Abstract
Background
Cartilage endplate (CEP) degeneration plays a vital role in the pathological process of intervertebral disc degeneration. It has been previously reported that microRNAs may participate in the occurrence and development of intervertebral disc degeneration through regulating its target genes directly. The regulatory roles of miR-142-3p/HMGB1 in some orthopedic diseases have been determined successively, but there was no report about the degeneration of CEP. Therefore, we aimed to determine the regulation of miR-142-3p/HMGB1 or potential molecular mechanisms on proliferation, apoptosis, migration, and autophagy of CEP cells.
Methods
The target gene of miR-142-3p was determined by double luciferase assay. We selected ATDC5 cell lines. CCK-8 method was used to detect cell proliferation. Real-time fluorescence quantitative polymerase chain reaction was used to determine gene expression levels, and western blot analysis was used to determine protein expression levels. We chose flow cytometry to measure cell apoptosis and cell cycle.
Results
The result of luciferase detection showed that the target gene of miR-142-3p in CEP cells was HMGB1. Knockdown of the miR-142-3p inhibited the expression level of HMGB1, the proliferation and migration of CEP cells, but it promoted apoptosis of CEP cells. In addition, the detection results of the proteins related to apoptosis or autophagy showed that knockdown of miR-142-3p promoted apoptosis and autophagy.
Conclusion
The negative regulation of miR-142-3p/HMGB1 can affect the proliferation, apoptosis, migration, and autophagy of CEP cells. Our results provide a new idea for the targeted treatment of CEP degeneration by inhibiting the expression of HMGB1.
Keywords: endplate chondrocytes, miR-142-3p, HMGB1, proliferation, apoptosis, autophagy
Introduction
Degenerative disc disease is the main cause and initiating factor of intervertebral disc protrusion, degenerative vertebral canal stenosis, lumbar spondylolisthesis, and other kinds of low back pain diseases. 1 Although the exact molecular mechanism of intervertebral disc degeneration is not clear, current studies suggested that apoptosis due to the various nonphysiological reasons may be the main cause of intervertebral disc cell decline.2-4 Studies have shown that oxidative stress-induced ferroptosis of annulus fibroblasts and nucleus pulposus cells is related to the pathogenesis of intervertebral disc degeneration. 5 In the process of intervertebral disc degeneration, fibrous annulus rupture, inflammation, or cartilage endplate (CEP) degradation will induce the production of reactive oxygen species and aggravate the oxidative stress of nucleus pulposus cells. 6 There have been many studies on the molecular mechanism of annulus fibrosus and nucleus pulposus degeneration affecting intervertebral disc degeneration for a long time.7-9 However, there are relatively few studies on the molecular mechanism of CEP degeneration affecting intervertebral disc degeneration. Studies have shown that intervertebral discs are the largest hypoxic tissue in the human body.10-12 However, the degeneration of the CEP at a certain level may lead to a decline in diffusion function, which will prevent the nucleus marrow from obtaining necessary nutrients and inhibit the excretion of metabolic wastes; thus, degeneration of intervertebral disc is accelerated.13,14 It is obvious that CEP cell degeneration may play a vital role in the pathological process of intervertebral disc degeneration, and prevention or inhibition of CEP cell degeneration may be a potential target for intervertebral disc degeneration. Therefore, it is necessary to explore the molecular mechanism of CEP degeneration.
MicroRNA (miRNA) is a single-stranded RNA molecule consisting of about 20 to 23 nucleotides in length. In recent years, miRNAs have attracted more and more attention in the study of disc degeneration. miRNAs are involved in the process of intervertebral disc cell proliferation, apoptosis, and extracellular matrix synthesis by directly regulating the target genes, which may play an important role in occurrence and development of intervertebral disc degeneration.15,16 Some miRNAs associated with various orthopedic diseases have been reported in previous studies, such as miR-34a,17,18 miR-181a,19,20 and miR-142-3p. 21 More important, Wang et al. found that miR-142-3p inhibited the inflammatory response and apoptosis of chondrocytes in osteoarthritis by inhibiting the NF-κβ pathway mediated by HMGB1. 21
High Mobility Group Protein (HMGB1) is a highly conserved nuclear protein that can be released into the extracellular environment to mediate inflammatory response after cell injury. 22 Andersson and Harris 23 found that the expression level of HMGB1 was significantly increased in synovial membrane and serum of patients with rheumatoid arthritis, and the expression level of HMGB1 and its mRNA was most significant in the pannus area of articular cartilage invaded by synovial fluid. Studies have confirmed that the expression level of HMGB1 is significantly increased during the process of intervertebral disc degeneration. 24 More important, some scholars speculated that blocking the expression of HMGB1 to delay the CEP degeneration might become an important target for the treatment of intervertebral disc degeneration after relevant experiments. 25 These studies have all shown that HMGB1 plays an important role in the degeneration of CEP.
Subsequently, through literature review, we found that there were no reports on the regulation of miR-142-3p/HMGB1 in CEP cells. Therefore, in this study, we studied the expression regulation mechanism of miR-142-3p/HMGB1 in CEP cells to further understand the potential mechanism of action of miR-142-3p/HMGB1 in CEP degeneration.
Materials and Methods
Cell Culture
Our study used ATDC5 cell line (BeNa Culture Collection: BNCC350793), which are derived from the mouse teratocarcinoma strain AT805.
After the primary cell attachment have reached 85% to 90%, the cell passage begins. We add digestion solution containing 0.25% trypsin and 0.01% EDTA (1:1) for 1 minute. Wait until the cells have shrunk and become rounded and the gap has increased before adding complete medium to stop the digestion. After the cells have shrunk, become rounded, and gaps have been increased, complete medium is added to stop the digestion. We collected the cell suspension and centrifuged for 4 minutes, leaving the bottom cell cluster. Then add fresh complete medium and mix well by pipetting. The cells were inoculated into multiple petri dishes at 2 × 105/mL, and cultured in a cell incubator with a constant temperature and saturated humidity (5% CO2 at 37 °C).
Cell Transfection
Seed the cells in a 6-well plate. When the cells are about 80% confluent, Lipofectamine 2000 transfection reagent (Thermofisher, 11668019) was used, and the experiment was carried out according to the manufacturer’s instructions. After 48 hours of transfection, the cells were collected and used for further analysis.
Plasmid Construction
The recovered and purified target fragment HMGB1-3UTR (XhoI/NotI) was linked with the pYr-MirTarget (XhoI/NotI) vector. The product was named pYr-MirTarget-HMGB1-3UTR.
Detection of Gene Expression Level by RT-PCR Method
Total RNA was extracted from the cells using TRIzol (Sigma, T9424-100ML). In our study, the expression level detection mainly included miRNA, HMGB1, Bcl-2, Bax, P62, and Beclin1.
The real-time fluorescent quantitative polymerase chain reaction (RT-PCR) kits used in this study include Takara’s TB Green Premix ExTaq II (Tli RNaseH Plus) and PrimeScript RT Master Mix (Perfect Real Time), and TIANGEN’s miRcute enhanced miRNA cDNA first-strand synthesis kit and miRcute enhanced miRNA fluorescence quantitative detection kit (SYBR Green). The specific experimental steps are carried out according to the instructions. All primers required for RT-PCR in this study are listed in Table 1 .
Table 1.
All Primers Used for RT-PCR.
| Gene/miRNA | Primer Sequence (5′-3′) | |
|---|---|---|
| M-miR-142-3p | F | GCGCGTGTAGTGTTTCCTACTTTATGGA |
| BAX | F | AGACAGGGGCCTTTTTGCTAC |
| R | AATTCGCCGGAGACACTCG | |
| Bcl2 | F | GTGGATGACTGAGTACCTGAACC |
| R | AGCCAGGAGAAATCAAACAGAG | |
| P62 | F | GTGGGACAGCCAGAGGAACA |
| R | GCCCTTCCGATTCTGGCAT | |
| Beclin | F | GGGTCACCATCCAGGAACTCA |
| R | CACCATCCTGGCGAGTTTCA | |
| HMGB1 | F | CAAGGACCCCAATGCACCCAAG |
| R | AAGCCAGGATGCTCGCCTTTG | |
| HMGB1 3′ UTR-mut | F | ATAGTTAACAGAGTTCCGAATGTGTCTTTAGATAGC |
| R | ACACATTCGGAACTCTGTTAACTATACAAAAAAAGA | |
F = forward; R = reverse; RT-PCR = real-time polymerase chain reaction.
Protein Detection (Western Blot)
The main proteins tested include HMGB1, the apoptosis-related protein Bcl-2/Bax, autophagy-related protein P62, and Beclin1. All antibodies used in this study are from Proteintech.
The cells were washed with precooled PBS (phosphate-buffered saline), then 50 μL cell lysate (RIPA) was add to each well. We transferred the cell-containing lysate to a 1.5 mL EP tube, sonicated it on ice for 1 hour, and then centrifuged at low temperature for 30 minutes. Finally, the supernatant (cell protein lysate) was transferred to a new EP tube.
Specific steps: Polyacrylamide gel for electrophoresis, transfer membrane (wet transfer method: transfer protein on the gel to the nitrocellulose membrane), wash membrane (0.05% TBST), milk blocking (5% skimmed milk powder, room temperature 1 hour), incubate primary antibody, wash membrane again, incubate secondary antibody (HRP-labeled antibody), and color development (Clarity Westren ECL Substrate, Bio-RAD).
Dual Luciferase Detection Report Related to the Regulation of HMGB1 and miRNA
We used bioinformatics related tools (TargetScan) to predict the target genes of miR-142-3p. In this study, primers were designed based on the predicted binding sites of miR-142-3p and target genes or mutations containing binding sites (target gene 3′UTR; Table 1 ). We amplified the 3′UTR sequence and constructed a 3′UTR luciferase reporter gene vector. Plasmid and miRNA were co-transfected into 293T cells, and the fluorescence intensity was measured by Promega GloMax 20/20 Luminometer.
Cell Proliferation Assay
CCK-8: Cells were inoculated to 96-well cell culture plates in the form of 1 × 105/mL. When the cells adhered to the wall and grew to about 90%, the cells were transfected according to different test groups. Change to complete medium after 6 hours. Discard the supernatant after 24 hours, then add the CCK-8 solution (dilute the medium and the CCK-8 stock solution at a ratio of 1:9), and react for 4 hours. Finally, the OD value was detected at 450 nm in the microplate reader.
Apoptosis Detection
FITC-Anexin-V/PI double-staining method: Cells are inoculated to 6-well cell culture plates. When the cells adhere to the wall and grow to about 105 to 106, the cells are transfected according to different groups. Change to complete medium after 6 hours, digest with trypsin after 24 hours, and wash with PBS. Then discard the supernatant and add 500 μL Binding Buffer to resuspend the cells.
Finally, according to the manufacturer’s instructions (Keygen BioTBCH: KGA108-2), add 5 μL Annexin V-FITC and 5 μL propidium iodide staining solution, respectively, and test on the flow cytometer.
Cell Migration and Cell Cycle Detection
The cell scratch method was used to detect the cell migration rate. The cell cycle is detected using a cell cycle detection kit (Keygen BioTBCH). The experimental operation was performed according to the manufacturer’s instructions. Finally, use flow cytometry for detection and analysis. The data were sorted and analyzed using the cell cycle fitting software ModiFit.
Statistical Analysis
All experimental data in our study were obtained through 3 repeated independent experiments, and the results are expressed as mean ± standard deviation. According to the experimental conditions, t test or ANOVA was selected to evaluate whether it is statistically significant (P < 0.05 indicates statistically significant).
Results
Transfection Efficiency
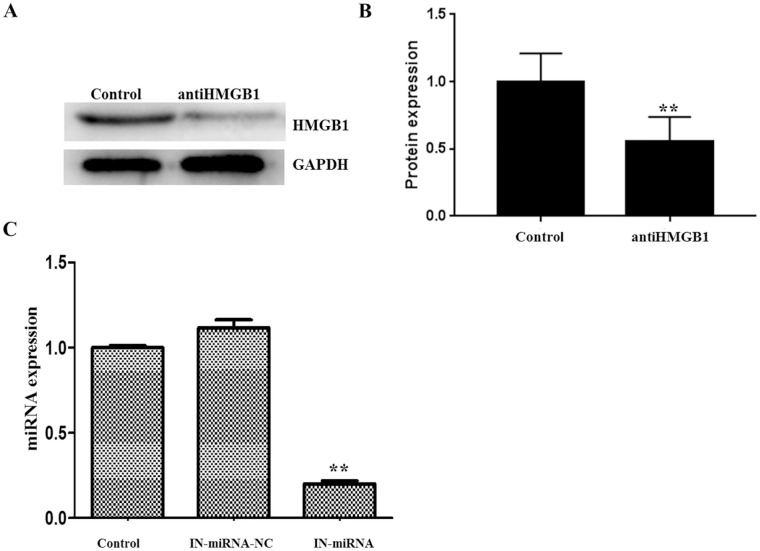
In this study, we conducted a series of experiments such as RNA extraction, fluorescent quantitative PCR (relevant primers are summarized in Table 1 ), western blot, and so on. The results showed that after we added HMGB1 inhibitor (100 μM), the expression level of HMGB1 protein was significantly reduced ( Fig. 1A and B ). After cell transfection, we tested the transfection efficiency. The results showed that the knockdown miR-142-3p group could significantly reduce the expression level compared with the control group, and the transfection efficiency reached 82% ( Fig. 1C ).
Figure 1.
Selection of HMGB1 inhibitor (A: RT-PCR, B: western blot; 100 μM HMGB1 inhibitor can effectively inhibit the expression level of HMGB1) and verification of transfection efficiency (C: the transfection efficiency was 82%; “**” indicates that P < 0.01).
HMGB1 Is miR-142-3p Target Gene
Results of double luciferase assay: First, we found that miR-142-3p may target HMGB1 through bioinformatics related tools (TargetScan) in the early stage. And mutants of mut-HMGB1 appeared in the predicted binding region (miR-142-3p and HMGB1 3’UTR) in this study ( Fig. 2A ). Then, we successfully constructed 2 vectors, which are wild-type wt-HMGB1 and mut-HMGB1 luciferase reporter vectors. The results ( Fig. 2B ) showed that in the cells transfected with miR-142-3p mimic, the luciferase activity of wt-HMGB1 vector was significantly reduced (P < 0.05). However, miR-142-3p mimic has no significant effect on mut-HMGB1. In short, we can conclude that the target gene of miR-142-3p is HMGB1.
Figure 2.
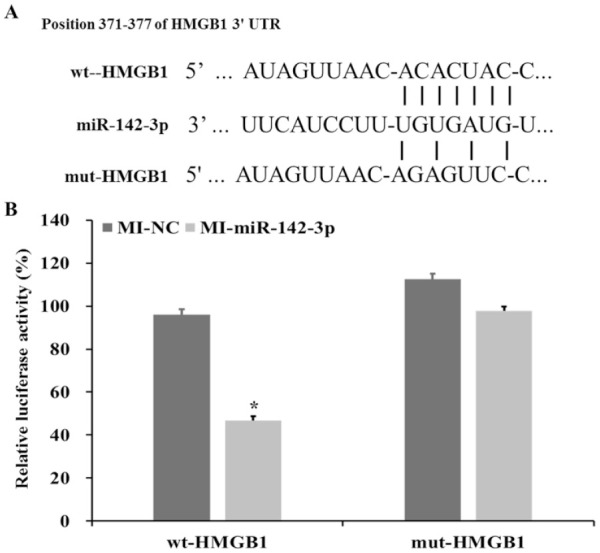
(A) Predict the target gene of miR-142-3p through bioinformatics (TargetScan) and mut-HMGB1 set according to the binding site (mutation is in the predicted binding site of miR-142-3p and HMGB1). (B) The luciferase activities of wt-HMGB1 and mut-HMGB1 were monitored in different experimental groups.
Cell Proliferation and Apoptosis
Cell Proliferation
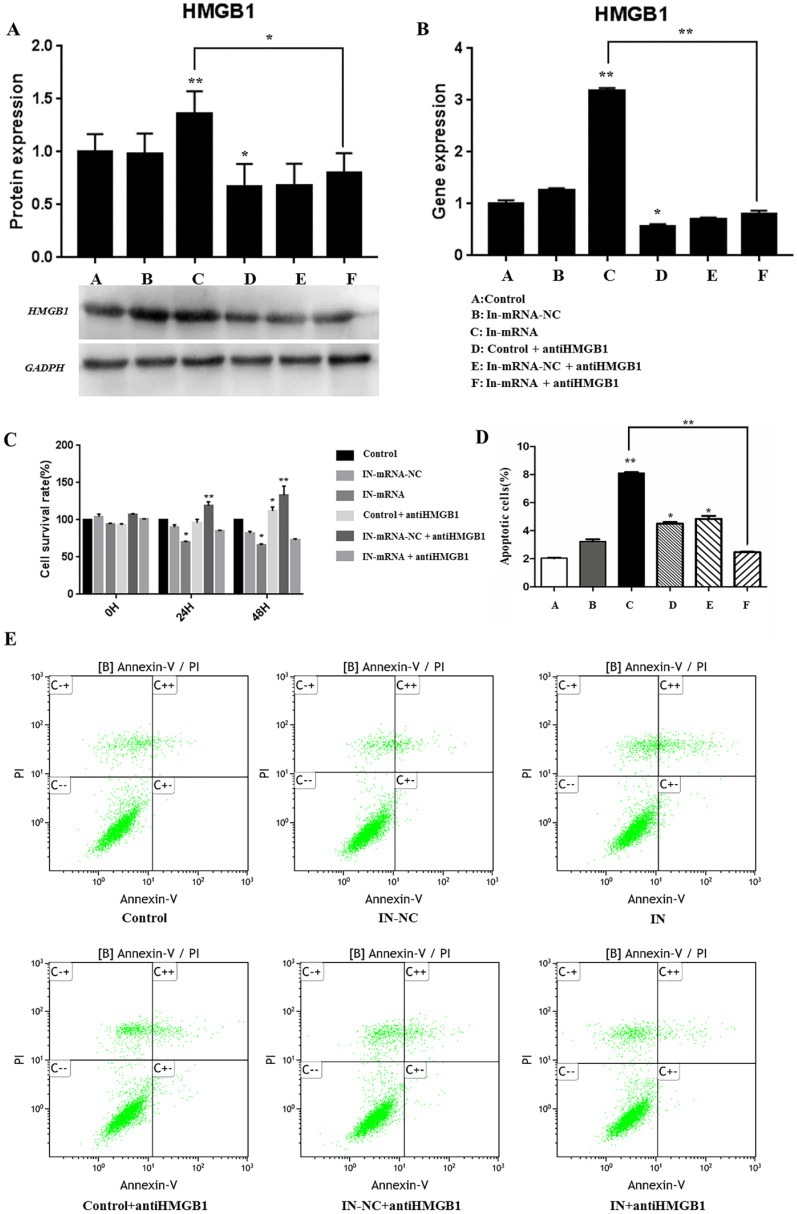
The results of protein detection ( Fig. 3A ) and RT-PCR ( Fig. 3B ) showed that the group of miR-142-3p inhibitor could significantly increase the gene and protein expression levels (HMGB1). However, when we added HMGB1 inhibitor, the expression level was significantly reduced. The above results strongly indicate that miR-142-3p has a negative regulatory effect on HMGB1. As shown in Fig. 3C , miR-142-3p inhibitor significantly inhibited the proliferation of cartilage end plate cells (P < 0.01), and the cell proliferation ability was restored after adding HMGB1 inhibitor.
Figure 3.
Detection of the regulatory effect of miR-142-3p on HMGB1 by western blot (A) and RT-PCR (B). (C) Cell proliferation detection (monitoring at 0 hours, 24 hours, and 48 hours, respectively). (D) Determination of cell apoptosis rate. (E) Flow cytometry to detect cell apoptosis rate.
Flow Cytometry to Detect Apoptosis
The results showed ( Fig. 3D and E ) that compared with the control group, knocking down of miR-142-3p significantly promoted the apoptosis of CEP cells, and the apoptosis rate reached 8.10%. But when the HMGB1 inhibitor was added, it significantly improved the effect of knocking down miR-142-3p on CEP apoptosis, and the apoptosis rate dropped to 2.47%.
Detection Results of Proteins Related Apoptotic
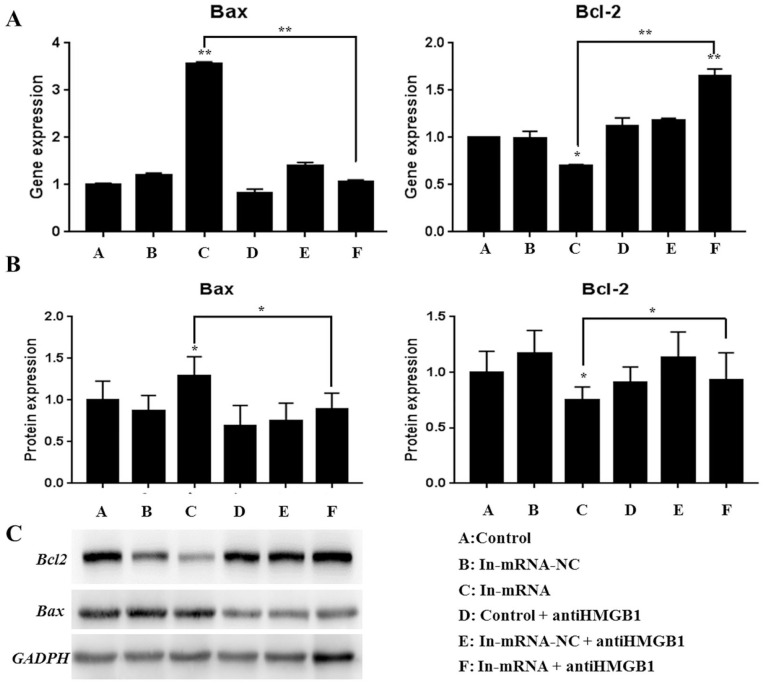
The results of RT-PCR ( Fig. 4A ) and western blot ( Fig. 4B and C ) showed that knocking down of miR-142-3p significantly increased the gene expression level of the apoptosis gene Bax (P < 0.01), and the protein expression also showed a significant increase (P < 0.05). However, gene and protein expression levels of Bax were significantly restored after adding the HMGB1 inhibitor. Conversely, knocking down miR-142-3p will significantly reduce the gene expression level of the Bcl-2 related to anti-apoptotic (P < 0.01), and the protein expression also showed a significantly reduction (P < 0.05). However, Bcl-2 gene and protein expression levels were restored after adding HMGB1 inhibitor. The apoptotic protein test results also showed that knocking down miR-142-3p can promote the apoptosis of cartilage end plate cells, and the apoptosis is improved after adding HMGB1 inhibitor. The above results also indicated that miR-142-3p knockdown promoted apoptosis of CEP cells, and the apoptosis may be improved after the addition of HMGB1 inhibitor.
Figure 4.
Detection of apoptosis-related proteins (A: RT-PCR to detect gene expression level; B and C: western blot to detect protein expression level).
Cell Cycle and Cell Migration
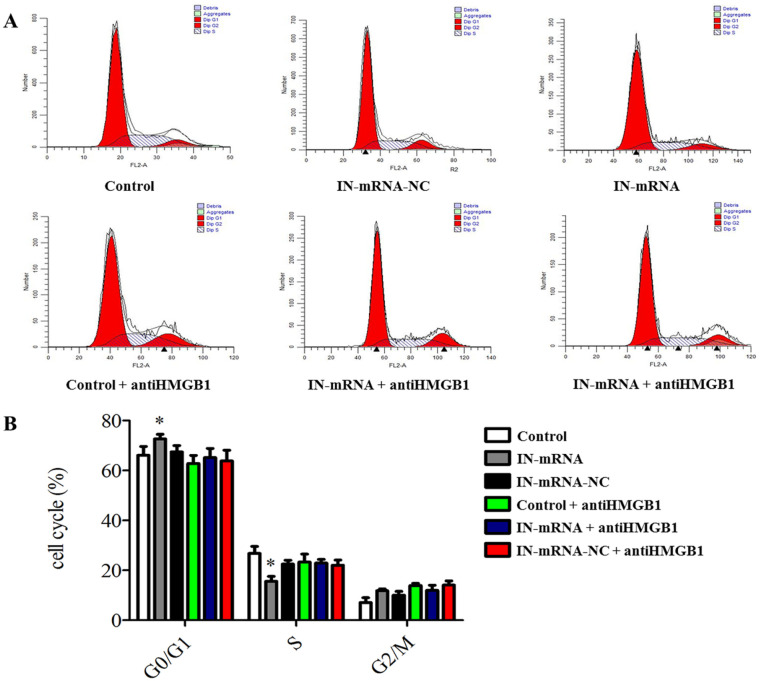
Flow cytometry monitoring results showed ( Fig. 5A and B ) that compared with the control group, knocking down miR-142-3p significantly increased the number of degenerative CEP cells in G0/G1 phase, but significantly reduced in S phase. Our results indicated that knocking down miR-142-3p may inhibit the cell cycle of CEP degeneration entering the S phase from G0/G1. And we found no other statistically significant results in other experimental groups.
Figure 5.
Cell cycle detection (A: flow cytometry to detect cell cycle in different experimental groups; B: knockdown of miR-142-3p can inhibit the degeneration of cartilage endplate cells from entering S phase from G0/G1 phase).
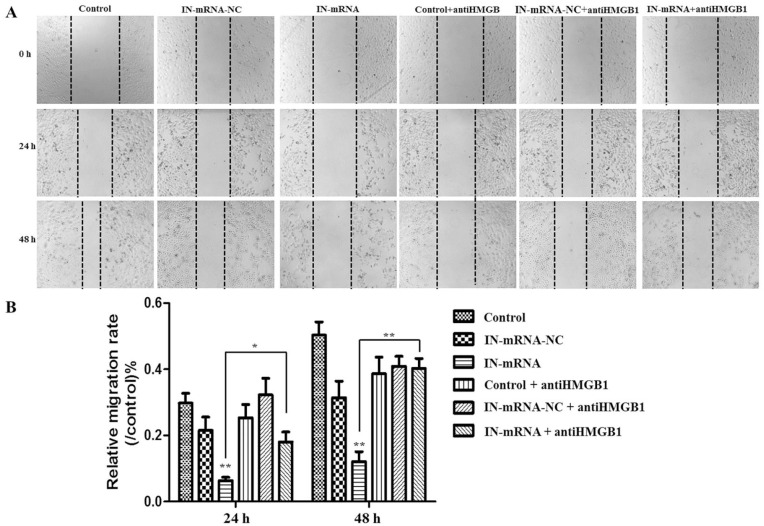
The cell scratch test results showed ( Fig. 6A and B ) that knocking down miR-142-3p significantly inhibited the migration of CEP cells, but when the HMGB1 inhibitor was added, the cell migration ability was restored.
Figure 6.
Detection of cell migration (A: use cell scratch method to detect cell migration in different experimental groups; B: Knockdown of miR-142-3p significantly inhibits the migration of cartilage degenerative cells, after adding HMGB1 inhibitor, the cell migration ability can be restored).
Autophagy
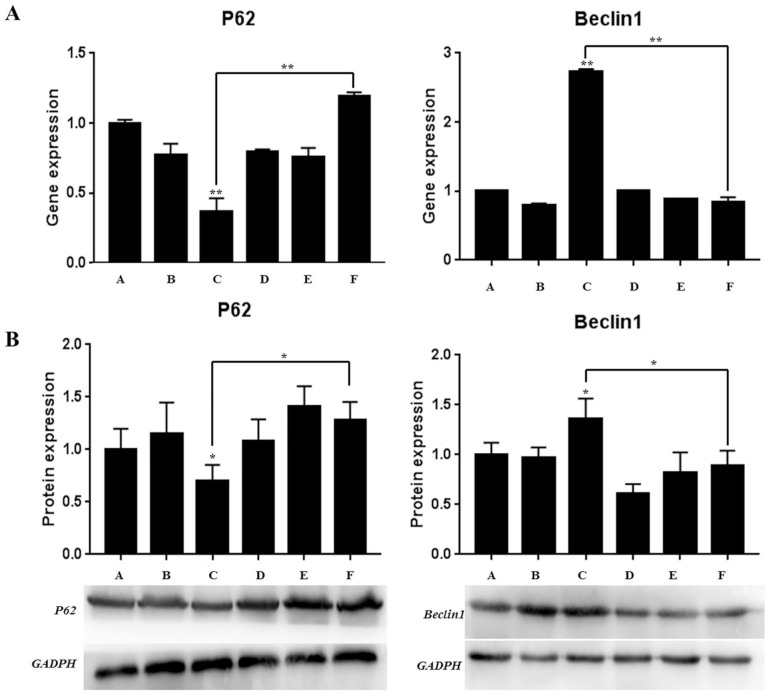
In our study, the autophagy-related proteins P62 and Beclin 1 were detected. The results of RT-PCR showed that ( Fig. 7A ), on the one hand, knocking down miR-142-3p significantly reduced the gene expression level of autophagy marker P62, while significantly increasing the gene expression level of Beclin 1. On the other hand, when the HMGB1 inhibitor was added, the gene expression levels of P62 and Beclin 1 were significantly increased and decreased, respectively.
Figure 7.
The detection of autophagy-related protein (A: detection of gene expression level by RT-PCR method; B: detection of protein expression level by western blot method).
Western blot obtained similar results to RT-PCR ( Fig. 7B ): knocking down miR-142-3p significantly reduced and increased the protein expression levels of P62 and Beclin 1, respectively. When HMGB1 inhibitor was added, the protein expression levels of P62 and Beclin 1 increased and decreased significantly. The above results indicated that the knockdown of miR-142-3p promotes autophagy of CEP cells, and this promotion effect may be restored after adding HMGB1 inhibitor.
Discussion
The mechanism of disc degeneration is complicated. Previous studies have found that the degeneration of CEP will cause a series of chain reactions to further increase the degree of degeneration. 26 Therefore, CEP degeneration may be an important cause of intervertebral disc degeneration. Gruber et al. found that HMGB1 gene expression levels have increased during the process of intervertebral disc degeneration. 24 More important, previous studies have shown that the expression of HMGB1 is regulated by microRNA in many diseases. 27 When we searched the literature, we found that miR-142-3p can inhibit the inflammatory response and apoptosis of chondrocytes in osteoarthritis by inhibiting the NF-κβ signaling pathway mediated by HMGB1. 21 These studies have suggested that it is very meaningful to investigate the regulatory effects of miR-142-3p/HMGB1 in the degeneration of the intervertebral disc CEPs and their regulatory mechanisms.
We have previously learned that HMGB1 is highly expressed in CEP cells, and that miR-142-3p can target and regulate HMGB1 in cartilage end plate cells has been verified in our dual luciferase assay. We verified that miR-142-3p can target and regulate HMGB1 in CEP cells in the results of dual luciferase detection. In this study, we discovered for the first time that miR-142-3p may have a negative regulatory effect on the expression of HMGB1 in the proliferation, apoptosis, migration, autophagy, and cell cycle of CEP degeneration cells. Specifically, we found that compared with control group (normal cells) or IN-miRNA-NC group (the negative control group that knocked down miRNA), knocking down miR-142-3p reduced the proliferation rate and migration rate of CEP cells, but the apoptosis rate was significantly increased. It also inhibits the cell cycle from G0/G1 phase into the S phase. However, when the HMGB1 inhibitor was added, it can reduce the inhibitory effect of miR-142-3p knockdown on cell proliferation and migration to a certain extent, and improve the apoptosis rate of CEP cells. The results of our study were similar to those previous studies. Schwickert et al. 28 and Lei et al. 29 found that miR-142-3p can inhibit breast cancer and non–small cell lung cancer, respectively, and miR-142-3p was considered to be a microRNA that can inhibit tumors. Moreover, previous studies have also reported that miR-142-3p may inhibit chondrocyte apoptosis and inflammation in osteoarthritis by regulating HMGB1. 21 The results of above research and our study can suggest that the negative regulation between miR-142-3p and HMGB1 affects the proliferation, apoptosis, and migration of CEP cells.
In addition, we also found that miR-142-3p and HMGB1 are negatively correlated in the expression of apoptosis-related proteins or autophagy-related proteins of CEP cells, and the results are statistically significant. The above suggests that the regulatory relationship between miR-142-3p and HMGB1 may be involved in related apoptosis or autophagy pathways, but the specific molecular mechanism is still unclear. We need to do more necessary, rigorous, and comprehensive experiments to verify our results.
What is more worthy of our attention is that studies have confirmed that autophagy is involved in various processes such as the regulation of biological development and metabolism, apoptosis, and aging. 30 At the same time, autophagy was also considered to be involved in the occurrence and development of a variety of degenerative diseases, 31 including the pathogenesis of intervertebral disc degeneration. 32 Gruber et al. 33 found that the expression of autophagy-related Beclin 1 in degenerated annulus fibrosus tissue was more significantly upregulated when compared with normal human annulus fibrosus tissue. Combined with our results, we speculated that the regulation of miR-142-3p/HMGB1 in CEP cells may be involved in the process of CEP degeneration. Our results suggest that it may be possible to prevent CEP degeneration by promoting high expression of miR-142-3p to inhibit the expression level of HMGB1. These results suggested that it may be possible to prevent CEP degeneration by promoting the high expression of miR-142-3p to inhibit the expression level of HMGB1.
Conclusion
In summary, our study is the first to find a negative correlation between miR-142-3p and HMGB1 in CEP cells. The regulation between them can affect the proliferation, apoptosis, and migration of cartilage end plate cells. Although the specific molecular mechanism or the specific involved are not yet clear, our results provide a new idea for the targeted treatment of CEP degeneration, that is, it may be possible to inhibit the expression of HMGB1 through the regulation of miR-142-3p.
Footnotes
Author Contributions: Conceptualization: Xuejun Yang and Yong Zhu; methodology: Bo Wang and Demin Ji; software: Wenhua Xing, Feng Li, and Zhi Huang; data curation: Wenhua Xing, Wenkai Zheng, and Jianmin Xue; writing, review, and editing: Bo Wang and Demin Ji.
Acknowledgments and Funding: We thank everyone who helped us in this research. We also thank all authors for their contributions and support. The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported by the National Natural Science Foundation of China (81960406).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: This study was conducted under the standard approved by the Second Affiliated Hospital of Inner Mongolia Medical University.
Informed Consent: Not applicable
ORCID iD: Xuejun Yang  https://orcid.org/0000-0002-5004-7435
https://orcid.org/0000-0002-5004-7435
References
- 1. Neidlinger-Wilke C, Boldt A, Brochhausen C, Galbusera F, Carstens C, Copf F, et al. Molecular interactions between human cartilaginous endplates and nucleus pulposus cells: a preliminary investigation. Spine (Phila Pa 1976). 2014;39(17):1355-64. [DOI] [PubMed] [Google Scholar]
- 2. Xu WN, Zheng HL, Yang RZ, Liu T, Yu W, Zheng XF, et al. Mitochondrial NDUFA4L2 attenuates the apoptosis of nucleus pulposus cells induced by oxidative stress via the inhibition of mitophagy. Exp Mol Med. 2019;51(11):1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He R, Cui M, Lin H, Zhao L, Wang J, Chen S, et al. Melatonin resists oxidative stress-induced apoptosis in nucleus pulposus cells. Life Sci. 2018;199:122-30. [DOI] [PubMed] [Google Scholar]
- 4. Yang CH, Qi WL, Zhao CW, Cai WJ, Gong Q, Niu JY, et al. Parecoxib prevents nucleus pulposus cells apoptosis by suppressing endoplasmic reticulum stress. Eur Rev Med Pharmacol Sci. 2020;24(21):11295-304. [DOI] [PubMed] [Google Scholar]
- 5. Yang RZ, Xu WN, Zheng HL, Zheng XF, Li B, Jiang LS, et al. Involvement of oxidative stress-induced annulus fibrosus cell and nucleus pulposus cell ferroptosis in intervertebral disc degeneration pathogenesis. J Cell Physiol. 2021;236(4):2725-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shamji MF, Setton LA, Jarvis W, So S, Chen J, Jing L, et al. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010;62(7):1974-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jimbo K, Park JS, Yokosuka K, Sato K, Nagata K. Positive feedback loop of interleukin-1beta upregulating production of inflammatory mediators in human intervertebral disc cells in vitro. J Neurosurg Spine. 2005;2(5):589-95. [DOI] [PubMed] [Google Scholar]
- 8. Cs-Szabo G, Ragasa-San Juan D, Turumella V, Masuda K, Thonar EJMA, An HS. Changes in mRNA and protein levels of proteoglycans of the anulus fibrosus and nucleus pulposus during intervertebral disc degeneration. Spine (Phila Pa 1976). 2002;27(20):2212-9. [DOI] [PubMed] [Google Scholar]
- 9. Dong W, Liu J, Lv Y, Wang F, Liu T, Sun S, et al. miR-640 aggravates intervertebral disc degeneration via NF-κB and WNT signalling pathway. Cell Prolif. 2019;52(5):e12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. González Martínez E, García-Cosamalón J, Cosamalón-Gan I, Blanco ME, García-Suarez O, Vega JA. Biology and mechanobiology of the intervertebral disc [in Spanish]. Neurocirugia (Astur). 2017;28(3):135-40. [DOI] [PubMed] [Google Scholar]
- 11. Chen JW, Li B, Yang YH, Jiang SD, Jiang LS. Significance of hypoxia in the physiological function of intervertebral disc cells. Crit Rev Eukaryot Gene Expr. 2014;24(3):193-204. [DOI] [PubMed] [Google Scholar]
- 12. Fujita N, Imai J, Suzuki T, Yamada M, Ninomiya K, Miyamoto K, et al. Vascular endothelial growth factor-A is a survival factor for nucleus pulposus cells in the intervertebral disc. Biochem Biophys Res Commun. 2008;372(2_suppl):367-72. [DOI] [PubMed] [Google Scholar]
- 13. Yuasa T, Otani T, Koike T, Iwamoto M, Enomoto-Iwamoto M. Wnt/beta-catenin signaling stimulates matrix catabolic genes and activity in articular chondrocytes: its possible role in joint degeneration. Lab Invest. 2008;88(3):264-74. [DOI] [PubMed] [Google Scholar]
- 14. Hwang SG, Ryu JH, Kim IC, Jho EH, Jung HC, Kim K, et al. Wnt-7a causes loss of differentiated phenotype and inhibits apoptosis of articular chondrocytes via different mechanisms. J Biol Chem. 2004;279(25):26597-604. [DOI] [PubMed] [Google Scholar]
- 15. Wang C, Wang WJ, Yan YG, Xiang YX, Zhang J, Tang ZH, et al. MicroRNAs: new players in intervertebral disc degeneration. Clin Chim Acta. 2015;450:333-41. [DOI] [PubMed] [Google Scholar]
- 16. Li Z, Yu X, Shen J, Chan MT, Wu WK. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48(3):278-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512(7515):431-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Kurowska-Stolarska M, Alivernini S, Melchor EG, Elmesmari A, Tolusso B, Tange C, et al. MicroRNA-34a dependent regulation of AXL controls the activation of dendritic cells in inflammatory arthritis. Nat Commun. 2017;8:15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shao B, Liao L, Yu Y, Shuai Y, Su X, Jing H, et al. Estrogen preserves Fas ligand levels by inhibiting microRNA-181a in bone marrow-derived mesenchymal stem cells to maintain bone remodeling balance. FASEB J. 2015;29(9):3935-44. [DOI] [PubMed] [Google Scholar]
- 20. Sun M, Zhou X, Chen L, Huang S, Leung V, Wu N, et al. The regulatory roles of microRNAs in bone remodeling and perspectives as biomarkers in osteoporosis. Biomed Res Int. 2016;2016:1652417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Guo Y, Wang C, Yu H, Yu X, Yu H. MicroRNA-142-3p inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting HMGB1. Inflammation. 2016;39(5):1718-28. [DOI] [PubMed] [Google Scholar]
- 22. Zhu Y, Ohba T, Ando T, Fujita K, Koyama K, Nakamura Y, et al. Endogenous TGF-β activity limits TSLP expression in the intervertebral disc tissue by suppressing NF-κB activation. J Orthop Res. 2013;31(7):1144-9. [DOI] [PubMed] [Google Scholar]
- 23. Andersson U, Harris HE. The role of HMGB1 in the pathogenesis of rheumatic disease. Biochim Biophys Acta. 2010;1799(1-2):141-8. [DOI] [PubMed] [Google Scholar]
- 24. Gruber HE, Hoelscher GL, Bethea S, Ingram J, Cox M, Hanley EN., Jr. High-mobility group box-1 gene, a potent proinflammatory mediators, is upregulated in more degenerated human discs in vivo and its receptor upregulated by TNF-α exposure in vitro. Exp Mol Pathol. 2015;98(3):427-30. [DOI] [PubMed] [Google Scholar]
- 25. Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8(4):195-202. [DOI] [PubMed] [Google Scholar]
- 26. Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976). 1995;20(11): 1307-14. [DOI] [PubMed] [Google Scholar]
- 27. Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72(1):230-8. [DOI] [PubMed] [Google Scholar]
- 28. Schwickert A, Weghake E, Brüggemann K, Engbers A, Brinkmann BF, Kemper B, et al. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha V, and additional cytoskeletal elements. PLoS One. 2015;10(12):e0143993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J, et al. MiR-142-3p represses TGF-β-induced growth inhibition through repression of TGFβR1 in non-small cell lung cancer. FASEB J. 2014;28(6):2696-704. [DOI] [PubMed] [Google Scholar]
- 30. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1-2):11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20(12):1338-48. [DOI] [PubMed] [Google Scholar]
- 32. Chen K, Lv X, Li W, Yu F, Lin J, Ma J, et al. Autophagy is a protective response to the oxidative damage to endplate chondrocytes in intervertebral disc: implications for the treatment of degenerative lumbar disc. Oxid Med Cell Longev, 2017;2017:4041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gruber HE, Hoelscher GL, Ingram JA, Bethea S, Hanley EN., Jr. Autophagy in the degenerating human intervertebral disc: in vivo molecular and morphological evidence, and induction of autophagy in cultured annulus cells exposed to proinflammatory cytokines-implications for disc degeneration. Spine (Phila Pa 1976). 2015;40(11):773-82. [DOI] [PubMed] [Google Scholar]