Chronic viral antigen stimulation may underlie CD8+ T-cell expansion and T-cell large granular lymphocyte leukemia (T-LGLL).1 In T-LGLL, CD8+ T-cell expansions are associated with somatic mutations that solidify clonal dominance, such as in the case of activating STAT3 mutations which are found in 40% of patients.2 However, whether chronic exposure to viral antigens are associated with somatic mutations in expanding CD8+ T cells among individuals without clinically detectable lymphoproliferations is currently not known.
Human T-cell leukemia virus type 2 (HTLV-2) preferentially targets CD8+ T cells, causing strong expansion of the infected CD8+ T-cell clones.3 Whereas HTLV-1 is the causative agent of adult T-cell leukemia/ lymphomas, 4 the etiologic role of HTLV-2 in lymphoproliferative diseases is less clear. In 1992 an incidental case of LGL leukemia with HTLV-2 seropositivity was described.5 Later, Thomas et al.6 reported anti-HTLV antibody positivity in 44% of T-LGLL patients. While some cross-reactivity cannot be excluded, 7.5% (4 of 53) of T-LGLL patients tested positive for HTLV-2 by both western blotting and polymerase chain reaction (PCR),6 suggesting that HTLV-2 may participate in T-LGLL pathogenesis in a minority of cases. In this study, we examined whether CD8+ T cells from healthy, asymptomatic blood donors with chronic HTLV-2 infection harbor somatic mutations in STAT3 or other immune-associated genes, potentially identifying invidiuals at risk of subsequent lymphoproliferative diseases.
This study was conducted with samples from the HTLV Outcomes Study (HOST) which includes samples from subjects recruited from five major US blood donation centers.7 HTLV status was analyzed with an ezymelinked immunosorbant (ELISA) assay, followed by western blot confirmation and HTLV-1 versus HTLV-2 typing by either real-time PCR or a type specific ELISA.7 At each visit, cohort participants were interviewed in detail for symptoms, followed by a physical and neurological examination. Informed consent was obtained from all participants. The study and sample collection were approved by the University of California San Francisco committee on human research and other Institutional Review Boards. We obtained frozen peripheral blood mononuclear cells (PBMC) of 30 HTLV-2 infected and 35 HTLV-2 uninfected blood donors from University of California San Francisco and Vitalant Research Institute (CA, USA). The PBMC samples collected between 2000 and 2008 were randomly selected from HOST.
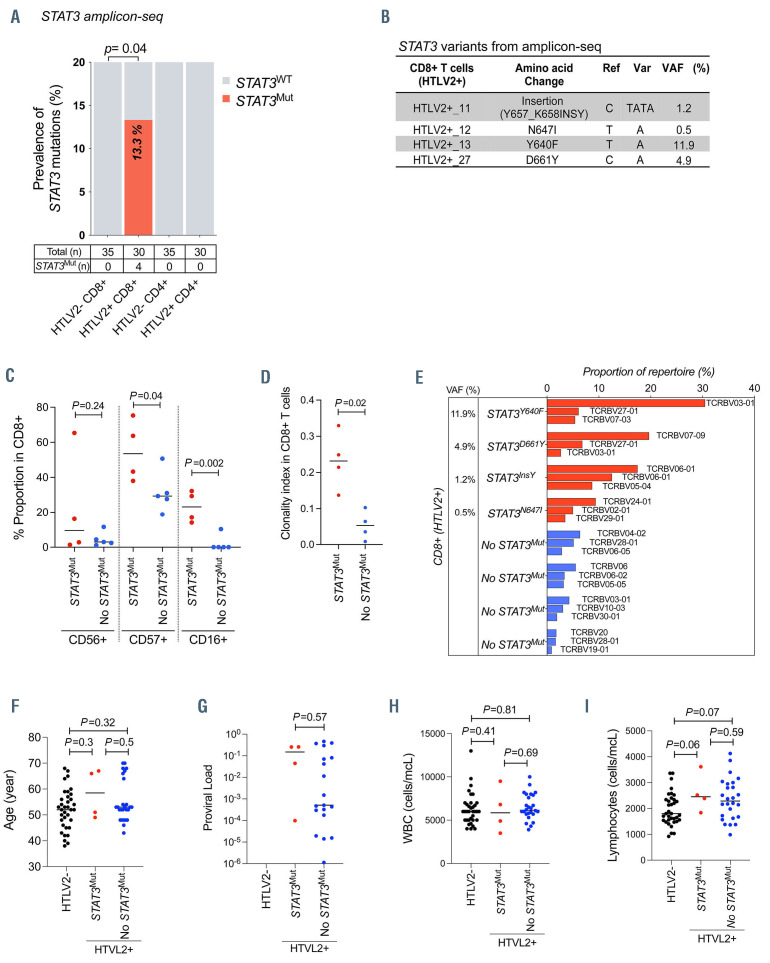
We separated CD4+ and CD8+ T cells from PBMC samples of HTLV-2 positive (n=30) and negative (n=35) healthy blood donors. The presence of STAT3 mutations in the sorted fractions was analyzed by ultra-deep targeted amplicon sequencing, covering hotspot regions of STAT3 gene (median coverage =7,472, sensitivity =0.5% variant allele frequency [VAF]). The sequencing was performed with Illumina Miseq System (Online Supplementary Figure S1A), and variant calling was performed as previously reported.8 Somatic nonsynonymous STAT3 mutations were discovered in CD8+ T cells from four of 30 (13.3%) HTLV-2 positive subjects, whereas no STAT3 mutations were discovered in HTLV- 2 negative subjects using deep amplicon sequencing of STAT3 (Figure 1A; Fisher’s exact test P=0.04). Furthermore, no STAT3 mutations were discovered in CD4+ T cells, indicating that the mutations were specific for the CD8+ T-cell subset. Among the four STAT3 mutations detected, three were missense STAT3 mutations (Y640F, N647I and D661Y) and one was a nonframeshift insertion (Y657_K658insY), with VAF of 11.9%, 0.5%, 4.9%, and 1.2%, respectively (Figure 1B). All the mutations identified in CD8+ T cells were located in the SH2 domain of STAT3 and have been previously reported in T-LGLL (Online Supplementary Figure S1A).2 The proportion of differentiated, putatively cytotoxic CD57+, CD16+ CD8+ T cells were higher in STAT3 mutated compared to STAT3 unmuted HTLV-2 positive individuals (Figure 1C). In addition, higher level of the cytotoxic marker perforin was noted in CD8+ T cells of STAT3 mutated, HTLV-2 positive individuals (Online Supplementary Figure S1B and C). TCRb deep sequencing9 of sorted CD8+ T cells revealed higher clonality index in the STAT3 mutated compared to STAT3 unmuted individuals (Figure 1D). The VAF of the STAT3 Y640F mutation was consistent with clonal event in the largest TCRBV03-01 clone; the other three STAT3 mutations with smaller VAF likely occurred as subclonal events or in the smaller CD8+ T-cell clones (Figure 1E).
The age distribution was similar between HTLV-2 negative subjects (median age 52 year), HTLV-2 positive subjects without STAT3 mutations (median age 53 years), and HTLV-2 positive subjects with STAT3 mutations (median age 58.5 years) (P-value between HTLV-2 positive subjects with and without STAT3 mutations, P=0.50; Mann–Whitney U test) (Figure 1F). No statistical difference in viral load was detected between STAT3 mutated (median =0.149) and unmuted HTLV-2 positive subjects (median =0.0005) (P=0.5; Mann–Whitney U test) (Figure 1G). No serial HTLV-2 viral load measurements were available; however, HTLV-2 viral load has been reported to be stable over time.10 There was no difference in total white blood cell and lymphocyte counts between STAT3 mutated and unmuted cases (Figure 1H and I).
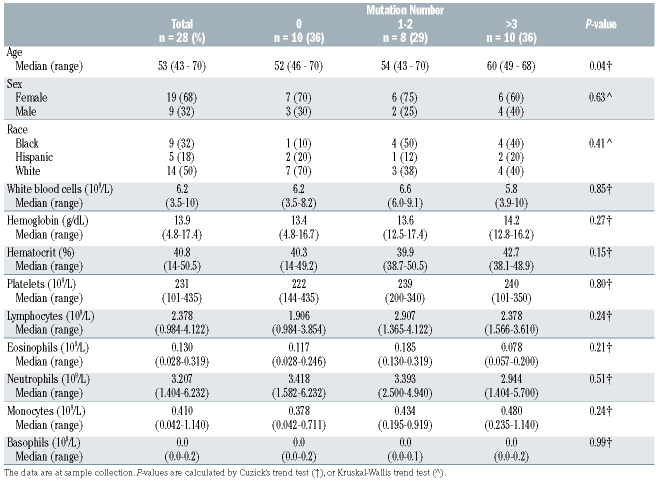
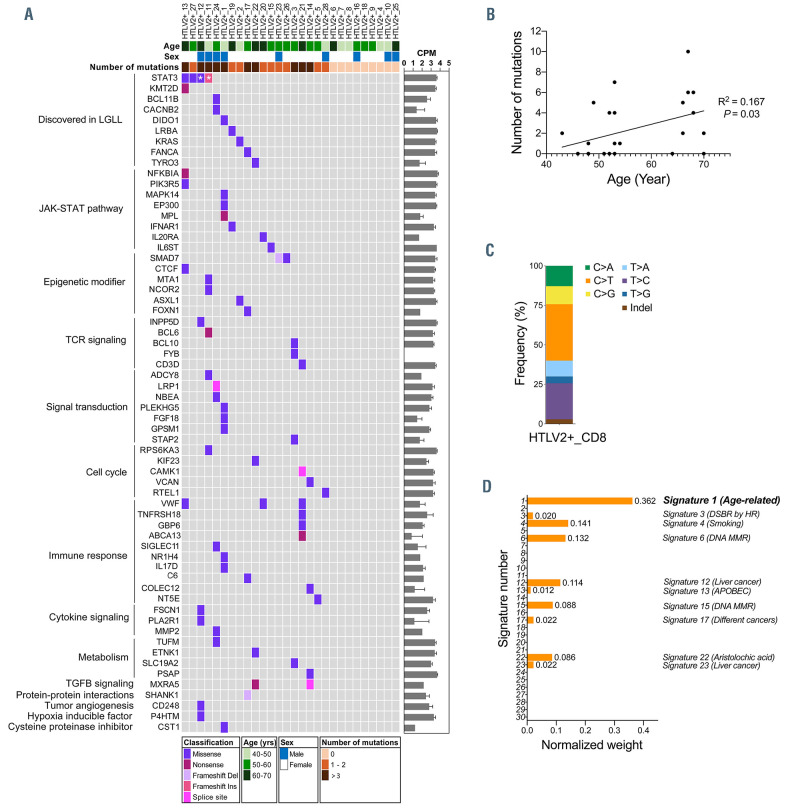
In order to characterize a larger spectrum of somatic variants in genes linked to immune regulation, we analyzed CD8+ T cells from 28 HTLV-2 positive subjects using a custom next generation sequencing panel covering the coding regions of 2,533 immune-related genes.11 Samples were sequenced with Illumina HiSeq or NovaSeq 6000 system (Online Supplementary Figure S2), and somatic variant calling followed a previously described approach.11 Variants were filtered using population based filtering, MuTect2 filters and against CD4+ and CD8+ panels of normals from 21 healthy controls (Online Supplementary Figure S2). Variants only found in CD8+ T cells, using sorted CD4+ T cells as matched normals, were evaluated further. Sequencing coverages are presented in the Online Supplementary Figure S2. Two STAT3 mutations (Y640F and D661Y) were detected in two of four STAT3 mutant cases. N647I and Y657_K658insY variants did not pass filtering due to lower sequencing coverage compared to amplicon sequencing; their presence was confirmed with visual inspection of the sequencing data using Integrative Genomics Viewer.12 In addition to STAT3 mutations, we identified a total of 66 coding somatic variants in 61 genes in CD8+ T cells (Figure 2A). Nineteen (68%) of the subjects had at least one variant, and the median number of variants was one per subject.
Eight subjects (29%) harbored variants in the genes previously discovered in LGLL (STAT3, KMT2D, TYRO3, DIDO1, BCL11B, CACNB2, KRAS, LRBA and FANCA),13 and five subjects (18%) harbored genes involved in JAKSTAT signaling pathway (NFKBIA, PIK3R5, MAPK14, EP300, MPL, IFNAR1, IL6ST and IL20RA) according to Uniprot identifier. Three genes had more than one variant: VWF (3 mutations), SMAD7 and MXRA5 (2 mutations each) (median VAF: 7%, 2% and 4%, respectively). Subject 13 who harbored a STAT3 Y640F mutation also had mutations in KMT2D, NFKBIA, PIK3R5, CTCF, and VWF. In this subject with multiple somatic mutations, STAT3 had the highest VAF (16.2%, Online Supplementary Table S1), suggesting that other variants may be subclonal. Subject 12 with a STAT3 N647I mutation also harbored variants in INPP5D, FSCN1 and PLA2R1, CD248, and P4HTM. Subject 11 with STAT3 Y657_K658insY harbored variants in MTA1, NCOR2, BCL6, ADCY8, RPS6KA3. Subject 27 with a STAT3 D661Y mutation had no additional variants. The overall number of mutations was higher in HTLV-2 positive blood donors harboring STAT3 mutations (median =6) compared to HTLV-2 positive blood donors without STAT3 mutations (median =1; P=0.061; Mann–Whitney U test), although the difference was not statistically significant. The complete list of variants and VAF can be found in the Online Supplementary Table S1.
Figure 1.
STAT3 mutations discovered in CD8+ T cells of HTLV-2 positive subjects. (A) Prevalence of STAT3 mutations in CD4+ and CD8+ T cells of human Tcell leukemia virus type 2 (HTLV-2) positive subjects (n=30) and HTLV-2 negative subjects (n=35). Four (13.3%; Fisher’s exact test P=0.04) of 30 HTLV-2 positive individuals had STAT3 mutations in CD8+ T cells. (B) STAT3 mutations found in HTLV-2 positive subjects by amplicon sequencing. STAT3 mutations (insertion, N647I, Y640F and D661Y) were discovered in CD8+ T cells from 4 HTLV-2 positive subjects. (C) Flow cytometry based immunophenotyping was performed to identify the proportion of differentiated, putatively cytotoxic (CD56+, CD57+, and CD16+) CD8+ T cells in 9 HTLV-2 positive blood donors. For the immunophenotyping, anti-CD3 APC, -CD45 V500, -CD4 APC-H7, -CD8 PE-Cy7, -CD16 PerCP-Cy5.5, -CD56 FITC, and -CD57 PE, were used. Each dot represents one individual, and horizontal lines indicate median values. Statistically significant difference was evaluated using Mann-Whitney U test. (D) The CD8+ T-cell clonality index by STAT3 mutation status in 8 HTLV-2 positive blood donors. The clonality index was calculated using ImmnoSEQ Analyzer software (Adaptive Biotechnologies, WA, USA) as 1 minus Shannon entropy normalized by the logarithm of the number of productive T-cell receptor (TCR) sequences. Each dot represents 1 individual, and horizontal lines indicate median values. P-values were evaluated using Mann-Whitney U test. (E) CD8+ T-cell repertoire analyzed with TCRb deep sequencing (Adaptive Biotechnologies). Sorted CD8+ T cells of HTLV-2 positive cases bearing STAT3 mutations (n=4) and without STAT3 mutations (n=4) were used. Variant allele frequency (VAF) was analyzed by amplicon sequencing. The graph shows top 3 TCR clones in each sample. (F-I) (F) Age distribution, (G) HTLV-2 proviral load in copies per peripheral blood mononuclear cells (PBMC), (H) white blood cells count and (I) lymphocytes count within HTLV-2 negative subjects (HTLV2-), HTLV-2 positive subjects without STAT3 mutations (No STAT3Mut) and HTLV-2 positive subjects harboring STAT3 mutations (STAT3Mut). Each dot represents one individual. P-values were calculated using Mann–Whitney U test (No STAT3Mut vs. STAT3Mut). Horizontal lines indicate median values. Ref: reference base; Var: variant base.
Table 1.
Clinical characteristics of HTLV-2 positive subjects analyzed in immunogene panel sequencing.
Somatic mutations can accumulate in tissues with aging.14 Accordingly, the total number of coding variants was associated with older age among HTLV-2 positive blood donors (no variants, median age 51 years; 1-2 variants, 53 years; 3 or more variants, 59 years; P=0.04) (Table 1; Figure 2B). The most frequent single nucleotide transition was C>T involved in age-associated mutational signature 1 (Figure 2C). The high prevalence of signature 1, revealed by mutational signature analysis (Figure 2D), further supported age-dependent accumulation of somatic variants in CD8+ T cells among HTLV-2 positive subjects. No association between peripheral blood counts and number of variants was observed (Table 1).
In summary, our results highlight the presence of STAT3 mutations in CD8+ T cells of healthy blood donors harboring HTLV-2 without clinical history of lymphoproliferative disease. HTLV-2 positive subjects with STAT3 mutations showed variable clonal expansion of CD8+ T cells, suggesting that HTLV-2 infection may promote lymphoproliferation and STAT3 mutagenesis outside the clinical context of T-LGLL. We identified additional mutations in CD8+ T cells of HTLV-2 positive subjects in genes involved in JAKSTAT signaling, immune regulation and lymphoproliferation. In addition to T-LGLL, somatic STAT3 mutations have been detected in LGLL associated diseases such as aplastic anemia, hypoplastic myelodysplastic syndrome and Felty’s syndrome, and in some patients with multiple sclerosis and rheumatoid arthritis.15 Although CD8+ T cell expansions can also be detected in other diseases such as in rheumatoid arthritis, somatic STAT3 mutations are not common in these conditions.8 STAT3 mutations and a history of HTLV-2 infection may highlight a subset of blood donors who are at risk of subsequent diagnosis of lymphoproliferative diseases. However, no clinical follow-up information is available from our study participants, and future studies are needed to elucidate whether HTLV-2 positive subjects carrying STAT3 and other mutations are at increased risk of T-LGLL or other lymphoproliferative diseases.
Figure 2.
Somatic mutations identified in CD8+ T cells of HTLV-2 positive subjects. (A) Mutation landscape in CD8+ T cells of human T-cell leukemia virus type 2 (HTLV-2) positive subjects (n=28). Coding variants identified by immunogene panel sequencing are presented, together with age, sex, number of mutations and mutation types. * (white asterisk), STAT3 variants detected in the deep amplicon sequencing but filtered out in the immunogene panel sequencing due to lower coverage but confirmed with visual inspection with the Integrative Genomics Viewer (Broad Institute, USA). The average expression of mutated genes in healthy controls are shown on the right, presented as counts per millions reads mapped (CPM) with mean ± standard deviation (n =5). The complete list of variants and variant allele frequencies (VAF) can be found in the Online Supplementary Table S1. (B) Correlation plot of age vs. mutation number in HTLV-2 positive blood donors. (C) Percentages of somatic base substitutions and indel identified by immunogene panel sequencing in CD8+ T cells of HTLV-2 positive blood donors. (D) Normalized weights of COSMIC signatures contributions. Signature 1 (weight: 0.362) was highly related signature.
Supplementary Material
Acknowledgment
Amplicon sequencing and Immunogene panel sequencing were performed at the Institute for Molecular Medicine Finland (FIMM) Technology Center, which is supported by Biocenter Finland. The authors also acknowledge IT Center for Science Ltd for computational resources.
Funding Statement
Funding: the research was funded by European Research Council (M-IMM project), ERAPerMed consortium JAKSTAT-TARGET, Academy of Finland, Finnish special governmental subsidy for health sciences, research and training, Sigrid Juselius Foundation, Helsinki Institute of Life Sciences Fellow funding, and Cancer Foundation Finland. The HOST study was supported by the US National Heart, Lung and Blood Institute research grant 2R01-HL- 62235 and Signe and Ane Gyllenberg Fundation.
References
- 1.Lamy T, Moignet A, Loughran TP. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129(9):1082-1094. [DOI] [PubMed] [Google Scholar]
- 2.Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melamed A, Witkover AD, Laydon DJ, et al. Clonality of HTLV-2 in natural infection. PLoS Pathog. 2014;10(3):e1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304-1315. [DOI] [PubMed] [Google Scholar]
- 5.Loughran TP, Coyle T, Sherman MP, et al. Detection of human T-cell leukemia/lymphoma virus, type II, in a patient with large granular lymphocyte leukemia. Blood. 1992;80(5):1116-1119. [PubMed] [Google Scholar]
- 6.Thomas A, Perzova R, Abbott L, et al. LGL leukemia and HTLV. AIDS Res Hum Retroviruses. 2010;26(1):33-40. [DOI] [PubMed] [Google Scholar]
- 7.Lee TH, Chafets DM, Busch MP, Murphy EL. Quantitation of HTLV-I and II proviral load using real-time quantitative PCR with SYBR Green chemistry. J Clin Virol. 2004;31(4):275-282. [DOI] [PubMed] [Google Scholar]
- 8.Savola P, Kelkka T, Rajala HL, et al. Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat Commun. 2017;8:15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D, Park G, Huuhtanen J, et al. Somatic mTOR mutation in clonally expanded T lymphocytes associated with chronic graft versus host disease. Nat Commun. 2020;11(1):2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwaan N, Lee TH, Chafets DM, et al. Long-term variations in human T lymphotropic virus (HTLV)-I and HTLV-II proviral loads and association with clinical data. J Infect Dis. 2006;194(11):1557-1564. [DOI] [PubMed] [Google Scholar]
- 11.Savola P, Martelius T, Kankainen M, et al. Somatic mutations and T-cell clonality in patients with immunodeficiency. Haematologica. 2020;105(12):2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppe A, Andersson EI, Binatti A, et al. Genomic landscape characterization of large granular lymphocyte leukemia with a systems genetics approach. Leukemia. 2017;31(5):1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blokzijl F, de Ligt J, Jager M, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538(7624):260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mustjoki S, Young NS. Somatic mutations in "benign" disease. N Engl J Med. 2021;384(21):2039-2052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.