Abstract
Background
Targeted therapy of patients with non-small cell lung cancer (NSCLC) who harbour sensitising mutations by tyrosine kinase inhibitors (TKIs) has been found more effective than traditional chemotherapies. However, target genes status (eg, epidermal growth factor receptor (EGFR) TKIs sensitising and resistant mutations) need to be tested for choosing appropriate TKIs. This study is to investigate the performance of a liquid biopsy-based targeted capture sequencing assay on the molecular analysis of NSCLC.
Methods
Plasma samples from patients with NSCLC who showed resistance to the first/second-generation EGFR TKIs treatment were collected. The AVENIO ctDNA Expanded Kit is a 77 pan-cancer genes detection assay that was used for detecting EGFR TKIs resistance-associated gene mutations. Through comparison of the EGFR gene testing results from the Cobas EGFR Mutation Test v2, and UltraSEEK Lung Panel, the effectiveness of the targeted capture sequencing assay was verified.
Results
A total of 24 plasma cell-free DNA (cfDNA) samples were tested by the targeted capture sequencing assay. 33.3% (8/24) cfDNA samples were positive for EGFR exon 20 p.T790M which leads to EGFR dependent TKIs resistance. 8.3% (2/24) and 4.2% (1/24) samples were positive for mesenchymal-epithelial transition gene amplification and B‐Raf proto‐oncogene, serine/threonine kinase exon 15 p.V600E mutations which lead to EGFR independent TKIs resistance. The median value of the p.T790M variant allele fraction and variant copy numbers was 2% and 36.10 copies/mL plasma, respectively. The next-generation sequencing test showed higher than 90% concordance with either MassArray or qPCR-based methods for detecting either EGFR TKIs sensitising or resistance mutations.
Conclusion
The targeted capture sequencing test can support comprehensive molecular analysis needed for TKIs treatment, which is promising to be clinically applied for the improved precision treatment of NSCLC.
Keywords: non-small cell lung cancer, lung cancer, drug reactions
Key messages.
What is already known on this topic
Liquid biopsy offers a non-invasive alternative to tissue biopsy for the molecular analysis-based targeted therapy of non-small cell lung cancer (NSCLC), but a highly sensitive detection method is needed for efficient testing extremely low levels of target genes variants in cell-free DNA (cfDNA) for the precision treatment of NSCLC.
What this study adds
The performance of the target capture sequencing-based assay on the systematic analysis of EGFR tyrosine kinase inhibitors (TKIs) sensitising and resistance mutations in the cfDNA of NSCLC were investigated, and compared with other leading techniques (real-time qPCR and MassArray).
How this study might affect research, practice or policy
This study confirmed that the target capture sequencing test has the highest sensitivity and throughput in the systematic analysis of EGFR TKIs therapy related genes mutations in cfDNA, and the clinical application of this test is expected to improve the precision treatment of NSCLC by EGFR TKIs.
Introduction
Lung cancer is the most commonly diagnosed primary malignancy in Canada and is currently the leading cause of cancer-related deaths in the country.1 Non-small cell lung cancer (NSCLC) is the major type of lung cancer, which accounts for more than 80% of all lung cancers.2 As lung cancer patients are often found to have advanced disease at the time of initial diagnosis, palliative therapies with the alleviation of symptoms and extension of survival are the main objectives of clinical treatments.1 Molecular analysis of tumour biomarker genes has become the standard of practice in the treatment of NSCLC. Patients with NSCLC who harbour sensitising mutations in epidermal growth factor receptor (EGFR) genes can be offered small-molecule tyrosine kinase inhibitors (TKIs), which are significantly more effective than traditional chemotherapies.3 Besides, other genes targeted therapies, specifically against sensitising mutations in ALK, ROS1 and B‐Raf proto‐oncogene, serine/threonine kinase (BRAF) also have a demonstrable clinical improvement in the outcome of NSCLC.2–5
However, after the median treatment duration (8–10 months), the majority of patients with NSCLC (50%–60%) will acquire a novel EGFR mutation in exon 20 known as p.T790M, which confers resistance to the first-generation and second-generation TKIs’ treatment.6 Osimertinib can be alternatively used to treat patients with the acquired EGFR exon 20 p.T790M mutation.7 Acquired EGFR exon 20 p.C797S mutation which mediates resistance to the third-generation EGFR TKIs treatment may also develop in EGFR exon 20 p.T790M-positive patients with NSCLC.8 Non-EGFR-dependent mechanisms of acquired TKIs resistance, such as gene copy number amplification of mesenchymal-epithelial transition (MET) gene and human epidermal growth factor receptor 2 gene, mutations of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha and BRAF genes have been reported as well.9–12 It is critical to systematically monitor the TKIs-based targeted therapy-associated gene mutations in patients with NSCLC for the improved outcome of targeted therapy.
Liquid biopsy offers a cost-effective and low-risk alternative to tissue biopsy for testing tumour biomarker genes in patients with NSCLC.13 14 The concordance between the testing of EGFR mutations in tumour DNA and cell-free DNA (cfDNA) of patients with NSCLC has been reported as 79%, though the mutation rates detected in plasma cfDNA reflect approximately 20% of that detected from tumour tissues.15 16 Liquid biopsy has been clinically used as a complementary assay to tissue biopsy for testing EGFR mutations in patients with NSCLC.17 Next-generation sequencing (NGS) was found to be able to detect EGFR exon 20 p.T790M in the liquid biopsy samples of patients with NSCLC with percentages of 0.3%–21.0%.18 More importantly, NGS technology-based assays are superior to currently used PCR-based assays with the capacity to simultaneously test multiple genes in one assay.19 20 Thus, more target genes can be tested by NGS to match patients with appropriate NSCLC treatment drugs for improved precision treatment.21–23
This research project aims to investigate the performance of an optimised hybridisation capture-based NGS assay (AVENIO ctDNA expanded kit, Roche Diagnostics, Canada) on testing somatic mutations in the plasma cfDNA for targeted treatment of patients with NSCLC. Based on the EGFR testing results, the concordance of the NGS test with clinically validated MassArray (UltraSEEK Lung Panel, Agena Bioscience, USA) and qPCR-based (Cobas EGFR Mutation Test v2) assays was evaluated.
Materials and methods
Patient and public involvement
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Patients and samples collection
NSCLC patients in advanced stages who experienced disease progression after the targeted treatment with first or second-generation EGFR TKIs were randomly included in this study. Ten millilitre peripheral blood was collected into Streck tubes. The plasma specimen was separated within 4 hours from peripheral blood collection by two rounds of centrifugation at 20°C, with 10 min at 1600× g and 3000× g respectively, and stored at −80°C before DNA extraction. Twenty-four plasma samples were collected and tested by the NGS test. Due to the availability of sample amount, 23 samples were tested by the MassArray-base assay as a replicate detection and 12 samples were tested by the qPCR-based assay as a second replicate detection.
Testing pan-cancer genes mutations in cfDNA by NGS
The AVENIO ctDNA expanded kit (Roche Diagnostics) is a hybridisation capture sequencing-based 77 genes pan-cancer assay (online supplemental table S1). AVENIO cfDNA Isolation Kit (Roche Diagnostics) was used to extract cfDNA from plasma according to the user’s manual. The extracted cfDNA was analysed by Agilent High Sensitivity DNA Analysis Kit (Agilent Technologies, California, USA) and Qubit dsDNA HS Assay Kit (ThermoFisher Scientific, California, USA) for quality control, and then used for library preparation with 10–50 ng cfDNA input. Prepared libraries were sequenced on the NextSeq 500 500/550 High Output Kit V2 (300 cycles) on Illumina NextSeq sequencing platform (Illumina, California, USA) and analysed by the AVENIO oncology analysis software V.2.0.0 (Roche Diagnostics) according to manufacturer’s instructions.24 25
bmjresp-2021-001154supp001.pdf (293.5KB, pdf)
Testing EGFR variants in cfDNA by qPCR and MassArray
Cobas EGFR Mutation Test v2 (Roche Diagnostics), a real-time qPCR-based in vitro diagnostics assay, was used for testing EGFR variants in 2 mL plasma of patients with NSCLC. A clinically validated UltraSEEK Lung Cancer Panel (Agena Bioscience) was also used as a reference method for testing EGFR mutations in 10 ng input cfDNA. Detailed information about these testing methods is provided in online supplemental file 1.
Statistical analysis
AVENIO ctDNA Analysis Software (Roche Diagnostics) was used for sequencing data analysis and generating diagnostic reports.26 With the data of abundance for different biomarker gene variants, the distribution of gene variants’ abundance is shown in box plots and the median value was calculated by using an R-based statistical data analysis tool (BoxPlotR).27
Results
EGFR gene testing by NGS
Plasma cfDNA is composed of fragmented double-stranded DNA circulating in the blood, which is mainly generated from the apoptotic and necrotic cells.28 29 The quality of extracted cfDNA samples was checked on the Bioanalyzer (Agilent Technologies). The major peak of the extracted cfDNA sample was typically located at 160–200 bp (online supplemental figure S1A, B). Several distinguishable cfDNA peaks with a size shift were observed, confirming the successful ligation of adapters to target gene fragments (online supplemental figure S1C, D). The library prepared by the AVENIO ctDNA Expanded Kit (Roche Diagnostics) typically showed the primary cfDNA peak at ~300 bp with additional small cfDNA peaks of larger size present (online supplemental figure S1C, D).
In this study, a total of 24 plasma cfDNA samples were tested by the NGS assay. The metrics of the NGS testing are shown in online supplemental table S2. After alignment analysis, the average ratio of mapped reads, and the on-target rate were 94.31% and 71.62% (online supplemental figure S2A). The typical median value of coverage uniformity was higher than 99% (online supplemental figure S2B). It can be observed that the high-quality sequencing data showed a very low error rate, excellent sequencing depth for target genes and high coverage uniformity, which also lead to a high theoretical sensitivity with a median value of 98.71% (online supplemental figure S2A).
The sequencing detection showed that 66.7% (16/24) ctDNA samples were positive for EGFR sensitising mutations, and 50.0% (8/16) of which were also positive for the first-generation and second-generation TKIs resistance mutation (p.T790M) (online supplemental table S3). The percentage of different EGFR sensitising mutations is shown in figure 1. The range of variant allele fraction for EGFR sensitising mutations was 0.11%–45.16% with the median value of 4% (figure 1A). It can be observed that exon 21 p.L858R and exon 19 dels are the main EGFR sensitising mutations detected in the plasma cfDNA, the percentages of which were 81.25% (13/16). And p.E746_A750del was the main exon 19 dels variant detected which compromise 62.5% (5/8) of all detected exon 19 dels variants (online supplemental table S3). And the variant allele fraction for EGFR exon 20 p.T790M was 0.08%–8.28% with the median value of 2% (figure 1A). The range of variant copy numbers for EGFR sensitive mutations was 1.79–4950 with the median value of 135.00 copies/mL plasma (figure 1B). And the range of variant copy numbers for EGFR exon 20 p.T790M was 0.68–561 with the median value of 36.10 copies/ mL plasma (figure 1B).
Figure 1.
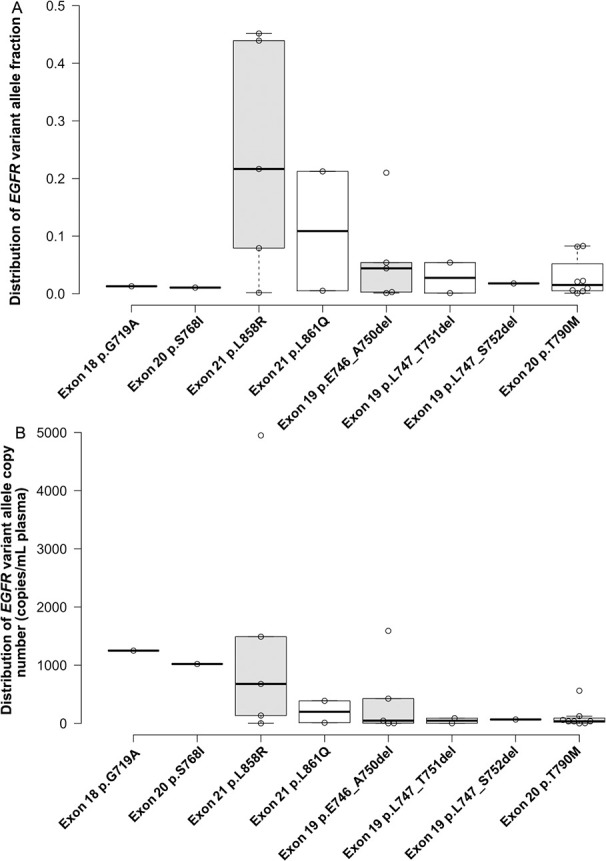
Detection of EGFR sensitising and TKIs resistance variants in plasma cell-free DNA (cfDNA) of patients with NSCLC by the NGS-based Avenio panel. (A) Variant allele fraction of the EGFR gene in plasma cfDNA; (B) molecular concentration of different EGFR variants in the plasma of patients with NSCLC. EGFR, epidermal growth factor receptor; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; TKI, tyrosine kinase inhibitor.
Concordance of NGS, qPCR and MassArray on EGFR gene testing
The qPCR detection showed 75.0% (9/12) and 33.3% (3/9) cfDNA samples were positive for EGFR sensitising mutations and EGFR exon 20 p.T790M (online supplemental table S3). Except for one inconclusive sample, the MassArray-based assay detected 56.5% (13/23) cfDNA samples that were positive for EGFR sensitising mutations, and 61.5% (8/13) of which were also positive for the EGFR exon 20 p.T790M (online supplemental table S3). The NGS-based assay showed 91.3% (21/23) concordance with the MassArray-based test for detecting either EGFR sensitising or EGFR TKIs resistant mutation (table 1). The qPCR assay showed 83.3% (10/12) and 100% (12/12) concordance with the MassArray-based test for testing EGFR sensitising mutations and exon 20 p.T790M variant, respectively (table 1). The concordance of the three assays on testing EGFR variants in the plasma cfDNA from patients with NSCLC was 83.3% (10/12) (table 1).
Table 1.
Concordance of EGFR testing results from NGS, MassArray and qPCR
| Mutations | Variants | NGS vs MassArray | NGS vs qPCR | MassArray vs qPCR |
| EGFR sensitising mutations | Exon 18 p.G719A | 100% (23/23) | 100% (12/12) | 100% (12/12) |
| Exon 20 p.S768I | 100% (23/23) | 100% (12/12) | 100% (12/12) | |
| Exon 21 p.L858R | 95.7% (22/23) | 100% (12/12) | 91.7% (11/12) | |
| Exon 21 p.L861Q | 100% (23/23) | 100% (12/12) | 100% (12/12) | |
| Exon 19 deletions | 95.7% (22/23) | 91.7% (11/12) | 91.7% (11/12) | |
| TKIs resistant EGFR mutation | Exon 20 p.T790M | 91.3% (21/23) | 91.7% (11/12) | 100% (12/12) |
EGFR, epidermal growth factor receptor; NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor.
Testing pan-cancer genes for improved TKIs resistance analysis
Of the cfDNA samples tested by the sequencing assay,66.7% (16/24) cfDNA samples were positive for EGFR mutations, 33.3% (8/24) were positive for TP53 mutations, 12.5% (3/24) were positive for PTCH1 and PTEN mutations, 8.3% (2/24) were positive for PMS2 mutations, 4.2% (1/24) were positive for NTRK1, RB1, ROS1, BRAF, GNAS, ERBB2, CTNNB1 genes mutations (table 2) (figure 2). The range of variant allele fraction for EGFR was 0.08%–45.16% with the median value of 2% (figures 1A and 2A). And the range of variant allele fraction for TP53 was 0.23%–12.65% with a median value of 4% (figures 2A and 3A). The range of variant copy number for EGFR was 0.679 copies/mL to 4950 copies/mL with the median value of 66.70 copies/mL plasma (figures 1B and 2B). And the range of variant copy numbers for TP53 was 12.2 copies/mL to 1110 copies/mL with the median value of 133.00 copies/mL plasma (figures 2B and 3B). In addition, the median value of variant allele fraction for NTRK1, RB1, PMS2, PTCH1, GNAS was 3%, 8%, 46%, 2% and 1%, respectively. And the median value of variant allele fraction for ROS1, BRAF, ERBB2, CTNNB1, PTEN was less than 1% (figure 2A) (table 2). The median value of variant copy number for NTRK1, RB1, PMS2, PTCH1, GNAS, CTNNB1 was 225.00, 598.00, 465.00, 22.50, 21.20, 63.00 copies/mL plasma (figure 2B) (table 2). And the median value of variant copy numbers for ROS1, BRAF, ERBB2, PTEN was less than 10 copies/ mL plasma (table 2).
Table 2.
Testing tumour biomarker genes (other than epidermal growth factor receptor and TP53) variants in cell-free DNA by the next-generation sequencing test
| SNV gene | Variant | Coding change | Variant description | Allele fraction (%) | Variant depth | Molecular mutant per mL (n) |
| NTRK1 | Exon 17 p.I737I | c.2211C>T | Synonymous variant | 2.97 | 119/4007 | 225 |
| RB1 | Exon 19 p.V654M | c.1960G>A | Missense variant | 7.91 | 286/3616 | 598 |
| ROS1 | Exon 41 p.L2157V | c.6469C>G | Missense variant | 0.28 | 4/1417 | 2.1 |
| PMS2 | Exon 11 p.A660A | c.1980C>T | Synonymous variant | 44.08 | 901/2044 | 425 |
| PMS2 | Exon 3 p.D70G | c.209A>G | Missense variant | 47.13 | 345/732 | 505 |
| PTCH1 | Exon 17 p.N929N | c.2787C>T | Synonymous variant | 46.50 | 1789/3847 | 4490 |
| PTCH1 | Exon 14 p.R665C | c.1993C>T | Missense variant | 1.32 | 44/3332 | 12.7 |
| PTCH1 | Exon 22 p.A1247A | c.3741G>A | Synonymous variant | 1.57 | 32/2034 | 22.5 |
| BRAF | Exon 15 p.V600E | c.1799T>A | Missense variant | 0.08 | 2/2561 | 1.3 |
| GNAS | Exon 8 p.R202C | c.604C>T | Missense variant | 1.32 | 35/2655 | 21.2 |
| ERBB2 | Exon 23 p.K937R | c.2810A>G | Missense variant | 0.21 | 5/2390 | 4.5 |
| ERBB2 | Exon 26 p.R1111Q | c.3332G>A | Missense variant | 0.61 | 12/1961 | 13.1 |
| CTNNB1 | Exon 3 p.S33Y | c.98C>A | Missense variant | 0.07 | 3/4597 | 63 |
| PTEN | Exon 5 p.H93Y | c.277C>T | Missense variant | 4.32 | 157/3633 | 162 |
| PTEN | Exon 8 p.N323fs | c.968dupA | Frameshift variant | 0.12 | 1/841 | 0.9 |
| PTEN | Exon 8 p.N323fs | c.968dupA | Frameshift variant | 0.11 | 1/892 | 1.2 |
Figure 2.
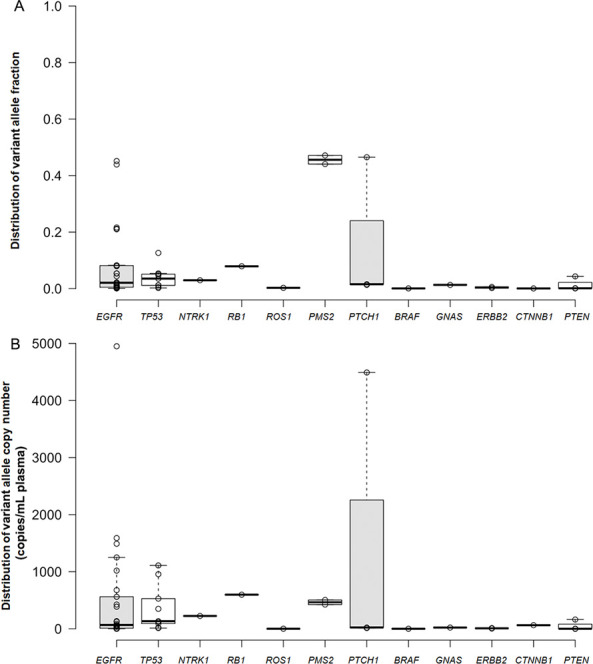
Percentages of tumour biomarker genes variant alleles located in different ranges of fraction ratio and copy numbers. (A) Distribution of tumour biomarker genes variant alleles in different ranges of fraction ratio; (B) distribution of tumour biomarker genes variant alleles in different ranges of copy numbers.
Figure 3.
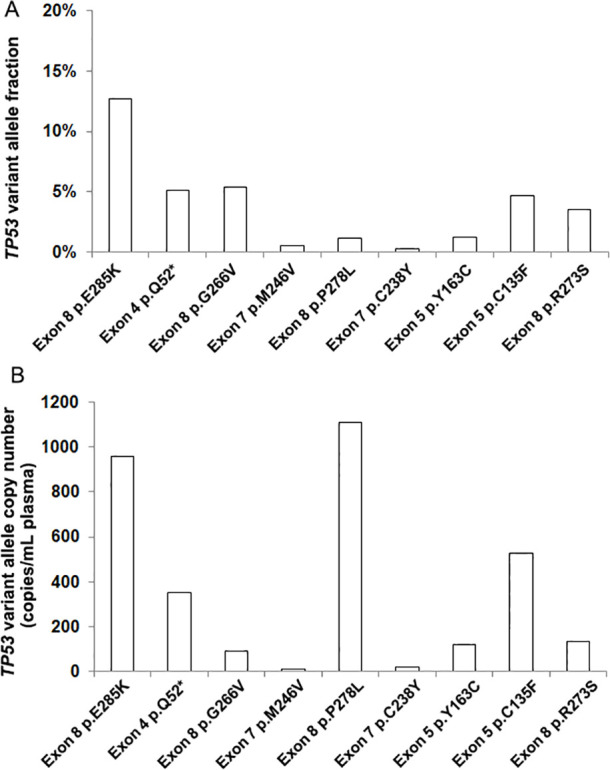
Testing of TP53 gene in the plasma cell-free DNA (cfDNA) of non-small cell lung cancer (NSCLC) patients by next-generation sequencing-based Avenio panel. (A) TP53 gene’s variant allele fraction in plasma cfDNA of NSCLC; (B) molecular concentration of TP53 gene variants in the plasma of NSCLC.
According to the sequencing data analysis process for the Avenio ctDNA kit (Roche Diagnostics), the copy number variant (CNV) score is statistically calculated by accounting for the log2 ratio to normal gene copy number, and the higher CNV score indicates the higher confidence value. The NGS detection showed that the copy number amplification variants were detected on EGFR and MET genes. Statistically, the range of the CNV scores for EGFR gene variants was from 4.14 to 29.06 with a median value of 14.45. And the range of the CNV scores for MET gene variants was 9.19–20.35 with a median value of 18.31 (figure 4). Twenty-five per cent (6/24) cfDNA samples were positive for the EGFR copy number amplification mutation, including 50% (3/6) cfDNA samples were also positive for MET amplification mutation. Fifty per cent (3/6) of EGFR amplification positive cfDNA samples and 66.7% (2/3) MET amplification positive cfDNA samples showed EGFR p.T790M negative results.
Figure 4.
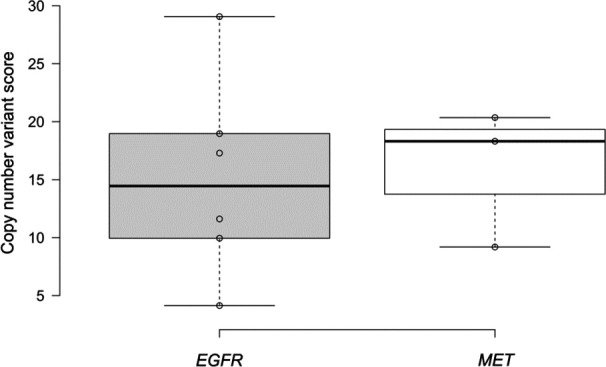
Testing of gene copy number variants in the plasma cell-free DNA of patients with non-small cell lung cancer by using next-generation sequencing-based Avenio panel. EGFR, epidermal growth factor receptor; MET, mesenchymal-epithelial transition.
Discussion
EGFR gene is the most important tumour biomarker to be tested for the targeted treatment of NSCLC because EGFR-TKIs treatment is the first-line therapy method for patients with NSCLC with sensitising EGFR mutations. It can be observed that some cfDNA samples showed a low EGFR variant allele fraction which can be lower than 0.1%, the testing of baseline EGFR p.T790M variant is critical for clinical NSCLC treatment.30 31 EGFR testing method with LOD lower than 0.1% should be applied for sensitive detection of EGFR variants in the plasma cfDNA of patients with NSCLC.
As shown in online supplemental table S4, the cfDNA samples which showed mismatch results from different assays were investigated. EGFR sensitising mutations with variant allele fractions less than 0.2% were easily missed in the MassArray-based test. And EGFR exon 20 p.T790M mutation with variant allele fraction less than 5% were missed by both the MassArray and qPCR-based assays. Though the variant allele fraction (EGFR exon 20 p.T790M) at 0.42% normally can still be caught by the MassArray and qPCR-based assays, the low copy number of variant alleles (4.45 copies/mL plasma) may be the reason for missing detected (online supplemental table S4). Besides the variant allele fraction, the copy number of gene variants can also be an important factor that can impact the sensitivity of assays on testing tumour biomarker genes in plasma cfDNA.
As a high-throughput testing method, the NGS assay has the advantages of simultaneous testing of multiple tumour biomarker genes in one test, interrogating both known and unknown gene variants. This makes it superior to qPCR and MassArray-based assays on gene detection efficiency.19 32 It was observed that the NGS-based assay showed high sensitivity in testing tumour biomarker genes in plasma cfDNA as gene allele variant fraction of less than 0.1% and the copy number of gene allele variant of less than 1 copy/mL plasma were also successfully detected (figures 1 and 3). This confirms other studies in which the deep sequencing approach showed higher detection sensitivity in testing tumour biomarker gene variants in cfDNA than qPCR and MassArray-based assays.30 32 33
Although EGFR exon 20 p.T790M mutation is the most possible reason for the resistance of NSCLC to the first-generation and second-generation EGFR TKIs, approximately 50% of resistance cases are caused by other molecular mechanisms such as the amplification of ERBB2, MET, and the mutation of BRAF, PI3K and KRAS.34–36 Also, other EGFR mutations such as exon 20 p.C797S, oncogenic fusions such as FGFR3-TACC3, and oncogene amplification such as MET and ERBB2 can also cause the resistance of NSCLC to the third generation TKIs.35 This study found that the detection of oncogenic amplification mutations can help clarify the resistance of NSCLC cases to the EGFR-TKIs by 12.5% (figure 4). Besides monitoring the EGFR status, it is necessary to comprehensively test other TKIs treatment-associated genes for choosing the most appropriate targeted treatment method. In this study, 4.2% (1/24) sample showed BRAF exon 15 p.V600E positive but EGFR p.T790M negative which helped clarify the EGFR independent TKIs resistance in NSCLC (table 2).37 12.5% (3/24) and 4.2% (1/24) samples showed mutations in PTEN and ERBB2, respectively (table 2), which may contribute to the resistance of NSCLC to TKIs as well, because ERBB2 amplification and PTEN loss have been found to activate PI3K/Akt pathway that is involved NSCLC progression.34 It was observed that the PTEN mutation (exon 8 p.N323fs) was only detected in patients with NSCLC without EGFR mutations. However, further studies need to be performed to confirm the relationship between PTEN and ERBB2 mutations and TKIs resistance.
Conclusion
All three methods (NGS, MassArray and qPCR-based assays) can effectively detect EGFR variants in the plasma cfDNA of patients with NSCLC and the concordance of these assays on testing EGFR variants was 83.3% (10/12). Based on the findings from this study, cfDNA testing methods with the LOD (lower than 2% and less than 102 copies/mL plasma) for target gene variants can detect more than 50% of EGFR mutation-positive plasma samples from patients with NSCLC. Compared with the MassArray and qPCR tests, the AVENIO ctDNA expanded kit (Roche Diagnostics) possesses the highest sensitivity and throughput on testing tumour biomarker genes in plasma cfDNA which shows high competitiveness to be clinically used for targeted therapy of NSCLC. The targeted capture sequencing assay is an excellent tool for comprehensive molecular analysis of NSCLC targeted therapy associated genes, which can help improve the precision treatment of NSCLC by TKIs. Considering the sample size used in this study is limited, further studies are needed to confirm the findings.
Footnotes
Contributors: LZ: methodology, project administration, writing original draft and editing. JC: resources and supervision. KF: methodology and resources. QC: resources, writing original draft and editing, responsible for the overall content as a guarantor. II: conceptualisation, funding acquisition, methodology, project administration, resources, supervision, writing original draft and editing, responsible for the overall content as a guarantor.
Funding: This study was supported by a research grant from Hoffmann - La Roche and AstraZeneca.
Competing interests: II has received honorarium for consultation from AstraZeneca, Roche, Bayer. The other authors of this study declare that they have no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the Ethics Committee of the Health Research Ethics Board of Universities of Alberta (HREBA.CC-18-0092). Participants gave informed consent to participate in the study before taking part.
References
- 1.Canadian-Cancer-Statistics-Advisory-Committee . Canadian cancer statistics 2018. Canadian Cancer Society, 2018. [Google Scholar]
- 2.Zugazagoitia J, Molina-Pinelo S, Lopez-Rios F, et al. Biological therapies in nonsmall cell lung cancer. Eur Respir J 2017;49:1601520. 10.1183/13993003.01520-2016 [DOI] [PubMed] [Google Scholar]
- 3.Kim C, Giaccone G. Precision oncology in non-small-cell lung cancer: opportunities and challenges. Nat Rev Clin Oncol 2018;15:348–9. 10.1038/s41571-018-0008-0 [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446–54. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 5.Saarenheimo J, Eigeliene N, Andersen H, et al. The value of liquid biopsies for guiding therapy decisions in non-small cell lung cancer. Front Oncol 2019;9:129. 10.3389/fonc.2019.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auliac JB, Pérol M, Planchard D, et al. Real-Life efficacy of osimertinib in pretreated patients with advanced non-small cell lung cancer harboring EGFR T790M mutation. Lung Cancer 2019;127:96–102. 10.1016/j.lungcan.2018.11.037 [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi F, Kato E, Wakabayashi A, et al. Effect of osimertinib treatment on lung adenocarcinoma with squamous cell transformation harboring the T790M mutation: a case report and literature review. Mol Clin Oncol 2019;11:127–31. 10.3892/mco.2019.1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Tsui ST, Liu C, et al. Egfr C797S mutation mediates resistance to third-generation inhibitors in T790M-positive non-small cell lung cancer. J Hematol Oncol 2016;9:59. 10.1186/s13045-016-0290-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takezawa K, Pirazzoli V, Arcila ME, et al. Her2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922–33. 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S-G, Chang Y-L, Yu C-J, et al. The role of PIK3CA mutations among lung adenocarcinoma patients with primary and acquired resistance to EGFR tyrosine kinase inhibition. Sci Rep 2016;6:35249. 10.1038/srep35249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho C-C, Liao W-Y, Lin C-A, et al. Acquired BRAF V600E mutation as resistant mechanism after treatment with Osimertinib. J Thorac Oncol 2017;12:567–72. 10.1016/j.jtho.2016.11.2231 [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Yang S, Wang K, et al. Met inhibitors for targeted therapy of EGFR TKI-resistant lung cancer. J Hematol Oncol 2019;12:63. 10.1186/s13045-019-0759-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci 2019;40:172–86. 10.1016/j.tips.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 14.Oellerich M, Christenson RH, Beck J, et al. Plasma EGFR mutation testing in non-small cell lung cancer: a value proposition. Clin Chim Acta 2019;495:481–6. 10.1016/j.cca.2019.05.019 [DOI] [PubMed] [Google Scholar]
- 15.Shu Y, Wu X, Tong X, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep 2017;7:583. 10.1038/s41598-017-00520-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772–82. 10.1158/1078-0432.CCR-16-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stockley T, Souza CA, Cheema PK, et al. Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol 2018;25:163–9. 10.3747/co.25.4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartels S, Persing S, Hasemeier B, et al. Molecular analysis of circulating cell-free DNA from lung cancer patients in routine laboratory practice: a Cross-Platform comparison of three different molecular methods for mutation detection. J Mol Diagn 2017;19:722–32. 10.1016/j.jmoldx.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 19.Vollbrecht C, Lehmann A, Lenze D, Hummel M, et al. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget 2018;9:18529–39. 10.18632/oncotarget.24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton BC, Birse RT, Maggert K, et al. Assessment of common somatic mutations of EGFR, KRAS, BRAF, NRAS in pulmonary non-small cell carcinoma using iPLEX® HS, a new highly sensitive assay for the MassARRAY® system. PLoS One 2017;12:e0183715. 10.1371/journal.pone.0183715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med 2017;6:154–62. 10.1002/cam4.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahama T, Sakai K, Takeda M, Azuma K, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan Oncology Group 8014LTR study). Oncotarget 2016;7:58492–9. 10.18632/oncotarget.11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Normanno N, Denis MG, Thress KS, Ratcliffe M, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501–16. 10.18632/oncotarget.13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwahashi N, Sakai K, Noguchi T, et al. A comprehensive gene mutation analysis of liquid biopsy samples from patients with metastatic colorectal cancer to the ovary: a case report. Oncol Lett 2018;16:6431–6. 10.3892/ol.2018.9467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spitzer M, Wildenhain J, Rappsilber J, et al. BoxPlotR: a web tool for generation of box plots. Nat Methods 2014;11:121–2. 10.1038/nmeth.2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer 2018;88:1–9. 10.1016/j.ejca.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kustanovich A, Schwartz R, Peretz T, et al. Life and death of circulating cell-free DNA. Cancer Biol Ther 2019;20:1057–67. 10.1080/15384047.2019.1598759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lettig L, Sahnane N, Pepe F, et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019;8:584–92. 10.21037/tlcr.2019.09.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu HA, Arcila ME, Hellmann MD, et al. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol 2014;25:423–8. 10.1093/annonc/mdt573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dono M, De Luca G, Lastraioli S, et al. Tag-based next generation sequencing: a feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients. Mol Med 2019;25:15. 10.1186/s10020-019-0082-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y-W, Stefaniuk C, Jakubowski MA. Real-time PCR and targeted next-generation sequencing in the detection of low level EGFR mutations: Instructive case analyses. Respir Med Case Rep 2019;28:100901. 10.1016/j.rmcr.2019.100901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Q, Yu S, Zhao W, et al. Egfr-Tkis resistance via EGFR-independent signaling pathways. Mol Cancer 2018;17:53. 10.1186/s12943-018-0793-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells 2018;7:212. 10.3390/cells7110212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westover D, Zugazagoitia J, Cho BC, et al. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 2018;29:i10–19. 10.1093/annonc/mdx703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minari R, Bordi P, La Monica S, et al. Concurrent acquired BRAF V600E mutation and Met amplification as resistance mechanism of first-line Osimertinib treatment in a patient with EGFR-mutated NSCLC. J Thorac Oncol 2018;13:e89–91. 10.1016/j.jtho.2018.03.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjresp-2021-001154supp001.pdf (293.5KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.


