Abstract
Mycoplasma gallisepticum is an important pathogen of chickens and turkeys that causes considerable economic losses to the poultry industry worldwide. The reemergence of M. gallisepticum outbreaks among poultry, the increased use of live M. gallisepticum vaccines, and the detection of M. gallisepticum in game and free-flying song birds has strengthened the need for molecular diagnostic and strain differentiation tests. Molecular techniques, including restriction fragment length polymorphism of genomic DNA (RFLP) and PCR-based random amplification of polymorphic DNA (RAPD), have already been utilized as powerful tools to detect intraspecies variation. However, certain intrinsic drawbacks constrain the application of these methods. The main goal of this study was to determine the feasibility of using an M. gallisepticum-specific gene encoding a phase-variable putative adhesin protein (PvpA) as the target for molecular typing. This was accomplished using a pvpA PCR-RFLP assay. Size variations among PCR products and nucleotide divergence of the C-terminus-encoding region of the pvpA gene were the basis for strain differentiation. This method can be used for rapid differentiation of vaccine strains from field isolates by amplification directly from clinical samples without the need for isolation by culture. Moreover, molecular epidemiology of M. gallisepticum outbreaks can be performed using RFLP and/or sequence analysis of the pvpA gene.
Mycoplasma gallisepticum is one of the major pathogens of domestic poultry, causing significant economic losses to the commercial poultry industry. M. gallisepticum infection has a wide variety of clinical manifestations, of which chronic respiratory disease of chickens is the most significant. The pathology associated with this disease is characterized by severe air sac infection where M. gallisepticum is the primary pathogen followed by secondary infections with Escherichia coli and/or viruses (17). Control of M. gallisepticum has been based on eradication of the organism from primary breeder flocks and on maintenance of the mycoplasma-free status in the breeders and breeder progeny by biosecurity of the premises (15). Serological monitoring of a representative sample of the flock is performed periodically, and isolation or DNA-based detection methods (15, 17) confirm suspected infection by M. gallisepticum. In recent years, a reemergence of mycoplasma infection has been observed in poultry, possibly due to the practice of placing large poultry populations in small geographical areas under poor biosecurity. This has necessitated a reevaluation of control strategies for M. gallisepticum (12).
One of the options as an alternative control method is the use of live M. gallisepticum vaccines (12, 28). Three live M. gallisepticum vaccines are currently used in many countries worldwide: F (Schering Plough, Kenilworth, N.J.), ts-11 (Bioproperties, Inc. Australia, marketed in the United States by Merial Select Laboratories, Gainesville, Ga.), and 6/85 (Intervet America, Millsboro, Del.). The more widespread utilization of M. gallisepticum vaccines requires the development of improved detection and differentiation methods in order to assess the efficacy of vaccines in displacing wild-type isolates (18, 26). Currently, the most widely used method to differentiate M. gallisepticum strains is a PCR-based randomly amplified polymorphic DNA (RAPD), or arbitrarily primed PCR, analysis (3, 5, 7). This method has been used for identifying vaccine strains in M. gallisepticum-vaccinated flocks (18) and for epidemiological studies (19). Due to the random nature of the primer and the low-stringency conditions of the RAPD reaction, this assay requires the use of pure cultures of the target mycoplasma. Isolation of mycoplasmas is expensive, time-consuming, and technically complicated in cases in which nonpathogenic mycoplasma species may overgrow the virulent mycoplasmas. In addition, the isolation process itself may favor the growth of one strain where more than one M. gallisepticum subtype may be present. Furthermore, technical factors such as the target DNA/primer ratio may significantly impact the reproducibility of RAPD patterns (27). Progress in the molecular biology of mycoplasmas has been achieved in the last decade, and several surface proteins in virulent mycoplasmas have been described (1, 22). Mycoplasmas exhibit a high degree of phenotypic variation, which is considered a major factor in pathogenicity and chronic infection of the host (22, 23, 24, 29). Changes in the surface topology of M. gallisepticum during host infection and the molecular characteristics of several M. gallisepticum surface proteins have been described (1, 6, 14, 21), including the recently described putative cytadhesin protein PvpA (2). PvpA is a phase-variable protein recognized by the chicken immune system (14, 30). Structurally, PvpA is a nonlipid integral membrane protein with a surface-exposed C-terminal portion (30). The surface-exposed C terminus of PvpA protein has a high proline content (28%) and contains identical direct repeat sequences consisting of 52 amino acids each, designated DR-1 and DR-2 (2). Size variation of the pvpA gene was observed among M. gallisepticum strains as a result of deletions occurring in the segment encoding the the proline-rich C-terminal region of the protein. Deletions were located exactly within the two direct repeat sequences (2). The presence of proline-rich regions in the surface-exposed C-terminal domains of other pathogenic mycoplasma adhesins (4, 8, 9) suggests an important role of these domains in the function of PvpA as an adhesin (10). Moreover, molecular typing of M. gallisepticum strains utilizing the C-terminus-encoding region of the pvpA gene may be a relevant target for epidemiological tracking of M. gallisepticum isolates.
The main goal of our study was to determine the feasibility of using the variable pvpA gene as the target to differentiate M. gallisepticum strains through a PCR restriction fragment length polymorphism (PCR-RFLP) assay. To design consensus primers to amplify all M. gallisepticum strains, the nucleotide sequence of the C-terminus-encoding region of the pvpA gene was determined from 10 reference M. gallisepticum strains and 24 field isolates. Various deletions were observed within DR-1 and DR-2, as previously described by Boguslavsky et al. (2). Further differentiation among M. gallisepticum reference strains and field isolates was obtained by RFLP using three restriction enzymes. To enhance the sensitivity of the assay, a seminested set of primers was also designed. This assay was used to detect M. gallisepticum directly from clinical samples. Amplicons obtained from clinical samples were analyzed according to the RFLP patterns typed with restriction enzymes and further sequenced. This assay accelerates the diagnosis and characterization of M. gallisepticum isolates and can be used as a molecular typing tool in order to better understand the epidemiology of M. gallisepticum outbreaks.
MATERIALS AND METHODS
Mycoplasma reference strains and isolates.
Most mycoplasma strains were obtained from our depository at the Poultry Diagnostic and Research Center (PDRC) in Athens, Ga. The type strains of the avian species Mycoplasma synoviae, Mycoplasma meleagridis, Mycoplasma iowae, Mycoplasma gallinarum, Mycoplasma gallinaceum, Mycoplasma iners, Mycoplasma imitans, and Mycoplasma cloacale were used for testing the specificity of the PCR test. The origins and biological properties of M. gallisepticum strains R, S6, A5969, F, K503, K703, K730, and HF-51 are described elsewhere (31). Vaccine strain ts-11 was obtained from Merial Select (Gainesville, Ga.), and 6/85 was obtained from Intervet America (Millsboro, Del.). Twenty-two field isolates obtained during recent outbreaks (1995 to 1999) and two field isolates from 1973 and 1984 were collected from our depository. Among these, seven isolates were from chickens, eleven were from turkeys, and six were from house finches (Carpodacus mexicanus). Table 1 presents a list of the isolates used.
TABLE 1.
Field isolates
| Case | Isolatea |
|---|---|
| K435 | TK/GA/73/1 |
| K2101 | CK/CO/84/1 |
| K3839 | HF/MA/94/1 |
| K3944 | TK/NC/95/1 |
| K4013 | HF/PA/95/1 |
| K4029 | TK/NB/95/2 |
| K4043 | TK/NB/95/1 |
| K4094 | HF/TN/96/1 |
| K4236 | TK/VA/96/1 |
| K4355 | CK/CA/96/1 |
| K4366 | GF/SC/97/1 |
| K4409 | HF/TX/97/1 |
| K4421A | TK/MI/97/2 |
| K4423B | TK/MI/97/1 |
| K4465 | TK/OH/97/1 |
| K4593 | HF/MA/98/1 |
| K4649A | TK/CO/98/1 |
| K4649B | TK/CO/98/2 |
| K4657 | CK/GA/98/1 |
| K4669A | TK/CO/98/3 |
| K4688C | CK/NC/99/1 |
| K4688F | CK/NC/99/2 |
| K4688G | CK/NC/99/3 |
| K4705 | CK/AK/99/1 |
Isolate names are in the form avian species/state/year/isolate number. TK, turkey; CK, chicken; HF, house finch; GF, goldfinch.
Culture procedures and sample preparations.
All avian mycoplasma strains were propagated in Frey's medium (17) with 12% swine serum (FMS) as previously described (5, 6). DNA isolation was performed as follows. One milliliter of mycoplasma broth culture was centrifuged at 13,000 rpm (Eppendorf 5417R; Brinkmann Instruments, Westbury, N.Y.) for 10 min. The cell pellet was then washed twice with 1 ml of 150 mM phosphate-buffered saline (PBS, pH 7.2) and resuspended in a final volume of 20 μl of PBS. The cell suspension was heated in a dry block at 110°C for 10 min and placed on ice for at least 10 min. After boiling, the lysate was centrifuged at 13,000 rpm for 2 min to remove debris. The supernatant containing DNA was collected and stored at −20°C until used.
DNA extraction from clinical samples was prepared by suspending tracheal swabs in 1 ml of PBS (three tracheal swabs per sample) and spinning at 13,000 rpm for 30 min. The supernatant was carefully removed and the resultant cell pellet suspended in 25 μl of PCR-grade H2O and treated as described above.
Clinical samples.
Clinical samples were obtained from the United States and Israel and processed at the PDRC or at the Kimron Veterinary Institute (Bet Dagan, Israel), respectively. Twenty tracheas were obtained upon necropsy from two commercial egg layer flocks which were serologically positive for M. gallisepticum infection. Tracheas were opened longitudinally, and mucous material was collected with a swab. Trachea material was cultured on Frey's media following standard isolation procedures. Material was resuspended in 500 μl of PBS for DNA extraction and pvpA PCR-RFLP analysis.
Tracheal swab samples from four Israeli broiler breeder flocks suspected of having M. gallisepticum infection were collected from live birds. Swabs were cultured on Frey's medium by standard isolation procedure, and DNA extracts for PCR testing were prepared from tracheal swabs by the boiling method as described above.
Primer selection.
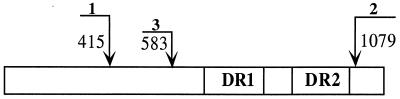
Initial primers were designed from the pvpA gene sequence of R strain (2). Seminested PCR primers were designed from conserved sequences of representative M. gallisepticum strains. These primers flank the direct repeat area within the C-terminus-encoding region of the pvpA gene. Primer positions are depicted in Fig. 1.
FIG. 1.
Schematic diagram of pvpA indicating the location of primers for seminested PCR. Primer positions are in relation to the R strain sequence (2). The first reaction used primer 1 (pvpA1F), located at nucleotide positions 415 to 437 (5′GCCAM TCCAACTCAACAAGCTGA3′), and primer 2 (pvpA2R), located at nucleotide positions 1059 to 1081 (5′GGACGTSGTCCTGGCT GGTTAGC3′). The seminested reaction was performed with primer 3 (pvpA3F), located at nucleotide positions 583 to 604 (5′GGTAGTCCTAAGTTATTAGGTC3′), and primer 2 (pvpA2R) as for the first amplification.
pvpA PCR.
Amplification reactions for pure culture samples were performed in a 50-μl reaction volume as follows. Five microliters of 10× PCR buffer containing 1.5 mM MgCl2, 2 μl of 1 mM deoxynucleoside triphosphate (dNTPs) (GIBCO BRL, Grand Island, N.Y.), 0.5 μl of outer primers pvpA1F and pvpA2R (50 μM), 0.25 μl of Taq DNA polymerase (5 U/μl) (Taq PCR; Qiagen Inc., Valencia, Calif.), 40.75 μl of distilled water, and 1 μl of template DNA. All amplifications were performed in a GeneAmp PCR system 2400 (PE Biosystems, Foster City, Calif.). Initially, annealing temperatures for primer extension of 50, 55, and 58°C were tested. The amplification reaction using 55°C as the annealing temperature produced the best amplification yield. This temperature was considered the optimal annealing temperature for pvpA primer extension. The thermocycler was programmed as follows: 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and 72°C for 10 min as the final extension step.
In order to increase sensitivity of detection in clinical samples, two amplifications were performed with seminested primers (Fig. 1). The first amplification was carried out in a 25-μl reaction volume consisting of 2.5 μl of 10× PCR buffer containing 1.5 mM MgCl2, 1 μl of 1 mM dNTPs, 0.25 μl of outer primers (pvpA1F and pvpA2R) (50 μM), 0.13 μl of Taq DNA polymerase (5 U/μl), 19.87 μl of distilled water, and 1 μl of template DNA. The first amplification was performed at 94°C for 3 min followed by 20 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and 72°C for 10 min was the final extension step. One microliter from the first amplification was used as the template for the second amplification. The second amplification was performed in 25 μl consisting of 2.5 μl of 10× PCR buffer containing 1.5 mM MgCl2, 1 μl of 1 mM dNTPs, 0.35 μl of seminested primer (pvpA3F) (50 μM), 0.35 μl of outer primer (pvpA2R) (50 μM), 0.13 μl of Taq DNA polymerase (5 U/μl), 19.80 μl of distilled water, and 1 μl of template DNA. The second amplification was performed at the same temperatures and times as the first amplification for 40 cycles.
The detection limit of seminested PCR and single PCR was determined by conducting nine 10-fold serial dilutions from R strain stock culture in FMS. Twenty microliters of each 10-fold dilution was plated on Frey's medium agar plates and incubated at 37°C until colonies were visible. Colonies were counted 5 days after inoculation of the plates. A 1-ml aliquot from each dilution was collected for DNA extraction followed by PCR amplification as described above.
The PCR products were detected by electrophoresis in ethidium bromide-stained agarose gels. Ten microliters of each amplification reaction was loaded on a 1.5% agarose gel containing 0.8 μg of ethidium bromide/ml. Agarose gel electrophoresis was run for 25 min at 110 V, and the gel was visualized under UV light.
Sequence analysis.
Amplified products from 10 reference M. gallisepticum strains and 24 field isolates were purified with a QIAquick PCR purification kit (Qiagen, Inc.). Purified PCR products were sequenced at the Molecular Genetics Instrumentation Facility, University of Georgia, utilizing automated sequencing with a Prism DyeDeoxy terminator cycle sequencing kit (PE Biosystems). Assembly of sequence contigs and initial multiple-sequence alignments were performed with sequencing project management (SeqMan) and multiple-sequence alignment (MegAlign) programs, respectively (DNASTAR; Lasergene, Inc. Madison, Wis.). Phylogenetic analysis was performed with PAUP 4.0b2 and 4a (Sinauer Associates, Inc., Sunderland, Mass.)
RFLP analysis.
Restriction enzyme analysis of sequences was performed with MapDraw (DNASTAR; Lasergene, Inc.). PvuII (Boehringer Mannheim, Indianapolis, Ind.), AccI (New England Biolabs, Inc., Beverly, Mass.), and ScrFI (New England Biolabs, Inc.) were identified as suitable for RFLP analysis and utilized for digestion of PCR products. The restriction enzyme digestions were performed in 10-μl reaction mixtures (6.6 μl of H2O, 1 μl of 10× buffer, 0.4 μl of restriction enzyme, 2 μl of PCR product), and mixtures were incubated at 37°C for 2 h. The digested PCR products were electrophoresed on a 10% Tris buffer–EDTA gel (Novex, San Diego, Calif.) at a constant 100 V for 50 min. Gels were visualized by silver staining according to the manufacturer's recommendation (Amersham Pharmacia Biotech, Piscataway, N.J.).
16S rRNA gene PCR.
An M. gallisepticum PCR test was performed with the 16S rRNA primers MG13 and MG14 (13), using 5 μl of DNA extracted as described above. In addition to serological testing and isolation, this PCR test is used routinely in the laboratory at the Kimron Veterinary Institute and at the PDRC to determine the M. gallisepticum status of chicken and turkey breeder flocks.
RESULTS
Amplification of the pvpA C-terminus-encoding region.
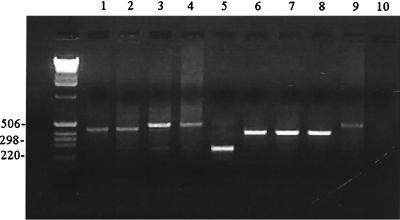
Amplification of the pvpA C-terminus-encoding region demonstrated size differences of PCR products among M. gallisepticum reference strains, as previously reported by Boguslavsky et al. (2). Amplification product size polymorphism was also observed within field isolates analyzed. Size variation of PCR products for reference strains and field isolates was confirmed by nucleotide sequencing analysis. Amplification product sizes for M. gallisepticum reference strains ranged from 267 to 497 bp (Fig. 2; Table 2). A PCR product with the expected size of 497 bp was observed for strains R, ts-11, and A5969 (Fig. 2, lanes 3, 4, and 9, respectively); a 437-bp PCR product was obtained for vaccine strain 6/85 and a field isolate (CK/CA/96/1) (Fig. 2, lanes 1 and 2) and for challenge strain S6 and finch isolate HF-51 (data not shown). Atypical strains K503, K703, and K730 produced an amplicon of 410 bp (Fig. 2, lanes 7 and 8). A 267-bp amplicon was obtained for strain F (Fig. 2, lane 5).
FIG. 2.
PCR of M. gallisepticum reference with outer primers pvpA 5′ and pvpA 3′. Molecular weight markers (Life Technologies, Rockville, Md.) are in the unmarked lane on the left (sizes on the left are in base pairs). Lane 1, 6/85; lane 2, field isolate (CK/CA/96/1); lane 3, R; lane 4, ts-11; lane 5, F; lane 6, K503; lane 7, K703; lane 8, K730; lane 9, A5969; lane 10, negative control. Size variation of amplification products from 497 bp (lanes 3, 4, and 9) to 267 bp (lane 5) was observed.
TABLE 2.
RFLP patterns of M. gallisepticum reference strains
| Strain | PCR sizea | Fragment sizes
|
RFLP pattern | ||
|---|---|---|---|---|---|
| PvuII | AccI | ScrFI | |||
| R | 497 | 231 + 217 + 49 | 231 + 156 + 110 | 240 + 183 + 74 | A |
| ts-11 | 497 | 231 + 217 + 49 | 341 + 156 | 240 + 183 + 74 | B |
| A5969 | 497 | 448 + 49 | 341 + 156 | 240 + 183 + 74 | C |
| 6/85 | 437 | 217 + 120 + 51 + 49 | Not cut | 363 + 74 | D |
| S6 | 437 | 217 + 120 + 51 + 49 | Not cut | 228 + 135 + 74 | E |
| HF-51 | 437 | 217 + 171 + 49 | Not cut | 228 + 135 + 74 | F |
| K503 | 410 | 231 + 130 + 49 | Not cut | Not cut | G |
| K703 | 410 | 231 + 130 + 49 | Not cut | Not cut | G |
| K730 | 410 | 231 + 130 + 49 | Not cut | Not cut | G |
| F | 267 | 217 + 50 | Not cut | 193 + 74 | H |
Size in base pairs, estimated from molecular weight marker and sequence analysis.
Size variation of PCR products was also observed among field isolates (Table 3). Like 6/85 and S6, 18 of the 24 field isolates analyzed produced a 437-bp amplicon; five isolates produced a 497-bp amplicon, and isolates CK/GA/98/1 and HF/TX/97/1 produced amplicons of 488 and 266 bp, respectively. Size variation of PCR products was confirmed by nucleotide sequencing analysis.
TABLE 3.
RFLP and sequencing analysis of the C-terminus-encoding region of the pvpA gene from field isolates
| Field strain | PCR product sizea | RFLP groupb | % Sequence identityc
|
|
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| TK/GA/73/1 | 497 | A | 99.8 | 99.4 |
| CK/CO/84/1 | 497 | A | 100 | 100 |
| CK/GA/98/1 | 488 | B | 98.5 | 96.9 |
| CK/NC/99/1 | 497 | B | 100 | 100 |
| CK/NC/99/2 | 497 | B | 100 | 100 |
| CK/NC/99/3 | 497 | B | 100 | 100 |
| TK/MI/97/1 | 437 | D | 100 | 100 |
| TK/MI/97/2 | 437 | D | 100 | 100 |
| TK/NB/95/1 | 437 | D | 100 | 100 |
| TK/NB/95/2 | 437 | D | 100 | 100 |
| TK/OH/97/1 | 437 | D | 100 | 100 |
| TK/NC/95/1 | 437 | D | 100 | 100 |
| CK/CA/96/1 | 437 | E | 100 | 100 |
| TK/VA/96/1 | 437 | F | 98.8 | 97.9 |
| TK/CO/98/1 | 437 | F | 98.1 | 96.5 |
| TK/CO/98/2 | 437 | F | 98.1 | 96.5 |
| TK/CO/98/3 | 437 | F | 98.1 | 98.8 |
| HF/MA/94/1 | 437 | F | 100 | 100 |
| HF/PA/95/1 | 437 | F | 100 | 100 |
| HF/TN/96/1 | 437 | F | 100 | 100 |
| GF/SC/97/1 | 437 | F | 100 | 100 |
| HF/MA/98/1 | 437 | F | 100 | 100 |
| HF/TX/97/1 | 266 | H | 96.1 | 91.9 |
| CK/AK/99/1 | 437 | I | 98.4 | 96.5 |
Size in base pairs, estimated from molecular weight marker and sequence analysis.
RFLP groups correspond to M. gallisepticum reference strains as follows: A, R; B, ts-11; D, 6185; E, S6; F, HF-51; H, F.
Sequence identity with corresponding RFLP group M. gallisepticum reference strain.
RFLP and sequencing analysis of M. gallisepticum reference strains.
The C-terminus-encoding region of the pvpA gene was sequenced for 10 M. gallisepticum reference strains. Multiple alignments of M. gallisepticum reference strain sequences indicated that variation among PCR products' relative mobilities was attributed to different size deletions at the end of the pvpA gene corresponding to the C terminus, as previously described by Boguslavsky et al. (2). The precise location of the deletions allowed the design of three conserved primers (pvpA1F, pvpA2R, and pvpA3F) (Fig. 1) utilized in a seminested PCR amplifying all the strains tested. In addition to size variation of PCR products, nucleotide sequence differences among M. gallisepticum reference strains produced different RFLP patterns. Digestion of M. gallisepticum reference strains with AccI, PvuII, and ScrFI produced eight distinct pattern combinations. The exact sizes of the fragments generated from digested PCR products are listed in Table 2. M. gallisepticum R, ts-11, A5969, HF-51, atypical strains (K503, K703, and K730), and F were readily differentiated into six distinct patterns designated A, B, C, F, G, and H, respectively, with PvuII and AccI. A third restriction enzyme, ScrFI, was used to differentiate 6/85 and S6, and the patterns were identified as D and E, respectively (Table 2).
RFLP and sequence analysis of field isolates.
The C-terminus-encoding regions of pvpA for 24 field isolates were compared by RFLP and sequence analysis. RFLP analysis classified the field isolates into seven of the eight RFLP groups described in Table 2 for M. gallisepticum reference strains. Field isolates were grouped as follows (Table 3): two isolates were classified as RFLP group A, represented by challenge strain R; four chicken isolates had RFLP pattern combinations similar to group B, represented by vaccine strain ts-11; six isolates were classified as RFLP group D, represented by vaccine strain 6/85; one isolate was placed within RFLP group E, represented by strain S6; nine isolates, four from turkeys and five from finches, were characterized as RFLP group F, represented by finch strain HF-51; one isolate showed RFLP patterns similar to vaccine strain F, classified as group H. Isolate CK/AK/99/1 produced a unique RFLP pattern (I), a combination not observed for any of the M. gallisepticum reference strains or field isolates analyzed.
A perfect correlation was observed between RFLP and sequencing analysis of the amplicon produced by the seminested PCR for 16 field isolates in which a 100% sequence identity was observed between the field isolate and its corresponding M. gallisepticum reference strain RFLP group (Tables 2 and 3). One isolate in RFLP group A showed 100% sequence identity to challenge strain R. Three of the four isolates identified as RFLP group B showed 100% identity to vaccine strain ts-11. Six of the seven isolates identified as RFLP group D showed 100% sequence identity to vaccine strain 6/85. One isolate in RFLP group E had 100% sequence identity to S6, in agreement with the results of ScrFI RFLP analysis. Five of the nine isolates within RFLP group F, isolated from finches, showed 100% identity to house finch isolate HF-51.
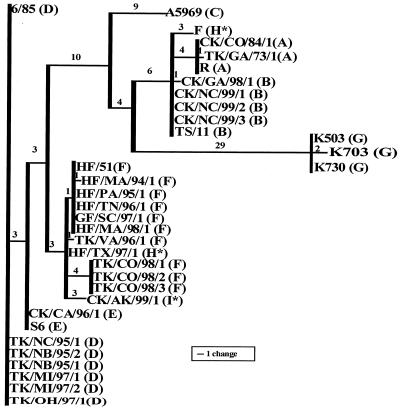
For the remaining eight field isolates, it was observed that the sequence of pvpA corresponding to the C-terminal region was not identical to that of the corresponding RFLP group. Sequence analysis of these isolates showed sequence divergences ranging from 99.8 to 96.1% for nucleotides and 99.4 to 91.9% for deduced amino acids (Table 3). Molecular grouping of M. gallisepticum isolates by RFLP was further confirmed by phylogenetic analysis of the pvpA gene sequences. Figure 3 shows a dendrogram constructed from the C-terminus-encoding end of the pvpA gene. Sequences were distributed in seven distinctive groups that correspond to the RFLP groups A, B, C, D, E, F, and G (Fig. 3). A discrepancy between phylogenetic and RFLP analysis was observed for the finch isolate HF/TX/97/1. This isolate, in contrast to the other finch isolates analyzed, produced a PCR product of the same size as vaccine strain F and was grouped by RFLP with this strain (RFLP group H). However, nucleotide sequence comparison indicated that HF/TX/97/1 was most closely related to other finch isolates rather than to F strain (Fig. 3). Further comparison of the deduced amino acid sequences of finch isolate HF/TX/97/1 and vaccine strain F indicated four amino acid substitutions (data not shown), resulting in amino acid sequence homology of only 91.9%. between the two strains (Table 3). Comparison of the deduced amino acid sequences of finch isolate HF/TX/97/1 and finch strain HF-51 indicated an amino acid sequence homology of 96.3% (data not shown).
FIG. 3.
Phylogentic analysis of the C-terminus-encoding portion of pvpA of M. gallisepticum reference strains and field isolates. The dendrogram was constructed using parsimony analysis in a full heuristic search with 500 bootstrap resampling replicates. The RFLP group for each respective isolate is indicated in parentheses. Phylogenetic analysis produced seven distinct groups, confirming RFLP grouping: A, B, C, D, E, F, and G. Discrepancies between phylogenetic and RFLP analysis were observed for isolates in groups H* and I*.
Specificity and sensitivity of pvpA PCR.
Single and seminested amplifications with the three pvpA primers were performed against DNA from eight avian mycoplasma species, as indicated in Materials and Methods, including M. gallinarum, and M. gallinaceum, common normal flora mycoplasmas of the upper respiratory track of chickens. M. gallisepticum was the only mycoplasma amplified by pvpA primers (data not shown).
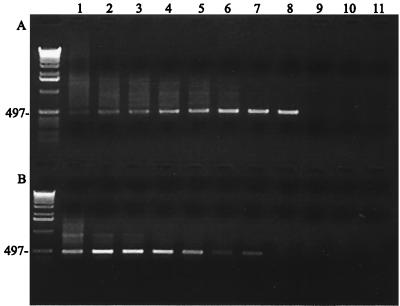
Sensitivity of single PCR versus seminested PCR was evaluated by comparing detection limits of both assays. Tenfold serial dilutions were performed from R strain stock culture with an initial titer of 5 × 108 CFU/ml (Fig. 4). A single amplification with primers pvpA3F and pvpA2R detected 500 CFU/ml (Fig. 4B), whereas the detection limit after a second amplification with primers pvpA3F and pvpA2R reached 50 CFU/ml (Fig. 4A), achieving a 10-fold increase in the detection limit.
FIG. 4.
Sensitivity of seminested PCR versus single PCR. (A) Seminested PCR. Serial dilutions of R strain amplified with seminested primers. Lanes 1 to 10 represent dilutions from 10−1 through 10−10. Lane 11 is the negative control. Lane 1, 5 × 108 CFU; lane 2, 5 × 107 CFU; lane 3, 5 × 106 CFU; lane 4, 5 × 105 CFU; lane 5, 5 × 104 CFU; lane 6, 5 × 103 CFU; lane 7, 5 × 102 CFU; lane 8, 5 × 101 CFU. (B) Single PCR with inner primers following the same serial dilutions. Both PCR systems produced a 497-bp band as expected.
Clinical samples.
Because of the increase in sensitivity achieved by the seminested PCR, this assay was used to analyze clinical samples. Six of 20 samples from two U.S. layer flocks from a common operation tested positive. One positive sample was from flock A, and the remaining five positive samples were from flock B. None of the PCR products were sensitive to AccI digestion, and RFLP group E (Table 3) was observed for the six amplicons when they were digested with PvuII. Sequences obtained from these amplicons showed 100% identity with field isolate TK/VA/96/1 (Table 3). Culture results from tracheal samples were negative for M. gallisepticum as well as saprophytic mycoplasma species, which may interfere with the isolation attempt. Since tracheal levels of M. gallisepticum in chronically infected birds are often very low (11), the number of viable cells removed from the trachea by swabbing may have been too low to be detected by isolation.
To further compare the sensitivity of pvpA PCR in clinical samples, tracheal swabs from three commercial breeder flocks from Israel were tested by three methods: pvpA seminested PCR, rRNA-based M. gallisepticum PCR (13), and isolation. All four flocks were positive for M. gallisepticum by both PCR tests, although the number of individual samples showing a positive reaction differed for the two tests (Table 4). M. gallisepticum, M. synoviae, and M. gallinarum were isolated from two flocks tested. Testing of three other chicken and turkey breeder flocks containing M. synoviae, M. gallinarum, and/or M. meleagridis but negative for M. gallisepticum were negative in both PCR tests (data not shown).
TABLE 4.
Comparative analysis of clinical samples from commercial broiler-breeder flocks
| Flock age (wk) | No. of positive samples/no. tested by:
|
Isolation | |
|---|---|---|---|
| M. gallisepticum PCRa | pvpA PCRb | ||
| 50 | 8/10 | 4/10 | M. gallisepticum, M. synoviae, M. gallinarum |
| 58 | 10/10 | 1/10 | NTc |
| 58 | 4/4 | 4/4 | NT |
| 40 | 5/5 | 2/5 | M. gallisepticum, M. synoviae, M. gallinarum |
PCR using primers MG13 and MG14, directed to the rRNA gene of M. gallisepticum.
Seminested pvpA PCR.
NT, not tested.
DISCUSSION
Sequence analysis of the whole pvpA gene of reference M. gallisepticum strains revealed that different-size deletions are located within DR-1 and DR-2 of the C-terminus-encoding region of pvpA (2). In this study we demonstrated by sequencing of the genomic region amplified by the seminested pvpA PCR primers that the pvpA C-terminal deletions are present also among M. gallisepticum strains currently circulating in the field. The largest deletion observed was that of a 230-bp fragment in vaccine strain F. An amplicon of the same size was observed for finch isolate HF/TX/97/1 (Table 3). Differing from isolates with this large deletion were atypical M. gallisepticum strains (K503, K703, and K730), with an amplicon of 410 bp. This amplicon had six small deletions scattered in the region of pvpA corresponding to the C terminus (2; additional data not shown). More significant yet, 16 of the 24 field isolates analyzed that produced a 437-bp amplicon shared a similar 60-bp deletion with M. gallisepticum representative strains 6/85, S6, and HF-51 (Table 2). Based on the deduced amino acid sequence, the deletion extends from amino acid position 88, located outside DR-2, to amino acid position 107, located within DR-2 (data not shown). However, not all field isolates had deletions within the pvpA gene. In common with representative strains R, A5969, and vaccine strain ts-11, six field isolates produced the expected 497-bp amplicon where no deletions were observed within the C-terminus-encoding pvpA region. In addition to PCR product length polymorphism, nucleotide differences allowed differentiation of representative M. gallisepticum strains and field isolates by RFLP of the amplification products. M. gallisepticum strains were characterized in seven RFLP groups, whereas field isolates were categorized within five of these seven groups and an additional unique group (Tables 2 and 3).
An important application of this method will be to identify specific M. gallisepticum strains and, in particular, to detect the presence of vaccine strains and to monitor their capacity to displace field strains during M. gallisepticum vaccination programs. Among analyzed field isolates, six turkey and four chicken isolates were grouped by RFLP as vaccine strains 6/85 and ts-11, respectively (Table 3). These results were confirmed by sequence analysis. A 100% sequence identity was observed among the turkey isolates and vaccine strain 6/85 and between three of the four chicken isolates and vaccine strain ts-11.
The identification of 13 vaccine strains among the 24 field isolates tested in this study was not surprising, since these samples were specifically selected from flocks with previous vaccination history or previous exposure to other vaccinated birds. The presence of vaccine strains in these flocks was suggested on the basis of clinical data and the identification of vaccine strains by the RAPD molecular typing used in our laboratory (S. H. Kleven and V. Leiting, unpublished data). RAPD analyses have been utilized previously to identify and determine the prevalence of vaccine strains ts-11 and 6/85 in experimental (18) and commercial (26) vaccinated flocks.
An outbreak of conjunctivitis in house finches (Carpodacus mexicanus) caused by M. gallisepticum was first reported during early 1994 (16, 20). Since the beginning of this outbreak, other wild bird species, such as American goldfinches (Carduelis tristis) and blue jays (Cyanocitta cristata), have been infected (16). RAPD typing of representative isolates from different bird species and geographical regions indicated the presence of a single type (19). In this study, RFLP and sequence analysis of finch isolates from different states demonstrated that sequences among isolates were identical with the exception of HF/TX/97/1. Although isolate HF/TX/97/1 was previously characterized as having an RAPD pattern identical to those of the other finch isolates (S. H. Kleven, unpublished data), this isolate had a 230-bp deletion, and based on RFLP, it was grouped with vaccine strain F. However, sequence analysis demonstrated that this isolate is more closely related to finch isolate HF-51 than to vaccine strain F. The pvpA gene divergence among HF/TX/97/1 and other finch isolates suggests that more than one strain of M. gallisepticum has been circulating among the finch population during the outbreak. Although free-flying birds have not been strongly implicated in the transmission of pathogenic mycoplasmas to commercial poultry, the relationship between poultry and finch isolates needs to be further analyzed (25).
Characterization of other chicken and turkey isolates was also more reliably obtained by sequencing than by size and RFLP grouping alone. For instance, the isolates TK/VA/96/1 and TK/CO/98/1, -2, and -3 (Table 3) have a pvpA C-terminus-encoding region closely related to that of finch isolates, suggesting that finch and poultry isolates may have a common ancestor. However, complete sequencing of pvpA and other surface protein genes will be required to better elucidate relationships among these strains.
A desired application of the seminested PCR-RFLP procedure is to detect and rapidly differentiate M. gallisepticum strains present in infection of commercial poultry flocks directly from tracheal samples, thus eliminating the need for culture. Detection of M. gallisepticum directly from tracheal scrapings with pvpA PCR was demonstrated in samples from commercial layer hens. Probably due to the low tracheal levels of M gallisepticum in chronically infected chickens (11), attempts to isolate the organism were not successful and the only evidence of infection was weak plate agglutination reactions. M. gallisepticum infection in acutely infected broiler-breeder flocks in Israel was diagnosed by the pvpA PCR test, by M. gallisepticum PCR, and by isolation. The lower percentage of positive samples in the pvpA PCR test, seen in Table 4, suggests that the sensitivity of this test may be less than that of the M. gallisepticum PCR test, which is <50 CFU (S. Levisohn, unpublished data). Nonetheless, this does not affect the diagnostic results in these cases, since these are based on the flock status rather than individual samples. However, further optimization of the PCR test may be necessary in order to improve the sensitivity as needed for testing of clinical samples.
Analysis of clinical isolates from Israel by pvpA PCR-RFLP showed that all samples have a 400-bp amplicon. In addition, an identical and unique RFLP pattern was observed for three Israel isolates, which had been previously characterized and differentiated by RAPD analysis (data not shown). Our findings suggest that the pvpA gene present in Israeli outbreak-related isolates is more conserved than the pvpA gene of U.S. isolates. However, these strains are readily differentiable from the vaccine strains.
The data presented here indicate that the pvpA gene of M. gallisepticum is a reliable target gene for differentiation of vaccine strains from field isolates based on either size variation, RFLP analysis, or sequencing of PCR products.
Moreover, it was demonstrated that sequence analysis of the pvpA gene could be utilized for epidemiology studies of M. gallisepticum outbreaks. This is the first study where a surface protein gene of M. gallisepticum has been utilized for molecular epidemiological analysis of a significant number of field isolates. Increasing sequence analysis of the pvpA and other genes will allow a better understanding on the origin of M. gallisepticum outbreaks and therefore will facilitate better strategies to control the disease. In addition, with further optimization, the pvpA PCR assay can be utilized as a tool to detect and identify specific M. gallisepticum strains directly from clinical material without the need for isolation.
ACKNOWLEDGMENTS
We thank Sylva Riblet, Bill Hall, Victoria Leiting, Lisa Griffeth, Lynn Luna, and Irina Gerchman for their technical assistance.
This work was supported by grant IS-3126-99 from BARD, United States-Israel Binational Agricultural Research and Development.
REFERENCES
- 1.Bencina D, Kleven S H, Elfaki M G, Snoj A, Dovc P, Dorrer D, Russ I. Variable expression of epitopes on the surface of Mycoplasma gallisepticum demonstrated with monoclonal antibodies. Avian Pathol. 1994;23:19–36. doi: 10.1080/03079459408418972. [DOI] [PubMed] [Google Scholar]
- 2.Boguslavsky S, Menaker D, Lysnyansky I, Liu T, Levisohn S, Rosengarten R, García M, Yogev D. Molecular characterization of the Mycoplasma gallisepticum pvpA gene which encodes a putative variable cytadhesin protein. Infect Immun. 2000;68:3956–3964. doi: 10.1128/iai.68.7.3956-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlton B R, Bickford A A, Walker R L, Yamamoto R. Complementary randomly amplified polymorphic DNA (RAPD) analysis patterns and primer sets to differentiate Mycoplasma gallisepticum strains. J Vet Diagn Investig. 1999;11:158–161. doi: 10.1177/104063879901100209. [DOI] [PubMed] [Google Scholar]
- 4.Dallo S F, Lazzell A L, Chavoya A, Reddy S P, Baseman J B. Bifunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan H H, Kleven S H, Jackwood M W. Application of polymerase chain reaction with arbitrary primers to strain identification of Mycoplasma gallisepticum. Avian Dis. 1995;39:729–735. [PubMed] [Google Scholar]
- 6.García M, Elfaki M G, Kleven S H. Analysis of the variability in expression of Mycoplasma gallisepticum surface antigens. Vet Microbiol. 1994;42:147–158. doi: 10.1016/0378-1135(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 7.Geary S J, Forsyth M H, Saoud S A, Wang G, Berg D E, Berg C M. Mycoplasma gallisepticum strain differentiation by arbitrary primer PCR (RAPD) fingerprinting. Mol Cell Probes. 1994;8:311–316. doi: 10.1006/mcpr.1994.1042. [DOI] [PubMed] [Google Scholar]
- 8.Hnatow L L, Keleer C L, Jr, Tessmer L L, Czymmek K, Dohms J E. Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae 30-kilodalton protein P30 and Mycoplasma genitalium P32. Infect Immun. 1998;66:3436–3442. doi: 10.1128/iai.66.7.3436-3442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeler C L, Jr, Hnatow L L, Whetzel P L, Dohms J E. Cloning and characterization of a putative cytadhesin gene (mgc1) from Mycoplasma gallisepticum. Infect Immun. 1996;64:1541–1547. doi: 10.1128/iai.64.5.1541-1547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehoe M A. Cell-wall-associated proteins in gram-positive bacteria. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1994. pp. 217–261. [Google Scholar]
- 11.Kleven S H. Tracheal populations of Mycoplasma gallisepticum after challenge of bacterin-vaccinated chickens. Avian Dis. 1985;29:1012–1017. [PubMed] [Google Scholar]
- 12.Kleven S H. Changing expectations in the control of Mycoplasma gallisepticum. Acta Vet Hung. 1997;45:299–305. [PubMed] [Google Scholar]
- 13.Lauerman L H. Mycoplasma PCR assays. In: Lauerman L H, editor. Nucleic acid amplification assays for diagnosis of animal diseases. Turlock, Calif: American Association of Veterinary Laboratory Diagnosticians; 1998. pp. 41–42. [Google Scholar]
- 14.Levisohn S, Rosengarten R, Yogev D. In vivo variation of Mycoplasma gallisepticum antigen expression in experimentally infected chickens. Vet Microbiol. 1995;45:219–231. doi: 10.1016/0378-1135(95)00039-d. [DOI] [PubMed] [Google Scholar]
- 15.Levisohn S, Kleven S H. Avian mycoplasmosis (Mycoplasma gallisepticum) Rev Sci Tech Off Int Epizoot. 2000;19:425–442. [PubMed] [Google Scholar]
- 16.Ley D H, Berkhoff J E, McLaren J M. Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis. Avian Dis. 1996;40:480–483. [PubMed] [Google Scholar]
- 17.Ley D H, Yoder H W., Jr . Mycoplasma gallisepticum infection. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. 10th ed. Ames: Iowa State University Press; 1997. pp. 194–207. [Google Scholar]
- 18.Ley D H, McLaren J M, Miles A M, Barnes H J, Franz G. Transmissibility of live Mycoplasma gallisepticum vaccine strains ts-11 and 6/85 from vaccinated layer pullets to sentinel poultry, and identification by random amplified polymorphic DNA (RAPD) analysis. Avian Dis. 1997;41:187–194. [PubMed] [Google Scholar]
- 19.Ley D H, Berkhoff J E, Levisohn S. Molecular epidemiological investigations of Mycoplasma gallisepticum (MG) conjunctivitis in songbirds by random amplified polymorphic DNA (RAPD) analyses. Emerg Infect Dis. 1997;3:375–380. doi: 10.3201/eid0303.970318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luttrell P M, Fischer J R, Stalknecht D E, Kleven S H. Field investigation of Mycoplasma gallisepticum infections in house finches (Carpodacus mexicanus) from Maryland and Georgia. Avian Dis. 1996;40:335–341. [PubMed] [Google Scholar]
- 21.Markham P F, Glew M D, Browning G F, Whithear K G, Walker I D. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect Immun. 1998;66:2845–2853. doi: 10.1128/iai.66.6.2845-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razin S, Yogev D, Naot Y. Molecular biology and pathogenesis of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun. 1994;62:5066–5074. doi: 10.1128/iai.62.11.5066-5074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosengarten R, Yogev D. Variant colony surface antigenic phenotypes within mycoplasma strain populations: implications for species diagnosis and strain standardization. J Clin Microbiol. 1996;34:149–158. doi: 10.1128/jcm.34.1.149-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stallknecht D E, Luttrell M P, Fischer J R, Kleven S H. Potential for transmission of the finch strain of Mycoplasma gallisepticum between house finches and chickens. Avian Dis. 1998;42:352–358. [PubMed] [Google Scholar]
- 26.Turner K S, Kleven S H. Eradication of live F strain Mycoplasma gallisepticum vaccine using live ts/11 on a multiage commercial layer farm. Avian Dis. 1998;42:404–407. [PubMed] [Google Scholar]
- 27.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whithear K G. Control of avian mycoplasmoses by vaccination. Rev Sci Tech Off Int Epizoot. 1996;15:1527–1533. doi: 10.20506/rst.15.4.985. [DOI] [PubMed] [Google Scholar]
- 29.Yogev D, Rosengarten R, Watson-McKown R, Wise K S. Molecular basis of mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yogev D, Menaker D, Strutzberg K, Levisohn S, Kirchhoff H, Hinz K-H, Rosengarten R. A surface epitope undergoing high-frequency phase variation is shared by Mycoplasma gallisepticum and Mycoplasma bovis. Infect Immun. 1994;62:4962–4968. doi: 10.1128/iai.62.11.4962-4968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yogev D, Levisohn S, Kleven S H, Halachmi D, Razin S. Ribosomal RNA gene probes to detect intraspecies heterogeneity in Mycoplasma gallisepticum and M. synoviae. Avian Dis. 1988;32:220–231. [PubMed] [Google Scholar]