Abstract
Convolutional neural networks (CNN) are widely used in computer vision and medical image analysis as the state-of-the-art technique. In CNN, pooling layers are included mainly for downsampling the feature maps by aggregating features from local regions. Pooling can help CNN to learn invariant features and reduce computational complexity. Although the max and the average pooling are the widely used ones, various other pooling techniques are also proposed for different purposes, which include techniques to reduce overfitting, to capture higher-order information such as correlation between features, to capture spatial or structural information, etc. As not all of these pooling techniques are well-explored for medical image analysis, this paper provides a comprehensive review of various pooling techniques proposed in the literature of computer vision and medical image analysis. In addition, an extensive set of experiments are conducted to compare a selected set of pooling techniques on two different medical image classification problems, namely HEp-2 cells and diabetic retinopathy image classification. Experiments suggest that the most appropriate pooling mechanism for a particular classification task is related to the scale of the class-specific features with respect to the image size. As this is the first work focusing on pooling techniques for the application of medical image analysis, we believe that this review and the comparative study will provide a guideline to the choice of pooling mechanisms for various medical image analysis tasks. In addition, by carefully choosing the pooling operations with the standard ResNet architecture, we show new state-of-the-art results on both HEp-2 cells and diabetic retinopathy image datasets.
Keywords: Medical image analysis, Pooling, Convolutional neural networks, HEp-2 cell image classification, Retinopathy image classification
Introduction
Convolutional neural networks (CNNs) are the state-of-the-art methods for various computer vision and medical image analysis tasks such as image classification [55, 95, 98, 109, 124, 126] and segmentation [35, 109, 118]. CNN often consists of multiple convolutional layers followed by one or more fully connected layers, where each convolutional layer often includes convolution, nonlinear activation and optionally pooling operators.
The purpose of pooling is mainly to down-sample the feature maps and to learn larger-scale image features that are invariant to small local transformations (e.g., translation, scaling, and rotation). It is a process of aggregating the features from each spatial region, e.g., averaging the values in each region at each feature channel.
Pooling does not only increase the size of the receptive field of convolutional kernels (neurons) over layers, but also reduces the computational complexity and the memory requirements as it reduces the resolution of the feature maps while preserving important features that are needed for processing by the subsequent layers. In medical image analysis, pooling can help to handle variance in lesion sizes [3] and positions [94].
Various pooling methods have been proposed for different purposes. For example, soft pooling (e.g., [42, 55, 90, 124, 129]) is proposed to take advantages of both the widely used max and average pooling; stochastic pooling (e.g., [39, 101, 132, 142, 143]) is proposed to overcome the overfitting issue in CNN training; spatial pyramid pooling and its variants are to capture spatial or structural information in the images (e.g., [45, 91, 130]); higher-order pooling (e.g., [25, 31, 34, 69–71, 141]) is to capture higher-order statistical information of the feature maps, etc.
However, most of these approaches were proposed for and evaluated on computer vision image datasets (e.g., PASCAL VOC 2012 [29], Cityscapes [23], CIFAR-10 [58]) and their applicability for medical image classification has not been well-investigated.
In this work, we review different pooling methods proposed in computer vision and medical imaging literature, and report examples of medical imaging applications where some of these pooling methods are used (refer Table 2). In addition, we conduct an experimental study to compare the performance of pooling methods on two different medical image classification tasks, i.e., classifications of HEp-2 cells and diabetic retinopathy images.
Table 2.
Overview of different pooling methods used for different medical imaging tasks
| Name of the pooling | Example applications in computer vision | Example applications in medical imaging | |
|---|---|---|---|
| Type of application | Modality | ||
| Max and average pooling | |||
| Max and/or average pooling | Classification [46] | Image classification and localization of lesions [93, 126] | Retina |
| Segmentation [90] | Cell image classification [77] | HEp-2 cells | |
| Image classification and detection of pneumonia [95] | X-Ray (chest) | ||
| Weakly supervised learning [55] | X-Ray (chest) | ||
| Multiple sclerosis identification [122] | MRI (brain) | ||
| Object localization [111] | – | – | |
| Linear combination of max and average pooling | |||
| Mixed max-average pooling [63] | Classification [63] | – | – |
| Gated max-average pooling [63] | Classification [63] | – | – |
| Dynamic correlation pooling [11] | Classification [11] | – | – |
| Soft pooling | |||
| Generalized max pooling | Segmentation [129], Classification [7] | Multiple Instance Learning [135] | Histopathology |
| Root-mean-square pooling [53] | Classification [53] | – | – |
| Log-sum-exp pooling [90] | Segmentation [90] | Weakly supervised classification and localization: thorax diseases [124] | X-Ray (chest) |
| Proximal femur fractures [55] | X-Ray (bone) | ||
| Histopathology cancer image classification [135] | Histopathology | ||
| Polynomial pooling [129] | Segmentation [129] | – | – |
| Learned-norm pooling [42] | Classification [42] | – | – |
| pooling [7] | Classification [7] | – | – |
| Rank-based pooling [101] | Classification [101] | Cerebral micro-bleed detection [120] | MRI (brain) |
| Multipartite pooling [99] | Classification [99] | ||
| Ordinal pooling [60] | Classification [60] | – | – |
| Multi-activation pooling [151] | Classification [151] | – | – |
| pooling [28] | Classification [28] | – | – |
| Global feature guided local pooling [57] | Classification [57] | – | – |
| SQUare-root (SQU) pooling [15] | Image instance retrieval [15] | – | – |
| Dynamic pooling [84] | – | Chronic kidney disease detection [84] | Saliva |
| Smooth-Maximum-Pooling [5] | Classification [5] | – | – |
| SoftPool [107] | Classification, Action recognition [107] | – | – |
| RunPool [54] | Classification [54] | – | – |
| Maxfun pooling [26] | Classification, Convolutional sparse coding [26] | – | – |
| Stochastic pooling to handle overfitting | |||
| Stochastic pooling [142] | Classification [142] | Multiple sclerosis identification [122] | MRI (brain) |
| Alcoholism Detection [121] | MRI (brain) | ||
| COVID-19 diagnosis [149] | CT (chest) | ||
| Rank-based stochastic pooling [101] | Classification [101] | Abnormal breast identification [148] | Breast |
| Mixed pooling [139] | Classification [139] | Brain tumor segmentation [10] | MRI (brain) |
| Hybrid pooling [112] | Classification [82, 112, 113] | – | – |
| Max pooling dropout [132] | Classification [132] | – | – |
| S3 pooling [143] | Classification [143] | – | – |
| Fractional max pooling [39] | Classification [39] | Retinopathy image classification [40] | Retina |
| Sparsity-based stochastic pooling [104] | Classification [104] | – | – |
| EasyConvPooling (ECP) [100] | Classification [100] | – | – |
| PatchShuffle stochastic pooling [123] | – | Diagnosis of COVID-19 [123] | CT (chest) |
| Pooling to encode spatial or structural information | |||
| Spatial pyramid pooling [45] | Classification, Detection [45] Hand gesture recognition [110], Image steganalysis [146] | Brain image segmentation [118] Prostate image segmentation [35] Tumor segmentation for rectal cancer radiotherapy [79] | MRI (brain) MRI (prostate) MRI, CT (rectum) |
| Concentric circle pooling [91] | Remote sensing scene classification [91] | – | – |
| Polycentric circle pooling [92] | Remote sensing image recognition. [92] | – | – |
| Pose pooling kernels [145] | Fine-grained image classification [145] | – | – |
| Geometric norm pooling [30] | Classification [30] | – | – |
| Cell pyramid matching [130] (non CNN) | – | Cell image classification [77, 130] | HEp-2 cells |
| Multi-pooling [117] | – | Brain tumor segmentation [117] | MRI (brain) |
| Donut-shaped spatial pooling [62] | – | Cell image classification [62] | HEp-2 cells |
| Structure based graph pooling [14] | Action recognition [14] | – | – |
| Atrous Spatial Pyramid Pooling (ASPP) [12] | Segmentation [12] | Multi-scale retinal vessel segmentation [134] | Retina |
| Higher-order pooling | |||
| Second oder pooling [9] | Classification, Segmentation [9] | – | – |
| Bilinear pooling [71] | Fine-grained classification [71] | – | – |
| Improved bilinear pooling [70] | Fine-grained classification [70] | – | – |
| -pooling [102] | Fine-grained classification [102] | – | – |
| Statistically-motivated second-order pooling [141] | Classification, Fine-grained classification [141] | – | – |
| Global second order pooling [34] | Classification [34] | – | – |
| Kernel pooling [25] | Classification [25] | – | – |
| Global covariance pooling [68] | Classification [68] | – | – |
| Global gated Mixture of Second-Order Pooling (GM-SOP) [119] | Classification [119] | – | – |
| Second-order temporal pooling [18] | Action recognition [18] | – | – |
| Graph pooling [125] | Graph classification [125] | – | – |
| Hierarchical adaptive pooling [74] | Graph classification, Graph Matching, Graph Similarity Learning [74] | – | – |
| Higher-order pooling [19] | Action recognition [19] | – | – |
| Detachable second-order pooling [66] | Classification [66] | – | – |
| Approaches that aim to keep important information when pooling | |||
| Detail preserving pooling [96] | Classification [96] | – | – |
| Local importance-based pooling [32] | Classification, Detection [32] | – | – |
| RNNPool [97] | Classification, Visual wake words, Face Detection [97] | – | – |
| Attention weighted pooling | |||
| Double-attention network (-network) [13] | Classification [13] | – | – |
| Convolutional Block Attention Module (CBAM) [131] | Classification [131] | Diabetic retinopathy grading [44] | Retina |
| Global learnable pooling [147] | Classification [147] | – | – |
| Zoom-in-Net [127] | – | Diabetic retinopathy grading [127] | Retina |
| Recurrent attention model [89] | – | Detection of pulmonary lesions [89] | X-Ray (chest) |
| Attention based CNN models | – | Glaucoma detection [67] | Retina |
| Thorax disease classification [41] | X-Ray (chest) | ||
| Implicit pooling mechanisms | |||
| Generalized max pooling [83] | Classification [83] | – | – |
| Task-driven feature pooling [133] | Classification [133] | – | – |
| Deep generalized max pooling [20] | Witter identification and document classification [20] | – | – |
| Adaptive spatial pooling [75] | Classification [75] | Retrieving brain tumors [17] | CE-MRI (brain) |
| Deep Adaptive Temporal Pooling (DATP) [103] | Human activity recognition [103] | – | – |
| Dynamic temporal pooling [64] | Time series classification [64] | – | – |
| Clustering-based aggregation schemes | |||
| Learnable Pooling Module (LPM) [86] | Full-face gaze estimation [86] | Brain surface analysis [38] | MRI (brain) |
| Video tagging [80] | |||
| Other approaches | |||
| Transformation Invariant Pooling (TI-Pooling) [61] | Classification [61] | Neuronal structures segmentation [61] | Microscopy |
| Hierarchical mix pooling [78] | - | HEp-2 cell image classification [78] X-Ray image classification [61] | HEp-2 cells X-Ray |
| Tree pooling [63] | Classification [63] | – | – |
| Virtual Pooling (ViP) [16] | Classification, Object Detection [16] | – | – |
| Kernelized subspace pooling [128] | Image patch matching [128] | – | – |
| LiftPool [150] | Classification, Segmentation [150] | – | – |
Selection of identified papers for review: An initial selection of papers was done by the aid of Google scholar. Different keywords related to pooling (e.g., pooling, pooling in CNN, pooling in medical imaging, attention weighted pooling, feature aggregation, etc.) were used to identify relevant papers. As the majority of the identified papers use existing pooling techniques, the papers which propose novel pooling approaches were mainly identified, and selected for review. This gave us around 121 papers in total, among them 87 papers proposed different pooling techniques and in 34 papers different pooling techniques are applied for different tasks. Among the selected papers, 90 and 31 papers, respectively, discuss pooling methods in computer vision and medical imaging.
The main contributions of this work include:
To our best knowledge, this is the first work to review various pooling methods in deep learning particularly for medical imaging applications.
As many of the pooling methods (e.g., higher-order pooling [25, 31, 34, 69–71, 141]) have not been explored for medical imaging, we perform an extensive set of comparative experiments on selected pooling methods to investigate their performance on two public medical image datasets.
The rest of this paper is organized as follows. Section 2 reviews the work related to different pooling methods proposed in computer vision and medical image analysis. Section 3 summarizes the dataset and the experimental settings. Results are reported and discussed in Sect. 4. A detail discussion about our work is given in Sect. 5 and Sect. 6 concludes this paper.
Pooling methods
There are two groups of pooling generally used in CNNs. The first one is local pooling, where the pooling is performed from small local regions (e.g., ) to downsample the feature maps. The second one is global pooling, which is performed from each of the entire feature map to get a scalar value of a feature vector for image representation. This representation is then passed to the fully connected layers for classification. For example, there are four local pooling and one global pooling layers included in the well-known DenseNet [51].
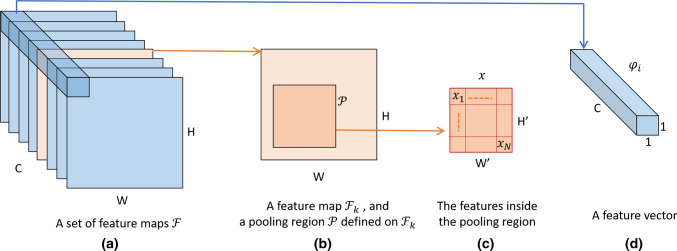
Notations: Consider a set of feature maps and a pooling region defined on one of these feature map, , as in Fig. 1. Assume that represents the features that are inside the pooling region on . For example, in the local pooling case and and in the global pooling case, where W and H represent the width and the height of the feature map, respectively. In the following, we assume that is vectorized to simplify the math operations, i.e., , where is the number of elements in . Let be the element of , where .
Table 1.
Notations used throughout this paper (please refer Fig. 1 for more information)
| Notation | Dimension | Detail |
|---|---|---|
| A set of feature maps | ||
| W | 1 | Width of |
| H | 1 | Height of |
| C | 1 | Number of feature maps (channels) in |
| A pooling region for each feature map | ||
| 1 | Width of | |
| 1 | Height of | |
| The part of a feature map (channel) within the pooling region . The channel index is omitted for simplicity. | ||
| 1 | The element or activation at the position in , | |
| N | 1 | The number of elements in , |
| A feature vector across channels at the position | ||
Fig. 1.
Demonstration of relevant notations. a A set of feature maps . b An example feature map from (or the channel of ), and a pooling region defined on . c The features inside the pooling region of the selected feature map . d The feature vector, , obtained across channels at the i-th position of the feature maps
Average pooling and max pooling
The average pooling and the max pooling [6] are widely used in CNNs [46, 49, 51, 59] because of their simplicity—they do not have any parameters to tune. The average pooling summarizes all the features in the pooling region and can be defined as
| 1 |
On the other hand, max pooling selects only the strongest activation in the pooling region, i.e.,
| 2 |
The average pooling and the max pooling have their own merits and disadvantages.
Averaging reduces the effect of noisy features. But as it gives equal importance to all the elements in the pooling region, background regions may dominate in the pooled representation, and hence, may reduce the discriminative power. In contrast, max pooling selects the largest value in each pooling region, and hence can avoid the effect of unwanted background features. However, as it selects only the maximum element, the pooled representation may capture noisy features.
The average and max pooling can be applied in different scenarios. Consider a situation in medical image analysis where lesion appears only in a small part of the image. In this case, average pooling may not be a good choice as the elements of the pooling region corresponding to background pixels will tend to dominate the pooled representation. However, average pooling may be more appropriate for some other scenarios, e.g., classification of abnormal images from normal ones where abnormality spread all over the abnormal image. Unlike average pooling, max pooling is a nonlinear operator1 which increases the nonlinearity of the network. In the training stage of a network, all the neurons that are connected to the average pooling layer will be updated via backpropagation as the output of all the neurons contribute to the output of average pooling. In contrast, as max pooling selects only the strongest activation, only the neurons which are connected to the neuron outputting the strongest activation will be allowed to learn.
Note that in addition to CNNs, max and average pooling also have been well-explored in traditional feature encoding approaches such as, bag-of-words [24] and its variations such as sparse coding [53], vector of locally aggregated descriptors [52] and Fisher vectors [88] (discussed in Sect. 2.10). Average pooling is widely used in all of these methods except sparse coding, where max pooling is widely used. As listed in Table 2, max and average pooling are very well-explored in medical image analysis for different problems, including HEp-2 cell image classification [77], retinopathy image classification [93, 126], multiple sclerosis identification from MRI images [122], etc.
Since neither max pooling nor average pooling consistently performs better than the other [6], approaches have been proposed to take advantages of both. This line of research includes a direct combination of max and average pooling with weights (Sect. 2.2) and soft pooling (Sect. 2.3). However, unlike max and average poolings, new parameters are introduced in these approaches, causing additional overhead in parameter learning or tuning.
Linear combination of max and average pooling
To overcome the problems associated with the max and the average pooling (discussed in Sect. 2.1), in mixed max-average pooling [63], the max pooling and the average pooling are simply added together with weights to take advantage of both, i.e.,
| 3 |
where is a learnable parameter that determines the mixing proportion. There are multiple options available here when choosing this parameter. The same a could be used for the entire network, or a set of a’s could be used, one for each pooling layer (i.e., , where L is the number of layers), or even different regions of different pooling layers may use different mixing proportions.
The mixing proportion, a, in Eq. (3) is a parameter which does not depend on the individual characteristics of a given image, although it can be learned in the network training process. The images from the same dataset could have different characteristics. For example in medical images, for some images, the lesions could be localized (appear only in some parts), but for some other images, lesions could be spread all over the image. In that case the mixing proportion should depend on the characteristics of each image than the characteristics of the dataset, and therefore it should be determined for each image separately. This is the motivation behind the Gated Mix-Average pooling [63].
The gated mix-average pooling can be defined as:
| 4 |
where is a weight vector (called the gating mask in [63]) to be learned when training the network, and is a sigmoid function which converts the transformed input () to a value between 0 and 1. This value is then used to weight the contribution of the max and the average pooled results as shown in Eq. (4). As with mixed max-average pooling (Eq. (3)), the new parameters () can also be learned in different ways, e.g., separately for each layer or separately for each of the channels in each layer of the network.
Both in mixed max-average and gated mix-average pooling, each pooling region (of a particular feature map) is considered independently from each other. Dynamic Correlation pooling [11] also uses the same formulation as in Eq. (3); however, the weighting proportion for each pooling region is determined based on the correlation between that region and its adjacent regions; average pooling gets higher weight if the correlation is high, and max pooling on the other hand.
To the best of our knowledge, as listed in Table 2, soft pooling approaches are widely used in medical imaging than using linear combination of max and average pooling techniques.
Soft pooling approaches
Soft pooling is used as an intermediate form between max and average pooling. Unlike simply adding the max and average pooling as in Sect. 2.2, in soft pooling, a smooth differentiable function is used to approximate the max and the average pooling for different parameter settings. For example, in the Generalized Mean (GM) [135] function,
| 5 |
the parameter r controls the softness, i.e., when this function is equivalent to average pooling, and when this approximates max pooling.
Various such approximations are used, including Log-Sum-Exp pooling (LSE) [55, 90, 124], Polynomial pooling [129], Learned-Norm pooling [42], pooling [7], Integration () pooling [28], Rank-based pooling [99, 101], Dynamic pooling [84], Smooth-Maximum pooling [5], Soft pooling [107], Maxfun pooling [26], Ordinal pooling [60]. As most these functions are differential approximation of max pooling, they are widely explored in (non-CNN based) Multiple Instance Learning approaches in computer vision [8] and medical image analysis [76, 135] (Table 2), in addition to CNN-based image classification [28, 42, 124, 144] and segmentation [90, 129].
The learned-norm pooling [42] and pooling [7] use similar formulation as in Eq. (5). The Root-Mean-Square pooling [53] is a special case () of GM. The pooling [28] introduces a formulation, where different statistics such as arithmetic mean, harmonic mean, maximum and minimum are special cases. pooling is given as
| 6 |
where
| 7 |
This pooling shows marginal improvements over the max pooling, pooling (Sect. 2.6) and pooling on some computer vision datasets in [28].
In the rank-based pooling [101], first the elements in each pooling region are ordered (ranked) and then the top-k elements (elements with highest activations) are averaged together as the pooled representation. When and this pooling is equivalent to max and average pooling, respectively. Ordinal pooling [60] and multi-activation pooling [151] are similar to rank-based pooling, which also use the rank of the elements when applying pooling.
The free parameter(s) in the above soft pooling functions could be the same for the entire network, or could be different for different layers and either could be fixed [90] or learned [42, 129]. For example, in pooling [28], the parameters (’s) are learned for each layer separately via back-propagation, and in polynomial pooling [129], a side-branch net is used to determine the parameters of each pooling region.
In all the above soft pooling approaches, the result of the pooling is just based on the characteristics of the pooling region of a particular feature map itself. But differently from these approaches, in Global Feature Guided Local pooling (GFGP) [57], the pooled result of a particular region is not only based on that region itself, but also it depends on some global statistics of the feature map. The GFGP is formulated as
| 8 |
where
| 9 |
The weights2 determine the type of pooling and are learned through an optimization process, and is channel (particular feature map)-dependent parameter, determined (learned) based on the statistics of the global features of that channel. Note that average and max pooling can be obtained when and , respectively.
Stochastic pooling approaches to handle overfitting
One of the main issue when training CNNs with limited data is overfitting. Mixed-pooling [139], Hybrid pooling [112], Stochastic pooling [142], Rank-Based Stochastic pooling [101], Max pooling dropout [132], Stochastic Spatial Sampling (S3 pooling) [143] and Fractional Max pooling [39] are proposed to reduce overfitting by introducing various forms of randomness in pooling configurations and/or the way the pooling is performed in the training process. Because of this randomness in training, the trained model can be thought as an ensemble of similar networks, with each random pooling configuration defining a different member of the ensemble.
As listed in Table 2, these stochastic pooling approaches are widely used in medical imaging (e.g., COVID-19 diagnosis [149], abnormal breast identification [148], brain tumor segmentation [10]) as usually the models in medical imaging are trained with small amount of training data, and these pooling approaches can help to handle the issues with overfitting.
Mixed pooling [139] and hybrid pooling [112] introduce randomness in training by randomly selecting either max or average operations for pooling, i.e.,
| 10 |
where is a random value to be either 0 or 1 that determines which pooling to be selected, i.e., when the max pooling and when the average pooling is selected, respectively. This randomness cannot be used in the testing time. Therefore, in [139], the statistics about how many times the max and the average operations are selected for pooling for each feature map in the training phase are recorded. Based on this statistics whatever pooling used frequently for each layer in the training phase is selected to use at the testing phase.
Stochastic pooling [142] introduces randomness in training by randomly selecting an activation (instead of selecting either maximum as in max pooling or all the elements as in average pooling) within each pooling region according to a multinomial distribution given by the values within that pooling region. Here, the values in each pooling region are first converted into probability values by dividing each of the value by the sum of all the values in that pooling region, i.e.,
| 11 |
Then, a location l within each pooling region is sampled based on the corresponding probability values to get the pooled representation of that region. The locations to sample for each pooling region in each layer for each training example are drawn independently to one another. In testing time, a probabilistic weighting scheme was used, where the pooled representation of a pooling region is calculated as follows:
| 12 |
This can be seen as a weighted average pooling, where the probability values are used to weight the corresponding elements in the pooling regions.
In stochastic pooling, still over-fitting may happen particularly when the training data are limited. This is because strong activations will always have the highest probability to be sampled. Therefore, rank-based stochastic pooling [101] suggests a different way to calculate the probabilities based on the ranks of the activations inside each pooling region.
Instead of sampling only one value from each pooling region as stochastic pooling does, a set of values could be randomly sampled first and then pooling could be applied on these random sampled activations as in max pooling dropout [132]. Max pooling dropout first applies dropout on the feature maps to drop of the features and then applies max pooling on the retaining features, and show better performance than stochastic pooling for particular values of p.
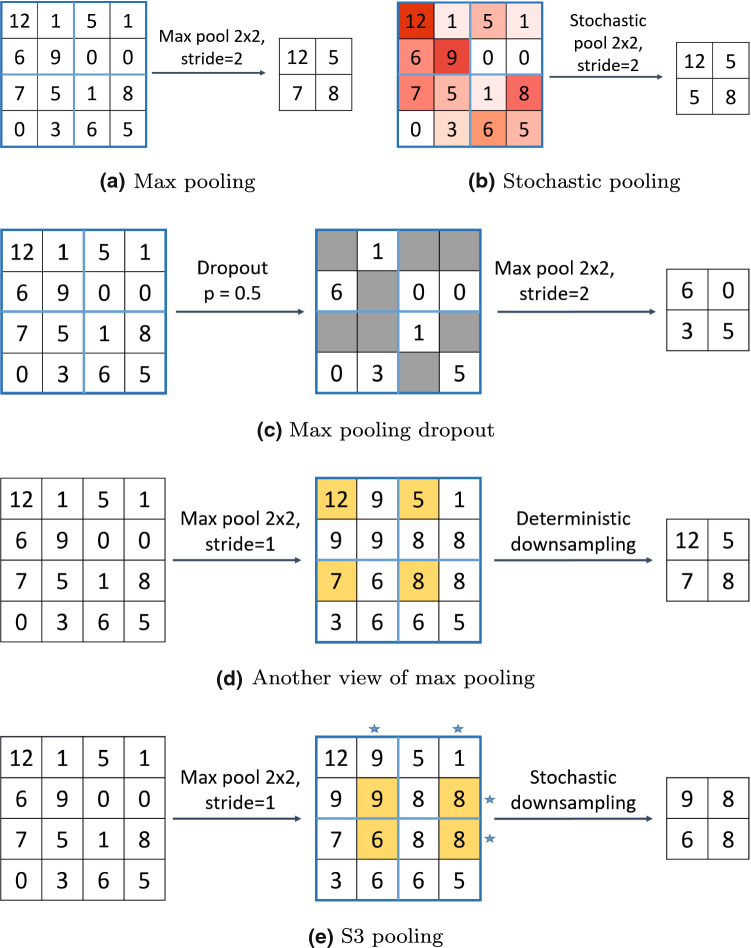
Unlike the above approaches where randomness is introduced in the pooling stage, in S3 pooling [143] and fractional max pooling [39], randomness is introduced in the spatial sampling stage. The standard max pooling can be viewed as a two-step procedure (Fig. 2d). In the first step, max pooling is performed from the feature map with a stride of 1. Then in the second step, spatial downsampling is performed uniformly on the resultant map by extracting the top-left corner element of each disjoint window, resulting in a feature map with s times smaller spatial dimensions. S3 pooling differs from the traditional max pooling in the second step. Instead of the uniform sampling used by max pooling, S3 pooling proposes non-uniform sampling to downsample the pooled feature maps.
Fig. 2.
Max pooling and different Stochastic pooling approaches: a the standard max pooling, b stochastic pooling, c max pooling dropout, d another view of max pooling with stride = 2, and e S3 pooling. For all the above, downsampling is performed with a filter size of with a step size of 2. In b, colors corresponding to probability values. High values of red correspond to high probability values and vice versa. In c, the values in the shaded squares are dropped. In e, ‘*’ corresponds to the selected rows and columns (Color figure online)
Max pooling reduces the size of the feature maps by an integer multiplicative factor s (the value of stride). Usually, s is set to two in most architectures (e.g., ResNet [46]), and therefore reducing the size of the feature maps by half of its original size every time pooling is applied, and hence, limiting the number of pooling layers used. In fractional max pooling [39], s is allowed to take a non-integer value, i.e., , to allow the use of larger number of pooling layers.
Because of this non-uniform nature of downsampling used in S3 pooling and fractional max pooling the downsampled feature maps get distorted. This distortion provides a way for data augmentation to improve the generality of the network.
Pooling approaches to encode spatial structure information
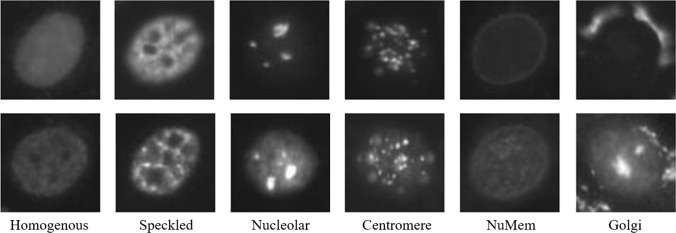
For some problems, encoding spatial information is necessary, for example, in natural images sky is always in the upper part of the image. Encoding such information may lead to more informative and discriminative feature representation. Similarly in some medical images, this kind of information is very useful. For example, the Golgi class in Fig. 4 has a unique ring like structure around the cells. Encoding that structure in the feature representation may help to easily discriminate that class from others. Various approaches [30, 43, 45, 91, 92, 115, 117, 130, 136, 145] have been proposed to encode local structure information in the pooled representation.
Fig. 4.
Example images from different classes of the HEp-2 cell image dataset
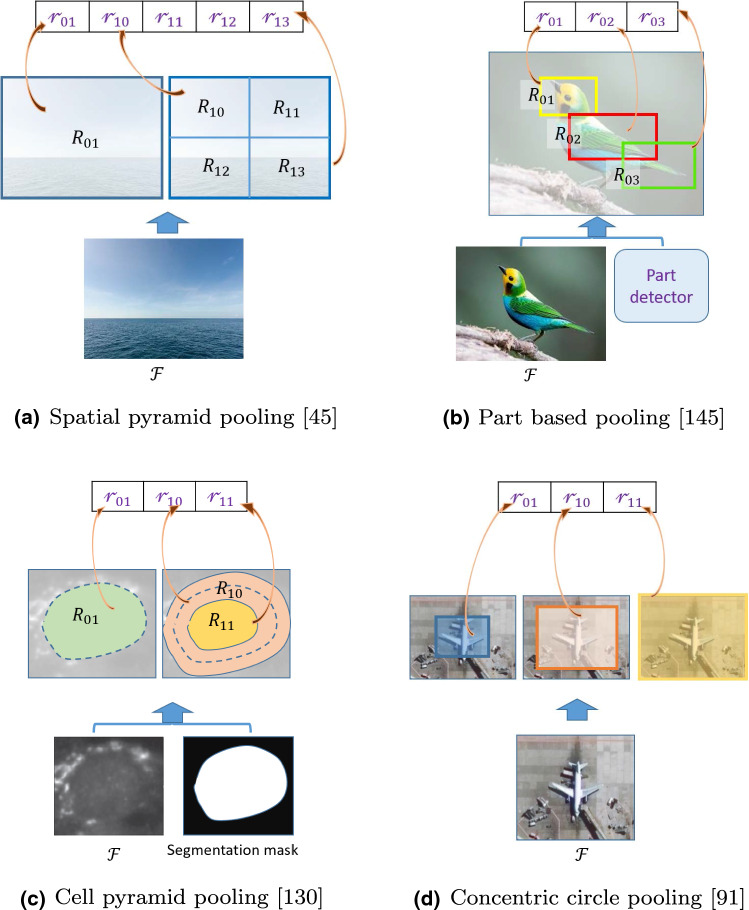
Spatial Pyramid pooling (SPP) [45] (Fig. 3a) is a popular way to include spatial structure information in the pooled representation. It divides the feature map into grids of cells and applies the standard max or average pooling from each cell separately. Then, these cell-based pooled representations are concatenated together as the image representation.
Fig. 3.
Different pooling techniques to capture information about spatial structures. represents feature maps. represents different pooling regions specified by different techniques. is the pooled representation of the region . Each of the pooled representation from an individual pooling region will have a dimension of , where C is the number of channels in . The final image representation will have a dimension of , where M is the total number of pooling regions specified by the pooling algorithm (images best viewed in color) (Color figure online)
SPP is very useful for rigid structures, but it may not be appropriate for images containing objects with different poses, e.g., birds with different poses. To overcome this, in [145] a part-based pooling strategy is proposed for fine-grained image classification. Here, from each image, different parts (e.g., head, tail, body of a bird) are detected first. Then, the features from each detected parts are pooled and concatenated together as the final image representation (Fig. 3b).
Both SPP and the part-based pooling strategies may not be very useful for the images with rotated objects. To capture rotationally invariant spatial structure, representations with CNNs Concentric Circle pooling [91] and Polycentric Circle pooling [92] were proposed and applied for recognizing remote sensing images, where the pooling regions are defined as concentric circles (Fig. 3d). A similar approach, Multi-pooling [117], was proposed to cope with lesions (brain tumors) with different sizes, where features extracted from different sized concentric regions are concatenated together as representations.
Cell Pyramid Matching (CPM) [130] is another approach to capture spatial structure information, specifically for cell image classification. In CPM, the segmentation mask of each cell is used to define the pooling regions as shown in Fig. 3c. CPM also adopted in [77] for the same purpose. Both in [130] and [77] CPM was used with traditional feature representations such as bag-of-words and not with CNNs. Note that CPM requires additional input in the form of segmentation masks to identify the border of each cell.
All the above approaches are meant to capture large-scale spatial structure information. On the other hand, Geometric -Norm pooling [30] aims to capture local structure information (e.g., from image regions of size ) for the sparse coding-based (non-CNN) representations by applying weights to different locations of the pooling region. However, with CNN, this pooling is equivalent to first applying a nonlinear transformation on the feature maps and then applying a convolution for aggregation.
Pooling approaches that capture higher-order information
Average pooling only captures the first-order statistics (i.e., mean) of each pooling region, by pooling from each channel (feature map) separately. This pooling, hence, neither captures the interaction between different feature maps, nor the interaction between the features from different regions of the same feature map. This interaction may capture additional details such as object co-occurrence [137]. Therefore, capturing higher-order statistical information via covariance matrices can improve the ability of CNNs to learn complex nonlinear class boundaries. Recently, incorporating higher-order statistical pooling approaches with CNNs got attention [18, 25, 31, 34, 69–71, 141] and have achieved state-of-the-art results on a variety of tasks including object recognition, fine-grained visual categorization, and object detection.
Second-Order pooling was initially proposed in [9] for aggregating SIFT descriptors (non-CNN). The max and the average second-order pooling are defined in [9] as follows:
| 13 |
| 14 |
where is the feature descriptor (e.g., SIFT [72]) from region , d is the dimensionality of and is the outer product between descriptor with itself, capturing the pairwise correlations between the elements of . The pooled representation (matrix of size ) was then passed through a nonlinear transformation and a normalization process before giving it to a linear classifier.
This idea is then extended with CNN features in [25, 34, 69–71, 141]. For example, in Improved Bilinear pooling [70] is a feature from the last layer of a CNN model (Fig. 1). The pooled features were then passed through a normalization layer before performing fine-grained classification. Both in [70, 71], second-order pooling is applied only at the end of the network; in contrast, in [34], second-order pooling is applied throughout the network (from lower to higher layers) and shows improved performance than applying them at the end of the network. Extensions of this pooling include compact [31, 141] and kernelized [25] versions. In addition, the association between second-order pooling and Attention-Based pooling is analyzed in [36]. The formulation of the -pooling [102] allows for a continuous transition between average and bilinear pooling by the introduction of a trainable parameter .
Approaches that aim to keep important information when pooling
Discriminative details could be lost due to improper pooling mechanisms, particularly, in the early stage of the networks. This information loss may hinder the learning process and result in sub-optimal models [32]. Detail Preserving pooling (DPP) [96] and Local Importance-Based pooling (LIP) [32] aim to reduce this information loss by preserving important features when pooling.
Because some activations are important than others, both of these approaches weight the contribution of activations in the pooling region as given in Eq. (8). However, they differ from each other (and from [57] discussed in Sect. 2.3) in the way the weights are determined. In DPP, higher weights are given to the activations which are different from the activation at the center of the pooling region as those activations are assumed to carry more information, i.e.,
| 15 |
where is the activation at the center of the preprocessed pooling region. The parameters and are learned together with other parameters in network training.
But in LIP, the weights are determined using a subnetwork attached to each pooling layer. Therefore, LIP can also be considered as an attention-based pooling approach (Sect. 2.8) as the subnetwork learns a saliency map to weight each element of the feature map. LIP shows improved recognition rate on the ImageNet dataset over DPP in [32].
In addition, these approaches also can be considered as soft pooling approaches; for particular parameter settings, they approximate the standard average and the max pooling. For example, when in Eq. (15), DPP becomes average pooling.
Larger networks cannot be deployed in resource constrained devices as they have large memory requirements. One way is to handle this problem is by reducing the number of layers of the network by rapid downsampling. Rapid downsampling of the feature maps by a large factor can simply lead to information loss, and hence reduced performance. RNNPool [97] tries to alleviate this problem by incorporating recurrent nets for downsampling, where two recurrent nets were used, the first one summarizes the feature maps horizontally and vertically, and the second one summarizes the outputs of the first one as the pooled results.
Attention-weighted pooling
In these kind of approaches, each element of the feature map is weighted by the corresponding weight from the attention/saliency map and then pooling is performed on this weighted feature map as a weighted average pooling [27]. Attention map highlights discriminative regions in the feature maps by giving higher weights to them compared to the non-discriminative regions. Therefore, one can expect to get a discriminative pooled representation when pooling from attention weighted feature maps than pooling directly from the original feature maps.
Attention-based pooling [36, 41, 50, 67, 73, 89, 127] has received much focus recently. Different attention models differ from the way the attention maps are generated. For example, in Cross Convolutional Layer pooling [73], the feature vectors from the feature map of a particular layer are weighted by each of feature maps from its subsequent layer. In [67], a separate subnetwork is used to learn the attention maps. Double-attention network (-network) [13] uses a double attention mechanism, where the first attention step uses a second-order attention pooling to aggregate the features from the entire feature map, and the second attention step distributes the key features. Convolutional Block Attention Module (CBAM) [131] contains two attention mechanisms: channel attention module followed by spatial attention module, where the channel attention module aims to capture the inter-channel relationship of features; on the other hand, the spatial attention module aims to capture the inter-spatial relationship of features. Global Learnable Pooling (GLPool) [147] can also be considered as an attention mechanism, where the weight of each pooling location is considered as a parameter and learned together with other network parameters in an end-to-end manner. [36] mathematically shows that the attention weighted pooling is equivalent to a low-rank approximation of second-order pooling.
Attention mechanisms also have been investigated in medical imaging (Table 2); for example, a separate branch of the network was used to get the attention maps for Glaucoma detection in [67]. A reinforcement learning-based recurrent attention model for pulmonary lesion detection from chest X-Rays was proposed in [89]. An attention-guided CNN was proposed for thorax disease classification in [41], where the regions identified by the global branch are further analyzed by the local branch, and then the outputs from both branches are fused for the final classification.
Implicit pooling mechanisms
The Generalized Max pooling (GMP) [83] does not explicitly specify the pooling function, but it implicitly learns the ‘pooled’ representation using an optimization framework which equalizes the similarity between the local descriptors of an image () and their ‘pooled’ representation (), i.e.,
| 16 |
where denotes a N dimensional vector of all ones. By doing so, the ‘pooled’ representation will capture the properties of the max pooling for the bag-of-words-based hard-encoded local descriptors (binary representation). However, for other descriptors such as features from the last layer CNN, this ‘pooled’ representation can be affected by the frequent descriptors, and hence, may not be similar to max pooling. It is shown in [83] that this ‘pooled’ representation of an image is equivalent to weighted average of its local descriptors, i.e.,
| 17 |
where is the vector of weights.
Since GMP is an unsupervised representation learning, in Task-Driven Feature pooling [133] GMP was extended to supervised learning, where the ‘pooled’ representations are learned jointly with a classifier to maximize the classification accuracy and showed improved accuracy over the traditional max and average pooling with fixed feature representations. Deep Generalized Max pooling [20] integrates the idea of GMP in a deep learning framework.
Clustering-based aggregation schemes
Bag-of-words (BoW) [24] and its variants such as Vector of Locally Aggregated Descriptors (VLAD) [52] and Fisher Vectors (FV) [88] are well-known (non-CNN-based) feature encoding and aggregation techniques for order-less representation of handcrafted local descriptors, and have been widely used in Computer Vision [24, 52, 81, 88] and Medical Imaging [77, 114] community. In these approaches, first the local features from all the training images are clustered into a set of clusters and then the local features from each image falling inside each cluster are aggregated using different statistics; BoW uses count statistics, VLAD aggregates gradients and FV uses second-order statistical information in addition to the statistics used by BoW and VLAD. The aggregated statistics from each cluster are then concatenated as the final feature representation of an image.
Methods also proposed to integrate these approaches with CNN as feature aggregation techniques by either using them with features extracted from pre-trained CNN [21, 37, 138], or learning CNNs together with the parameters of BoW [80, 87], VLAD [2, 80, 140] and FV [80] in an end-to-end manner. A recent work [80] reports significant performance improvement by the learned aggregation schemes (BoW, VLAD and FV) over average pooling for video classification. To the best of our knowledge, these techniques are only used at the end of the network for feature aggregation.
Other approaches for feature aggregation
Various other approaches based on the max, average, and their variants also proposed for different reasons. For example, Transformation Invariant pooling (TI-pooling) [61] applies max pooling on the CNN features extracted from the transformed versions of an image to represent that image, so that the representation will capture transformation invariant features. The Hierarchical Mix-pooling [78] applies max pooling on the average pooled feature maps (and vice versa) to reduce information loss and shows improved performance than applying either max or average pooling alone on the Sparse Coding-based representations. Most of the pooling techniques for downsampling the feature maps are not invertible due to information loss, i.e., upsampling a downsampled feature map cannot recover the lost information in the downsampling. LiftPool [150] is a recently proposed pooling technique, which aims to build pooling layers that are invertible. Kernelized subspace pooling [128] was proposed to obtain a highly invariant description (invariant to flipping, rotation, etc.) from the CNN for image patch matching, where, the principal components of the feature maps from the last layer of the CNN are considered as the pooled output, and showed that descriptors obtained in this way are discriminative and highly invariant for image patch matching.
Convolutions also can be viewed as weighted average pooling, where the filters are learned in the training process. Tree pooling [63] learns a set of filters and ways to combine them. In [63], Tree pooling shows improved performance over max, average, mixed max-average (Sect. 2.2), and gated pooling (Sect. 2.2). However, Tree pooling contains more parameters to learn than mixed max-average and gated pooling. Strided Convolutions [105], on the other hand, are convolutions, use larger strides () for downsampling the feature maps. Unlike the traditional max and average pooling, where pooling is performed from each input feature map (channel) independently, in strided convolutions all the set of input feature channels are used to generate each output feature map/channel. Therefore, they need to learn many extra parameters. In Learning pooling (LEAP) [108] strided convolutions are applied independently from each channel of the feature maps to reduce the number of parameters required with strided convolutions. As discussed in Sect. 1, pooling makes the features robust to local transformation invariance. In contrary, strided convolutions capture local structures or positional information.
Experiment setup
In this section, we explain the datasets, evaluation criteria, network architecture, experimental settings, and the considered pooling strategies.
Datasets and evaluation criteria
We use the following two public medical image datasets for comparing different pooling strategies: (1) Human Epithelial type 2 (HEp-2) cells dataset3 and (2) Diabetic Retinopathy (DR) dataset.4 In the HEp-2 cells dataset, each cell covers the entire image as shown in Fig. 4. The lesions in the DR dataset (Fig. 5) covers only small parts of the images. In both datasets, the task is to classify each image into one of the predefined classes.
Fig. 5.
Example images from different classes of the DR image dataset
HEp-2 cells dataset
This dataset3 is from the I3A HEp-2 (Indirect Immunofluorescence Image Analysis—Human Epithelial Type-II) Cell and Specimen image classification competition organized by the International Conference on Pattern Recognition (ICPR), 2014. There were two tasks in this competition: Task 1 is to classify individual cell images into one of the six classes (Homogeneous, Speckled, Nucleolar membrane, Centromere, and Golgi), and the Task 2 is to classify specimen images into one of the seven classes (Homogeneous, Speckled, Nucleolar Membrane, Centromere, Golgi, and Mitotic Spindle). Each specimen image contains a large number of cell images of same type. In this work we focus on Task 1 - cell image classification. The training set of both tasks were released to the participants of the competition and the test sets were kept private by the organizers of the competition. As the training set of Task 1 dataset contains a smaller number (13, 596) of images and its test set is inaccessible, we used the Task 2 dataset to extract 26, 078 cell images as explained in [77]. We sample of these images from each class and use them as our training set, and use the rest of the images as the test set. When sampling, we make sure that the training data and the test data contain cell images from disjoint set of specimen images. The number of images in the training and test sets from each class of this dataset is given in Table 3.
Table 3.
HEp-2 cells image dataset
| Class | Training | Testing |
|---|---|---|
| Homogeneous | 3435 | 2363 |
| Speckled | 3498 | 2403 |
| Nucleolar | 3322 | 2253 |
| Centromere | 3339 | 2419 |
| Nuclear membrane | 1169 | 930 |
| Golgi | 571 | 396 |
| Total | 15,314 | 10,764 |
Note that all of these images are in gray scale, and the size of each image is approximately pixels. We resize each image into pixels of size . Images are normalized (zero mean and unit variance) before giving them to the CNN. Data augmentation, such as random mirroring, rotations (), and random cropping of size pixels were used at the training time. In the testing time, images were cropped at the center and no augmentation were used.
Mean Class Accuracy (MCA) was used as the evaluation measure, as it is the required metric by the competition. It is defined as:
| 18 |
where is the correct classification rate for class k, and is the number of classes.
Diabetic retinopathy dataset
This dataset is from the Kaggle Diabetic Retinopathy Detection challenge.5 It contains five classes from the scale of 0 to 4, which rates the presence of diabetic retinopathy (DR) in each image, where, 0—No DR, 1—Mild, 2—Moderate, 3—Severe, 4—Proliferative DR. The training set of this dataset contains 35, 126 images. To reduce the computational time required to run the experiments, we randomly sample 3, 500 images from the classes which contain over 3, 500 images, and fix this as the training set for all of our experiments. The images from the public leader–board were used as the test set. The number of images from the training and testing set from different classes is given in Table 4. Example images from each class are shown in Fig. 5.
Table 4.
DR image dataset
| Class | Training | Testing |
|---|---|---|
| No DR | 3500 | 8130 |
| Mild | 2443 | 720 |
| Moderate | 3500 | 1579 |
| Severe | 873 | 237 |
| Proliferative DR | 708 | 240 |
| Total | 11,024 | 10,906 |
Each image is preprocessed as explained in [40] as follows: First, the images were rescaled to have the same radius, then the local average color is subtracted from each channel and are mapped to of gray level (intensity value of 128).
In training, each image is first resized to pixels. Each channel of the image is normalized (zero mean and unit variance) before it is used by the CNN. Data augmentation, such as random mirroring, rotations (), and random cropping of size pixels, was used at the training time. In the testing time, images were cropped at the center, and no augmentation was used.
We used the Quadratic Weighted Kappa (QK) as the evaluation measure as it is used by the competition. QK measures the level of agreement between the predictions made by the system (A), and the annotator (B), and can be defined as
| 19 |
where W, O and E are matrices of size . O is the confusion matrix, each element in O, i.e., indicates how many times an image received the rating i by A, and rating j by B. The expected outcomes, E, is calculated as the outer product between the actual histogram vector of outcomes and the predicted histogram vector. E is normalized such that E and O have the same sum. The element, , of the weight matrix, W, is given as
| 20 |
Network architecture, initialization and training
For both datasets, we use a ResNet [46]-based CNN architecture. Table 5 illustrates the components of our selected CNN, which contains an input layer and three residual blocks. The input layer and each of the first two residual blocks are followed by transition layers to downsample the feature maps by half of its original sizes. Two approaches were considered for the transition layers. In the first case, a convolution with a stride of 2 is applied, and in the second case this convolution is replaced by a pooling operation. Global pooling is applied at the end of the network to get an image representation, which is then passed to a linear classification layer to get the classification scores. Note that as the images from the HEp-2 cells dataset are small in size (), a stride of one is used in the first convolutional layer for this dataset.
Table 5.
Network architecture used for both HEp-2 cells and the DR datasets
| Description | HEp-2 cells dataset | DR dataset |
|---|---|---|
| Convolution layer | Conv 33, 64, stride 1 | Conv 77, 64, stride 2 |
| Transition layer | Pooling/convolution | |
| Residual blocks | Conv | |
| Transition layer | Pooling/convolution | |
| Residual blocks | Conv | |
| Transition layer | Pooling/convolution | |
| Residual blocks | Conv | |
| Global pooling layer | Pooling | |
| Fully connected layer | FC–6 | FC–5 |
Conv indicates the convolutional layers, FC represents fully connected layer
The below settings were used unless otherwise specified.
HEp-2 cells dataset
For this dataset, the network was trained from scratch as we have a larger number of training images and the sizes of them are small. The initial learning rate was set to 0.01, which was then divided by a factor of 10 at the end of the and then at the end of the epochs, respectively. The total number of epochs were set to 80. We used weighted Cross-Entropy loss to handle imbalanced classes, where the weights are set to the inverse class frequencies. The network is optimized using Stochastic Gradient Decent (SGD) with the Nesterov momentum of 0.9 and a weight decay of . The batch size was set to 64.
Diabetic retinopathy dataset
For this dataset, the weights of the CNN were initialized with the weights of an ImageNet pre-trained model as recommended in [109]. The initial learning rate was set to 0.005, and it was divided by a factor of 10 at the end of the 90th and 120th epochs, while the total number of epochs was set to 130. Following [4], we directly use Quadratic Weighted Kappa as the loss function. The network was optimized using SGD with the Nesterov momentum of 0.9 and a weight decay . The batch size was set to 32. But for the bilinear pooling, we found that the above selected initial learning was quite large, and therefore, we set the learning rate to 0.001 and the batch size to 18 due to memory constraints.
Considered pooling techniques for comparison
As it is infeasible to experiment with all the pooling techniques proposed in the literature, we selected the following techniques for comparison: average and max pooling (Sect. 2.1), mixed max-average pooling [63] (Sect. 2.2), Generalized Mean (GM) pooling [135] (Sect. 2.3), improved bilinear pooling [70] (Sect. 2.6), stochastic pooling [142], S3-pooling [143], and max-pooling dropout [132] (Sect. 2.4). In addition, we considered two attention-based pooling (Sect. 2.8): Double-Attention Block (-block) [13] and Convolutional Block Attention Module (CBAM) [131]. Some of the pooling techniques such as clustering-based pooling (Sect. 2.10) and implicit pooling mechanisms (Sect. 2.9) need significant changes in the CNN architecture, and therefore, make it difficult to have a direct comparison with simple mechanisms such as max or average. Therefore, not considered for comparison in this work.
Results and discussion
All the reported experiments in this work are iterated ten times and the mean and the standard deviation of the MCA for HEp-2 cells and QK for the DR datasets over these iterations are reported as the evaluation measures. In addition, for each experiment, the independent samples t test was used to calculate the p values to get the statistical significance of the obtained results compared to the top performing method in that experiment.
Comparison of max, average, combination of max-average and soft pooling approaches
Table 6 compares the performance of max pooling, average pooling, mixed max-average pooling [63], and GM pooling [7]. Here, each of these pooling was applied to all the transition and the global pooling layers.
Table 6.
Comparison of different pooling approaches on the HEp-2 cells and DR datasets
| Pooling method | HEp-2 Cells | DR | ||
|---|---|---|---|---|
| MCA | p value | QK | p value | |
| Max | 0.0000 | 0.0004 | ||
| Average | – | 0.0199 | ||
| Mixed max-average | 0.0020 | 0.0258 | ||
| GM pooling | 0.0019 | – | ||
Here, pooling is applied to all the transition layers of the CNN
On the HEp-2 cells dataset, average pooling gives significantly better performance () than max pooling ( vs ). The performance of mixed max-average and GM pooling are in between the performance obtained by the average and max pooling. This result is expected as in most of the cases each image of the HEp-2 cells dataset contains exactly one cell and it covers almost all the image regions. Therefore, averaging will help to capture the property of each cell, and hence, gives better performance than max pooling.
On the other hand, GM pooling gives significantly improved performance () than the max and the average pooling on the DR dataset. This result is also expected, as the lesions in the DR datasets are small (do not cover the entire image). Max pooling may capture noisy features as it focus on the top activated elements of each feature map, and the average pooling averaging out all the activations, and therefore, the background features will dominate in the image representation. Therefore, GM pooling is a balance between max and average pooling—it considers not only the top activated element, but also other elements which have high activations.
Table 7 reports the effect of the value r in GM pooling. On the HEp-2 cells dataset larger values of r () lead to a significant drop () in performance. This aligns with the results obtained with the average and max pooling, as corresponding to average and corresponding to max pooling, respectively. On the DR dataset gives better QK values than others. Larger values of r () give a significant drop in performance () as they approximate max pooling, and hence, may capture noisy features. However, although gives the best kappa scores, we observed that smaller r values (i.e., ) give statistically similar () performance.
Table 7.
Effect of the value r in GM pooling
| r | HEp-2 cells (MCA) | DR (QK) |
|---|---|---|
| 2 | ||
| 3 | ||
| 5 | ||
| 7 |
Table 8 reports effect of the mixing proportion, a, for the mixed max-average pooling. Small value of a give better performance than large values.
Table 8.
Effect of mixing proportion a in mixed max-average pooling
| a | HEp-2 Cells (MCA) | DR (QK) |
|---|---|---|
| 0.2 | ||
| 0.4 | ||
| 0.6 | ||
| 0.8 |
Convolution versus pooling for downsampling the feature maps
As explained in Sect. 3.2, the transition layers for downsampling the featuremaps could be either pooling layers or convolutional layers. In Sect. 4.1, pooling was used for downsampling in all the transition layers. This section investigates the effect in performance when using convolutions for downsampling the feature maps instead of pooling. Table 9 reports the results.
Table 9.
Comparison of max, avg and GM pooling
| Pooling method | HEp-2 cells | DR | |||
|---|---|---|---|---|---|
| First transition layer | Global pooling layer | MCA | p value | QK | p value |
| Average | Average | – | 0.0080 | ||
| Max | Average | 0.0000 | 0.0918 | ||
| Conv | Average | 0.0061 | 0.0001 | ||
| Max | Max | 0.0000 | 0.0038 | ||
| Average | Max | 0.0000 | 0.0002 | ||
| Conv | Max | 0.0001 | 0.0001 | ||
| GM | GM | 0.0000 | – | ||
Here pooling is applied at the first transition layer and at the global pooling layer only. Convolution is applied for downsampling at other transition layers
The top scores are highlighed in bold
For the HEp-2 cells, dataset applying average pooling at the first transition layer gives significantly better performance () than applying max pooling. We believe that this is due to the size of the images. As the image sizes are small (), applying max pooling at the early stage of the network easily discards many valuable information, and leads to drop in performance. Here, applying average pooling at the first layer generally gives significantly better performance () regardless of the global pooling operation used. Applying average pooling at both the first transition layer and the global pooling layer leads to significantly better performance than any other combination ().
For the DR dataset, applying max or average pooling at the first transition layer gives similar performance when fixing the global pooling operation. But applying average pooling as the global pooling operator gives improved QK values than applying max pooling as the global pooling operator. Applying GM pooling on both the first transition and global pooling layers, on the other hand, gives the best QK values (the reason is discussed in Sect. 4.1) compared to most of the considered combinations (). However, this (GM pooling) gives statistically similar performance () compared to applying max and average pooling at the first and the global pooling layers respectively.
When comparing Tables 6 and 9, applying convolution at the intermediate transition layers for downsampling the feature maps or applying pooling for downsampling give similar performance () on both datasets.
Can higher-order information help? Improved bilinear pooling as the global pooling operator
This section investigates whether higher-order information give improved performance than other pooling approaches considered in Sects. 4.1 and 4.2. Here, we used convolution as the downsampling operation in all the transition layers except the first one. The improved bilinear pooling [70] was used to capture higher-order statistical information between feature channels at the global pooling layer.
Table 10 reports the results. On both datasets bilinear pooling does not show significant improvement in performance than applying average pooling as the global pooling operator. We conduct another experiment to investigate whether the higher-order information obtained by the bilinear pooling can add complementary information to the feature representation obtained by other pooling approaches, e.g., global average pooling. The results in Table 10 does not show any considerable improvement () when combining bilinear pooling with global average pooling.
Table 10.
Bilinear pooling as the global pooling operator
| Pooling method | HEp-2 Cells | DR | |||
|---|---|---|---|---|---|
| First transition layer | Global pooling layer | MCA | p value | QK | p value |
| Max | Bilinear | 0.0000 | 0.6932 | ||
| Average | Bilinear | 0.7421 | 0.0000 | ||
| Max | Average + bilinear | 0.0000 | 0.1813 | ||
| Average | Average + bilinear | – | 0.0002 | ||
| Average | Average | 0.1625 | – | ||
Convolution is applied at all the transition layers except the first one
The top scores are highlighed in bold
Experiments with stochastic pooling
The CNN trained on the HEp-2 cell images dataset may overfit to the training data as we got a high training MCA (). In this experiment, we investigate different stochastic pooling approaches to reduce overfitting, such as stochastic pooling [142], max pooling dropout [132], and S3 pooling [143]. Here, we applied stochastic pooling/dropout only at the global pooling layer.
As expected, stochastic pooling and max pooling dropout give lower MCA (Table 11), as they are based on max pooling. Remember that stochastic pooling selects an (only one) activation from each pooling region based on the multinomial distribution given by the values inside it, and the max-pooling dropout randomly drop (in our experiments we set ) of the elements (make them equal to zero) and then apply max pooling on this new pooling region. We also considered average-pooling dropout, where we randomly drop of the elements (make them equal to zero) and then apply average pooling instead of max pooling, which gives improvement compared to max pooling dropout.
Table 11.
Effect of stochastic pooling: convolution is applied for downsampling transition layers except in the first one, where average pooling is used
| Global pooling layer | HEp-2 cells | |
|---|---|---|
| MCA | p value | |
| Stochastic pooling [142] | 0.0200 | |
| S3 pooling [143] | 0.0003 | |
| Max pooling dropout [132] | 0.0009 | |
| Average pooling dropout [132] | – | |
| Global average pooling with no stochasticity/dropout | 0.4585 | |
The top scores are highlighed in bold
S3 pooling gives the lowest MCA compared to all the approaches we considered. We found that none of these stochastic pooling approaches give significant improvements compared to our baseline—average pooling without any stochastic pooling/dropout ( from Table 9).
Experiments with attention weighted pooling
Here, we experiment with two different attention mechanisms (explained under the Sect. 2.8): Double Attention (-block) [13] and Convolutional Block Attention Module (CBAM) [131]. These blocks were added before the global pooling layer. Table 12 reports the results of these attention weighted blocks with different pooling operations applied at the first and the last pooling layers. Results show significant performance improvement () compared to the experiments which do not have attention layers. Both attention mechanisms gives similar performance regardless of the first and the last pooling operations.
Table 12.
Effect of attention weighted blocks with different pooling operations at the first and the last transition layers
| Method | DR (QK) | p value | ||
|---|---|---|---|---|
| First transition layer | Attention | Global pooling layer | ||
| Max | – | Average | 0.0006 | |
| Max | CBAM | Average | 0.4051 | |
| Max | Average | – | ||
| GM | – | GM | 0.0219 | |
| GM | CBAM | GM | 0.0961 | |
| GM | GM | – | ||
The top scores are highlighed in bold
Comparison with the state-of-the-art
Note that the focus of this work is to compare different pooling mechanisms to find out which one is better under some scenarios, and we are not particularly focused on building a state-of-the-art system. However, based on the findings from our previous experiments reported in this paper, in this section, we compare our results with the state-of-the-art and show that on both datasets our approach leads to new state-of-the-art results.
DR dataset
Most of the existing work [1, 40, 56, 93] for DR image analysis focus on building custom CNN architectures. For example, multiple filter sizes and different color spaces were explored for fine-grained classification of DR lesions in [116]. Attention-based mechanisms [127] were also explored. A recent work [85] analyses different loss functions for optimizing Kappa as the evaluation measure for DR image analysis.
As explained in Sect. 3.1.2 all the experiments on the DR dataset reported previously in this paper are based on a subset of the entire training set. To compare with the state-of-the-art, in this section we use all the images from the training set (35, 126 images) to train the CNN, and test it separately on the validation (10, 906 images) and the test (42, 670 images) sets respectively. Table 13 reports the results. Our result beats the state-of-the-art methods, and establishes a new state-of-the-art.
Table 13.
Comparison of our approach with the state-of-the-art methods on the DR dataset with different evaluation measures (QK, accuracy, and weighted F1 score)
| Method | Validation | Testing | ||||
|---|---|---|---|---|---|---|
| QK | Accuracy | F1-score | QK | Accuracy | F1-score | |
| Single models | ||||||
| MobileNet-Dense [22] | – | – | – | 0.825 | – | – |
| MobileNetV2 [22] | – | – | – | 0.822 | – | – |
| M-Net [127] | 0.832 | – | – | 0.825 | – | – |
| Ours: max-avg | 0.858 | 84.25 | 0.844 | 0.849 | 83.17 | 0.833 |
| Ours: GM-GM | 0.852 | 83.60 | 0.841 | 0.850 | 82.63 | 0.831 |
| Ours: max-A-avg | 0.854 | 83.83 | 0.841 | 0.851 | 82.77 | 0.831 |
| Ours: GM-A-GM | 0.850 | 83.93 | 0.842 | 0.847 | 82.80 | 0.831 |
| Ensemble | ||||||
| Model ensemble [22] | – | – | – | 0.852 | – | – |
| Min-pooling [40] | 0.860 | – | – | 0.849 | – | – |
| Zoom-in-Net [127] | 0.865 | – | – | 0.854 | – | – |
| o_O [1] | 0.854 | – | – | 0.844 | – | – |
| Reformed gamblers [56] | 0.851 | – | – | 0.839 | – | – |
| Ours | 83.37 | 0.840 | 82.34 | 0.830 | ||
The top scores are highlighed in bold
In this experiment, we select four different pooling settings from the previous experiment (Table 12) and train four separate ResNet-18 models based on each of these pooling settings. The pooling settings considered here are: (1) max-avg: max and the average pooling are applied to the first and the global pooling layers respectively, (2) GM-GM: GM pooling is applied at the first and the global pooling layers, (3) max-A-avg: max and the average pooling are applied to the first and the global pooling layers, respectively, and A attention layer is added before the global pooling layer, (4) GM-A-GM: GM pooling is applied to the first and the global pooling layers, and A attention layer is added before the global pooling layer. As most of the state-of-the-art methods (e.g., [40, 127]) make use of the features from both eyes (left and right, as they have high correlation) for the classification of a particular eye, we also combine the features from both eyes before pass it to the classification layer of each model. To make it consistent with other state-of-the-art methods, we use mean squared error as our loss function. Adam optimizer with an initial learning rate of was used to optimize the network parameters. The number of epochs and the batch size was set to 60 and 16, respectively.
From Table 13, we can observe that all the different pooling settings (our single models) give similar QK values compared to each other, and compared to the state-of-the-art methods. We believe that this is because the results are almost saturated at a QK value of . We can also observe that the ensemble of our four models improves the overall QK values and leads to the state-of-the-art results on both the validation and the test sets. Note that compared to Zoom-in-net [127], our method is not only simple, but also make use of a standard network architecture (ResNet-18) with different pooling mechanisms.
HEp-2 cells dataset
A significant amount of work has been done for HEp-2 cell image classification, and can be categorized into handcrafted features-based approaches, and deep learning-based approaches. Various handcrafted features such as multi-resolution local patterns [77], shape index histograms [62], gray-level histogram statistics [48], co-occurrence matrix features [48], Local Binary Patterns [47], and SIFT [47, 77] features have been explored. Recently, CNN [33, 65]-based approaches also became popular for HEp-2 cell image classification.
In this literature of HEp-2 cell image classification, different methods use different test sets for comparison as the test set of this dataset is not publicly available (explained in Sect. 3.1.1). Some methods completely discard the specimen information when constructing the test set. It is observed in [77] that when the specimen information is discarded, a very high MCA () can be easily obtained even with handcrafted features. As explained in Sect. 3.1.1, we considered specimen information when splitting the dataset, and compare our method with the methods which also consider specimen information when constructing the test set.
Table 14 compares our results with the state-of-the-art results on the HEp-2 cells image dataset, and show that our results are the new state-of-the-art. We can observe from Table 14 that our method beats other methods with a ResNet architecture with carefully chosen pooling layers. It also can be noted that we achieve the new state-of-the-art results with a small amount of training data (15, 314 images) compared to other methods, for example, the work of [65] uses a training set which contains over 100, 000 images.
Table 14.
Comparison with the state-of-the-art methods on the HEp-2 cells dataset
| Method | MCA | Accuracy | F1 score |
|---|---|---|---|
| LeNet-based CNN [33] | 71.88 | – | – |
| Deep CNN [65] | 74.67 | – | – |
| Shape index histograms with donut-shaped spatial pooling [62] | 78.70 | – | – |
| Multi-resolution patterns with ensemble SVMs [77] | 87.10 | – | – |
| Ours | 87.95 | 0.88 |
The top scores are highlighed in bold
Discussion
The following section summarizes the work of this paper based on the pooling techniques reviewed and the findings of the experiments.
As discussed, pooling can help to learn invariant features, reduces overfitting, and reduces computational complexity by downsampling the feature maps. There are two types of pooling operations used in CNNs, they are: local pooling and global pooling. Local pooling is applied from small image regions (e.g., ) at the early stages of the CNNs to capture local features, and the global pooling is applied at the end of the network from the entire feature map to get a feature representation, which will be then used by the fully connected layer for classification.
The max and the average pooling are the widely used pooling techniques. They are used both in local and the global pooling layers, and their applicability depends on the application. Max pooling considers only the mostly activated elements in each feature map, and discards all the other activations as irrelevant. This activated element could be a noisy one. Our experiments suggest that max pooling is appropriate in situations where the class specific features (e.g., abnormal regions in medical images) are smaller in size compared to the image size. In the learning stage of the network, the network nodes which are connected only to this maximum activated element will be updated, which makes the learning of the network slow. Usually maximum pooling is applied at the early stages of the network to capture the important local image features. This is appropriate when the size of the images are large enough. However, our experiments show that when the size of the images are small, applying max pooling at the early stages of the network leads to information loss, and hence, drop in classification performance compared to applying average pooling at the early layers. The average pooling, on the other hand, gives equal weights to all the activations, regardless of their importance. Therefore, the class specific information in the feature maps could be downgraded and the features correspond to the background could dominate in the pooled representation. Usually average pooling is used as the global pooling operator to capture the contribution of all the features (e.g., Resnet [46]). In addition, the network may converge faster as all the network nodes are updated in the learning stage. Our experiments also prove that applying average pooling is a better choice than max pooling as the the global pooling operator.
As discussed, the max or the average pooling cannot be applicable in all the scenarios. Each of them have their own merits and demerits. To overcome this, and to take the advantage of both, mix max-average pooling (Sect. 2.2) and the soft pooling techniques are proposed (Sect. 2.3). The max and the average pooling are combined with weights in the mix max-average pooling. In soft pooling, the pooled representation is obtained as the weighted sum of the local features. In this way, all the elements in a feature map will contribute to the pooled representation, and their contributions are determined based on their activation values - larger weights for high activations, and the lower weights on the other hand. There are various ways proposed to determine these weights (refer Sect. 2.3). Rank-based pooling (explained in Sect. 2.3) also can be considered as softpooling techniques, but they differ in the way how the local features are weighted in the pooled representations. In rank-based pooling, top k activations receive a weight of one, and the others receive zero weights. However, unlike the max and the average pooling, these approaches (mix max-average, soft pooling and the rank based pooling) introduce new free parameters which need to be selected carefully for improved classification performance. We show by experiments that these approaches are applicable in situations where the class specific features are small in size compared to the size of the images. In this scenarios, softpooling gives improved performance compared to max and average pooling. In addition, we also show by experiments that when the class specific features spread all over the images (e.g., each abnormal image in the medical domain contains more abnormal regions than the normal ones), the average pooling is the better choice.
One of the main problem with training CNNs is overfitting, particularly when the CNN is trained with small amount of data. To reduce overfitting, various pooling techniques are investigated, which tries to apply some stochasticity in the pooling process (These techniques are explained in Sect. 2.4). Mainly, two types of stochasticity are generally used. The first type is focusing on the pooling stage itself, and the second type is focusing on the spatial sampling stage of the pooling. For example, the Stochastic pooling [142] introduces randomness in the pooling stage of the network training by randomly selecting an activation within each pooling region according to a multinomial distribution given by the values within that pooling region. In S3 pooling [143] and Fractional Max Pooling [39] randomness is applied at the spatial sampling stage of the pooling. Note that, dropout [106] is another way to reduce overfitting by randomly dropping some network nodes at the training stage, and it is a computationally efficient approach than most of the above mentioned approaches for reducing overfitting.
The global pooling operation helps to get an orderless representation of the local features. This orderless representation is very useful to capture discriminative features regardless of where they appear in images. However, for some classification problems this orderless representation may fail to capture some very important information as it completely discards the location information of the local features. In natural images, the sky is always in the upper part of the images. Similarly, for some medical image classification problems such as, classifying cell images, the location information may be useful. The Golgi class (refer Fig. 4) has a ring-like structure, capturing this information is very useful to discriminate the Golgi class from the others. The orderless representation may fail to capture such information. To capture this local structure information there are various approaches such as Spatial Pyramid Pooling [45], Cell Pyramid Pooling [130], etc. are proposed. These approaches are discussed in detail in Sect. 2.5.
It has been reported in the literature that for fine-grained image classification capturing the interaction between different features is very much useful to improve the classification performance [71]. The widely used average pooling uses a first order statistics, i.e., mean, to aggregate the features. This statistics is not designed to capture the interaction between different local features, such as object co-occurrence, and may not be useful for find-grained image classification. Higher-oder statistical pooling techniques such as bilinear pooling [31, 70, 71] are proposed for this purpose, and reportedly show improved performance over max and the average pooling for fine-grained image classification. We discuss these techniques in detail in Sect. 2.6. However, our experiments hardly show any improvement by bilinear pooling compared to the traditional max and the average pooling as the global pooling operators.
All the features in a particular feature map cannot be considered equally. Some feature are important than others. Attention based pooling (discussed in Sect. 2.8) recently received much attention. These approaches weights the importance of the local image features by the use of attention maps generated in the training process. In the attention maps, the ‘important’ regions receive higher weights than the ‘unimportant’ regions. The pooled representation is then obtained as the attention weighted aggregation of the local features. Our experiments on the DR dataset shows improved performance when applying attention based pooling compared to without using them.
Most of the pooling approaches discussed above (including, max, average, mixed max-average [63]) only consider the statistics of the features that are inside the considered pooling region of a particular feature map when applying pooling. Here, pooling regions in each feature map are considered independently from each other. However, some approaches also consider the statistics of the entire feature map (e.g., Global Feature Guided Local pooling [57]), or some statistics from the adjacent pooling regions of the considered pooling region (e.g., Dynamic Correlation pooling [11]) to calculate the output or to determine the type of pooling to be applied on the considered pooling region. Usually, pooling is performed separately from each feature map (or channel), and therefore, the number of channels in the pooled representation is same as the number of input channels. Examples of such approaches include, the average and max pooling, linear combination of them, soft pooling, and stochastic pooling. However, different channels of the same set of feature maps are highly correlated and should be treated jointly [83]. Second-order pooling (Sect. 2.6), implicit pooling mechanisms (Sect. 2.9), clustering-based aggregation schemes (Sect. 2.10), and strided convolutions (Sect. 2.11) do not consider feature channels independently. Therefore, the number of channels in the output is not necessarily the same as the number of channels in the input feature maps.For example, in strided convolution [105] the number of channels in the output feature maps is equivalent to the number of convolutions used. To take the advantage of different types of pooling mechanisms, their combinations were also considered, e.g., in [96] DPP is combined with S3 pooling to retain the important information in the feature maps and at the same time learning representations which are less prune to overfitting.
It should be noted that there is no single pooling technique which can work in all the scenarios, and the selection of it usually depends on the characteristics of the application. One of the limitation of our study is only two datasets were considered, but note that they are different from each other in terms of their modalities and characteristics.
Conclusion
In this paper, we reviewed different kinds of pooling techniques proposed in the literature of computer vision, together with the medical imaging domains where these techniques are used (refer Table 2). The advantages, disadvantages and their applicability in different scenarios are discussed in detail. In addition, a comprehensive set of experiments are conducted on a selected set of pooling techniques tested on two public medical image datasets.
Our experimental results suggest that the pooling technique for a particular classification task should be selected by considering the scale of the class specific features that appear in the images. We found that global average pooling generally gives better results than global max pooling. In addition, applying max pooling at the earlier stages of the network, particularly for the dataset with smaller sized images may lead to drop in performance due to information loss by max pooling. Higher-order statistics in terms of bilinear pooling to capture the interaction between different feature channels do not seem to provide significant improvement compared to some simple approaches such as max and average pooling on the two datasets we considered. Adding attention layers improve the classification performance compared to a system where no attention layers are used.
We believe that this review and the comparative study will provide a guideline to the choice of pooling mechanisms for various medical image analysis tasks.
Acknowledgements
NR was partially supported by the NSF Sri Lanka grant NSF/RPHS/2016/D02. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Lets assume that , where is the input, y is the output, and f represents the pooling function. If f is a linear function, this pooling is a linear pooling; otherwise, it is a nonlinear pooling. For example, max is a nonlinear pooling as it is a nonlinear function.
In [57] position priors are also added to determine the value of the weights. Here, in Eq. (9), we omit those priors to make it simple.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rajendran Nirthika and Siyamalan Manivannan have contributed equally to this work.
References
- 1.Antony M, Brggemann S (2015) Kaggle diabetic retinopathy detection: team o\_O solution. Technical report
- 2.Arandjelovic R, Gronat P, Torii A, Pajdla T, Sivic J (2016) NetVLAD: CNN architecture for weakly supervised place recognition. In: IEEE conference on computer vision and pattern recognition, pp 5297–5307 [DOI] [PubMed]
- 3.B Z, Crawford R, Dogdas B, Goldmacher G, Chen A (2019) A progressively-trained scale-invariant and boundary-aware deep neural network for the automatic 3D segmentation of lung lesions. In: IEEE winter conference on applications of computer vision, pp 1–10
- 4.Beckham C, Pal C (2016) A simple squared-error reformulation for ordinal classification. abs/1612.00775
- 5.Bieder F, Sandkühler R, Cattin PC (2021) Comparison of methods generalizing max-and average-pooling. arXiv:2103.01746
- 6.Boureau YL, Ponce J, LeCun Y (2010) A theoretical analysis of feature pooling in visual recognition. In: 27th international conference on machine learning, pp 111–118
- 7.Bruna J, Szlam A, Lecun Y (2014) Signal recovery from pooling representations. In: 31st international conference on machine learning, pp 1585–1598
- 8.Carbonneau MA, Cheplygina V, Granger E, Gagnon G. Multiple instance learning: a survey of problem characteristics and applications. Pattern Recognit. 2018;77:329–353. [Google Scholar]
- 9.Carreira J, Caseiro R, Batista J, Sminchisescu C (2012) Semantic segmentation with second-order pooling. In: European conference on computer vision. Springer, pp 430–443
- 10.Chang J, Zhang L, Gu N, Zhang X, Ye M, Yin R, Meng Q. A mix-pooling CNN architecture with FCRF for brain tumor segmentation. J Vis Commun Image Represent. 2019;58:316–322. [Google Scholar]
- 11.Chen J, Hua Z, Wang J, Cheng S (2017) A convolutional neural network with dynamic correlation pooling. In: International conference on computational intelligence and security, pp 496–499
- 12.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2017;40(4):834–848. doi: 10.1109/TPAMI.2017.2699184. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Kalantidis Y, Li J, Yan S, Feng J (2018) -Nets: double attention networks. In: Advances in neural information processing systems, vol 31
- 14.Chen Y, Ma G, Yuan C, Li B, Zhang H, Wang F, Hu W. Graph convolutional network with structure pooling and joint-wise channel attention for action recognition. Pattern Recogn. 2020;103:107321. [Google Scholar]
- 15.Chen Z, Lin J, Chandrasekhar V, Duan LY (2018) Gated square-root pooling for image instance retrieval. In: 25th IEEE international conference on image processing, pp 1982–1986
- 16.Chen Z, Zhang J, Ding R, Marculescu D (2020) ViP: virtual pooling for accelerating CNN-based image classification and object detection. In: IEEE/CVF winter conference on applications of computer vision, pp 1180–1189
- 17.Cheng J, Yang W, Huang M, Huang W, Jiang J, Zhou Y, Yang R, Zhao J, Feng Y, Feng Q, et al. Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. Public Library Sci One. 2016;11(6):e0157112. doi: 10.1371/journal.pone.0157112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherian A, Gould S. Second-order temporal pooling for action recognition. Int J Comput Vis. 2019;127(4):340–362. [Google Scholar]
- 19.Cherian A, Koniusz P, Gould S (2017) Higher-order pooling of CNN features via kernel linearization for action recognition. In: IEEE winter conference on applications of computer vision, pp 130–138
- 20.Christlein V, Spranger L, Seuret M, Nicolaou A, Král P, Maier A (2019) Deep generalized max pooling. In: International conference on document analysis and recognition, pp 1090–1096
- 21.Cimpoi M, Maji S, Vedaldi A (2015) Deep filter banks for texture recognition and segmentation. In: IEEE conference on computer vision and pattern recognition, pp 3828–3836 [DOI] [PMC free article] [PubMed]
- 22.Gao J, Leung C, Miao C (2019) Diabetic retinopathy classification using an efficient convolutional neural network. In: IEEE international conference on agents, pp 80–85
- 23.Cordts M, Omran M, Ramos S, Rehfeld T, Enzweiler M, Benenson R, Franke U, Roth S, Schiele B (2016) The cityscapes dataset for semantic urban scene understanding. In: IEEE conference on computer vision and pattern recognition, pp 3213–3223
- 24.Csurka G, Dance C, Fan L, Willamowski J, Bray C (2004) Visual categorization with bags of keypoints. In: Workshop on statistical learning in computer vision, European conference on computer vision, vol 1. Prague, pp 1–2
- 25.Cui Y, Zhou F, Wang J, Liu X, Lin Y, Belongie S (2017) Kernel pooling for convolutional neural networks. In: IEEE conference on computer vision and pattern recognition, pp 3049–3058
- 26.Czaja W, Li W, Li Y, Pekala M (2021) Maximal function pooling with applications. arXiv:2103.01292
- 27.De Campos T, Csurka G, Perronnin F. Images as sets of locally weighted features. Comput Vis Image Underst. 2012;116(1):68–85. [Google Scholar]
- 28.Eom H, Choi H (2018) Alpha-pooling for convolutional neural networks. arXiv:1811.03436
- 29.Everingham M, Van Gool L, Williams CK, Winn J, Zisserman A. The pascal visual object classes (voc) challenge. Int J Comput Vis. 2010;88(2):303–338. [Google Scholar]
- 30.Feng J, Ni B, Tian Q, Yan S (2011) Geometric lp-norm feature pooling for image classification. In: IEEE conference on computer vision and pattern recognition, pp 2609–2704
- 31.Gao Y, Beijbom O, Zhang N, Darrell T (2016) Compact bilinear pooling. In: IEEE conference on computer vision and pattern recognition, pp 317–326
- 32.Gao Z, Wang L, Wu G (2019) LIP: Local importance-based pooling. In: IEEE international conference on computer vision, pp 3355–3364
- 33.Gao Z, Wang L, Zhou L, Zhang J. HEp-2 cell image classification with deep convolutional neural networks. IEEE J Biomed Health Inform. 2016;21(2):416–428. doi: 10.1109/JBHI.2016.2526603. [DOI] [PubMed] [Google Scholar]
- 34.Gao Z, Xie J, Wang Q, Li P (2019) Global second-order pooling convolutional networks. In: IEEE conference on computer vision and pattern recognition, pp 3024–3033
- 35.Geng L, Wang J, Xiao Z, Tong J, Zhang F, Wu J. Encoder–decoder with dense dilated spatial pyramid pooling for prostate MR images segmentation. Comput Assist Surv. 2019;24(sup2):13–19. doi: 10.1080/24699322.2019.1649069. [DOI] [PubMed] [Google Scholar]
- 36.Girdhar R, Ramanan D (2017) Attentional pooling for action recognition. In: Advances in neural information processing systems, vol 30. pp 34–45
- 37.Gong Y, Wang L, Guo R, Lazebnik S (2014) Multi-scale orderless pooling of deep convolutional activation features. In: European conference in computer vision, vol. 8695. pp 392–407
- 38.Gopinath K, Desrosiers C, Lombaert H (2019) Learnable pooling in graph convolution networks for brain surface analysis. arXiv:1911.10129 [DOI] [PubMed]
- 39.Graham B (2014) Fractional max-pooling. arXiv:1412.6071
- 40.Graham B (2015) Kaggle diabetic retinopathy detection competition report. Technical report
- 41.Guan Q, Huang Y, Zhong Z, Zheng Z, Zheng L, Yang Y (2018) Diagnose like a Radiologist: attention guided convolutional neural network for thorax disease classification. abs/1801.09927
- 42.Gulcehre C, Cho K, Pascanu R, Bengio Y (2014) Learned-norm pooling for deep feedforward and recurrent neural networks. In: Joint European conference on machine learning and knowledge discovery in databases. Springer, pp 530–546
- 43.Han XH, Lei J, Chen YW (2016) HEp-2 cell classification using K-support spatial pooling in deep CNNs. In: Deep learning and data labeling for medical applications, pp 3–11
- 44.He A, Li T, Li N, Wang K, Fu H. CABNet: category attention block for imbalanced diabetic retinopathy grading. IEEE Trans Med Imaging. 2021;40(1):143:153. doi: 10.1109/TMI.2020.3023463. [DOI] [PubMed] [Google Scholar]
- 45.He K, Zhang X, Ren S, Sun J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2015;37(9):1904–1916. doi: 10.1109/TPAMI.2015.2389824. [DOI] [PubMed] [Google Scholar]
- 46.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition, pp 770–778
- 47.Hobson P, Percannella G, Vento M, Wiliem A (2013) International competition on cells classification by fluorescent image analysis. Technical report, international conference on image processing
- 48.Hsieh TY, Huang YC, Chung CW, Huang YL (2009) HEp-2 cell classification in indirect immunofluorescence images. In: 7th International conference on information, communications and signal processing, pp 1–4
- 49.Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: IEEE conference on computer vision and pattern recognition, pp 7132–7141
- 50.Hu Y, Wang B, Lin S (2017) Fc4: fully convolutional color constancy with confidence-weighted pooling. In: IEEE conference on computer vision and pattern recognition, pp 4085–4094
- 51.Huang G, Liu Z, Weinberger KQ (2017) Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition, pp 2261–2269
- 52.Jégou H, Douze M, Schmid C, Pérez P (2010) Aggregating local descriptors into a compact image representation. In: IEEE computer society conference on computer vision and pattern recognition, pp 3304–3311
- 53.Jianchao Y, Yu K, Gong Y, Huang T (2009) Linear spatial pyramid matching using sparse coding for image classification. In: IEEE conference on computer vision and pattern recognition, pp 1794–1801
- 54.Jie HJ, Wanda P. RunPool: a dynamic pooling layer for convolution neural network. Int J Comput Intell Syst. 2020;13(1):66–76. [Google Scholar]
- 55.Jiménez-Sánchez A, Kazi A, Albarqouni S, Kirchhoff S, Sträter A, Biberthaler P, Mateus D, Navab N (2018) Weakly-supervised localization and classification of proximal femur fractures. arXiv:1809.10692 [DOI] [PubMed]
- 56.John Dunavent JX, Dunavent RK (2015) Kaggle diabetic retinopathy detection: 3rd place solution report. Technical report
- 57.Kobayashi T (2019) Global feature guided local pooling. In: IEEE international conference on computer vision, pp 3365–3374
- 58.Krizhevsky A (2009) Learning multiple layers of features from tiny images. Technical report, University of Toronto
- 59.Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: advances in neural information processing systems, vol 25. pp 1097–1105
- 60.Kumar A (2018) Ordinal pooling networks: for preserving information over shrinking feature maps. abs/1804.02702
- 61.Laptev D, Savinov N, Buhmann JM, Pollefeys M (2016) TI-POOLING: transformation-invariant pooling for feature learning in convolutional neural networks. In: IEEE conference on computer vision and pattern recognition, pp 289–297
- 62.Larsen ABL, Vestergaard JS, Larsen R. HEp-2 cell classification using shape index histograms with donut-shaped spatial pooling. IEEE Trans Med Imaging. 2014;33(7):1573–1580. doi: 10.1109/TMI.2014.2318434. [DOI] [PubMed] [Google Scholar]
- 63.Lee CY, Gallagher PW, Tu Z (2016) Generalizing pooling functions in convolutional neural networks: Mixed, gated, and tree. In: Artificial intelligence and statistics, pp 464–472
- 64.Lee D, Lee S, Yu H (2021) Learnable dynamic temporal pooling for time series classification. arXiv:2104.02577
- 65.Li H, Zheng WS, Zhang J (2016) Deep CNNs for HEp-2 cells classification: a cross-specimen analysis. CoRR abs/1604.05816
- 66.Li L, Xie J, Li P, Zhang L (2021) Detachable second-order pooling: Toward high-performance first-order networks. IEEE Trans Neural Netw Learn Syst, 1–15. 10.1109/TNNLS.2021.3052829 [DOI] [PubMed]
- 67.Li L, Xu M, Liu H, Li Y, Wang X, Jiang L, Wang Z, Fan X, Wang N. A large-scale database and a CNN model for attention-based glaucoma detection. IEEE Trans Med Imaging. 2020;39(2):413–424. doi: 10.1109/TMI.2019.2927226. [DOI] [PubMed] [Google Scholar]
- 68.Li P, Xie J, Wang Q, Gao Z (2018) Towards faster training of global covariance pooling networks by iterative matrix square root normalization. In: IEEE conference on computer vision and pattern recognition, pp 947–955
- 69.Li P, Xie J, Wang Q, Zuo W (2017) Is second-order information helpful for large-scale visual recognition? In: IEEE international conference on computer vision
- 70.Lin TY, Maji S (2017) Improved bilinear pooling with CNNs. arXiv:1707.06772
- 71.Lin TY, RoyChowdhury A, Maji S (2015) Bilinear CNNs for fine-grained visual recognition. arXiv:1504.07889 [DOI] [PubMed]
- 72.Lowe DG. Object recognition from local scale-invariant features. IEEE Int Conf Comput Vision. 1999;2:1150–1157. [Google Scholar]
- 73.Liu L, Shen C, Hengel A. Cross-convolutional-layer pooling for image recognition. IEEE Trans Pattern Ana Mach Intell. 2017;39(11):2305–2313. doi: 10.1109/TPAMI.2016.2637921. [DOI] [PubMed] [Google Scholar]
- 74.Liu N, Jian S, Li D, Zhang Y, Lai Z, Xu H (2021) Hierarchical adaptive pooling by capturing high-order dependency for graph representation learning. arXiv:2104.05960
- 75.Liu Y, Zhang YM, Zhang XY, Liu CL. Adaptive spatial pooling for image classification. Pattern Recognit. 2016;55:58–67. [Google Scholar]
- 76.Manivannan S, Cobb C, Burgess S, Trucco E. Subcategory classifiers for multiple-instance learning and its application to retinal nerve fiber layer visibility classification. IEEE Trans Med Imaging. 2017;36(5):1140–1150. doi: 10.1109/TMI.2017.2653623. [DOI] [PubMed] [Google Scholar]
- 77.Manivannan S, Li W, Akbar S, Wang R, Zhang J, McKenna SJ. An automated pattern recognition system for classifying indirect immunofluorescence images of HEp-2 cells and specimens. Pattern Recognit. 2016;51:12–26. [Google Scholar]
- 78.Manivannan S, Wang R, Trucco E (2016) Hierarchical mix-pooling and its applications to biomedical image classification. In: IEEE 13th international symposium on biomedical imaging, pp 541–544
- 79.Men K, Boimel P, Janopaul-Naylor J, Zhong H, Huang M, Geng H, Cheng C, Fan Y, Plastaras JP, Ben-Josef E, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol. 2018;63(18):185016. doi: 10.1088/1361-6560/aada6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miech A, Laptev I, Sivic J (2017) Learnable pooling with Context Gating for video classification. ArXiv abs/1706.06905
- 81.Mohedano E, McGuinness K, O’Connor NE, Salvador A, Marques F, Giro-i Nieto X (2016) Bags of local convolutional features for scalable instance search. In: ACM on international conference on multimedia retrieval, pp 327–331
- 82.Momeny M, Jahanbakhshi A, Jafarnezhad K, Zhang YD. Accurate classification of cherry fruit using deep CNN based on hybrid pooling approach. Postharvest Biol Technol. 2020;166:111204. [Google Scholar]
- 83.Murray N, Perronnin F (2014) Generalized max pooling. In: IEEE conference on computer vision and pattern recognition, pp 2473–2480
- 84.Navaneeth B, Suchetha M. A dynamic pooling based convolutional neural network approach to detect chronic kidney disease. Biomed Signal Process Control. 2020;62:102068. [Google Scholar]
- 85.Nirthika R, Manivannan S, Ramanan A (2020) Loss functions for optimizing kappa as the evaluation measure for classifying diabetic retinopathy and prostate cancer images. In: IEEE 15th international conference on industrial and information systems, pp 144–149
- 86.Ogusu R, Yamanaka T (2019) LPM: learnable pooling module for efficient full-face gaze estimation. In: 14th IEEE international conference on automatic face and gesture recognition, pp 1–5
- 87.Passalis N, Tefas A (2017) Learning bag-of-features pooling for deep convolutional neural networks. In: IEEE international conference on computer vision, pp 5766–5774
- 88.Perronnin F, Dance C (2007) Fisher kernels on visual vocabularies for image categorization. In: 2007 IEEE conference on computer vision and pattern recognition, pp. 1–8
- 89.Pesce E, Withey SJ, Ypsilantis PP, Bakewell R, Goh V, Montana G. Learning to detect chest radiographs containing pulmonary lesions using visual attention networks. Med Image Anal. 2019;53:26–38. doi: 10.1016/j.media.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 90.Pinheiro PO, Collobert R (2015) From image-level to pixel-level labeling with convolutional networks. In: IEEE conference on computer vision and pattern recognition, pp 1713–1721
- 91.Qi K, Guan Q, Yang C, Peng F, Shen S, Wu H. Concentric circle pooling in deep convolutional networks for remote sensing scene classification. Remote Sens. 2018;10(6):934. [Google Scholar]
- 92.Qi K, Yang C, Hu C, Guan Q, Tian W, Shen S, Peng F. Polycentric circle pooling in deep convolutional networks for high-resolution remote sensing image recognition. IEEE J Sel Top Appl Earth Observ Remote Sens. 2020;13:632–641. [Google Scholar]
- 93.Quellec G, Charrière K, Boudi Y, Cochener B, Lamard M. Deep image mining for diabetic retinopathy screening. Med Image Anal. 2017;39:178–193. doi: 10.1016/j.media.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 94.Rikiya Y, Nishio M, Do RKG, Togashi K . Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K et al (2017) CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv:1711.05225
- 96.Saeedan F, Weber N, Goesele M, Roth S (2018) Detail-preserving pooling in deep networks. In: IEEE conference on computer vision and pattern recognition, pp 9108–9116
- 97.Saha O, Kusupati A, Simhadri HV, Varma M, Jain P (2020) RNNPool: efficient non-linear pooling for ram constrained inference. arXiv:2002.11921
- 98.Scherer D, Müller A, Behnke S (2010) Evaluation of pooling operations in convolutional architectures for object recognition. In: International conference on artificial neural networks, pp 92–101
- 99.Shahriari A, Porikli F (2017) Multipartite pooling for deep convolutional neural networks. arXiv:1710.07435
- 100.Sheng J, Chen C, Fu C, Xue CJ (2018) EasyConvPooling: random pooling with easy convolution for accelerating training and testing. arXiv:1806.01729
- 101.Shi Z, Ye Y, Wu Y. Rank-based pooling for deep convolutional neural networks. Neural Netw. 2016;83:21–31. doi: 10.1016/j.neunet.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 102.Simon M, Gao Y, Darrell T, Denzler J, Rodner E (2017) Generalized orderless pooling performs implicit salient matching. In: IEEE international conference on computer vision, pp 4960–4969
- 103.Song S, Cheung NM, Chandrasekhar V, Mandal B (2018) Deep adaptive temporal pooling for activity recognition. In: 26th ACM international conference on Multimedia, pp 1829–1837
- 104.Song Z, Liu Y, Song R, Chen Z, Yang J, Zhang C, Jiang Q. A sparsity-based stochastic pooling mechanism for deep convolutional neural networks. Neural Netw. 2018;105:340–345. doi: 10.1016/j.neunet.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 105.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M (2014) Striving for simplicity: the all convolutional net. arXiv:1412.6806
- 106.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(56):1929–1958. [Google Scholar]
- 107.Stergiou A, Poppe R, Kalliatakis G (2021) Refining activation downsampling with SoftPool. arXiv:2101.00440
- 108.Sun M, Song Z, Jiang X, Pan J, Pang Y. Learning pooling for convolutional neural network. Neurocomputing. 2017;224:96–104. [Google Scholar]
- 109.Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 110.Tan YS, Lim KM, Tee C, Lee CP, Low CY. Convolutional neual network with spatial pyramid pooling for hand gesture recognition. Neural Comput Appl. 2021;33(10):5339–5351. [Google Scholar]
- 111.Tolias G, Sicre R, Jégou H (2015) Particular object retrieval with integral max-pooling of CNN activations. arXiv:1511.05879
- 112.Tong Z, Aihara K, Tanaka G (2016) A hybrid pooling method for convolutional neural networks. In: International conference on neural information processing, pp 454–461
- 113.Tong Z, Tanaka G. Hybrid pooling for enhancement of generalization ability in deep convolutional neural networks. Neurocomputing. 2019;333:76–85. [Google Scholar]
- 114.Tsai CF (2012) Bag-of-words representation in image annotation: a review. Int Scholarly Res Not 1–19
- 115.Tsai YH, Hamsici OC, Yang MH (2015) Adaptive region pooling for object detection. In: IEEE conference on computer vision and pattern recognition, pp 731–739
- 116.Vo HH, Verma A (2016) New deep neural nets for fine-grained diabetic retinopathy recognition on hybrid color space. In: IEEE international symposium on multimedia, pp 209–215
- 117.Wang F, Huang S, Shi L, Fan W. The application of series multi-pooling convolutional neural networks for medical image segmentation. Int J Distrib Sensor Netw. 2017;13:1–10. [Google Scholar]
- 118.Wang L, Xie C, Zeng N. RP-Net: a 3D convolutional neural network for brain segmentation from magnetic resonance imaging. IEEE Access. 2019;7:39670–39679. [Google Scholar]
- 119.Wang Q, Gao Z, Xie J, Zuo W, Li P (2018) Global gated mixture of second-order pooling for improving deep convolutional neural networks. In: Advances in neural information processing systems, pp 1277–1286
- 120.Wang S, Jiang Y, Hou X, Cheng H, Du S. Cerebral micro-bleed detection based on the convolution neural network with rank based average pooling. IEEE Access. 2017;5:16576–16583. [Google Scholar]
- 121.Wang SH, Lv YD, Sui Y, Liu S, Wang SJ, Zhang YD. Alcoholism detection by data augmentation and convolutional neural network with stochastic pooling. J Med Syst. 2018;42(1):2. doi: 10.1007/s10916-017-0845-x. [DOI] [PubMed] [Google Scholar]
- 122.Wang SH, Tang C, Sun J, Yang J, Huang C, Phillips P, Zhang YD. Multiple sclerosis identification by 14-layer convolutional neural network with batch normalization, dropout, and stochastic pooling. Front Neurosci. 2018;12:818. doi: 10.3389/fnins.2018.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang SH, Zhang Y, Cheng X, Zhang X. Zhang YD (2021) PSSPNN: PatchShuffle stochastic pooling neural network for an explainable diagnosis of COVID-19 with multiple-way data augmentation. Comput Math Methods Med. 2021;6633755:1–18. doi: 10.1155/2021/6633755. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 125.Wang Z, Ji S. Second-order pooling for graph neural networks. IEEE Trans Pattern Anal Mach Intell. 2020 doi: 10.1109/TPAMI.2020.2999032. [DOI] [PubMed] [Google Scholar]
- 126.Wang Z, Yang J (2017) Diabetic retinopathy detection via deep convolutional networks for discriminative localization and visual explanation. arXiv:1703.10757
- 127.Wang Z, Yin Y, Shi J, Fang W, Li H, Wang X (2017) Zoom-in-net: deep mining lesions for diabetic retinopathy detection. In: International conference on medical image computing and computer assisted intervention, pp. 267–275
- 128.Wei X, Zhang Y, Gong Y, Zheng N (2018) Kernelized subspace pooling for deep local descriptors. In: IEEE conference on computer vision and pattern recognition, pp 1867–1875
- 129.Wei Z, Zhang J, Liu L, Zhu F, Shen F, Zhou Y, Liu S, Sun Y, Shao L (2019) Building detail-sensitive semantic segmentation networks with polynomial pooling. In: IEEE conference on computer vision and pattern recognition, pp 7115–7123
- 130.Wiliem A, Sanderson C, Wong Y, Hobson P, Minchin RF, Lovell BC. Automatic classification of human epithelial type 2 cell indirect immunofluorescence images using cell pyramid matching. Pattern Recogn. 2014;47(7):2315–2324. [Google Scholar]
- 131.Woo S, Park J, Lee JY, Kweon IS (2018) CBAM: convolutional block attention module. In: European conference on computer vision, pp 3–19
- 132.Wu H, Gu X (2015) Max-pooling dropout for regularization of convolutional neural networks. abs/1512.01400
- 133.Xie G, Zhang X, Shu X, Yan S, Liu C (2015) Task-driven feature pooling for image classification. In: IEEE international conference on computer vision, pp 1179–1187
- 134.Xie H, Tang C, Zhang W, Shen Y, Lei Z. Multi-scale retinal vessel segmentation using encoder-decoder network with squeeze-and-excitation connection and atrous spatial pyramid pooling. Appl Opt. 2021;60(2):239–249. doi: 10.1364/AO.409512. [DOI] [PubMed] [Google Scholar]
- 135.Xu Y, Zhu J, Chang E, Tu Z (2012) Multiple clustered instance learning for histopathology cancer image classification, segmentation and clustering. In: IEEE conference on computer vision and pattern recognition, pp 964–971
- 136.Yang F, Choi W, Lin Y (2016) Exploit all the layers: fast and accurate CNN object detector with scale dependent pooling and cascaded rejection classifiers. In: IEEE conference on computer vision and pattern recognition, pp 2129–2137
- 137.Yang Y, Newsam S (2011) Spatial pyramid co-occurrence for image classification. In: International conference on computer vision, pp 1465–1472
- 138.Yoo D, Park S, Lee J (2015) In So Kweon: Multi-scale pyramid pooling for deep convolutional representation. In: IEEE conference on computer vision and pattern recognition workshops, pp 71–80
- 139.Yu D, Wang H, Chen P, Wei Z (2014) Mixed pooling for convolutional neural networks. In: International conference on rough sets and knowledge technology. Springer, pp 364–375
- 140.Yu J, Zhu C, Zhang J, Huang Q, Tao D. Spatial pyramid-enhanced NetVLAD with weighted triplet loss for place recognition. IEEE Trans Neural Netw Learn Syst. 2020;31(2):661–674. doi: 10.1109/TNNLS.2019.2908982. [DOI] [PubMed] [Google Scholar]
- 141.Yu K, Salzmann M (2018) Statistically-motivated second-order pooling. In: European conference on computer vision, pp 600–616
- 142.Zeiler MD, Fergus R (2013) Stochastic pooling for regularization of deep convolutional neural networks. arXiv:1301.3557
- 143.Zhai S, Wu H, Kumar A, Cheng Y, Lu Y, Zhang Z, Feris RS (2017) S3Pool: pooling with stochastic spatial sampling. In: IEEE conference on computer vision and pattern recognition, Honolulu, pp 4003–4011
- 144.Zhang B, Zhao Q, Feng W, Lyu S. AlphaMEX: a smarter global pooling method for convolutional neural networks. Neurocomputing. 2018;321:36–48. [Google Scholar]
- 145.Zhang N, Farrell R, Darrell T (2012) Pose pooling kernels for sub-category recognition. In: IEEE conference on computer vision and pattern recognition, pp 3665–3672
- 146.Zhang R, Zhu F, Liu J, Liu G. Depth-wise separable convolutions and multi-level pooling for an efficient spatial CNN-based steganalysis. IEEE Trans Inf Forensics Secur. 2019;15:1138–1150. [Google Scholar]
- 147.Zhang X, Zhang X. Global learnable pooling with enhancing distinctive feature for image classification. IEEE Access. 2020;8:98539–98547. [Google Scholar]
- 148.Zhang YD, Pan C, Chen X, Wang F. Abnormal breast identification by nine-layer convolutional neural network with parametric rectified linear unit and rank-based stochastic pooling. J Comput Sci. 2018;27:57–68. [Google Scholar]
- 149.Zhang YD, Satapathy SC, Liu S, Li GR. A five-layer deep convolutional neural network with stochastic pooling for chest CT-based COVID-19 diagnosis. Mach Vis Appl. 2021;32(1):1–13. doi: 10.1007/s00138-020-01128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao J, Snoek CG (2021) Liftpool: Bidirectional convnet pooling. arXiv:2104.00996
- 151.Zhao Q, Lyu S, Zhang B, Feng W. Multiactivation pooling method in convolutional neural networks for image recognition. Wirel Commun Mob Comput. 2018;2018:1–15. [Google Scholar]