Abstract
We developed a rapid pulsed-field gel electrophoresis (PFGE) protocol for subtyping Campylobacter isolates based on the standardized protocols used by PulseNet laboratories for the subtyping of other food-borne bacterial pathogens. Various combinations of buffers, reagents, reaction conditions (e.g., cell suspension concentration, lysis time, lysis temperature, and restriction enzyme concentration), and electrophoretic parameters were evaluated in an effort to devise a protocol that is simple, rapid, and robust. PFGE analysis of Campylobacter isolates can be completed in 24 to 30 h using this protocol, whereas the most widely used current protocols require 3 to 4 days to complete. Comparison of PFGE patterns obtained in six laboratories showed that subtyping results obtained using this protocol are highly reproducible.
Campylobacter jejuni is the most common bacterial cause of diarrheal illness in the United States (1). It is estimated that Campylobacter infections affect over 2.5 million persons, with 80% of infections attributable to food-borne transmission (10). The majority of Campylobacter infections occur as sporadic events and not as part of outbreaks. Cases of campylobacteriosis are often associated with handling raw poultry or eating raw or undercooked poultry meat, though large outbreaks have most often been associated with the consumption of unpasteurized milk or contaminated water (7, 14, 20, 21). Strain differentiation is necessary for the identification of sources of contamination and determination of routes of transmission; this could in turn enable us to more accurately detect outbreaks and limit the spread of Campylobacter infections. A wide range of phenotypic and genotypic subtyping techniques has been applied to Campylobacter species to improve our understanding of the epidemiology of infection (15, 22). Macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) has been used successfully for inter- and intraspecies differentiation of campylobacters (6, 13, 18, 23). Although the sensitivity of PFGE is dependent on the choice of restriction enzyme, it is generally accepted that PFGE is one of the most discriminatory genotypic typing methods currently available for subtyping of Campylobacter species (11; C. Fitzgerald, L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields, submitted for publication).
A number of Campylobacter PFGE protocols have been described in the literature (8, 9, 12, 17, 23). However, individual laboratories use different procedures for plug preparation, restriction digestions, and electrophoretic separation of DNA fragments. This makes interlaboratory comparisons of Campylobacter PFGE profiles a challenging task. Several studies have investigated the interlaboratory reproducibility of PFGE analysis (3, 19). These studies highlight the importance of using standardized protocols in instances where the data to be compared will be generated in different laboratories. These issues were addressed by PulseNet, the National Molecular Subtyping Network for Foodborne Disease Surveillance, established by the National Center for Infectious Diseases, Centers for Disease Control and Prevention (CDC), in 1996. The use of standardized PFGE protocols by the PulseNet system allows for rapid comparison of DNA fingerprints from pathogens such as Escherichia coli O157:H7, Salmonella spp., and Shigella spp. between different laboratories to enhance food-borne disease surveillance. While there is international agreement that a standardized method is needed for Campylobacter species, until now, no significant advances have been made towards achieving this goal. Here we describe a rapid and robust PFGE protocol for the molecular subtyping of Campylobacter jejuni and other Campylobacter species. This protocol is based on the standardized PFGE procedure used by PulseNet (http://www.cdc.gov/ncidod/dbmd/pulsenet/pulsenet.htm). The robustness and reproducibility of the results obtained with this protocol were demonstrated by a study conducted at the CDC and five independent laboratories. Analysis of the data from these laboratories showed perfect correlation of the PFGE patterns for the seven test strains that were compared.
MATERIALS AND METHODS
Bacterial strains.
A set of three isolates was used for the development of the PFGE protocol described below (Table 1). An additional 18 isolates of different Campylobacter spp., including C. jejuni, C. coli, and C. lari, were used for the evaluation and validation of the protocol at the CDC. Seven of these isolates (see Fig. 5) were sent to five independent (non-CDC) laboratories (the Department of Consolidated Laboratory Services in Virginia, the South Carolina Department of Health and Environmental Control, the Minnesota Department of Health, the Los Angeles County Public Health Laboratory [Los Angeles, Calif.], and the University of Alberta Hospital [Edmonton, Alberta, Canada]) for evaluation and reproducibility testing. The bacteria were grown at 42°C overnight on heart infusion agar with 5% (vol/vol) defibrinated rabbit blood (Becton Dickinson, Franklin Lakes, N.J.) under microaerobic conditions (10% CO2, 5% H2, and 85% N2). All isolates had been previously identified to species level by standard procedures (2) and characterized by somatic O serotyping (16) and PFGE using an established protocol (13).
TABLE 1.
List of Campylobacter isolates used in the evaluation of the rapid PFGE protocol
| Isolate no. | Species | Source |
|---|---|---|
| D125 | C. jejuni | Human |
| D224 | C. jejuni | Bovine |
| D226 | C. jejuni | Human |
| D473 | C. jejuni | Avian |
| D1108 | C. jejuni | Bovine |
| D1118 | C. jejuni | Milk |
| D2577 | C. jejuni | Human |
| D2807 | C. jejuni | Human |
| D4996 | C. jejuni | Human |
| D5497 | C. jejuni | Human |
| EDL3 | C. jejuni | Bovine |
| EDL4 | C. jejuni | Bovine |
| EDL18 | C. jejuni | Human |
| EDL22 | C. jejuni | Human |
| SSH9892 | C. jejuni | Bovine |
| SSH9896 | C. jejuni | Bovine |
| D2489 | C. coli | Unknown |
| D2596 | C. coli | Swine |
| D1014 | C. lari | Avian |
| D1758 | C. lari | Human |
| L35 | C. lari | Avian |
FIG. 5.
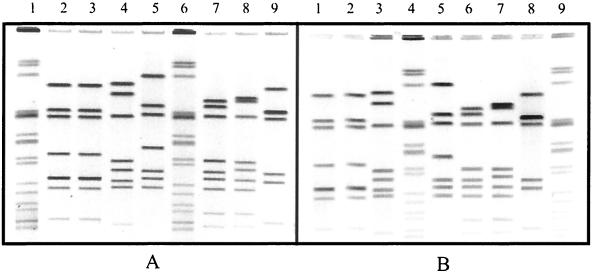
Gels showing the PFGE patterns of the seven isolates used in the validation study. (A) PFGE results obtained in our laboratory. (B) Results obtained in one of the independent laboratories. Lanes 2, 3, 4, 6, 7, 8, and 9 of panel A contain the PFGE profiles of isolates CDC1 to CDC7, respectively. These isolates are located in lanes 2, 3, 4, 5, 7, 8, and 9, respectively, in the gel shown in panel B. Each sample was digested with SmaI restriction enzyme. Lanes 1, 5, and 10 in panel A and lanes 1, 6, and 10 in panel B contain the strain used by PulseNet laboratories as the size standard for PFGE analysis of Salmonella enterica serovar Typhimurium isolates.
Plug preparation.
Cell suspensions were prepared by removing the cells from the surface of the culture plates using a cotton or polyester fiber applicator swab and suspending them in a polystyrene round-bottomed tube (Falcon; 12 by 75 mm; Becton Dickinson) containing 2 ml of phosphate-buffered saline (PBS; 0.01 M phosphate buffer [pH 7.4], 0.85% NaCl) or the cell suspension buffer (CSB;100 mM Tris, 100 mM EDTA, pH 8.0) recommended in the PulseNet protocol. Each cell suspension was adjusted to an optical density of 0.35 to 0.45 using a Dade MicroScan turbidity meter (Baxter Diagnostics, Inc., McGaw Park, Ill.). This corresponds to absorbance values of 0.570 to 0.820 at a wavelength of 610 nm when using a spectrophotometer. A 400-μl aliquot of adjusted cell suspensions was transferred to 1.5-ml microcentrifuge tubes containing 25 μl of proteinase K (20-mg/ml stock from Amresco, Solon, Ohio) and mixed gently by inverting the tubes two to four times. An equal volume (400 μl) of melted 1.0% SeaKem Gold (SKG) agarose in TE (10 mM Tris, 1 mM EDTA, pH 8.0) was added to the cell suspension, one sample at a time, and mixed gently by pipetting the mixture up and down two to three times. Plugs were also made using 1% SKG agarose containing 1% sodium dodecyl sulfate (SDS), the plug agarose used in the PulseNet protocol. The agarose-cell suspension mixture was dispensed immediately into the wells of reusable plug molds (catalog no. 170-3622; Bio-Rad Laboratories, Hercules, Calif.). The agarose plugs were allowed to solidify at room temperature for 10 to 15 min or at 4°C for 5 min.
Lysis of cells in plugs.
The plugs were transferred to 50-ml tubes (polypropylene tubes; 30 by 115 mm; Becton Dickinson) containing 5 ml of cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% sarcosine, 0.1 mg of proteinase K/ml). Lysis was allowed to proceed for 15 min at 54°C in an orbital shaker water bath with constant and vigorous agitation (150 to 200 rpm). We arrived at a 15-min lysis by testing different lysis times (0.25, 0.5, 1, 2, and 4 h).
Washes.
After lysis, the plugs were washed four times (15 to 20 min/wash) at 54°C (once with sterile ultrapure water and three times with TE, pH 8) in a shaking water bath. The water and TE were prewarmed at 54°C before each washing step. After the last wash, the TE was discarded and 5 ml of fresh TE (room temperature) was added to each tube.
Restriction digestion.
A 2-mm-wide slice from each plug was cut with a scalpel or single-edge razor blade and transferred to a tube containing 1× restriction buffer solution (SureCut buffer A; Roche Molecular Biochemicals, Indianapolis, Ind.). The plug slices were incubated in this restriction buffer at room temperature for 5 min. Then, the prerestriction mixture was removed, and 200 μl of the restriction enzyme mixture containing 40 U of SmaI (Roche) was added to each tube. The plug slices were incubated at room temperature (∼23 to 25°C) for 2 h. Prior to casting of the gel, the restriction mixture was removed from each tube and replaced with 200 μl of 0.5× TBE (10× TBE contains 0.89 M Tris borate and 0.02 M EDTA, pH 8.3; Sigma Chemical Co., St. Louis, Mo.). The plug slices were allowed to stand at room temperature for 5 min, after which they were loaded into the appropriate wells of a 1% SKG agarose gel.
Electrophoresis conditions.
The electrophoresis conditions used during the developmental stage of this PFGE protocol consisted of an initial switch time of 6.75 s and a final switch time of 38.35 s (gradient of 6 V/cm and an included angle of 120). These switch time values can be set using the AutoAlgorithim function of the CHEF Mapper (Bio-Rad) to separate fragments in the range of 50 to 400 kbp. The gels were electrophoresed for ∼18 h, depending on the equipment used (CHEF Mapper or GenePath; Bio-Rad), in 0.5× TBE. After the electrophoresis run was completed, the gels were stained with 400 ml of ethidium bromide solution (50 μg/ml), and the band pattern was observed under UV illumination.
Computer analysis of PFGE patterns.
The PFGE patterns were analyzed using the Molecular Analyst Fingerprinting Plus software package (version 1.2; Bio-Rad). The TIFF images were normalized by aligning the peaks of the size standard strain (Salmonella Newport strain AM01144), which was loaded on three lanes (lanes 1, 5, and 10 or lanes 1, 6, and 10) in each gel, with the database global standard. Matching and dendrogram UPGMA (unweighted pair group method with averages) analysis of the PFGE patterns was performed using the Dice coefficient with a 1.0 to 1.5% tolerance window.
RESULTS AND DISCUSSION
Preparation of agarose plugs.
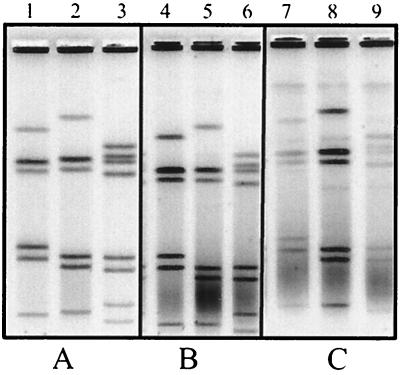
Two different CSBs were used in our evaluation: PBS and the modified TE buffer (CSB) described in the E. coli O157:H7 standardized protocol (4) used by PulseNet laboratories (Table 2 shows the buffer composition). While either buffer can be used to prepare the cell suspensions, we observed that occasionally plugs made with CSB showed some background. We chose PBS to prepare the cell suspensions because it provides an osmotically balanced environment that minimizes lysis of the cells prior to immobilization in the agarose. Adjusting the cell suspensions to an optical density of 0.35 to 0.45 worked best in our experiments. Suspensions adjusted to a higher cell concentration yielded DNA concentrations that were too high, resulting in poor resolution of the restriction fragments. Plugs made using SKG agarose containing 1% SDS consistently showed higher background than did those made with the agarose containing no SDS (Fig. 1). Additional evidence showing the effects of SDS on plug quality came from experiments where the concentration of detergent used was increased from 1 to 2%. As shown in Fig. 1C, the background observed in the plugs made with agarose containing 2% SDS was much higher than that in plugs made with 1% SDS. The presence of SDS in the plug agarose may cause the cells to lyse immediately after the melted agarose is added to the cell suspensions, resulting in the release of DNA while the agarose-cell suspension mixture is still in a liquid form. The pipetting force applied to the samples during the casting step might be causing shearing of the DNA, which leads to smearing. These results conclusively show that addition of SDS to the plug agarose is detrimental and unnecessary.
TABLE 2.
Comparison of reagents and conditions between the E. coli O157:H7 PFGE protocol and the rapid PFGE protocol for Campylobacter
| Reagent or conditions | Reagent and/or condition(s) for protocola:
|
|
|---|---|---|
| PulseNet protocol for E. coli O157:H7 | Campylobacter rapid PFGE protocol | |
| CSB | TE: 100 mM Tris, 100 mM EDTA, pH 8.0 | PBS: 0.01 M phosphate buffer (pH 7.4), 0.85% NaCl |
| Plug agarose | 1% SKG–1% SDS in TE | 1% SKG in TE |
| Lysis buffer | 50 mM Tris, 50 mM EDTA (pH 8.0), 1% sarcosine, 0.1 mg of proteinase K/ml | 50 mM Tris, 50 mM EDTA (pH 8.0), 1% sarcosine, 0.1 mg of proteinase K/ml |
| Washes (10 to 15 min each) | Six at 50°C (twice with sterile ultrapure water and four times with TE) | Four at 54°C (once with sterile ultrapure water and three times with TE) |
| Restriction digestion | 25 to 50 U (XbaI) at 37°C for 2 h | 40 U (SmaI) at 25°C for 2 h |
| Electrophoresis conditions | IST = 2.16 s; FST = 54.17 s; gradient = 6 V/cm; included angle = 120; running time = 18 to 19 h | IST = 6.75 s; FST = 38.35; gradient = 6 V/cm; included angle = 120; running time = 18 to 19 h |
IST, initial switch time; FST, final switch time.
FIG. 1.
PFGE gels showing a comparison of plugs made with 1% SKG agarose without SDS (A) with plugs made with 1% SKG containing 1 and 2% SDS (B and C, respectively). Each experiment was conducted using the same three isolates: D450 (lanes 1, 4, and 7), D4996 (lanes 2, 5, and 8), and D5497 (lanes 3, 6, and 9).
Lysis of cells.
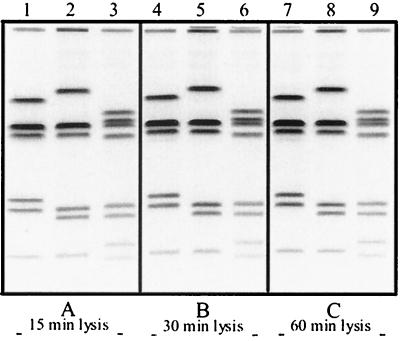
Several lysis incubation times (0.25, 0.5, 1, 2, and 4 h) were used to determine the shortest incubation time required for optimal lysis of cells in agarose plugs. The results showed that optimal lysis occurs within the first 15 to 30 min of incubation at 54°C (Fig. 2). This is in contrast to the much longer lysis incubation times (24 to 48 h of incubation at 56°C) recommended by other published PFGE protocols for Campylobacter (3, 8). Some PFGE protocols use a double-lysis method in which the lysis buffer is replaced halfway through the lysis incubation step (8). The need for extended or double lysis treatment of the plugs seemed unlikely to us, given the fact the Campylobacter is a fragile organism that can be lysed quite readily by standard methods, such as alkali lysis. Furthermore, PulseNet protocols for E. coli O157:H7 and Salmonella and Shigella species show that optimal lysis of cells in agarose plugs occurs within 1.5 to 2 h (at 54°C). Our data indicate that neither extended nor double lysis is required for efficient release of DNA from Campylobacter cells. While a 15-min lysis is sufficient, plugs can be lysed longer if needed. We observed no difference in the quality of the plugs when the lysis step was allow to proceed for up to 4 h (data not shown).
FIG. 2.
PFGE gels showing the results of different lysis incubation times. (A) Results of a 15-min lysis. (B) Results of lysis of plugs for 30 min. (C) Results of lysis of plugs for 60 min. Each experiment was conducted using the same three isolates as in Fig. 1: D450 (lanes 1, 4, and 7), D4996 (lanes 2, 5, and 8), and D5497 (lanes 3, 6, and 9).
Washing of agarose plugs.
Various washing schemes were used in an effort to determine the most efficient way to remove cellular debris from the lysed plugs. Plugs were washed three to six times (once with ultrapure water and two to five times with TE, pH 8.0, for 20 min/wash) at 54°C. We recommend that the lysed plugs be washed a total of four times (once with type 1 water and three times with TE, pH 8.0, for 20 min/wash) to ensure the removal of cell debris and proteinase K. Water is used in the first wash to increase the dialysis effect of the first wash (there is no significant amount of solute in water). Subsequent washes with TE allow for further removal of cell debris and lysis buffer while providing a stable environment for the DNA. The use of other chemicals, such as phenylmethanosulfonyl fluoride, was not evaluated, as such chemicals were not necessary to obtain adequate results for the strains tested in this study.
Restriction digestion.
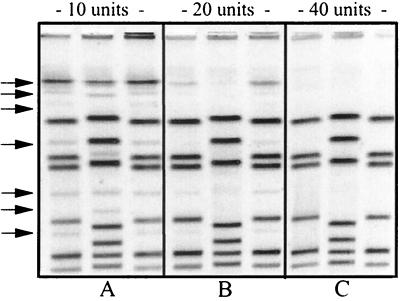
The restriction enzyme SmaI was used in this study because it is the most widely used restriction enzyme for PFGE analysis of Campylobacter (11, 13, 23). As shown in Fig. 3, the lanes containing the samples restricted with 10 and 20 U of SmaI show incomplete restriction. These lanes also illustrate the impact that incomplete restriction can have on the overall PFGE pattern. A total of 40 U of SmaI was needed to fully restrict Campylobacter DNA, in agarose plugs, in a 2-h incubation period. All the restriction digestion reactions were carried out at room temperature, which in our laboratory ranged from 23 to 26°C. Incomplete restriction was observed when restriction digestions were carried out at temperatures higher than 28°C (data not shown). We are currently evaluating different restriction endonucleases, such as SalI, SacII, BssHII, and KpnI, to be used as secondary enzymes for PFGE analysis. Preliminary data suggest that discriminatory potential, ease of use, and cost-effectiveness are enzyme specific (data not shown). However, the PFGE method described herein works well with all the enzymes mentioned above. The use of a secondary enzyme will enable us to improve the discrimination power of PFGE in cases where the results obtained with the primary enzyme, SmaI, are inconclusive.
FIG. 3.
PFGE gel showing the results of a 2-h restriction digestion using 10, 20, and 40 U of SmaI (A, B, and C, respectively). The numbers of units used in each reaction (three isolates per reaction) are indicated at the top of the gel. Arrows indicate incomplete restriction.
Electrophoresis conditions.
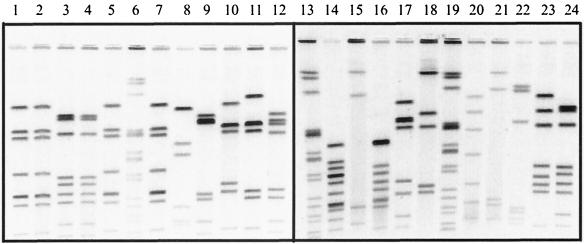
The electrophoresis conditions used were optimized for the analysis of C. jejuni isolates using the restriction enzyme SmaI. These conditions also provide adequate resolution of DNA fragments generated in PFGE analysis of other Campylobacter species (Fig. 4).
FIG. 4.
SmaI PFGE fingerprints of 21 different isolates tested with the protocol described in the text. Lanes 1 to 5 (EDL3, EDL4, D224, D226, and EDL18, respectively), 7 to 12 (EDL22, D473, D1108, SSH9892, D4996, and D5497, respectively), 17 and 18 (D2577 and D125, respectively), 20 (D2807), 23 (SSH9896), and 24 (D1118) contain the PFGE patterns from C. jejuni isolates. Lanes 14 and 16 contain patterns from C. coli isolates (D2596 and D2489, respectively). Lanes 15 (D1014), 21 (D1758), and 22 (L35) show the patterns from C. lari isolates. Lanes 6, 13, and 19 contain a Salmonella strain used as a size standard in our analysis.
Validation of the protocol.
Although the utility of PFGE for subtyping Campylobacter isolates has been demonstrated in several laboratories, data are often obtained using protocols unique to each laboratory, making the exchange and comparison of information virtually impossible. The first step in the validation process of our protocol was to assess its reproducibility. This was accomplished by analyzing the 21 isolates at least three times in our laboratory. No differences in the PFGE patterns were observed between different experiments (data not shown). The reproducibility of the protocol was further evaluated in a second laboratory at the CDC prior to the external evaluation. The PFGE patterns obtained in both laboratories matched perfectly (data not shown). The most important evaluation of this protocol was conducted in five external laboratories. A total of seven isolates were evaluated by these laboratories. The PFGE patterns obtained at the CDC for the seven isolates used in the external validation study are presented in Fig. 5A. Figure 5B shows the results obtained, using the same isolates, in one of the independent laboratories. The Molecular Analyst Fingerprinting Plus software was used to compare the PFGE patterns obtained at the CDC with the patterns obtained by five independent laboratories using this protocol. In each case, there was a perfect match between the PFGE patterns for each of the isolates (Fig. 6), indicating that the PFGE protocol described above is rapid, robust, and reproducible. Furthermore, this protocol is versatile, as demonstrated by the successful analysis of various Campylobacter species, including C. jejuni, C. coli, and C. lari (Fig. 4) as well as C. hyointestinalis, C. fetus, and C. concisus (data not shown).
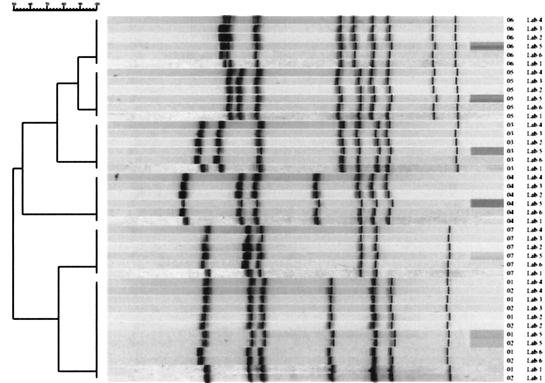
FIG. 6.
Dendrogram showing the results of the comparison between the PFGE patterns obtained at CDC and the patterns submitted by the five external laboratories. The dendrogram shows that each laboratory obtained the same pattern on matching isolates, indicating a high degree of reproducibility at the interlaboratory level.
In conclusion, standardization of protocols is crucial for the successful implementation of any molecular method as a practical epidemiologic tool. We chose the standardized protocols used by PulseNet as our model for the development of the Campylobacter protocol for several reasons. Since Campylobacter will be added to the PulseNet database, we wanted to maintain the reagents and equipment used in the protocol as close as possible to those listed in the standardized protocols already established for other pathogens (4). In addition, PulseNet laboratories must follow and comply with quality assurance and quality control measures when analyzing samples with the purpose of comparing them with patterns in the database (5). By following the Campylobacter protocol described above, PulseNet laboratories will not have to set and follow different quality assurance-quality control procedures for Campylobacter. Perhaps most importantly, the standardized protocols used by PulseNet have been validated through the testing of thousands of isolates every year by dozens of laboratories, nationally and internationally, and in outbreak situations, making them the most scrutinized PFGE protocols available. The protocol described here is the foundation for what will become the standardized PFGE protocol for C. jejuni to be used by PulseNet laboratories.
ACKNOWLEDGMENTS
We thank Mabel Ann Nicholson for assistance with the maintenance of the Campylobacter isolates used in this study and Susan B. Hunter for assistance with the computer analysis of the PFGE patterns. We are indebted to Denise Toney (Department of Consolidated Laboratory Services in Virginia), Lucy Scarborough (South Carolina Department of Health and Environmental Control), David Boxrud (Minnesota Department of Health), Eleanor Lehnkering (Los Angeles County Public Health Laboratory), and Linda Chui (University of Alberta Hospital, Canada) for their critical evaluation of the protocol. We thank Patricia Fields for reviewing the manuscript.
REFERENCES
- 1.Altekruse S F, Stern N J, Fields P I, Swerdlow D L. Campylobacter jejuni—an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T J, Patton C M, Morris G K. Differentiation of Campylobacter species using phenotypic characterization. Lab Med. 1988;19:96–102. [Google Scholar]
- 3.Bolton F J, Fox A J, Gibson J, Madden R H, Moore J E, Moran L, Murphy P, Owen R J, Pennington T H, Stanley T, Thompson-Carter F, Wareing D R A, Wilson T. A. multi-center study of methods for sub-typing Campylobacter jejuni. In: Newell D G, Ketley J M, Feldman R A, editors. Campylobacters, helicobacters, and related organisms. New York, N.Y: Plenum Press; 1996. pp. 1–35. [Google Scholar]
- 4.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: training manual. Atlanta, Ga: Centers for Disease Control and Prevention; 2000. [Google Scholar]
- 5.Centers for Disease Control and Prevention. Quality assurance (QA)/quality control (QC) manual for the standardized pulsed-field gel electrophoresis (PFGE) technique used by PulseNet Laboratories for foodborne disease surveillance. Training manual. Atlanta, Ga: Centers for Disease Control and Prevention; 2000. [Google Scholar]
- 6.Chang N, Taylor D E. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J Bacteriol. 1990;172:5211–5217. doi: 10.1128/jb.172.9.5211-5217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahey T, Morgan D, Gunneburg C, Adak G K, Majid F, Kaczmarski E. An outbreak of Campylobacter jejuni enteritis associated with failed milk pasteurisation. J Infect. 1995;31:137–143. doi: 10.1016/s0163-4453(95)92160-5. [DOI] [PubMed] [Google Scholar]
- 8.Gibson J R, Fitzgerald C, Owen R J. Comparison of PFGE, ribotyping and phage-typing in the epidemiological analysis of Campylobacter jejuni serotype HS2 infections. Epidemiol Infect. 1995;115:215–225. doi: 10.1017/s0950268800058349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–162. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 10.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.On S L, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.On S L W, Van Damme P. Identification and epidemiological typing of Campylobacter hyointestinalis subspecies by phenotypic and genotypic methods and description of novel groups. Syst Appl Microbiol. 1997;20:238–247. [Google Scholar]
- 13.Owen R J, Sutherland K, Fitzgerald C, Gibson J, Borman P, Stanley J. Molecular subtyping scheme for serotypes HS1 and HS4 of Campylobacter jejuni. J Clin Microbiol. 1995;33:872–877. doi: 10.1128/jcm.33.4.872-877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer S R, Gully P R, White J M, Pearson A D, Suckling W G, Jones D M, Rawes J C, Penner J L. Water-borne outbreak of Campylobacter gastroenteritis. Lancet. 1983;i:287–290. doi: 10.1016/s0140-6736(83)91698-7. [DOI] [PubMed] [Google Scholar]
- 15.Patton C M, Wachsmuth I K. Typing schemes: are current methods useful? In: Nachamkin L, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 110–128. [Google Scholar]
- 16.Penner J L, Hennessy J N. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salama S M, Tabor H, Richter M, Taylor D E. Pulsed-field gel electrophoresis for epidemiologic studies of Campylobacter hyointestinalis isolates. J Clin Microbiol. 1992;30:1982–1984. doi: 10.1128/jcm.30.8.1982-1984.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley J, Linton D, Sutherland K, Jones C, Owen R J. High-resolution genotyping of Campylobacter coli identifies clones of epidemiologic and evolutionary significance. J Infect Dis. 1995;172:1130–1134. doi: 10.1093/infdis/172.4.1130. [DOI] [PubMed] [Google Scholar]
- 19.van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Melchers J M, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrary primed PCR for typing of Staphylococcus aureus strain. J Clin Microbiol. 1995;31:406–409. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogt R L, Little A A, Patton C M, Barrett T J, Orciari L A. Serotyping and serology studies of campylobacteriosis associated with consumption of raw milk. J Clin Microbiol. 1984;20:998–1000. doi: 10.1128/jcm.20.5.998-1000.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt R L, Sours H E, Barrett T, Feldman R A, Dickinson R J, Witherell L. Campylobacter enteritis associated with contaminated water. Ann Intern Med. 1982;96:292–296. doi: 10.7326/0003-4819-96-3-292. [DOI] [PubMed] [Google Scholar]
- 22.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W, Chang N, Taylor D E. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J Infect Dis. 1991;163:1068–1072. doi: 10.1093/infdis/163.5.1068. [DOI] [PubMed] [Google Scholar]