper cells using a negative magnetic separation cocktail purchased from Miltenyi Biotec, we subdivided these cells using CD62L magnetic antibody provided in the same kit; after checking the purity of the sorted subpopulation was higher than 95%, we measured the ability of CD4+/CD62Llow T cells to interact with extracellular matrix in vitro using 96 transwells with 5 μm pores and Matrigel in a standard assay or polymerized with osteopontin (100 ng/μL) or type I collagen (4 mg/mL), upon different stimulations in the presence or absence of synthetic ligands for TLR2, such as PAM2/CSK4 (100 ng/mL), PAM3/CSK4 (100 ng/mL) or PPD (25 μg/mL), for TLR4, such as LPS (20 μg/mL), for TLR9, such as CpG (2 ng/mL) and of T cell activation microparticles for mouse (with αCD3/αCD28) and for human (αCD3/αCD28/αCD2). The assay was performed also in the presence of blocking, anti-pan CD44 mAb (clone Rat IgG2a, clone MJ-64), to assess the contribution of this molecule to modification of trafficking. T cells were cultured in this invasion test for 6, 12 and 24 h and at each time point have been harvested from the upper and lower chamber and recovered from the Matrigel and counted using flow cytometer (Cytoflex) and software (Kaluza 2.1.1.) and analyzed with a proliferation assay (CCK8) evaluating absorbance with microplates reader (Tecan, Switzerland).
In addition, we enriched CD4+CD62Llow T cells from the spleens of naÏve SJL/Jwt (n = 3) and C57Bl/6wt (n = 4) mice and cultured them in the absence of any stimulus, in the absence or presence of TLR-9,-4, -2/6 or -2/1 ligands (respectively CpG, LPS, PAM2/CSK4 or PAM3/CSK4, alone or combined with microparticles conjugated with αCD3/αCD28 (mouse T cell activation kit from Miltenyi Biotec). After 16 h, cells were collected, and the mRNA was extracted. RT-qPCR was performed as described below for CD44 splicing variants, splicing factors and housekeeping genes.
For human cell cultures we enriched CD4+ T cells (Miltenyi Biotec) from PBMC (separated with gradient of Lympholite, Cedarlane) of healthy donors (n = 3) and MS patients (n = 7) and cultured in the absence of any stimulus, in the absence or presence of TLR-9,-4, -2/6 or -2/1 ligands (respectively CpG, LPS, PAM2/CSK4 or PAM3/CSK4, alone or combined with microparticles conjugated with human T cell activation kit. After 24 h, cells were harvested, and the mRNA was extracted. RT-qPCR was performed as described below for CD44 splicing variants and housekeeping genes.
Flow cytometry
We examined the expression on the T cell surface of 4 markers known to play a relevant role in the interaction between leukocytes and endothelia, namely α4-integrin/VLA4 (CD49d), LFA1 (CD11a), L-selectin (CD62L), CD44, used at a concentration of 5 μg/106 cells. To examine the level of expression of these proteins only on activated ag-specific T cells by flowcytometry, we took advantage of a mouse SJL/J strain transgenic for the β-chain of a public TCR specific for PLP139-151 in the same strain (Nicolò et al., 2006, 2010). This strain was obtained by back crossing the C57Bl/6vβ10+ transgenic mouse described in three for more than 10 generations onto the SJL/J strain (SJL/Jvβ10+).Reflecting the general characteristics of the pathogenic PLP139-151-specific repertoire (Penitente et al., 2008).T cells expressing this transgenic β-chain and the appropriate β-chain are not spontaneously activated in vivo but become activated by PLP139-151upon immunization with the peptide.
SJL/Jvβ10+ mice were immunized with PLP139-151 in IFA containing 200 (fast mobilizer) or 50 (slow mobilizer) μg/20g of heat-killed Mtb, and F1(SJL/Jvβ10+xB6wt) (fast mobilizer) and F1(SJL/Jvβ10+xB6tlr2−) (slow mobilizer) were challenged with the same amount of PLP139-151 in IFA containing 50 μg/20g of heat-killed Mtb. Cells from popliteal draining lymph nodes were recovered at day 8 post-immunization, labeled with 1.5 μM of carboxy-fluorescein succinimidyl ester (CFSE, Invitrogen, Thermo Fisher Scientific) and stimulated in vitro with PLP139-151. Three days later, cells were recovered stained for TCR vβ10 PE-conjugated (BD Pharmigen, Thermo Fisher Scientific), CD49d, CD11a, CD62L and CD44 above described and combined with an APC-conjugated Goat anti-rat IgG (BD Pharmigen, Thermo Fisher Scientific), and the expression of these markers was compared on vβ10high/CFSElow cells, i.e. T cells that had proliferated in response to PLP139-151. Gating strategy and results are reported in Figure 1.
RT–qPCR assays
CNS mouse samples were collected as described above, while human cerebrospinal fluids (CSF) were centrifugated at 1,200 rpm for 10 min and pellet were stored in TRIzol™ reagent at −80°C. Total RNA was isolated from mice and human samples with Direct-zol Rna Micro-Prep (Zymoresaerch). RNA concentration was evaluated by spectrophotometric reading at 280 and 260 nm. Total RNA was used for first strand cDNA synthesis with cDNA Sensifast kit (Bioline). The quantification of gene expression was obtained from Biorad instruments and products (iQ5 Multicolor Real-Time PCR Detection System, SYBR Green Master Mix and semiskirted 96 well plates) according to the manufacturer′s recommendations. Primers were deduced from literature both for human (Sugano et al., 2011; Wang et al., 2017) (Table S1) and murine (Bader et al., 2003; Cao and Malon, 2018; Giampietro et al., 2015; Piermattei et al., 2016; Sen et al., 2013) (Table S2) sequences genes. Each gene target quantification reaction was performed separately with the respective primer sets. Conditions were set as follows: 50°C for 2 min, followed by 95°C for 10 min, forty cycles at 95°C for 15 s, followed by 60°C for 1 min. The melt standard curve was at 95°C for 15 s, followed by 60°C for 1 min, 95°C for 15 s, and finally, 60°C for 15 s. Gene expression results were analyzed using Biorad software. For the expression of murine CD44 variants (standard, v1-v10, v8-v10 and v9-v10) ready-to-use-assays have been purchased (Figure S6) following Applied Biosystems protocol of Taqman Universal Master Mix II with UNG. Relative mRNA expression levels were calculated by normalizing examined genes against β-actin using the 2−ΔCt method.
Model building and simulation of molecular dynamics
The molecular models of the CD44 isoforms were built using I-TASSER, a metaserver that automatically employs ten threading algorithms in combination with ab initio modeling to generate the tertiary structure of a protein as well as replica-exchange Monte Carlo dynamics (REMD) simulations for the atomic-level refinement (Pirolli et al., 2014; Roy et al., 2010). The quality of the models generated by I-TASSER are evaluated accordingly to the C-score value, provided by the same program. The C-score was calculated based on the significance of the threading alignments and the convergence of the I-TASSER simulations. C-scores typically range from −5 to 1, with higher scores reflecting a model of better quality. To take into account protein flexibility and solvent effects, the behavior of the model structures was studied in a dynamic context by molecular dynamics (MD) simulations and the solvent accessible surface area (SASA) was monitored (Righino et al., 2018). Average SASA values from MD simulations may therefore estimate the surface area that is available for contact with solvents and other molecules. Indeed, surface properties play a key role in determining the interactions between the protein and various other molecules and solvent and may provide important information on functional properties.
Molecular dynamics (MD) simulations of the model structures were executed using Desmond, version Build 13 (D. E. Shaw Research, New York) as implemented in the Schrodinger Suite 2020-1 (Schrödinger Inc.) employing the OPLS2005 force field (Shivakumar et al., 2010). The systems for the simulations were built with the “System Builder” tool of the Maestro suite (Schrödinger Inc.), drawing an orthorombic box around each CD44 isoform with a distance buffer of 10 Å between the protein and the side of the box, and filling it with SPC water molecules. A 0.15 M NaCl salt concentration was added, and the system was neutralized by additional Na+ ions. The Particle-Mesh Ewald method was used to calculate long-range electrostatic interactions and the SHAKE algorithm was used to constrain the bonds. van der Waals and short-range electrostatic interactions were smoothly truncated at 9.0 Å. The systems were equilibrated with the default protocol provided in Desmond. Subsequently, 200 ns MD simulations were carried for each system and the SASA calculated from the trajectories was analyzed using VMD (Humphrey et al., 1996).
Gene expression microarray
Total RNA (100ng/each sample or each control) was processed using the Ambion WT Expression kit; 5.5 μg of ss-cDNA were fragmented with GeneChip WT Terminal Labeling kit and labeled with biotin using terminal deoxynucleotidyl transferase, before hybridization (50-pM control oligonucleotide B2; 15, 5, 25, and 100 p.m., respectively, of BioB, BioC, BioD, and cre hybridization controls; 1× hybridization mix; 10% Dimethylsulfoxide; nuclease-free water): this mixture was heated to 99°C for 5min and to 45°C for another 5min. Then, it was injected into Affymetrix Mouse Exon 1.0 ST microarray chips, and the hybridization was performed under rotation at 45°C ± 16 h. The GeneChip Mouse Exon 1.0 ST Array is a single array with over 4.5 million unique 25-mer oligonucleotide features constituting approximately 1.2 million probe sets. For the gene-level analysis, the chip contains about 23,339 probe sets (Piermattei et al., 2016).
Western blot
Splenocytes were collected from naïve female SJL/J mice. CD4+CD62Llow T cells were enriched using CD4+ CD62L+ T cell Isolation Kit II. Cells were plated and stimulated in vitro with PAM2/CSK4, PAM3/CSK4 and αCD3/αCD28 T cell activation microparticles for mouse. After 1 and 6 h, cells were harvested, washed with PBS 1X. Cell pellets were stored at −80°C. Control and treated cells were lysate in ice-cold RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) completed with Halt Protease Inhibitor Cocktail and Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). The protein lysates (30 μg) were separated on Bolt 4–12% SDS–polyacrilamide gel (Thermo Fisher Scientific) and electroblotted onto PolyVinylidene-DiFluoride membrane (PVDF, Amersham, Merck, Germany). Nonspecific binding sites were blocked by SuperBlockTM (TBS) Blocking Buffer (Thermo Fisher Scientific) for 1 h at room temperature. Membranes were then incubated overnight at 4°C with the following primary antibodies, diluted in TBS-T and 5% non-fat dry milk or 5% BSA (Sigma-Aldrich, Merck): β-catenin 1:1000, P-β-catenin 1:1000, β-actin 1:1000. The day after, membranes were incubated with the secondary, HRP-conjugated anti-rabbit antibody (Thermo Fisher Scientific) for 2 h. Immunoreactive bands were visualized by PierceTM ECL Western Blotting Substrate (Thermo Fisher Scientific) according to manufacturer′s instructions. The densitometry analysis was determined and normalized to B-actin using Alliance Q9 Advanced UVITEC software.
Quantification and statistical analysis
Student′s t-test, one-way ANOVA or two-way ANOVA were performed to examine the effects and possible interaction of independent variables (GraphPad, Prism 9.3 software).When appropriate, post hoc comparisons were made using Tukey′s HSD, with a significance level of p< 0.05. For statistical analysis of qPCR data, the unpaired t-test was used to compare ΔCt values across the replicates, setting the pvalue cut-off at 0.05. A gene-level analysis was performed by TAC software: a total of 23,339 genes were tested to compare their expression between three F1 (SJL/Jwt × C57Bl/6Tlr2−) samples and three F1 (SJL/Jwt × C57Bl/6wt) samples. The ANOVA was used for the detection of differentially expressed genes (DE-Gs). Only the probe sets whose fold changes were higher than or equal to 1.5 and p value of 0.05 were selected as modulated (Piermattei et al., 2016).
Acknowledgments
This research was funded by linea D1 – Università Cattolica del Sacro Cuore and FISM (Fondazione Italiana Sclerosi Multipla) cod 2016/R/22 (FR and GDS) and co-financed with 5-per-mille public funding; European Research Council (ERC) advanced grant nr. 697 695714 IMMUNOALZHEIMER (GC).
Author contributions
Conceptualization, F.R., G.D.S.; methodology, G.D.S.; software, G.D.S., M.T., C.C., D.P., M.Fi., M.R., M.F., B.R., M.C.D.R.; validation; formal analysis: M.T., G.D.S., M.V., C.C., C.M., M.C.G., E.G.; investigation; data curation: G.D.S., M.T., M.L., M.M., M.C.G.,; writing—original draft preparation: F.R., G.D.S., M.F., G.C.; writing—review and editing, M.T., G.D.S., F.R., D.P., M.C.D.R., M.F., G.C., M.Fi., M.R., M.C.G., M.M.; visualization, F.R.; supervision, G.D.S., F.R; project administration, G.D.S.; funding acquisition, G.D.S., F.R. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103763.
Contributor Information
Francesco Ria, Email: francesco.ria@unicatt.it.
Gabriele Di Sante, Email: gabriele.disante@unipg.it.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
All additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baaten B.J., Li C.-R., Bradley L.M. Multifaceted regulation of T cells by CD44. Commun.Integr. Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader A.G., Felts K.A., Jiang N., Chang H.W., Vogt P.K. Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12384–12389. doi: 10.1073/pnas.2135336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D., Amor S. Publication guidelines for refereeing and reporting on animal use in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2012;242:78–83. doi: 10.1016/j.jneuroim.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Bordonaro M. Crosstalk between Wnt signaling and RNA processing in colorectal cancer. J. Cancer. 2013;4:96–103. doi: 10.7150/jca.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan F.R., Mikecz K., Glant T.T., Jobanputra P., Pinder S., Bavington C., Morrison P., Nuki G. CD44 expression by leucocytes in rheumatoid arthritis and modulation by specific antibody: implications for lymphocyte adhesion to endothelial cells and synoviocytes in vitro. Scand. J. Immunol. 1997;45:213–220. doi: 10.1046/j.1365-3083.1997.d01-382.x. [DOI] [PubMed] [Google Scholar]
- Brennan F.R., O’Neill J.K., Allen S.J., Butter C., Nuki G., Baker D. CD44 is involved in selective leucocyte extravasation during inflammatory central nervous system disease. Immunology. 1999;98:427–435. doi: 10.1046/j.1365-2567.1999.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvier L., Demuth G., Manouchehri N., Wong C., Sacharidou A., Mineo C., Shaul P.W., Monson N.L., Kounnas M.Z., Stüve O., et al. Reelin depletion protects against autoimmune encephalomyelitis by decreasing vascular adhesion of leukocytes. Sci. Transl.Med. 2020;12:eaay7675. doi: 10.1126/scitranslmed.aay7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camponeschi C., De Carluccio M., Amadio S., Clementi M.E., Sampaolese B., Volonté C., Tredicine M., Romano Spica V., Di Liddo R., Ria F., et al. S100B protein as a therapeutic target in multiple sclerosis: the S100B inhibitor arundic acid protects from chronic experimental autoimmune encephalomyelitis. IJMS. 2021;22:13558. doi: 10.3390/ijms222413558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Malon J.T. Anti-nociceptive role of CXCL1 in a murine model of peripheral nerve injury-induced neuropathic pain. Neuroscience. 2018;372:225–236. doi: 10.1016/j.neuroscience.2017.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sante G., Migliara G., Valentini M., Delogu G., Ria F. Regulation of and regulation by CD 44: a paradigm complex regulatory network. Int. Trends Immun. 2013;1:33–41. [Google Scholar]
- Fagone P., Mazzon E., Cavalli E., Bramanti A., Petralia M.C., Mangano K., Al-Abed Y., Bramati P., Nicoletti F. Contribution of the macrophage migration inhibitory factor superfamily of cytokines in the pathogenesis of preclinical and human multiple sclerosis: in silico and in vivo evidences. J. Neuroimmunol. 2018;322:46–56. doi: 10.1016/j.jneuroim.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Fallarino F., Gargaro M., Mondanell G., Downer E.J., Hossain M.J., Gran B. Delineating the role of Toll-like receptors in the neuro-inflammation model EAE. Methods Mol. Biol. 2016;1390:383–411. doi: 10.1007/978-1-4939-3335-8_23. [DOI] [PubMed] [Google Scholar]
- Filippov V., Filippova M., Duerksen-Hughes P.J. The early response to DNA damage can lead to activation of alternative splicing activity resulting in CD44 splice pattern changes. Cancer Res. 2007;67:7621–7630. doi: 10.1158/0008-5472.CAN-07-0145. [DOI] [PubMed] [Google Scholar]
- Galluzzo E., Albi N., Fiorucci S., Merigiola C., Ruggeri L., Tosti A., Grossi C.E., Velardi A. Involvement of CD44 variant isoforms in hyaluronate adhesion by human activated T cells. Eur. J. Immunol. 1995;25:2932–2939. doi: 10.1002/eji.1830251033. [DOI] [PubMed] [Google Scholar]
- Ghazi-Visser L., Laman J.D., Nagel S., van Meurs M., van Riel D., Tzankov A., Frank S., Adams H., Wolk K., Terracciano L., et al. CD44 variant isoforms control experimental autoimmune encephalomyelitis by affecting the lifespan of the pathogenic T cells. FASEB J. 2013;27:3683–3701. doi: 10.1096/fj.13-228809. [DOI] [PubMed] [Google Scholar]
- Giampietro C., Deflorian G., Gallo S., Di Matteo A., Pradella D., Bonomi S., Belloni E., Nyqvist D., Quaranta V., Confalonieri S., et al. The alternative splicing factor Nova2 regulates vascular development and lumen formation. Nat. Commun. 2015;6:8479. doi: 10.1038/ncomms9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V., Matos P., Jordan P. The -catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14:2538–2549. doi: 10.1261/rna.1253408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraju P., Todd L., Shetye S., Monslow J., Puré E. CD44-dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol. 2019;75:314–330. doi: 10.1016/j.matbio.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.C., Schulte-Mecklenbeck A., Hanning U., Posevitz-Fejfár A., Korsukewitz C., Schwab N., Meuth S.G., Wiendl H., Klotz L. Distinct pattern of lesion distribution in multiple sclerosis is associated with different circulating T-helper and helper-like innate lymphoid cell subsets. Mult.Scler. J. 2017;23:1025–1030. doi: 10.1177/1352458516662726. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- Idris M., Harmston N., Petretto E., Madan B., Virshup D.M. Broad regulation of gene isoform expression by Wnt signaling in cancer. RNA. 2019;25:1696–1713. doi: 10.1261/rna.071506.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Crossman D.K., Mitchell E.H., Sohn P., Crowley M.R., Serra R. WNT5A inhibits metastasis and alters splicing of Cd44 in breast cancer cells. PLoS ONE. 2013;8:e58329. doi: 10.1371/journal.pone.0058329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko R., Park J.H., Ha H., Choi Y., Lee S.Y. Glycogen synthase kinase 3β ubiquitination by TRAF6 regulates TLR3-mediated pro-inflammatory cytokine production. Nat. Commun. 2015;6:6765. doi: 10.1038/ncomms7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin B., Ben-Shaanan T.L., Schiller M., Dubovik T., Azulay-Debby H., Boshnak N.T., Koren T., Rolls A. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci. 2017;20:1300–1309. doi: 10.1038/nn.4610. [DOI] [PubMed] [Google Scholar]
- Latini A., Novelli L., Ceccarelli F., Barbati C., Perricone C., De Benedittis G., Conti F., Novelli G., Ciccacci C., Borgiani P. mRNA expression analysis confirms CD44 splicing impairment in systemic lupus erythematosus patients. Lupus. 2021;30:1086–1093. doi: 10.1177/09612033211004725. [DOI] [PubMed] [Google Scholar]
- Lindner M., Klotz L., Wiendl H. Mechanisms underlying lesion development and lesion distribution in CNS autoimmunity. J. Neurochem. 2018;146:122–132. doi: 10.1111/jnc.14339. [DOI] [PubMed] [Google Scholar]
- Loh T.J., Moon H., Cho S., Jang H., Liu Y.C., Tai H., Jung D.-W., Williams D.R., Kim H.-R., Shin M.-G., et al. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol. Rep. 2015;34:1231–1238. doi: 10.3892/or.2015.4110. [DOI] [PubMed] [Google Scholar]
- Luz A., Fainstein N., Einstein O., Ben-Hur T. The role of CNS TLR2 activation in mediating innate versus adaptive neuroinflammation. Exp. Neurol. 2015;273:234–242. doi: 10.1016/j.expneurol.2015.08.021. [DOI] [PubMed] [Google Scholar]
- Martino M.M., Maruyama K., Kuhn G.A., Satoh T., Takeuchi O., Müller R., Akira S. Inhibition of IL-1R1/MyD88 signalling promotes mesenchymal stem cell-driven tissue regeneration. Nat. Commun. 2016;7:11051. doi: 10.1038/ncomms11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., Kubes P. Interactions between CD44 and hyaluronan in leukocyte trafficking. Front.Immunol. 2015;6:68. doi: 10.3389/fimmu.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley A.C., Oltean S., Hascall V., Woods E.L., Steadman R., Phillips A.O., Meran S. Nuclear hyaluronidase 2 drives alternative splicing of CD44 pre-mRNA to determine profibrotic or antifibrotic cell phenotype. Sci. Signal. 2017;10:eaao1822. doi: 10.1126/scisignal.aao1822. [DOI] [PubMed] [Google Scholar]
- Miller S.D., Karpus W.J., Davidson T.S. Experimental autoimmune encephalomyelitis in the mouse. Current Protocols in Immunology. 2010;88:15.1.1–15.1.20. doi: 10.1002/0471142735.im1501s88. [DOI] [PubMed] [Google Scholar]
- Miyauchi E., Kim S.-W., Suda W., Kawasumi M., Onawa S., Taguchi-Atarashi N., Morita H., Taylor T.D., Hattori M., Ohno H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020;585:102–106. doi: 10.1038/s41586-020-2634-9. [DOI] [PubMed] [Google Scholar]
- Mousavi A. CXCL12/CXCR4 signal transduction in diseases and its molecular approaches in targeted-therapy. Immunol.Lett. 2020;217:91–115. doi: 10.1016/j.imlet.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Nicolò C., Di Sante G., Orsini M., Rolla S., Columba-Cabezas S., Romano Spica V., Ricciardi G., Chan B.M.C., Ria F. Mycobacterium tuberculosis in the adjuvant modulates the balance of Th immune response to self-antigen of the CNS without influencing a “core” repertoire of specific T cells. Int. Immunol. 2006;18:363–374. doi: 10.1093/intimm/dxh376. [DOI] [PubMed] [Google Scholar]
- Nicolò C., Sali M., Di Sante G., Geloso M.C., Signori E., Penitente R., Uniyal S., Rinaldi M., Ingrosso L., Fazio V.M., et al. Mycobacterium smegmatis expressing a chimeric protein MPT64-proteolipid protein (PLP) 139-151 reorganizes the PLP-specific T cell repertoire favoring a CD8-mediated response and induces a relapsing experimental autoimmune encephalomyelitis. J. Immunol. 2010;184:222–235. doi: 10.4049/jimmunol.0804263. [DOI] [PubMed] [Google Scholar]
- Nicolò C., Di Sante G., Procoli A., Migliara G., Piermattei A., Valentini M., Delogu G., Cittadini A., Constantin G., Ria F. M tuberculosis in the adjuvant modulates time of appearance of CNS-specific effector T cells in the spleen through a polymorphic site of TLR2. PLoS ONE. 2013;8:e55819. doi: 10.1371/journal.pone.0055819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli L., Barbati C., Ceccarelli F., Perricone C., Spinelli F.R., Alessandri C., Valesini G., Perricone R., Conti F. CD44v3 and CD44v6 isoforms on T cells are able to discriminate different disease activity degrees and phenotypes in systemic lupus erythematosus patients. Lupus. 2019;28:621–628. doi: 10.1177/0961203319838063. [DOI] [PubMed] [Google Scholar]
- Oukka M., Bettelli E. Regulation of lymphocyte trafficking in central nervous system autoimmunity. Curr.Opin.Immunol. 2018;55:38–43. doi: 10.1016/j.coi.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I., Akerman M., Dror I., Kosti I., Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38:W281–W285. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penitente R., Nicolò C., Van den Elzen P., Di Sante G., Agrati C., Aloisi F., Sercarz E.E., Ria F. Administration of PLP 139–151primes T cells distinct from those spontaneously responsive in vitro to this antigen. J. Immunol. 2008;180:6611–6622. doi: 10.4049/jimmunol.180.10.6611. [DOI] [PubMed] [Google Scholar]
- Perry V.H., Cunningham C., Holmes C. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 2007;7:161–167. doi: 10.1038/nri2015. [DOI] [PubMed] [Google Scholar]
- Piermattei A., Migliara G., Di Sante G., Foti M., Hayrabedyan S.B., Papagna A., Geloso M.C., Corbi M., Valentini M., Sgambato A., et al. Toll-like receptor 2 mediates in vivo pro- and anti-inflammatory effects of Mycobacterium tuberculosis and modulates autoimmune encephalomyelitis. Front.Immunol. 2016;7:191. doi: 10.3389/fimmu.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirolli D., Sciandra F., Bozzi M., Giardina B., Brancaccio A., De Rosa M.C. Insights from molecular dynamics simulations: structural basis for the V567D mutation-induced instability of zebrafish alpha-dystroglycan and comparison with the murine model. PLoS ONE. 2014;9:e103866. doi: 10.1371/journal.pone.0103866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H., Sherman L., Herrlich P.A. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Righino B., Galisson F., Pirolli D., Vitale S., Réty S., Gouet P., De Rosa M.C. Structural model of the full-length Ser/Thr protein kinase StkP from S. pneumoniae and its recognition of peptidoglycan fragments. J. Biomol.Struct.Dyn. 2018;36:3666–3679. doi: 10.1080/07391102.2017.1395767. [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor A.M., Jacobelli J., Friedman R.S. Immune cell trafficking to the islets during type 1 diabetes. Clin. Exp. Immunol. 2019;198:314–325. doi: 10.1111/cei.13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S., Jumaa H., Webster N.J.G. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat. Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theor. Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Strazza M., Azoulay-Alfaguter I., Silverman G.J., Mor A. T cell chemokine receptor patterns as pathogenic signatures in autoimmunity. Discov. Med. 2015;19:117–125. [PubMed] [Google Scholar]
- Sugano E., Isago H., Wang Z., Murayama N., Tamai M., Tomita H. Immune responses to adeno-associated virus type 2 encoding channelrhodopsin-2 in a genetically blind rat model for gene therapy. Gene Ther. 2011;18:266–274. doi: 10.1038/gt.2010.140. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., Correale J., Fazekas F., Filippi M., Freedman M.S., et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Thorsen K., Mansilla F., Schepeler T., Øster B., Rasmussen M.H., Dyrskjøt L., Karni R., Akerman M., Krainer A.R., Laurberg S., et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002998. M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijink B.M., Buter J., de Bree R., Giaccone G., Lang M.S., Staab A., Leemans C.R., van Dongen G.A.M.S. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin.Cancer Res. 2006;12:6064–6072. doi: 10.1158/1078-0432.CCR-06-0910. [DOI] [PubMed] [Google Scholar]
- Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., Apuzzo T., Sperduti I., Volpe S., Cocorullo G., et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Tripathi V., Zhang Y.E. Redirecting RNA splicing by SMAD3 turns TGF-β into a tumor promoter. Mol. Cell Oncol. 2017;4:e1265699. doi: 10.1080/23723556.2016.1265699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée A., Vallée J.-N., Guillevin R., Lecarpentier Y. Interactions between the canonical WNT/beta-catenin pathway and PPAR gamma on neuroinflammation, demyelination, and remyelination in multiple sclerosis. Cell. Mol. Neurobiol. 2018;38:783–795. doi: 10.1007/s10571-017-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D. How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 2015;15:692–704. doi: 10.1038/nri3908. [DOI] [PubMed] [Google Scholar]
- Wang X., Du Z., Liu X., Song Y., Zhang G., Wang Z., Wang Q., Gao Z., Wang Y., Wang W. Expression of CD44 standard form and variant isoforms in human bone marrow stromal cells. Saudi Pharm. J. 2017;25:488–491. doi: 10.1016/j.jsps.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle U.H., Maisel D., Klostermann S., Weiss E.H., Schmitt M. Differential splicing generates new transmembrane receptor and extracellular matrix-related targets for antibody-based therapy of cancer. Cancer Genom.Proteomics. 2011;8:211–226. [PubMed] [Google Scholar]
- Yang C., Liang H., Zhao H., Jiang X. CD44 variant isoforms are specifically expressed on peripheral blood lymphocytes from asthmatic patients. Exp. Ther. Med. 2012;4:79–83. doi: 10.3892/etm.2012.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qian J., Gu C., Yang Y. Alternative splicing and cancer: a systematic review. Sig. Transduct.Target.Ther. 2021;6:78. doi: 10.1038/s41392-021-00486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
All additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.