Abstract
Angioimmunoblastic T-cell lymphoma (AITL) and peripheral T-cell lymphoma with T follicular helper phenotype (PTCL-TFH) are a group of complex clinicopathological entities that originate from T follicular helper cells and share a similar mutation profile. Their diagnosis is often a challenge, particularly at an early stage, because of a lack of specific histological and immunophenotypic features, paucity of neoplastic T cells and prominent polymorphous infiltrate. We investigated whether the lymphoma-associated RHOA Gly17Val (c.50G>T) mutation, occurring in 60% of cases, is present in the early “reactive” lesions, and whether mutation analysis could help to advance the early diagnosis of lymphoma. The RHOA mutation was detected by quantitative polymerase chain reaction with a locked nucleic acid probe specific to the mutation, and a further peptide nucleic acid clamp oligonucleotide to suppress the amplification of the wild-type allele. The quantitative polymerase chain reaction assay was highly sensitive and specific, detecting RHOA Gly17Val at an allele frequency of 0.03%, but not other changes in Gly17, nor in 61 controls. Among the 37 cases of AITL and PTCL-TFH investigated, RHOA Gly17Val was detected in 62.2% (23/37) of which 19 had multiple biopsies including preceding biopsies in ten and follow-up biopsies in 11 cases. RHOA Gly17Val was present in each of these preceding or follow-up biopsies including 18 specimens that showed no evidence of lymphoma by combined histological, immunophenotypic and clonality analyses. The mutation was seen in biopsies 0-26.5 months (mean 7.87 months) prior to the lymphoma diagnosis. Our results show that RHOA Gly17Val mutation analysis is valuable in the early detection of AITL and PTCL-TFH.
Introduction
Angioimmunoblastic T-cell lymphoma (AITL), peripheral T-cell lymphoma with T follicular helper phenotype (PTCL-TFH) and follicular T-cell lymphoma are a group of complex clinicopathological entities that originate from T follicular helper (TFH) cells. These lymphomas are the most common among various T-cell malignancies in Western countries. Patients with these lymphomas commonly show an aggressive clinical course, with a median 3-year survival rate of only 30% (https://www.hmrn.org/ statistics/disorders/34). The poor clinical outcome is largely a consequence of the lack of understanding of their molecular mechanism and targeted therapy.
The diagnosis of these lymphoma entities, particularly at an early stage, is a challenge because of the lack of specific clinical and histological features, low tumor cell content and the presence of prominent polymorphous inflammatory infiltrates that often mask the neoplastic cells. To help the diagnosis, T-cell clonality analysis is commonly used, but this is frequently not helpful because of the paucity of malignant T cells. The diagnostic difficulty is further exacerbated by the increasing use of needle core biopsies to avoid more invasive surgical excision. When a lymphoproliferative lesion is suspected but uncertain on histological diagnosis, patients are commonly subjected to a “watch and wait” approach, and further biopsied when showing signs of disease progression.
There is a wide range of genetic changes in T-cell lymphomas with TFH phenotype, including early TET2 and DNMT3A mutations, which occur in hematopoietic stem cells, and late genetic changes, which are specifically seen in lymphoma cells.1-4 Among the latter, RHOA mutation is the most frequent,2,5-9 occurring in 60-70% of cases, with Gly17Val (c.50G>T) accounting for 95% of the changes. Interestingly, studies of mouse models have suggested that RHOA Gly17Val mutation induces TFH cell differentiation and, together with loss-of-function TET2 mutations, can promote the development of AITL-like lymphomas.10-12 Importantly, RHOA mutation is preferentially associated with the above lymphoma entities and has, so far, been shown only in the neoplastic T-cell population, but not in reactive B and T cells in these malignant conditions.2,5,13,14 Studies to date suggest that the RHOA Gly17Val mutation could be used as a marker for the diagnosis of AITL and PTCL-TFH.6,15 However, it is unclear whether the mutation is present in tissue biopsies prior to the diagnosis of AITL and whether mutation analysis could help in the early detection of these lymphomas. To investigate these issues, we established a highly sensitive quantitative polymerase chain reaction (qPCR) assay to detect the RHOA mutation, and screened a large cohort (n=37 cases) of AITL and PTCL-TFH with multiple sequential biopsies together with 61 controls.
Methods
Tissue materials and DNA extraction
The use of archival tissues for research was approved by the ethics committees of the institutions involved. In total, 78 tissue specimens from 37 patients (multiple consecutive specimens in 29 cases) with AITL (n=35) or PTCL-TFH (n=2), together with 61 controls (13 with reactive lymph nodes with paracortical expansion of T cells, 10 with classic Hodgkin lymphoma and 1 with marginal zone lymphoma with a prominent background of T cells) and peripheral blood DNA samples from 16 individuals (TET2 and DNMT3A mutations in 2, TET2 mutation in 4, DNMT3A mutation in 5) with clonal hematopoiesis of indeterminate potential (CHIP) (1 individual classed as having high-risk CHIP) were successfully investigated. DNA samples were prepared from a whole tissue section of each specimen. Purified DNA was obtained using a QIAamp DNA-Micro kit (Qiagen), while crude DNA was obtained by digesting samples at 37°C overnight with proteinase K and NP-40 buffer. Purified DNA samples from nine cases of AITL or PTCL-TFH with known RHOA mutations (Gly17Val, c.50G>T, n=6 with variant allele frequency [VAF] ranging from 2-32%; Gly17Leu, c.49-50GG>TT, n=2; Gly17Glu, c.50G>A, n=1; Ser26Arg, c.76A>C, n=1) were available from our previous study,2 while crude DNA samples were available from routine clonality analysis or similarly prepared. The histology of these specimens was reviewed by specialist hematopathologists.
Quantitative polymerase chain reaction with peptide nucleic acid clamp and locked nucleic acid probe
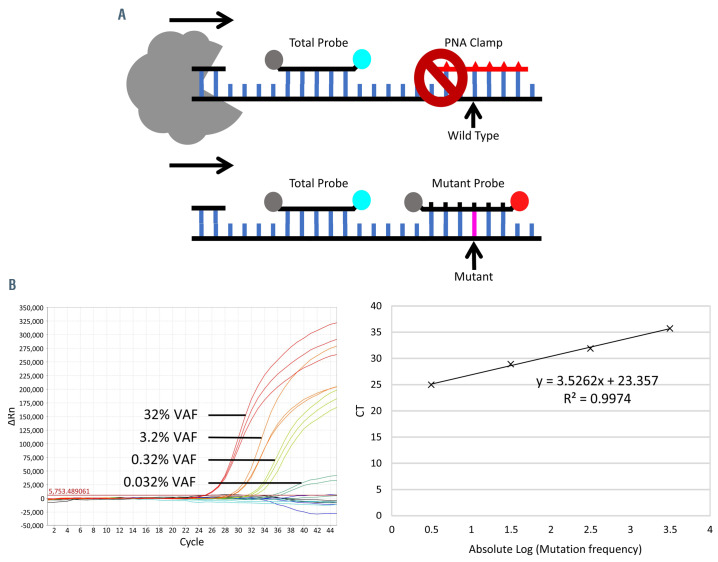
The qPCR with peptide nucleic acid (PNA) clamp and locked nucleic acid (LNA) probe was performed as previously described.16 The qPCR assay contained two probes and a PNA clamp (Figure 1A, Online Supplementary Table S1). The total probe served as an internal control to monitor the PCR performance. The PNA clamp specifically and strongly binds to the wild-type DNA sequence, resisting the 5’ nuclease activity of Taq DNA polymerase, and thus blocks the wild-type allele from PCR amplification. This results in preferential amplification of the mutant allele which is detected specifically by the LNA mutant probe.
PCR primers with various probes were first tested using purified DNA samples, with the optimized conditions outlined below (Online Supplementary Table S1). Briefly, the PNA-LNA PCR was carried out in a 20 μL reaction containing 10 mL Premix Ex Taq (Probe qPCR) Master Mix (Takara, Shiga, Japan), 0.2 mM of each forward and reverse primer, 0.1 μM of each total and mutant probe, 0.05 mM of the PNA clamp probe, and 2 mL of crude DNA or 25 ng of purified DNA. Real-time PCR was carried out in triplicate using Quantstudio 6 (Thermo Fisher Scientific, Waltham, MA, USA) with denaturation at 95°C for 30 s followed by 45 cycles at 95°C for 3 s, and at 62°C for 30 s.
Targeted sequencing using the Fluidigm Access Array and Illumina MiSeq
Targeted sequencing was performed on a selected case with consecutive tissue biopsies as described in our recent study.2 Each of the DNA samples was investigated in duplicate for mutations in TET2, DNMT3A, IDH2, RHOA, PLCG1, CCND3, CD28 and TNFRSF21 by Fluidigm PCR and Illumina MiSeq sequencing. Sequence reads alignment, variant calling, filtering to eliminate false positive and benign changes were carried out according to our previously established protocols.2 Only the reproducible variants that appeared in both replicates were regarded as true changes.
BaseScope in situ hybridization
This was performed on a selected case. The sequence of the clonal TRB rearrangement in a case of AITL was available from a recent study.2 Based on the unique VDJ junctional sequence, unique BaseScope probes were designed and used to identify the lymphoma T cells by in situ hybridization. The BaseScope in situ hybridization was carried out according to the manufacturer's instructions (Advanced Cell Diagnostics, Newark, CA, USA),2 with the conditions optimized using the AITL specimen from which the clonal TRB rearrangement was sequenced. Tonsils were used as a negative control.
Results
Peptide nucleic acid – locked nucleic acid quantitative polymerase chain reaction is highly sensitive and specific for the detection of RHOA Gly17Val (c.50G>T)
Upon the optimization of PCR conditions for RHOA mutation detection, we first determined the sensitivity of the qPCR assay using serial dilutions of three purified DNA samples with known VAF of Gly17Val (c.50G>T) into tonsil DNA. The PNA-LNA qPCR was highly sensitive, capable of detecting the mutation at a VAF of 0.032% (Figure 1B). As expected, the qPCR was highly specific to Gly17Val (c.50G>T) and showed no detectable signal of the LNA mutant probe with the DNA samples harboring other RHOA mutations including Gly17Leu (c.49-50GG>TT) and Gly17Glu (c.50G>A), or tonsillar DNA (Online Supplementary Figure S1A).
Figure 1.
Detection of RHOA p.Gly17Val (c.50G>T) by real time polymerase chain reaction with peptide nucleic acid/locked nucleic acid probes. (A) Schematic illustration of the real time polymerase chain reaction (PCR) design. The total probe is used as an internal control to monitor the PCR performance. The peptide nucleic acid (PNA) clamp binds to the wild-type sequence, resists the 5’ nuclease activity of Taq DNA polymerase and thus blocks the wild-type allele from PCR amplification. This results in preferential amplification of the mutant allele which is detected specifically by the locked nucleic acid (LNA) mutant probe. (B) The PCR assay is highly sensitive, capable of detecting the RHOA mutation at a variant allele frequency (VAF) of 0.03% based on serial dilutions of an angioimmunoblastic T-cell lymphoma sample with known mutation allele frequency by next-generation sequencing (left panel). For simplicity, only the LNA mutant but not total probe signals are shown. The PCR assay shows a linear correlation among the serial dilutions (right panel).
As crude DNA preparations were routinely used for clonality analysis in our clinical diagnostic laboratories, we tested whether such crude DNA samples were amenable to the above PNA-LNA qPCR (Online Supplementary Figure S1C). Of the ten crude DNA samples initially tested, nine yielded excellent amplification comparable with the results from purified DNA samples. We then used crude DNA preparations for the qPCR in the remaining investigations, with high quality results from 139 of the 144 samples investigated. The reason that the other five samples failed to support qPCR was essentially the poor quality of the DNA, as evidenced by quality control PCR.
RHOA Gly17Val (c.50G>T) is detected in initial biopsies not diagnostic for lymphoma by conventional approaches
To confirm that the RHOA mutation was lymphomaspecific, not associated with reactive conditions proliferations or other lymphoproliferative disorders with enriched T cells, we investigated 45 tissue biopsies including 27 from lymph nodes, for which T-cell clonality analysis was requested during routine histological diagnosis, but showed no evidence of a T-cell lymphoma. These included 13 specimens showing paracortical T-cell expansion or enriched CD4+ T cells, and ten classic Hodgkin lymphomas with a prominent background of T cells. The qPCR was successful for all these specimens, as indicated by the total probe control, but they were all negative for the RHOA mutation. Similarly, we also showed absence of the RHOA mutation in peripheral blood lymphocytes (Online Supplementary Figure S1D) from 16 individuals with CHIP (mutational information provided in Online Supplementary Table S2), in keeping with the findings from whole exome or panel sequencing.17,18
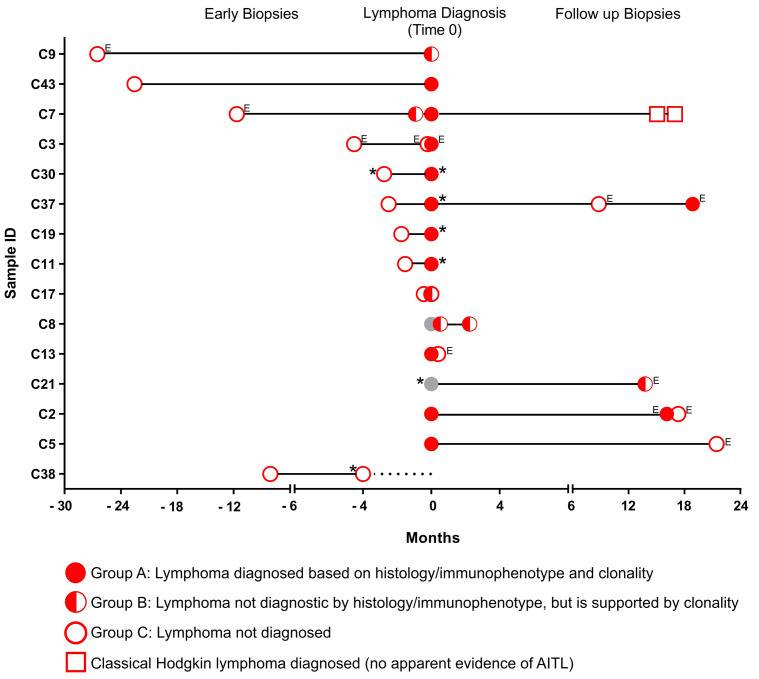
Figure 2.
Analysis of RHOA (c.50G>T; p.Gly17Val) mutation by quantitative polymerase chain reaction in the RHOA-positive cases with longitudinal biopsies in patients with angioimmunoblastic T-cell lymphoma (only including cases with at least one biopsy classed as not diagnostic [“group B” or “group C”]). The diagnoses of these specimens were reviewed, and categorized into three groups: group A (filled circles): lymphoma diagnosed by histology and immunophenotype, further supported by clonal TCR gene rearrangement; group B (half-filled circles): lymphoma not diagnostic by histology and immunophenotype alone, but ascertained by clonal TCR gene rearrangement; group C (open circles): lymphoma not diagnosed by combined analyses. Red (regardless of the symbol: including outline only, half-filled, and completely filled symbols) indicates RHOA p.Gly17Val mutant positive biopsy. Gray indicates RHOA p.Gly17Val status unknown. E denotes extranodal biopsies. *denotes lymph node excision biopsies and all others are core biopsy specimens. An open square denotes a diagnosis of classic Hodgkin lymphoma with no apparent evidence of angioimmunoblastic T-cell lymphoma (AITL). Case 38 lacks a final diagnosis as the patient died, indicated by a dashed line. Cases with only group A multiple biopsies are not included in this figure.
We then investigated 37 cases of AITL (n=35) or nodal PTCL-TFH (n=2) with unknown RHOA mutation status, of which 29 had multiple longitudinal biopsies (2-5) available for RHOA analysis. RHOA mutation was seen in 23 cases, but was negative in the remaining 14 cases. Among the 23 cases with RHOA mutation, 19 cases had multiple biopsies. We reviewed the original histological diagnosis in each of these specimens and categorized them into three groups. Group A specimens (n=26) showed clear evidence of lymphoma by histology and immunophenotype, further supported by clonal TCR rearrangement. Group B (n=6) had suspicious histological and immunophenotypic findings, although not entirely diagnostic for lymphoma, which was ascertained by detection of strong clonal TCR gene rearrangements. Finally, group C mutations (n=18) were not diagnostic for lymphoma by combined histological, immunophenotypic and clonality analyses (Online Supplementary Table S3). Interestingly, RHOA mutation was detected in each of these specimens, in the positive cases, including those not diagnostic for lymphoma by conventional integrated diagnostic investigations (Figure 2).
Characteristics of biopsies not diagnostic for lymphoma but positive for RHOA mutation
Of the 23 cases of AITL or PTCL-TFH with RHOA mutation, the initial biopsy was not diagnostic in ten cases including two with an excisional lymph node specimen. The time from the initial non-diagnostic biopsy to that establishing AITL or PTCL-TFH ranged from 0 to 26.5 months, with an average of 7.87 months. All these initial non-diagnostic biopsies showed polyclonal (n=4), weak clonal (n=5) or oligoclonal (n=2) TCR gene rearrangements or failed (n=1) by BIOMED-2 TRB and TRG clonality analysis. Seven of the initial non-diagnostic biopsies were also subjected to BIOMED-2 B-cell clonality analysis and six showed a clonal IG gene rearrangement. Epstein-Barr virus (EBV)-encoded small RNA (EBER) in situ hybridization was carried out in eight cases during the routine diagnostic workup, and four showed variable positivity from a few scattered to numerous EBER-positive cells, of which three displayed clonal IG gene rearrangement. In four cases, EBV-driven proliferation or classic Hodgkin lymphoma were considered in the initial diagnosis.
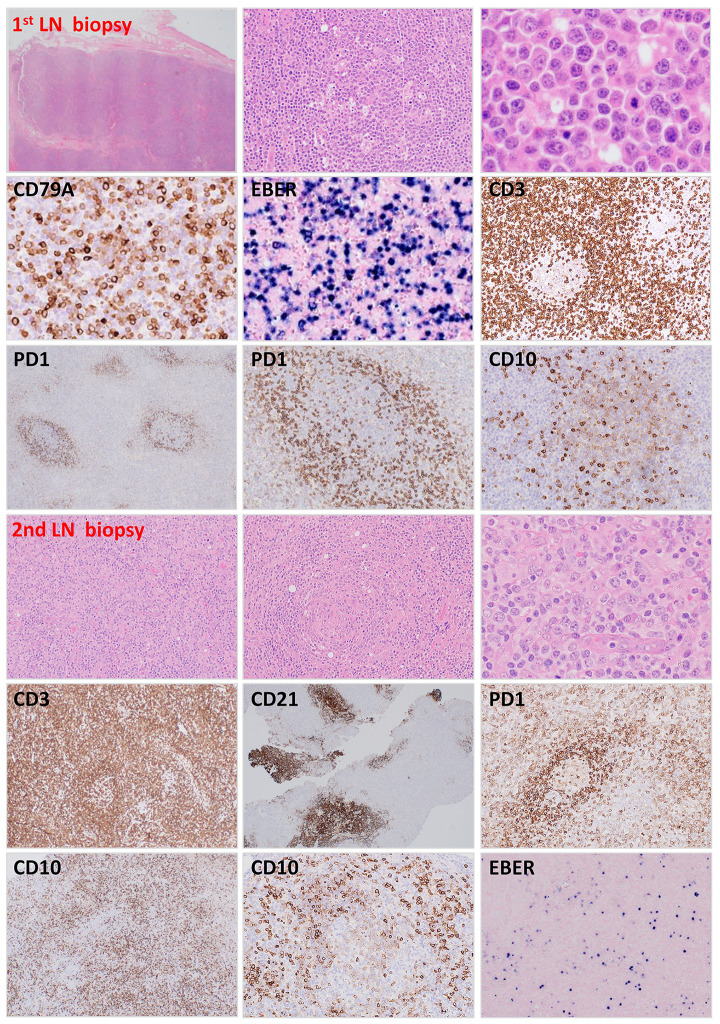
Figure 3.
Histological and immunophenotypic findings in case 30. The first lymph node excision biopsy shows partial effacement of the lymph node architecture by polymorphous infiltrates, particularly B cells with plasmacytoid differentiation. A large proportion of B cells were EBER-positive and showed IG κ light chain restriction (not shown). The lymphoid follicles appear to be reactive and shows no apparent expansion of T follicular helper (TFH) cells, with only a few CD10-positive cells spilling out of the germinal center, consistent with pattern-1 histology of angioimmunoblastic T-cell lymphoma (AITL). The second lymph node biopsy shows effacement of the lymph node architecture by medium-sized atypical lymphoid cells with regressed follicles. There is a prominent proliferation of follicular dendritic cell meshworks and high endothelial venules with atypical lymphoid cells clustered in their vicinity. The atypical lymphoid cells are T cells expressing TFH markers, spilling out of the germinal center to the interfollicular region. EBER in situ hybridization shows only scattered positive cells.
Figure 4.
Histological and immunophenotypic findings in case 7. The first biopsy shows a subcutaneous perivascular infiltrate of CD3+ T cells with vasculitic features. The third biopsy shows cardinal features of angioimmunoblastic T-cell lymphoma (AITL) and also a prominent pleomorphic infiltrate including an EBER-positive B-cell population with Hodgkin and Reed/Sternberg (HRS)-like morphology and immunophenotype. The fifth biopsy shows no apparent evidence of AITL, but a polymorphous infiltrate with a more prominent EBER-positive B-cell population that has HRS cell morphology and immunophenotype, rosetting by CD4+ T cells.
Table 1.
Summary of histological, immunohistochemical, clonality analysis and genetic findings in case 7.
Six follow-up biopsies were not diagnostic of involvement by AITL or PTCL-TFH by routine diagnostic investigations; these included two bone marrow and two skin specimens. In each specimen, a CD4+ T-cell infiltrate was noted, but a definite aberrant immunophenotype and expression of TFH markers could not be ascertained. Tcell clonality analysis showed weak polyclonality in two and a weak clonal or oligoclonal pattern in two.
Representative cases
Case 30. A 79-year-old man presented with bilateral tender neck lymph nodes, and had mild sweats, but no weight loss or fever. Clinical examination revealed multiple bilaterally enlarged neck and groin lymph nodes (up to 1.5 cm diameter), and palpable liver and spleen. Right level II neck lymph node excision biopsy showed partial effacement of the lymph node architecture and expansion of the interfollicular area by a polymorphous population of lymphoid cells, including B cells with plasmacytoid differentiation, and scattered large cells (Figure 3). These B cells expressed pan B-cell markers (CD20, CD79a, CD19), and MUM1, but were negative for CD10 and BCL6 (data not shown). A high proportion of the B cells were EBERpositive and showed IG k light chain restriction. The lymphoid follicles appeared to be reactive and showed no apparent expansion of TFH cells, with only a few CD10- positive cells spilling out of the germinal centers (Figure 3). BIOMED-2 clonality analyses showed clonal IGH and IGK gene rearrangements, but a weak oligoclonal pattern with TRG and TRB. The histological diagnosis was uncertain: a clonal EBV-positive polymorphous lymphoproliferation was considered. Close follow-up with a low threshold for re-biopsy was recommended.
Two months later, the patient presented with increasing fatigue, fever, maculopapular chest rash and increased size of peripheral lymphadenopathy. Positron-emission tomography (PET) scan revealed extensive bilateral cervical, mediastinal, bilateral iliac and groin lymphadenopathy. Left level V neck lymph node excision biopsy showed partial effacement of the lymph node architecture by an infiltrate of medium-sized atypical lymphoid cells with regressed follicles (Figure 3). There was hyperplasia of follicular dendritic cell meshworks and high endothelial venules with the atypical lymphoid cells clustered in their vicinity. The atypical lymphoid cells were positive for CD3, CD5 and TFH markers (PD1, CD10, ICOS, BCL6) (Figure 3). EBER in situ hybridization revealed only scattered positive cells. BIOMED-2 clonality analyses showed clonal TRB and TRG gene rearrangements, and also weak clonal IGH and IGK gene rearrangements which were different from those of the previous biopsy in the size of their amplified IG products. A diagnosis of AITL was made. The patient was initially treated with six cycles of CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone), then avelumab (cycle 12 at the most recent follow-up) under the AVAIL-T trial, and was well, showing no constitutional symptoms or palpable cervical lymph nodes, 20 months following the AITL diagnosis.
Retrospective analysis showed the presence of RHOA Gly17Val mutation in the above two specimens by qPCR and targeted sequencing with 3% and 23% VAF in the first and second biopsy, respectively. The targeted sequencing also revealed a pathogenic nonsense substitution in DNMT3A (c.2311C>T, p.R771*) in the first and second biopsy with 17% and 26% VAF, respectively.
Case 7. An 82-year-old man, with remission of a previous mantle cell lymphoma, presented with a skin rash on the right calf. A punch biopsy showed a mild perivascular infiltrate by CD3+ T cells, with a histological diagnosis of panniculitis (Figure 4, Table 1). To investigate potential lymphoma relapse, lymph node core biopsies were taken at 11 and 12 months of follow-up: neither specimen showed evidence of mantle cell lymphoma, but both revealed cardinal features of AITL and also a prominent pleomorphic infiltrate including an EBER-positive B-cell population with Hodgkin and Reed/Sternberg (HRS)-like morphology and immunophenotype (Figure 4, Table 1). T-cell clonality analysis demonstrated clonal rearrangements by TRG-A and TRB-A with identical sized amplified products between the two biopsies, and an additional clonal rearrangement by TRB-C in the second biopsy. Further follow-up biopsies were taken at 25 and 26 months, and neither showed apparent evidence of AITL, but both had polymorphous infiltrates with a more prominent EBER-positive B-cell population that had HRScell morphology and immunophenotype (Figure 4, Table 1). Neither specimen showed any evidence of the clonal TRG/TRB rearrangements seen in the early lymph node biopsies, although a fifth biopsy displayed an isolated clonal rearrangement by TRB-C. B-cell clonality analyses demonstrated polyclonal IG gene rearrangements in both specimens (Figure 4, Table 1). A classic Hodgkin lymphoma arising from the EBV-positive B-cell component of the AITL was considered.
In a retrospective study, the TRB-A and B PCR products from the second biopsy were sequenced using an Illumina MiSeq platform and a dominant TRBV5-J2 rearrangement (86%) was identified.2 Based on the unique VDJ junctional sequence, we designed unique BaseScope probes to identify the lymphoma T cells by in situ hybridization (Online Supplementary Figure S2). As expected, both the second and third biopsies with an AITL diagnosis showed diffuse positivity with the lymphoma clone-specific probe. Interestingly, the initial skin biopsy also displayed isolated positive cells, while the fifth biopsy with a diagnosis of EBV lymphoproliferative disease (LPD) gave a negative result. Both the second (AITL) and fifth (EBV-LPD) biopsies were investigated by panel sequencing for recurrent somatic mutations, and this identified five shared mutations – one DNMT3A, three TET2 and one RHOA changes – between the two specimens, and one further TET2 mutation only in the second specimen (Table 1). In general, the mutation load in each of the above shared changes was much higher in the second biopsy (AITL) than in the fifth biopsy (EBV-LPD). This was particularly striking for the RHOA Gly17Val mutation, with a VAF of 20% in the second biopsy but of only 1% in the fifth biopsy. As expected, qPCR for the RHOA mutation demonstrated its strong positivity in both the second and third biopsies showing AITL, but a weak positive signal in the initial skin biopsy, and the fourth and fifth biopsies displaying classic Hodgkin lymphoma-like EBV-LPD (Figure 4, Online Supplementary Figure S2).
Discussion
By using a highly sensitive qPCR assay, we confirmed that RHOA Gly17Val (c.50G>T) mutation is specifically associated with AITL or PTCL-TFH, but is not found in other lymphoproliferative conditions including those with florid infiltration of T-helper cells. More importantly, detection of RHOA Gly17Val (c.50G>T) mutation could help early detection of AITL and PTCL-TFH, and also diagnosis of their extranodal involvement.
The difficulty in making a histological diagnosis of AITL or PTCL-TFH is well recognized, particularly when the neoplastic cell content is low, inconspicuous by histological and immunophenotypic assessment and undetectable by analysis of TCR gene rearrangements. Apart from the paucity of neoplastic T cells, the polymorphous infiltrate, particularly the presence of numerous EBV-positive B cells, including HRS-like cells (EBV-positive or -negative), may lead to diagnostic consideration of an EBV-driven B-cell proliferation.19-21 Such misleading diagnostic features may also be reinforced by the frequent demonstration of clonal IG gene rearrangement. As shown in this study, EBV-driven B-cell proliferation was considered as a diagnosis in four of 11 of the initial biopsies. Indeed, it has been reported by several independent studies that EBV-associated lymphoproliferation is a frequent pitfall in the diagnosis of AITL.19-22
In the present study, the majority of the initial specimens that were not diagnostic were core biopsies. It is possible that these core biopsies were not totally representative, with characteristic lymphoma components missed because of sampling errors. Nevertheless, two initial nondiagnostic biopsies were excisional lymph node specimens, indicating that an absence of diagnostic features of AITL/PTCL-TFH in the initial biopsies was also a real issue. Interestingly, the time interval between the initial non-diagnostic biopsies and the follow-up biopsies that established the diagnosis varied considerably, ranging from 0 to 26.5 months. In the cases with a long interval, it is likely that the initial biopsy represented an early premalignant lesion, while the follow-up biopsy reflected more progressed disease, thus having more cardinal features for making the AITL/PTCL-TFH diagnosis. While in the cases with a short interval, it is possible that the enlarged lymph nodes were variably involved by AITL/PTCL-TFH and the initial non-diagnostic biopsies represented early involvement by the lymphoma.
Both the above possibilities may exist, without excluding the other. It is impossible to distinguish the two scenarios based on the analysis of a single biopsy, including RHOA mutation analysis. Nonetheless, detection of RHOA Gly17Val (c.50G>T) mutation can certainly raise the alarm to perform more in-depth histological and immunophenotypic investigations, for example a more careful search for evidence of TFH cell expansion, as shown in the first biopsy in case 30. It is important to emphasize that detection of a RHOA mutation is not equivalent to a diagnosis of lymphoma because mutational analysis by qPCR or targeted sequencing is highly sensitive, and the mutation is seen in biopsies without histological evidence of AITL. As discussed above, the RHOA mutation-positive non-diagnostic biopsy may represent a premalignant lesion or early involvement by the lymphoma. Therefore, RHOA mutation analysis should be used as an auxiliary tool with the results interpreted in the context of histological and immunophenotypic findings. If a histological diagnosis of AITL/PTCL-TFH cannot be made, an excision biopsy or a low threshold for an early follow up biopsy, when appropriate, is indicated.
In a few case studies, RHOA mutation was detected in circulating free DNA or peripheral blood lymphocytes from patients with AITL/PTCL-TFH,15 and the mutation burden appeared to correlate with the treatment outcome.23,24 It remains to be investigated whether the mutation is detectable in circulating free DNA or peripheral blood lymphocytes at the time of initial non-diagnostic biopsies. Given that the RHOA mutation burden in circulating free DNA and peripheral blood lymphocytes reflects, at least theoretically, the overall lymphoma load, simultaneously analyzing blood samples, in addition to tissue biopsy, could add further value in lymphoma diagnosis, particularly when a specimen is not representative.
Apart from the initial biopsies, the diagnosis of extranodal involvement by AITL/PTCL-TFH such as skin and bone marrow is also difficult for reasons similar to those discussed above, including low tumor cell content and polymorphous infiltrate.25 In particular, extranodal infiltrates may not harbor the cardinal features of AITL such as TFH marker expression in the neoplastic T cells and follicular dendritic cell meshworks. The DNA sample from these extranodal sites is often not informative for clonality analysis because of insufficient lymphoid cells and/or poor DNA quality. As shown in our study, RHOA mutation analysis is highly valuable for confirming AITL/PTCL-TFH involvement in follow-up biopsies. Similarly, RHOA mutation analysis is valuable in the diagnosis of AITL/PTCL-TFH from cytological specimens. 9
Aside from the RHOA mutation, there are several other genetic changes including IDH2, CD28, PLCG1, VAV1 and TNFRSF21 mutations, and VAV1-STAP2, CTLA4-CD28 and ITK-SYK fusions, which occur at variable frequencies in AITL and PTCL-TFH.2,26-28 Of note, mutations in VAV1, a signaling molecule downstream of TCR, occurs in 8.2% of AITL and appears to be mutually exclusive of the RHOA mutation.28 Detection of these additional lymphoma- associated genetic changes could also help the early detection of AITL/PTCL-TFH, together with RHOA mutation detection, potentially being valuable for diagnosis in up to 90% of these T-cell lymphomas. These diverse genetic changes could be readily investigated by targeted sequencing either alone or as part of a comprehensive lymphoma panel.
The detailed analyses of multiple biopsies in case 7 also revealed the mutation burden in non-malignant T cells. The fifth lymph node biopsy showed little involvement by AITL as the lymphoma clone was undetectable by BaseScope in situ hybridization and the VAF of the RHOA mutation was only 1%, but the specimen had a high TET2 mutation burden (20% VAF). This implies that the TET2 mutation must have been present in a large proportion of reactive B and T cells,1,5,29-31 probably up to 40% of the total cell population, which is well above the EBER-positive cell fraction (~5%). Hence, this enlarged lymph node was essentially caused by lymphoid proliferation driven by the TET2 mutation and/or EBV infection. These findings further highlight the markedly variable histological presentation of enlarged lymph nodes in patients with AITL and, hence, the danger of potential sampling errors in the diagnosis of AITL. In this context, it is pertinent to investigate RHOA and other lymphoma-associated mutations in patients with advanced age and biopsies showing EBV-positive polymorphous infiltrates, to explore how such mutation analyses can help to circumvent this diagnostic pitfall. At the genetic level, the high number of TET2 mutations and their frequent presence in non-neoplastic T cells suggest that the patient most likely had an underlying CHIP with TET2 and DNMT3A mutations that occurred in hematopoietic stem cells, consequently extending to the progenies of these cells.
In general, patients with AITL typically have an aggressive clinical course and respond poorly to currently available therapies. Case 7 appeared to be an exception to this rule, showing slow disease progression in the absence of any treatment, although it is not possible to rule out a response to the R-GCVP (rituximab, gemcitabine, cyclophosphamide, vincristine, prednisolone) that was aimed at treating the EBV-associated proliferations (Table 1). An indolent clinical course has been previously reported for some patients with AITL showing pattern-1 histology, including those treated only with steroids.22,32 It remains to be investigated how to identify such indolent cases at the time of diagnosis and stratify their clinical management accordingly, particularly in view of the advance in early detection of AITL.
In summary, we have shown that RHOA mutation is specifically associated with AITL, PTCL-TFH and their related lesions, and investigation of the mutation is highly valuable in early detection of these T-cell lymphomas and their extranodal involvement.
Supplementary Material
Acknowledgments
The authors thank Shubha Anand and Yuanxue Huang for their assistance with using TapeStation, Graeme Clark and Ezequiel Martin for their assistance with Illumina sequencing and Fangtian Wu for carrying out DNA extraction on a control case.
Funding Statement
Funding: The research in MQD's laboratory was supported by grants from Blood Cancer UK (13 006, 15 019), CRUK (C8333/A29707), and the Kay Kendall Leukaemia Fund (KKL582) UK. WY was supported by a research fellowship from the China Scholarship Council, and an International Collaborative Award from the Pathological Society of Great Britain and Ireland, UK. The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research Centre.
References
- 1.Nguyen TB, Sakata-Yanagimoto M, Asabe Y, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017;7(1):e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao WQ, Wu F, Zhang W, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J Pathol. 2020;250(3):346-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iqbal J, Amador C, McKeithan TW, Chan WC. Molecular and genomic landscape of peripheral T-cell lymphoma. Cancer Treat Res. 2019;176:31-68. [DOI] [PubMed] [Google Scholar]
- 4.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakata-Yanagimoto M, Enami T, Yoshida K, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171-175. [DOI] [PubMed] [Google Scholar]
- 6.Nakamoto-Matsubara R, Sakata- Yanagimoto M, Enami T, et al. Detection of the G17V RHOA mutation in angioimmunoblastic T-cell lymphoma and related lymphomas using quantitative allele-specific PCR. PLoS One. 2014;9(10):e109714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palomero T, Couronne L, Khiabanian H, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014; 46(2):166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ondrejka SL, Grzywacz B, Bodo J, et al. Angioimmunoblastic T-cell lymphomas with the RHOA p.Gly17Val mutation have classic clinical and pathologic features. Am J Surg Pathol. 2016;40(3):335-341. [DOI] [PubMed] [Google Scholar]
- 9.Lee PH, Weng SW, Liu TT, et al. RHOA G17V mutation in angioimmunoblastic Tcell lymphoma: a potential biomarker for cytological assessment. Exp Mol Pathol. 2019;110:104294. [DOI] [PubMed] [Google Scholar]
- 10.Zang S, Li J, Yang H, et al. Mutations in 5- methylcytosine oxidase TET2 and RhoA cooperatively disrupt T cell homeostasis. J Clin Invest. 2017;127(8):2998-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes JR, Ambesi-Impiombato A, Couronné L, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33(2):259-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng SY, Brown L, Stevenson K, et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132(9):935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobay MP, Lemonnier F, Missiaglia E, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102(4):e148-e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371-375. [DOI] [PubMed] [Google Scholar]
- 15.Hayashida M, Maekawa F, Chagi Y, et al. Combination of multicolor flow cytometry for circulating lymphoma cells and tests for the RHOA(G17V) and IDH2(R172) hotspot mutations in plasma cell-free DNA as liquid biopsy for the diagnosis of angioimmunoblastic T-cell lymphoma. Leuk Lymphoma. 2020;61(10):2389-2398. [DOI] [PubMed] [Google Scholar]
- 16.Tanzima Nuhat S, Sakata-Yanagimoto M, Komori D, et al. Droplet digital polymerase chain reaction assay and peptide nucleic acid-locked nucleic acid clamp method for RHOA mutation detection in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018;109(5):1682-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis NE, Petrova-Drus K, Huet S, et al. Clonal hematopoiesis in angioimmunoblastic T-cell lymphoma with divergent evolution to myeloid neoplasms. Blood Adv. 2020;4(10):2261-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steciuk MR, Massengill S, Banks PM. In immunocompromised patients, Epstein- Barr virus lymphadenitis can mimic angioimmunoblastic T-cell lymphoma morphologically, immunophenotypically, and genetically: a case report and review of the literature. Hum Pathol. 2012;43(1):127-133. [DOI] [PubMed] [Google Scholar]
- 20.Nicolae A, Pittaluga S, Venkataraman G, et al. Peripheral T-cell lymphomas of follicular T-helper cell derivation with Hodgkin/ Reed-Sternberg cells of B-cell lineage: both EBV-positive and EBV-negative variants exist. Am J Surg Pathol. 2013;37(6):816-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laforga JB, Gasent JM, Vaquero M. Potential misdiagnosis of angioimmunoblastic T-cell lymphoma with Hodgkin's lymphoma: a case report. Acta Cytol. 2010;54(5 Suppl):840-844. [PubMed] [Google Scholar]
- 22.Tan LH, Tan SY Tang T, et al. Angioimmunoblastic T-cell lymphoma with hyperplastic germinal centres (pattern 1) shows superior survival to patterns 2 and 3: a meta-analysis of 56 cases. Histopathology. 2012;60(4):570-585. [DOI] [PubMed] [Google Scholar]
- 23.Sakata-Yanagimoto M, Nakamoto-Matsubara R, Komori D, et al. Detection of the circulating tumor DNAs in angioimmunoblastic T- cell lymphoma. Ann Hematol. 2017;96(9):1471-1475. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen TB, Sakata-Yanagimoto M, Fujisawa M, et al. Dasatinib Is an effective treatment for angioimmunoblastic T-cell lymphoma. Cancer Res. 2020;80(9):1875-1884. [DOI] [PubMed] [Google Scholar]
- 25.Attygalle AD, Diss TC, Munson P, Isaacson PG, Du MQ, Dogan A. CD10 expression in extranodal dissemination of angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2004;28(1):54-61. [DOI] [PubMed] [Google Scholar]
- 26.Vallois D, Dobay MP, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490-1502. [DOI] [PubMed] [Google Scholar]
- 27.Rohr J, Guo S, Huo J, et al. Recurrent activating mutations of CD28 in peripheral Tcell lymphomas. Leukemia. 2016; 30(5):1062-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujisawa M, Sakata-Yanagimoto M, Nishizawa S, et al. Activation of RHOAVAV1 signaling in angioimmunoblastic Tcell lymphoma. Leukemia. 2018;32(3):694-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95-96. [DOI] [PubMed] [Google Scholar]
- 30.Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25-38. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz FH, Cai Q, Fellmann E, et al. TET2 mutations in B cells of patients affected by angioimmunoblastic T-cell lymphoma. J Pathol. 2017;242(2):129-133. [DOI] [PubMed] [Google Scholar]
- 32.Ch'ang HJ, Su IJ, Chen CL, et al. Angioimmunoblastic lymphadenopathy with dysproteinemia--lack of a prognostic value of clear cell morphology. Oncology. 1997;54(3):193-198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.