Abstract
Diamond-Blackfan anemia (DBA) is an inherited bone marrow failure disorder in which pure red blood cell aplasia is associated with physical malformations and a predisposition to cancer. Twentyfive percent of patients with DBA have mutations in a gene encoding ribosomal protein S19 (RPS19). Our previous proof-of-concept studies demonstrated that DBA phenotype could be successfully treated using lentiviral vectors in Rps19-deficient DBA mice. In our present study, we developed a clinically applicable single gene, self-inactivating lentiviral vector, containing the human RPS19 cDNA driven by the human elongation factor 1αshort promoter, which can be used for clinical gene therapy development for RPS19-deficient DBA. We examined the efficacy and safety of the vector in a Rps19-deficient DBA mouse model and in human primary RPS19- deficient CD34+ cord blood cells. We observed that transduced Rps19-deficient bone marrow cells could reconstitute mice long-term and rescue the bone marrow failure and severe anemia observed in Rps19-deficient mice, with a low risk of mutagenesis and a highly polyclonal insertion site pattern. More importantly, the vector can also rescue impaired erythroid differentiation in human primary RPS19-deficient CD34+ cord blood hematopoietic stem cells. Collectively, our results demonstrate the efficacy and safety of using a clinically applicable lentiviral vector for the successful treatment of Rps19-deficient DBA in a mouse model and in human primary CD34+ cord blood cells. These findings show that this vector can be used to develop clinical gene therapy for RPS19-deficient DBA patients.
Introduction
Diamond-Blackfan anemia (DBA) is a congenital bone marrow (BM) failure disorder with erythroid hypoplasia that presents early in infancy.1-3 The classic hematologic profile of DBA consists of macrocytic anemia with reticulocytopenia, normal or decreased levels of neutrophils, and a variable platelet count.3,4 Additionally, patients with DBA can also manifest with non-hematologic symptoms such as physical abnormalities and a predisposition to cancer.2,5
The majority of DBA cases (60-70%) are caused by heterozygous loss-of-function mutations in genes coding for ribosomal proteins (RP), resulting in functional RP haploinsufficiency.2 Recent studies have also identified mutations in erythroid transcriptional factors GATA1 and TSR2 (a direct binding partner of RPS26) as a cause of the DBA phenotype.6-8 Mutations in RPS19 are the most common alterations among patients with putative causal mutations, contributing to over 25% of cases.2 The main therapeutic option for DBA patients is corticosteroids, with more than 80% of the subjects responding well during early stages of treatment. However, half of these patients become non-responsive to corticosteroid therapy over prolonged treatment and have to be given blood transfusions.9 Importantly none of the currently available treatments is curative, and the treatments are often accompanied by serious complications. 1,2,10 Hematopoietic stem cell transplantation is currently the sole curative option for the treatment of DBA. This treatment is, however, limited by the availability of suitable donors and the potential for serious immunological complications.11 Gene therapy, using gene-corrected hematopoietic stem cells, would be a potential alternative therapeutic strategy, as highlighted in our previous studies.12-16 For clinical applications using this approach, the efficiency of transgene expression and safety aspects, including potential insertional mutagenesis, should be addressed.17-19 Our group recently demonstrated that correction of Rps19-deficient BM cells using lentiviral vectors containing a clinically relevant promoter could rescue BM failure and defects in erythroid development, while exhibiting limited risk of insertional mutagenesis. 12
In our previous study, we utilized a lentiviral vector containing the RPS19 gene as well as a GFP marker. We have subsequently designed a clinically applicable single gene lentiviral self-inactivating (SIN) vector for the clinical development of gene therapy for Rps19-deficient DBA patients. This vector harbors a codon-optimized human RPS19 cDNA driven by the short human elongation factor 1a promoter and lacks a fluorescent marker. In this study, we demonstrate that this vector can rescue the anemia and lethal BM failure observed in mouse models of Rps19-deficient DBA, with a low-risk insertion profile and no evidence of clonal expansion associated with vector integration near cancer-associated genes. We also observed the rescue of impaired erythroid differentiation in human RPS19-deficient CD34+ cord blood cells treated with this vector. Our results demonstrate the feasibility and preclinical efficacy for treatment of RPS19-deficient DBA using a clinically applicable SIN lentiviral vector, which opens the possibility for the development of clinical gene therapy for RPS19-deficient DBA patients.
Methods
Lentiviral vector constructs
The SIN lentiviral vector is derived from the pRRL.PPT.PGK. vector backbone.20 A codon-optimized human RPS19 cDNA was designed as described previously21 and inserted downstream of the EFS promoter. Lentiviral vectors were produced by the Vector Unit at Lund University as previously described.12
Mice and transplantations
Mice were maintained at the Lund University animal facility and all animal experiments were approved by the Lund University animal ethics committee. The homozygous doxycycline- inducible Rps19-deficient mouse model used in the study was established as previously described.15 A detailed description of transplantations is provided in the Online Supplementary Methods.
Transduction
c-kit+ or lineage negative (Lin–) cells isolated from BM of transgenic mice were enriched by using CD117 or Lin– microbeads and magnetic-activated cell sorting separation columns (all from Miltenyi Biotec) according to the manufacturer’s protocol. After enrichment, cells were pre-stimulated for 24 h in StemSpan serum-free expansion medium (Stem Cell Technologies), supplemented with penicillin/streptomycin (Gibco), murine stem cell factor (100 ng/mL; PeproTech), and human thrombopoietin (50 ng/mL; PeproTech) in six-well plates at the concentration of 0.5x106 cells/mL. For transduction, retronectin-coated (20 ng/mL; Takara) 12-well plates were preloaded with the viral vectors (multiplicity of infection [MOI]=5-10), followed by seeding of 0.5x106 cells into each well filled with 1 mL pre-stimulation medium.
Flow cytometry
A complete description of all antibodies used is listed in the Online Supplementary Methods.
Human primary cord blood cells and erythroid differentiation
Human cord blood samples were obtained from the maternity wards of Helsingborg General Hospital and Skåne University Hospital in Lund and Malmö, Sweden, after informed, written consent according to guidelines approved by the regional ethical committee. Mononuclear cells were separated through densitygradient centrifugation. CD34+ cells were magnetically isolated according to the manufacturer’s description (Milteny Biotec, cat. n. 130-046-702). Cells were cultured in serum-free expansion medium (Stem Cell Technologies), supplemented with human stem cell factor, thrombopoietin, and FLT3-ligand at 100 ng/mL from Peprotech. Full descriptions of transduction and erythroid differentiation are provided in the Online Supplementary Methods.
Other experimental details
Full descriptions of the quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), determination of transduction efficiency and vector copy number measurements are provided in the Online Supplementary Methods.
Insertion site analysis
Whole BM cells were isolated at 16 weeks after transplantation. Genomic DNA was isolated from the BM of flushed femora using the DNA Blood & Tissue kit (Qiagen). The vectorgenome junction was amplified using the INtegration Site PIpeline for paIRed-End reaDs (INSPIIRED) workflow as described by Sherman and colleagues.24
Statistical analysis
t-tests and one-way analysis of variance with the Tukey multiple- comparison test were used to determine statistical significance. Computations were performed using GraphPad Prism (version 6; GraphPad Software).
Results
High transduction efficiency of the EFS-RPS19 vector
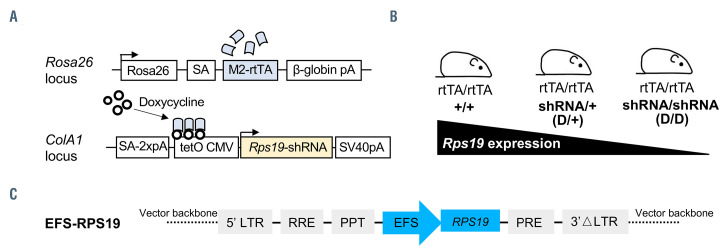
We first studied the transduction efficiency of the vector in BM progenitor cells isolated from our established Rps19-deficient DBA mouse model.15 This model contains the Rps19-targeting shRNA expressed under a doxycycline- responsive promoter located downstream of the collagen A1 gene (Figure 1A). Experimental animals were bred to be either heterozygous (D/+) or homozygous (D/D) for the shRNA to generate two models with intermediate or severe Rps19 deficiency (Figure 1B). Rps19 mRNA expression was reduced by approximately 50%, and a trend toward more efficient knockdown in D/D mice compared to D/+ mice was seen, as shown in our previous studies.15 Upon induction with doxycycline, transplanted recipients receiving D/D BM cells developed an acute and lethal BM failure, while recipients of D/+ BM cells developed a mild chronic anemia.15 As D/D mice develop lethal BM failure shortly after doxycycline induction, the more severely affected D/D mice were used in the present study to investigate whether the lethal phenotype could be fully rescued with the vector.
The clinically applicable single gene lentiviral vector was developed using a SIN lentiviral vector design harboring the codon-optimized human RPS19 cDNA driven by an internal EFS promoter (named EFS-RPS19) (Figure 1C). Compared to the human codon-optimized RPS19 cDNA, there are six mismatches in the shRNA construct for generating the mouse model. Because of this, gene expression derived from the human codon-optimized RPS19 cDNA is not affected by the shRNA.
We first examined the transduction efficiency of the vector. In our previous study, we transduced cells at a MOI of 10-20 and demonstrated the rescue of the anemia and BM failure with a reduced risk of insertional mutagenesis. 12 To investigate the therapeutic effects of a lower average vector copy number per cell, we decided to use an MOI of 5-10. As shown in Online Supplementary Figure S1, the transduction efficiency was 75% on average in ckit+ cells isolated from D/D mice. We next examined both endogenous Rps19 and vector-derived RPS19 mRNA expression in c--kit+ cells isolated from D/D and +/+ mice at a MOI of 5. As shown in Online Supplementary Figure S2, the endogenous Rps19 mRNA expression levels were significantly decreased in the cells isolated from D/D mice compared to the levels in cells from +/+ mice after doxycycline administration. Cells transduced with EFSRPS19 exhibited a 2.5-fold higher level of expression of human RPS19 mRNA compared to the endogenous Rps19 expression in the D/D group. Interestingly, transduced cells isolated from +/+ mice showed a significantly lower level of human RPS19 mRNA expression than the transduced cells isolated from D/D mice at the same MOI, indicating the internal physiological regulation of excess RPS19 production as reported by others.29,30 The overall results indicated that cells transduced with the EFS-RPS19 vector could successfully express the human RPS19 transgene
Gene-corrected bone marrow cells can rescue the Diamond-Blackfan anemia phenotype in vivo
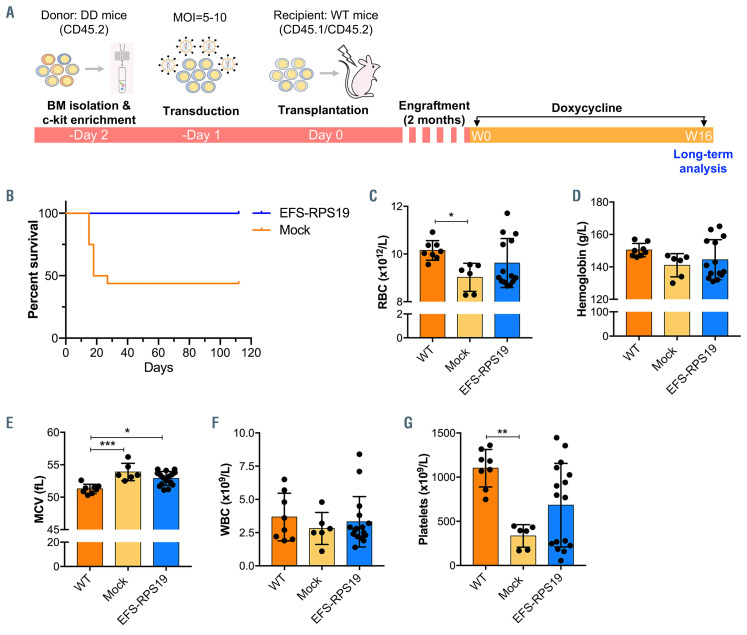
We next assessed the function of gene-corrected BM cells using the EFS-RSP19 vector in vivo. As shown in Figure 2A, uninduced (no doxycycline) c-kit+ BM cells from D/D mice (CD45.2) were transduced with the EFSRPS19 vector (MOI=5-10), and then transplanted into lethally irradiated wild-type B6SJL recipient mice (CD45.1/CD45.2, named the EFS-RPS19 group). Mice receiving uninduced c-kit+ BM cells without vector transduction were regarded as the mock group (negative control). Following engraftment and stable donor-derived reconstitution of the hematopoietic system, doxycycline was administrated to all recipients to induce the DBA phenotype. To determine whether the vector-treated cells could achieve a full correction, age-matched B6SJL wildtype (WT) mice receiving no irradiation and no transplantation but the same doxycycline administration were used as the control group.
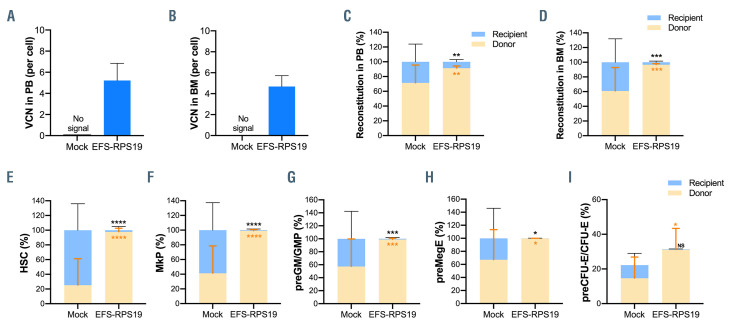
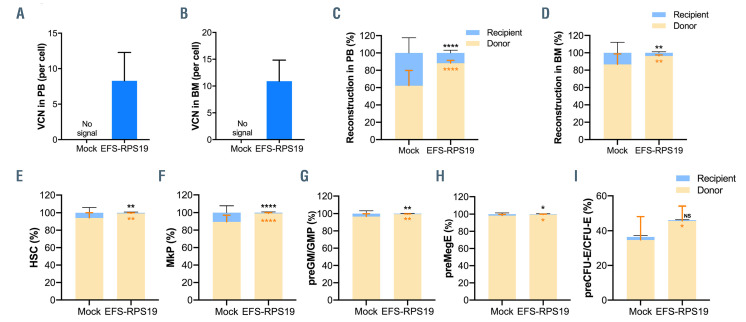
Before doxycycline administration, both the mock and EFS-RPS19 groups showed high overall donor reconstitution, indicating minimal to absent recipient-derived hematopoiesis (Online Supplementary Figure S3). After induction with doxycycline for 2 weeks, recipients in the mock group showed a dramatic decrease in red blood cell counts, mean corpuscular volume (MCV), and white blood cell and platelets counts, indicating that the mice developed BM failure shortly after doxycycline administration (Figure 2B-F). In contrast, recipients in the EFSRPS19 vector-treated group showed normal blood cellularity compared to the WT group. To assess the long-term therapeutic effects, recipients were administered doxycycline for 16 weeks (Figure 3A). As shown in Figure 3B, most of the recipients in the mock group died (9 out of 16) due to severe anemia or BM failure (data not shown) at 2-3 weeks after doxycycline administration. The few remaining recipients exhibited a macrocytic anemia phenotype with significantly reduced red blood cell counts and increased MCV at 16 weeks. The hemoglobin levels and platelet counts were also decreased compared to those in the WT group (Figure 3C-G). As expected, there was a significantly decreased expression of endogenous Rps19 in donor-derived BM cells from both the mock and EFS-RPS19 groups after 16 weeks of doxycycline administration (Online Supplementary Figure S4). Strikingly, all the mice in the EFS-RPS19 group survived without any signs of anemia and with normal BM cellularity compared to the WT group. These results indicate that the lethal BM failure can be prevented by the vector. The vector copy number at 16 weeks after transplantation was 5.2±1.6 and 4.7±1.0 on average in gene-corrected cells isolated from peripheral blood and BM, respectively (Figure 4A, B). We also analyzed the fraction of myeloiderythroid compartments by flow cytometry (Online Supplementary Figure S5). Donor-derived hematopoiesis was observed in the EFS-RPS19 group, and the mean percentage of donor cells (CD45.2) in every progenitor population was significantly higher in the EFS-RPS19 group than in the mock group (Figure 4C-I). Unlike in the EFSRPS19 group, the transplanted cells in the mock group had limited reconstituting ability. In addition, we observed a significantly higher reconstitution of resident recipient cells (CD45.1/CD45.2) in the few surviving mice in the mock group than in the EFS-RPS19 group, perhaps explaining why these animals did not develop severe BM failure.
Figure 1.
The inducible Rps19-deficient mouse model and structure of the EFS-RPS19 self-inactivating lentiviral vector. (A) Overview of modified loci. Black arrowheads indicate the transcriptional start sites. (B) Breeding strategy to adjust the level of Rps19 downregulation. Homozygous mice (D/D mice) are used in the project. (C) The self-inactivating lentiviral vector harboring a codon-optimized human RPS19 cDNA driven by human elongation factor 1a short (EFS) promoter. LTR: long terminal repeat; pA: polyadenylation signal; PPT: polypurine tract; RRE: Rev response element; SA: splice acceptor.
EFS-RPS19 vector-treated Rps19-deficient bone marrow cells provide long-term reconstitution
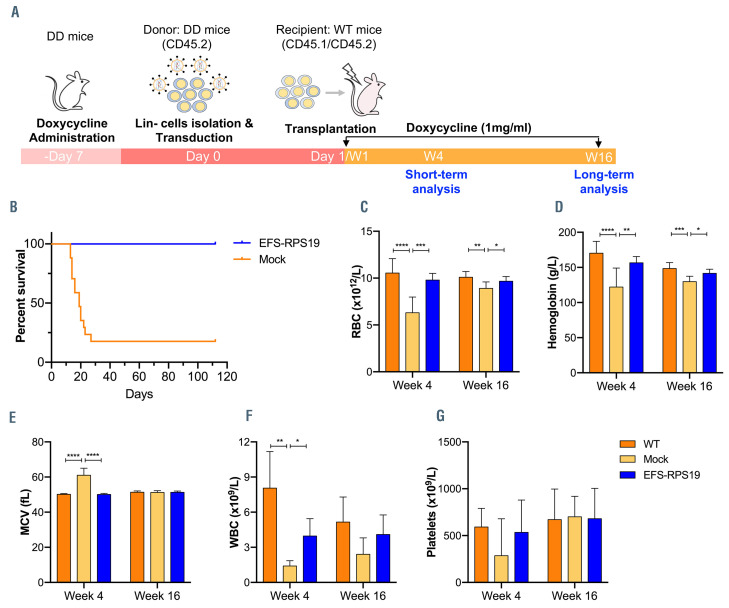
We next investigated whether the EFS-RPS19 vectortreated Rps19-deficient BM cells could generate long-term engraftment and reconstitution in doxycycline-induced lethally irradiated WT recipients (Figure 5A). To this end, D/D mice were induced with doxycycline for 1 week and red blood cell counts and hemoglobin levels were measured to confirm the DBA phenotype (Online Supplementary Figure S6). The isolated Lin– BM cells from doxycyclineinduced mice were transduced with the vector and transplanted into the doxycycline-induced lethally irradiated WT recipients (EFS-RPS19 group). Recipients receiving untransduced Rps19-deficient Lin– BM cells from doxycycline-induced D/D mice were regarded as the mock group. The setting of the WT group was the same as described above. Doxycycline was administrated to all the recipients directly after transplantation. After induction with doxycycline for 2-3 weeks, the majority of mice in the mock group died (13 out of 16 animals) due to severe anemia or BM failure (Figure 5B, Online Supplementary Figure S7). The few surviving mice exhibited significantly decreased levels of endogenous Rps19 expression in donor-derived BM cells at 16 weeks after doxycycline administration (Online Supplementary Figure S8) with the concomitant development of a severe anemia phenotype in the mock group (Figure 5C-G). In contrast, all recipients in the EFS-RPS19 group survived with normal blood cellularity compared to the WT group. The vector copy number was on average 8.3±4.0 and 10.9±3.9 in gene-corrected Rps19-deficient cells isolated from peripheral blood and BM, respectively (Figure 6A, B). By analyzing the fraction of myeloid-erythroid compartments at 16 weeks, we observed almost complete donor-derived hematopoiesis in the EFS-RPS19 group, which was significantly higher than in the mock group (Figure 6C-I). Taken together, our results demonstrate that the EFS-RPS19 vector-treated group obtained full correction of anemia and BM failure.
Figure 2.
Effective correction of anemia by the EFS-RPS19 vector at 2 weeks after induction of the Diamond-BLackfan phenotype. (A) The scheme of the uninduced genecorrected cell transplantation model and plan for examining short-term therapeutic effects. (B-F) Blood cellularity at 2 weeks after doxycycline induction (n=13-16, error bars represent the standard deviation, *P<0.05, **P<0.01, ***P<0.005 ****P<0.001 by one-way analysis of variance). BM: bone marrow; MOI: multiplicity of infection; WT: wild-type; RBC: red blood cells; MCV: mean corpuscular volume; WBC: white blood cells.
Gene-corrected bone marrow cells showed polyclonal hematopoiesis and had a typical lentiviral insertion profile
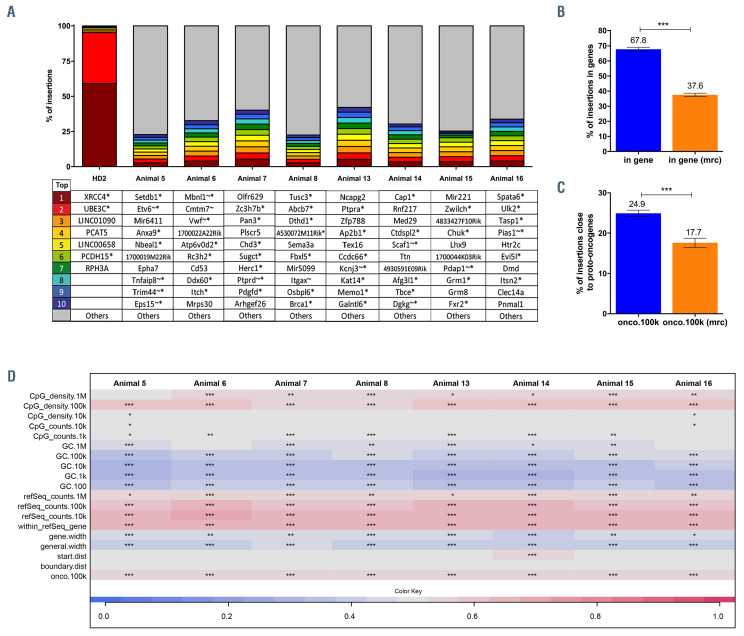
The risk of insertional mutagenesis is a major concern for future applications of gene therapy in the clinic. To assess the safety of the EFS-RPS19 vector integration profile, as well as the clonal dynamics of the transduced cells, insertion site analysis was performed using the INSPIIRED workflow.24 BM cells from uninduced donors (Figure 3A) of cohort 1 (animals 5-8) and cohort 2 (animals 13-16), or those from doxycycline-induced donors (Figure 5A) of cohorts 3 (animals 7-11) and cohort 4 (animals 17-20) were isolated 16 weeks after disease induction in the recipients. Detailed information on pool size estimation and sequence diversity in each sample is shown in Online Supplementary Tables S2 and S3. Particularly, the top ten integrations with the highest sequence contribution in each sample are depicted in Figure 7A and Online Supplementary Figure S9A. Overall, EFS-RPS19-transduced cells showed a highly polyclonal insertion site pattern, reflecting the overall integration preferences of lentiviral vectors. No integration site contributed with more than 5.37% in uninduced gene-corrected cells (Figure 7A) or 4.4% in gene-corrected Rps19- deficient cells (Online Supplementary Figure S9A) to the overall sequence pool, and there were no integrations in or close to known high-risk proto-oncogenes (Lmo2, Ccnd2 or Hmga2). However, one integration at 16.5 kb upstream of the high-risk locus Ikzf1 was detected (accounting for 1.65%), and another integration at 20.7 kb upstream of the high-risk gene Mecom was found (accounting for 0.04%) in recipients of gene-corrected Rps19-deficient BM cells. For the analysis of overlaps between integrations in or near the same genes among BM samples, we observed common lentiviral integration sites identified in previous integration site analysis (Online Supplementary Tables S4 and S5).31,32 We identified 11 integrations in or near the same refSeq genes between cohorts 1+2 and cohorts 3+4 (Online Supplementary Figure S10). Four of these shared common insertion sites in or near Hgf, Kdm6a, Lnpep and Mef2c also listed as protooncogenes in the All Onco database.25 The sole occurrence of integration sites in or near high-risk loci was not an indication of a higher risk of insertional mutagenesis if no dominant clones were detected. The detected integrations might simply reflect that lentiviral vectors were capable of integrating at these genomic sites. We performed analysis of insertion site profile including parameters of the integration site preferences close to CpG islands, GC-rich regions, in or near transcription units, the transcriptional start site of genes, gene boundaries or proto-oncogenes (Figure 7B-D, Online Supplementary Figure S9B-D). More integrations were detected in a distance of 100 kb relative to CpG islands, marking actively transcribed regions but not in the direct vicinity of CpG islands (1-10 kb), hence not close to the promoter region. Our data showed that GC rich regions (marking promoter regions of genes) and long intergenic regions were generally disfavored by the vector. The integrations inside transcriptional units and in or close to proto-oncogenes (within a 100 kb window = onco.100k) are displayed relative to the matched random controls (Figure 7B, Online Supplementary Figure S9B).
Figure 3.
Effective long-term correction of the anemia and bone marrow failure in mice treated with the EFS-RPS19 vector. (A) The scheme of the induced genecorrected cell transplantation model and the plan for examining long-term therapeutic effects. (B) Surviral rate analysis. (C-G) Blood cellularity at 16 weeks after doxycycline induction (n=13-16, error bars represent the standard deviation, *P<0.05, **P<0.01, ***P<0.005 by one-way analysis of variance). BM: bone marrow; MOI: multiplicity of infection; WT: wild-type; RBC: red blood cells; MCV: mean corpuscular volume; WBC: white blood cells.
The EFS-RPS19 vector rescued impaired erythroid differentiation of human RPS19-deficient CD34+ cord blood cells
We next examined the therapeutic effects of the EFSRPS19 vector using human primary CD34+ cord blood cells. Since primary CD34+ cells from DBA patients are difficult to obtain, we utilized previously validated lentiviral shRNA vectors that silence RPS19 expression in human CD34+ cells to induce a DBA phenotype.23
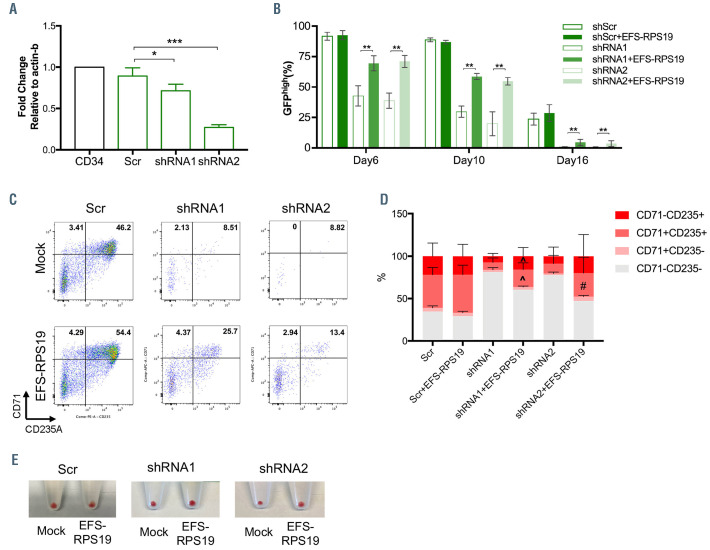
Two lentiviral vectors expressing shRNA (shRNA1 and shRNA2) targeting different regions of the human RPS19 mRNA sequence were used to induce the DBA phenotype, and a vector expressing a scrambled shRNA sequence (Scr) was used as a healthy control. To ensure that the human construct in the vector would not be degraded by the targeting shRNA, alignments were performed and there were two mismatches (out of 19 nucleotides) for shRNA1 and four mismatches (out of 19 nucleotides) for shRNA2. Thus, it is very unlikely that the gene expression derived from the human codon-optimized RPS19 cDNA would be affected by either shRNA. Since the shRNA vectors also contain a GFP marker gene, transduced GFP+ cells were sorted for further examination. As shown in Figure 8A, both shRNA1 and shRNA2 significantly decreased RPS19 mRNA expression, with slightly more efficient knockdown being obtained with shRNA1 than shRNA2, which resembles our previous observations in D/+ and D/D mouse models.15 In order to examine the function of EFS-RPS19, we transduced sorted GFP+ cells and cultured them in erythroid differentiation medium for 48 h after transduction. The PCR results indicated successful integration of the vector into human cells, as shown in Online Supplementary Figure S11. It is a well-known phenomenon that lentiviral vector-mediated transgene expression is silenced in a fraction of CD34+ cells, and that this fraction increases during differentiation of cells, likely due to changes in chromatin accessibility. As shown in Figure 8B, transgene silencing was evident in a fraction of the Scr-transduced CD34+ cells in which the fraction of GFPhigh shRNA expressing cells decreased from 100% in the sorted cells to around 90% on day 6 and day 10, with a further reduction to around 25% at day 16. However, in the RPS19-deficient groups, the loss of GFPhigh cells was evident from day 6, with an increased fraction of GFPlow and GFP– cells (Online Supplementary Figure S12A-B). By further analysis of cell distribution, significantly decreased outputs of CD71+CD235A– cells on day 6 and CD71– CD235A+ cells on day 10 were observed in GFPhigh populations of RPS19-deficient groups (Online Supplementary Figure S12C, D). The impaired differentiation was rescued by EFS-RPS19, with significantly increased GFPhigh populations (1.6-fold for shRNA1 and 1.8-fold for shRNA2) (Figure 8B) and progenitor populations (CD71+CD235A–) (Online Supplementary Figure S12C) compared to the RPS19-deficient groups on day 6. Particularly, during terminal erythropoiesis on day 16, cells in the RPS19-deficient groups showed reduced red blood cell production (especially in the shRNA2 group) and few GFPhigh cells (<1%) could be detected (Figure 8C-E). However, EFSRSP19 vector-treated groups produced more red blood cells and maintained significantly higher GFPhigh populations (~4%) than the RPS19-deficient groups. These results collectively demonstrate that the EFS-RPS19 vector can rescue the impaired erythroid differentiation in human primary RPS19-deficient CD34+ cord blood cells.
Figure 4.
Gene-corrected bone marrow cells show a competitive advantage in contributing to long-term hematopoiesis in vivo. (A, B) Vector copy number in peripheral blood (A) and bone marrow (B). (C, D) Donor reconstitution in peripheral blood (C) and bone marrow (D). (E–I) The percentage of transduced cells in hematopoietic stem cells (E), megakaryocyte progenitors (F), pre-granulocyte-macrophage and granulocyte-macrophage progenitors (G), pre-megakaryocyte-erythroid (H), and pre-colony-forming unit erythroid and colony-forming unit erythroid (I) (n=13-16, error bars represent the standard deviation, black asterisks indicate the statistical significance of the comparison of recipient-derived cells between the mock and EFS-RPS19 groups, orange asterisks indicate the statistical significance of the comparison of donor-derived cells between the mock and EFS-RPS19 groups. *P<0.05, **P<0.01, ***P<0.005, ****P<0.001 by one-way analysis of variance). VCN: vector copy number; PB: peripheral blood; BM: bone marrow; HSC: hematopoietic stem cells; MkP: megakaryocyte progenitors; pre-GM/GMP: pre-granulocyte macrophage and granulocyte macrophage progenitors; preMegE: pre-megakaryocyte-erythroid; preCFU-E/CFU-E: pre-colony-forming unit–erythroid (CFU-E)/CFU-E.
Figure 5.
Amelioration of disease phenotype in Rps19-deficient animals transplanted with gene-corrected cells. (A) Scheme of the gene-corrected Rps19-deficient cell transplantation model and plan for examining short-term and long-term therapeutic effects. (B) Survival rate analysis. (C-G) Blood cellularity at 4 and 16 weeks after doxycycline induction (n=14-16, error bars represent the standard deviation, *P<0.05, **P<0.01, ***P<0.005 ****P<0.001 by one-way analysis of variance). WT: wild-type; RBC: red blood cells; MCV: mean corpuscular volume; WBC: white blood cells.
Discussion
Currently, hematopoietic stem cell transplantation is the sole curative option for DBA patients, but suitable donors are often unavailable and there can be serious immunological complications.3,33 Gene therapy using gene-corrected hematopoietic stem cells has been shown to be a promising therapeutic strategy for genetic blood disorders in recent years.19,34,35 Our previous proof-of-concept studies also demonstrated the feasibility of applying gene therapy to cure DBA.12-16 In the present study, a clinically applicable lentiviral vector was used to investigate the efficacy and safety for treating anemia and lethal BM failure in Rps19-deficient mice and for ameliorating the impaired erythroid differentiation in human primary RPS19-deficient CD34+ cord blood cells. The next step towards clinical gene therapy will be to perform toxicology and biodistribution analyses, and thereafter proceed with submitting an application to regulatory authorities in order to initiate a phase I/II clinical trial with 6-12 patients focusing on safety.
For successful development of clinical gene therapy, vector efficacy, generating long-term therapeutic effects, is crucial. To examine the therapeutic effects of the vector with lower copies per cell, we decreased the MOI to 5-10 in the present study and demonstrated that the EFS-RPS19 vector has robust therapeutic effects with no evidence of clonal expansion associated with vector integration near cancer-associated genes, as we showed in our previous study.12 It has already been shown that ribosomal proteins are produced in excess of the needs of the ribosome assembly, and that the excess protein is subjected to proteasomal degradation.22,29,30 Similarly, in the present study, our transduced Rps19-deficient cells had physiological levels of expression of RPS19. Hence, it is unlikely that the ectopic expression of RPS19 would promote uncontrolled growth. Since single-copy insertion of the therapeutic gene in the target cells is suggested to avoid the risk of genotoxicity in clinical gene therapy manipulation,19 we plan to examine the therapeutic effects using transduced cells with a lower MOI (e.g. MOI=1) in future studies. Our results also demonstrated that the vector could rescue the impaired erythroid differentiation of RPS19-deficient cord blood cells by increasing red blood cell production. Overall, we showed that the EFS-RPS19 vector could rescue the anemia and BM failure of RPS19-deficient DBA.
Apart from efficacy, vector safety is the other essential factor to assess when applying gene therapy. The risk of insertional mutagenesis is a concern for future applications of gene therapy in the clinic. To prevent this risk, we utilized a third-generation SIN lentiviral vector that lacks potent enhancers in the long terminal repeat regions, since such vectors were shown to exhibit a safer integration profile in previous clinical trials19,34,36 and also in our previous animal studies.12,15 By using the state-of-the-art INSPIIRED workflow, which can provide better quantification of clonal abundance compared to a linear amplificationmediated PCR approach, we found that gene-corrected BM cells in both models exhibited a low risk of mutagenesis with no evidence of clonal expansion associated with vector integration near cancer-associated genes. In our study, no hematologic abnormalities were observed due to enforced expression of RPS19. The results collectively demonstrate the safety of the EFS-RPS19 vector for clinical gene therapy development. The bioinformatic pipeline of the INSPIIRED workflow is a more automated approach, making it well suited for monitoring patients in gene therapy trials in the future.
Increased MCV, due to macrocytic anemia, is a classic clinical observation in patients with DBA. It is, at least in part, caused by the stabilization of p53 and activation of p53 targets (e.g., p21, Bax), which are responsible for cell cycle arrest in G0/G1 phases leading to larger cell size.9,37 We also observed a significantly increased MCV in the mock group after induction with doxycycline for more than 4 weeks (Figures 3E and 5E). In particular, during the initial phase after doxycycline induction, the majority of red blood cells in the circulation were produced before the DBA phenotype was induced. Since old red blood cells are smaller than the newly produced cells, the MCV is decreased compared to normal for a short period during the first 2 weeks after doxycycline induction (Figure 2E). In addition to the severe anemia phenotype after induction of Rps19 deficiency, we observed decreased white blood cell and platelet counts in the mock group. Mild thrombocytopenia and neutropenia (low levels of neutrophilic granulocytes) have also been observed in about 25% of DBA patients during the course of the disease.4,37,38 In addition, several patients with RPS19-deficient DBA have developed myelodysplastic syndrome with multilineage cytopenia, which suggests a multilineage defect.37 This observation correlates with a reduction in the absolute numbers of hematopoietic stem and progenitor cells in BM due to Rps19 deficiency,15 which led to the lethal BM failure we observed in untreated animals shortly after doxycycline administration. This is also supported by the limited reconstitution ability of progenitor compartments in the mock group (Figure 4C-I) and impaired erythroid differentiation of human primary RPS19-deficient cord blood cells. RPS19-deficient patients who develop thrombocytopenia and neutropenia also experience similar progressive phenotypes of hypocellularity in the BM.38,39 Moreover, DBA is a very heterogeneous disease. It is unknown why family members with the same genetic mutation in RPS19 may have very different phenotypes, ranging from no anemia to severe anemia with progression to multilineage BM failure.40 The current understanding of phenotype-genotype correlations is far from comprehensive and needs to be studied further.
Figure 6.
EFS-RPS19 vector-treated Rps19-deficient cells show a competitive advantage in contributing to long-term hematopoiesis in vivo. (A, B) Vector copy number in peripheral blood (A) and bone marrow (B). (C, D) Donor reconstitution in peripheral blood (C) and bone marrow (D). (E–I) The percentage of transduced cells in hematopoietic stem cells (E), megakaryocyte progenitors (F), pre-granulocyte-macrophage and granulocyte-macrophage progenitors (G), pre-megakaryocyte-erythroid (H), and pre-colony-forming unit erythroid and colony-forming unit erythroid (I) (n=14-16, error bars represent the standard deviation, black asterisks indicate the statistical significance of the comparison of recipient-derived cells between the mock and EFS-RPS19 groups, orange asterisks indicate the statistical significance of the comparison of donor-derived cells between the mock and EFS-RPS19 groups. *P<0.05, **P<0.01, ***P<0.005, ****P<0.001 by one-way analysis of variance). VCN: vector copy number; PB: peripheral blood; BM: bone marrow; HSC: hematopoietic stem cells; MkP: megakaryocyte progenitors; pre-GM/GMP: pregranulocyte macrophage and granulocyte macrophage progenitors; preMegE: pre-megakaryocyte-erythroid; preCFU-E/CFU-E: pre-colony-forming unit –erythroid (CFUE)/ CFU-E.
Although the majority of mice in the mock group died, a few mice did survive until the planned endpoint at 16 weeks. One of the possible reasons for this may be the emergence of resident hematopoietic stem cells derived from the recipients, which contribute to the reconstitution of hematopoiesis as a protective mechanism against stress-induced exhaustion in the BM.41-43 In support of this concept, recipient-derived hematopoietic stem cells were observed in BM of the mock group at 16 weeks after transplantation, even though full lethal irradiation was performed prior to transplantation. Other unknown reasons may also contribute to this observation and the underlying mechanism is unknown.
Figure 7.
Gene-corrected bone marrow cells show a vector integration pattern that indicates low risk of mutagenesis and a highly polyclonal insertion site pattern. (A) The top ten integration sites in each sample (*indicates that the integration was within a transcription unit, ~ indicates that the insertion was within 50 kb of a cancer-related gene). (B, C) Percent of all integrations inside transcriptional units (B) and percent of integrations within 100 kb of proto-oncogenes compared to matched random control sites (C). (D) Genomic heatmap analysis of the insertion site profile. mrc: matched randon control. ***P<0.001 by an unpaired t-test.
Figure 8.
Impaired erythroid differentiation of RPS19-deficient CD34+ cord blood cells can be rescued by the EFS-RPS19 vector. (A) RPS19 mRNA expression in CD34+ cord blood cells transduced with shRNA. (B) Percentage of GFPhigh population in RPS19-deficient CD34+ cord blood cells treated or not with EFS-RPS19 during erythroid differentiation from stage I to stage III. (C) Fluorescnce activated cell sorting analysis of erythroid differentiation of RPS19-deficient cells treated or not with EFS-RPS19 on day 16. (D) Percentage of indicated cell outputs of GFPhigh populations on day 16. (E) Red blood cell pellets at the end of stage III initiated with equal numbers of CD34+ cord blood cells (data shown as mean ± standard deviation, ^P<0.05 compared to the shRNA1 group, #P<0.05 compared to the shRNA2 group, *P<0.05, **P<0.01, ***P<0.005 by a t-test, 3 independent experiments).
As mentioned before, the majority of DBA patients have mutations in genes coding for ribosomal proteins, and 25% of them are RPS19-deficient (mostly because of point mutations or small deletions). Our findings indicate the possibility of developing SIN lentiviral vectors also tailored for other DBA mutations (e.g., RPL5) in the future.
In conclusion, our data show the safety and efficacy of a clinically applicable SIN lentiviral vector for the successful treatment of Rps19-deficient DBA in our mouse model and in human primary CD34+ cord blood cells. We did not observe any hematologic abnormalities in vivo due to enforced expression of RPS19. Our present study suggests that the clinically applicable SIN lentiviral vector, EFSRPS19, has the potential to be employed in a clinical gene therapy strategy for RPS19-deficient DBA patients.
Supplementary Material
Acknowledgments
The authors thank Beata Lindqvist and Xiaojie Xian for lentivirus production, Zhi Ma for technical assistance, and Alexander Doyle for English language editing.
Funding Statement
Funding: This work was supported by a Hemato-Linne grant from the Swedish Research Council Linnaeus, project grants from the Swedish Research Council, the Swedish Cancer Society and the Swedish Children’s Cancer Society (to SK), the Tobias Prize awarded by the Royal Swedish Academy of Sciences financed by the Tobias Foundation, a clinical research grant from Lund University Hospital (to SK), European Union project grants STEMEXPAND and PERSIST (to SK), a grant from The Royal Physiographic Society of Lund, Sweden (to YL), and a grant from Stiftelsen Lars Hiertas Minne (to YL).
References
- 1.Lipton JM, Ellis SR. Diamond-Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematol Oncol Clin North Am. 2009;23(2):261-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulirsch JC, Verboon JM, Kazerounian S, et al. The genetic landscape of Diamond- Blackfan anemia. Am J Hum Genet. 2018; 103(6):930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlachos A, Muir E. How I treat Diamond- Blackfan anemia. Blood. 2010;116(19):3715-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willig TN, Niemeyer CM, Leblanc T, et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Societe d'Hematologie et d'Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI). Pediatr Res. 1999;46(5):553-561. [DOI] [PubMed] [Google Scholar]
- 5.Narla A, Vlachos A, Nathan DG. Diamond Blackfan anemia treatment: past, present, and future. Semin Hematol. 2011;48(2):117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gripp KW, Curry C, Olney AH, et al. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A(9):2240-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig LS, Gazda HT, Eng JC, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142(6):859-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan anemia: mutation and database update. Hum Mutat. 2010;31(12):1269-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers KC, Davies SM. Hematopoietic stem cell transplantation for bone marrow failure syndromes in children. Biol Blood Marrow Transplant. 2009;15(3):279-292. [DOI] [PubMed] [Google Scholar]
- 12.Debnath S, Jaako P, Siva K, et al. Lentiviral vectors with cellular promoters correct anemia and lethal bone marrow failure in a mouse model for Diamond-Blackfan anemia. Mol Ther. 2017;25(8):1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flygare J, Olsson K, Richter J, Karlsson S. Gene therapy of Diamond Blackfan anemia CD34(+) cells leads to improved erythroid development and engraftment following transplantation. Exp Hematol. 2008;36(11): 1428-1435. [DOI] [PubMed] [Google Scholar]
- 14.Hamaguchi I, Ooka A, Brun A, Richter J, Dahl N, Karlsson S. Gene transfer improves erythroid development in ribosomal protein S19-deficient Diamond-Blackfan anemia. Blood. 2002;100(8):2724-2731. [DOI] [PubMed] [Google Scholar]
- 15.Jaako P, Flygare J, Olsson K, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118(23):6087-6096. [DOI] [PubMed] [Google Scholar]
- 16.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat Rev Genet. 2011;12(5):301-315. [DOI] [PubMed] [Google Scholar]
- 17.Anguela XM, High KA. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273-288. [DOI] [PubMed] [Google Scholar]
- 18.Biffi A, Bartolomae CC, Cesana D, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117(20): 5332-5339. [DOI] [PubMed] [Google Scholar]
- 19.Cavazzana M, Bushman FD, Miccio A, Andre-Schmutz I, Six E. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov. 2019;18(6):447-462. [DOI] [PubMed] [Google Scholar]
- 20.Dull T, Zufferey R, Kelly M, et al. A thirdgeneration lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Carranza B, Gentsch M, Stein S, et al. Transgene optimization significantly improves SIN vector titers, gp91phox expression and reconstitution of superoxide production in X-CGD cells. Gene Ther. 2009;16(1):111-118. [DOI] [PubMed] [Google Scholar]
- 22.Jaako P, Debnath S, Olsson K, et al. Gene therapy cures the anemia and lethal bone marrow failure in a mouse model of RPS19- deficient Diamond-Blackfan anemia. Haematologica. 2014;99(12):1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flygare J, Kiefer T, Miyake K, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105(12):4627-4634. [DOI] [PubMed] [Google Scholar]
- 24.Sherman E, Nobles C, Berry CC, et al. INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol Ther Methods Clin Dev. 2017;4:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry CC, Nobles C, Six E, et al. INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol Ther Methods Clin Dev. 2017;4:17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao A, Chazdon RL, Colwell RK, Shen TJ. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics. 2006;62(2): 361-371. [DOI] [PubMed] [Google Scholar]
- 27.Haemmerle R, Phaltane R, Rothe M, et al. Clonal dominance with retroviral vector insertions near the ANGPT1 and ANGPT2 genes in a human xenotransplant mouse model. Mol Ther Nucleic Acids. 2014;3: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLury BD. On the estimation of biological populations. Biometrics. 1947;3(4):145-167. [PubMed] [Google Scholar]
- 29.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17(9):749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116(15):2826-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesana D, Ranzani M, Volpin M, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther. 2014;22(4):774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deichmann A, Brugman MH, Bartholomae CC, et al. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol Ther. 2011;19(11):2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Da Costa L, O'Donohue MF, van Dooijeweert B, et al. Molecular approaches to diagnose Diamond-Blackfan anemia: The EuroDBA experience. Eur J Med Genet. 2018;61(11):664-673. [DOI] [PubMed] [Google Scholar]
- 34.High KA, Roncarolo MG. Gene therapy. N Engl J Med. 2019;381(5):455-464. [DOI] [PubMed] [Google Scholar]
- 35.Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science. 2018;359(6372):eaan4672. [DOI] [PubMed] [Google Scholar]
- 36.Marktel S, Scaramuzza S, Cicalese MP, et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ss-thalassemia. Nat Med. 2019;25(2):234-241. [DOI] [PubMed] [Google Scholar]
- 37.Da Costa L, Leblanc T, Mohandas N. Diamond-Blackfan anemia. Blood. 2020;136(11):1262-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giri N, Kang E, Tisdale JF, et al. Clinical and laboratory evidence for a trilineage haematopoietic defect in patients with refractory Diamond-Blackfan anaemia. Br J Haematol. 2000;108(1):167-175. [DOI] [PubMed] [Google Scholar]
- 39.Casadevall N, Croisille L, Auffray I, Tchernia G, Coulombel L. Age-related alterations in erythroid and granulopoietic progenitors in Diamond-Blackfan anaemia. Br J Haematol. 1994;87(2):369-375. [DOI] [PubMed] [Google Scholar]
- 40.Engidaye G, Melku M, Enawgaw B. Diamond Blackfan anemia: genetics, pathogenesis, diagnosis and treatment. EJIFCC. 2019;30(1):67-81. [PMC free article] [PubMed] [Google Scholar]
- 41.Shi W, Vu T, Boucher D, et al. Ssb1 and Ssb2 cooperate to regulate mouse hematopoietic stem and progenitor cells by resolving replicative stress. Blood. 2017;129(18):2479-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgartner C, Toifl S, Farlik M, et al. An ERK-dependent feedback mechanism prevents hematopoietic stem cell exhaustion. Cell Stem Cell. 2018;22(6):879-892.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh SK, Singh S, Gadomski S, et al. Id1 ablation protects hematopoietic stem cells from stress-induced exhaustion and aging. Cell Stem Cell. 2018;23(2):252-265.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.