Continued advances in understanding the pathobiology of posttraumatic osteoarthritis (PTOA) and mesenchymal stromal cells (MSCs)-based therapies can be achieved by probing archived samples to ask a posteriori questions. Herein, we isolated RNA and performed 42-plex gene analysis on synovial membrane samples that were embedded in paraffin and stored at room temperature for more than a year. Samples were obtained as part of an in vivo study where intra-articular administration of integrin α10-selected MSCs was shown to protect cartilage and subchondral bone from traumatic injury in equine talocrural joints. 1 Interestingly, synovial membrane histology scores were significantly higher in treated joints at 6 months due to a mononuclear cellular infiltrate of macrophages and lymphocytes. To gain insight into possible mechanisms driving the chondroprotective effects of integrin α10-selected MSCs, RNA was isolated from the synovial membrane for gene expression analysis using the NanoString nCounter platform (NanoString Technologies Inc., Seattle, WA) and a custom-designed 42-gene equine panel. Six months after initiation of PTOA, there was decreased expression of TIMP2 (P = 0.028) and NF-κB (P = 0.031) in treated joints, suggesting that integrin α10-selected MSCs diminish long-term pro-inflammatory and catabolic states after injury. Treatment with integrin α10-selected MSCs also resulted in increased expression of CCL5 (P = 0.049). Through promoting macrophage reprogramming (M1/M2 transition), CCL5 may be aiding in the reparative response elicited by the integrin α10-selected MSCs. 2 This study demonstrates the ability to use archived synovial membrane samples to gain insight into mechanisms of action, and identification of potential targets to mitigate PTOA.
Methods, Results, and Discussion
Horses (n = 8) received bilaterally focal impact injuries to the talar articular surface and were treated intra-articularly with 20 × 106 allogeneic, adipose-derived integrin α10-selected MSCs (treated) or vehicle only (control), as previously described. 1 Synovial biopsies were obtained at the time of surgery (before injury), at 6-week second-look arthroscopy, and postmortem at 6 months. Samples were fixed in 10% neutral-buffered formalin for 24 hours, embedded in paraffin (herein referred to as FFPE samples), and stored at room temperature for 17 to 23 months prior to this study. RNA was extracted from the FFPE samples using High Pure FFPET RNA Isolation Kit (Roche, Indianapolis, IN) and according to NanoString’s recommendations, where a minimum tissue input of 48 mm2 is recommended to obtain about 10 ng/μL of RNA. For this study, ten 10-µm thick sections from FFPE blocks were required to obtain RNA of sufficient quantity and quality to perform gene expression analysis in most samples.
Quality control criteria to select samples for NanoString analysis, included (1) minimum RNA concentration (>10 ng/μL) and (2) RNA purity (A260/A280 ratio between 1.7 and 2.3) measured on a NanoDrop (Thermo Scientific, Waltham, MA). For samples that did not meet these criteria, the RNA extraction was repeated on 10 more sections. If criteria were still not met, the sample was excluded from the NanoString analysis. The minimum input of RNA from each sample to be used for the analysis was calculated based on the extent of RNA degradation, as estimated by smear analysis on the basis of the RNA Quality Number (RQN, <7) and amount of fragments sized 50 to 300 nucleotides (nt) (expressed as percentage of total RNA) measured using the AATI Fragment Analyzer (Advanced Analytical Technologies Inc., Ames, IA). Synovial membrane biopsies that were collected at the initial surgery and 6-week second-look arthroscopy were obtained with an arthroscopic biopsy punch, resulting in very small (<8 mm2) samples, and the RNA extracted from 21 of 28 samples did not meet the inclusion criteria for NanoString analysis. Therefore, only the results obtained from samples harvested during necropsy at 6 months after injury are reported, finally including synovial membrane from control and treated joints of 7 horses (n = 14).
A custom NanoString CodeSet was developed to measure expression of 39 genes associated with inflammation, immunoregulation, and cartilage matrix homeostasis in early PTOA ( Table 1 ).3,4 NanoString nSolver 4.0 Analysis Software was used to process raw data. Background threshold was calculated from the raw data as the mean ±2 SD mRNA count across all the negative controls. Data were normalized against 3 housekeeping genes (GAPDH, HPRT1, and UBC). Normalized log transform data of the gene counts were exported for further analysis in JMP Pro 13 (SAS, Cary, NC). The expression of each gene was compared between treated and control joints using a paired t test, to account for the dependency of the matched joints within the same horse, with P < 0.05 considered significant.
Table 1.
Custom CodeSet Panel: List of Genes Selected for the NanoString Gene Expression Analysis Based on Their Contributions to the Pathogenesis of Posttraumatic Osteoarthritis.
| Inflammation | Immunoregulation | Cartilage Matrix Homeostasis | Housekeeping Genes |
|---|---|---|---|
| CCL2 | ARG1 | MMP-1 | GAPDH |
| COX1 | CCL5 | MMP-3 | HPRT1 |
| COX2 | CD163 | MMP-13 | UBC |
| CXCL12 | CTLA-4 | TIMP1 | |
| CXCL8 | FOXP3 | TIMP2 | |
| IFN-γ | GATA-3 | ||
| IL-12 | IL-1RA | ||
| IL-17A | IL-4 | ||
| IL-23 | IL-10 | ||
| IL-1β | MRC1 | ||
| NF-κB | STAT6 | ||
| NOS-2 | TGF-β1 | ||
| PRG4 | |||
| RUNX2 | |||
| SOX9 | |||
| T-bet | |||
| TNF-α | |||
| IL-2 | |||
| IL-6 | |||
| IL-13 | |||
| IL-22 | |||
| PGE2 | |||
The concentration of the extracted RNA was 272.4 ± 127.5 ng/µL (range 50.9-468 ng/µL). On smear analysis, 30.5% ± 8.7% (range 17%-42.5%) of RNA was represented by 50 to 300 nt length fragments, from which we derived the percentage of RNA with length >300 nt (69.5% ± 8.7%, range 57.5%-83%). Based on the quality control analysis results, a volume was calculated for each sample so that 200 ng of RNA was run against the custom CodeSet on nCounter SPRINT profiler. Gene expression analysis revealed that treatment with integrin α10-selected MSCs, resulted in increase of CCL5 expression (P = 0.049) in treated joints 6 months after injury, and additionally revealed decreased expression of TIMP2 (P = 0.028) and NF-κB (P = 0.031) ( Fig. 1A-C ).
Figure 1.
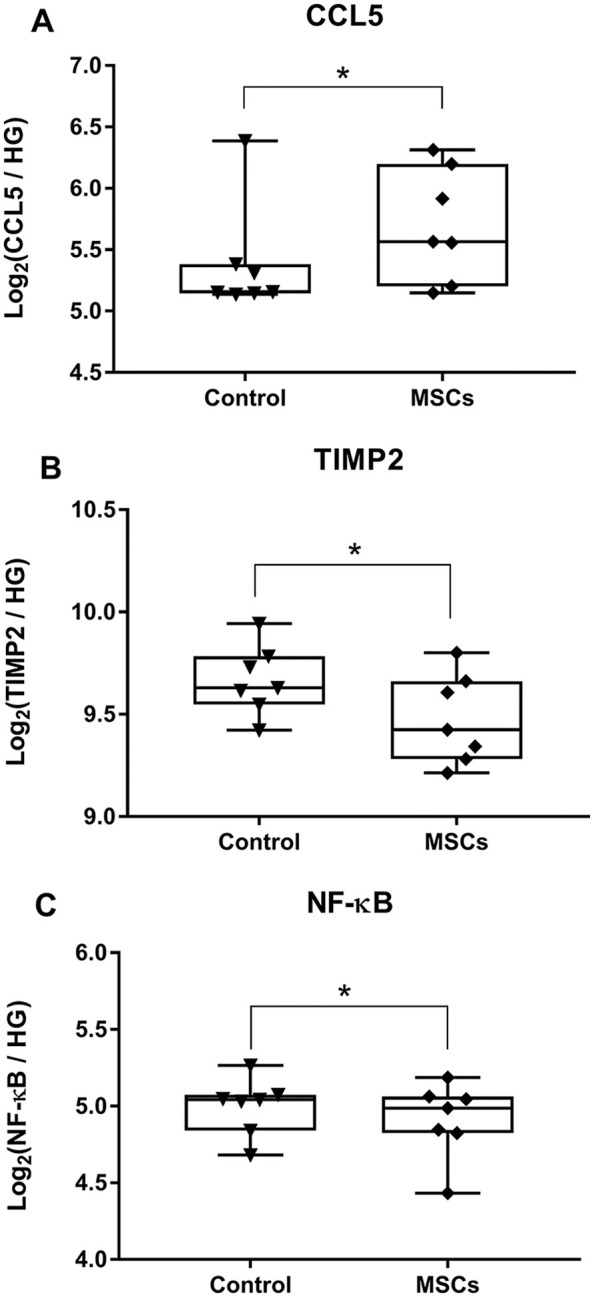
Intra-articular injection of integrin α10-selected mesenchymal stromal cells (MSCs) following acute injury results in long-term changes in gene expression of the synovial membrane including (A) increased expression of CCL5, (B) decreased expression of TIMP2, and (C) decreased expression of NF-κB. Data are expressed as log2 transformed gene counts relative to 3 housekeeping genes (GAPHD, HPRT1, UBC) detected using NanoString. Data are represented as single datapoints, with box-and-whiskers plot indicating median with interquartile range (n = 7 in each group) (*P < 0.05, paired t test). HG = housekeeping genes.
Although CCL5 is classically considered an inflammatory chemokine, recent evidence suggests that CCL5 is critical in the reprogramming of macrophages (M1/M2 transition) during the resolution phase of inflammation, and that CCL5 is actively secreted in high concentrations by resolution phase macrophages.2,5 A transition from M1 to M2 phenotype is also consistent with the decrease in NF-κB expression, a key driver of M1 phenotype, which is upregulated during activation of pro-inflammatory M1 macrophages. 6 Additionally, monocytes are capable of secreting TIMP2 in response to inflammation. 7 TIMP2 is critical for maintenance of tissue homeostasis through its role as a matrix metalloproteinases antagonist. Although counter intuitive, early suppression of TIMPs seems predictive of a positive therapeutic outcome, as normal physiological extracellular matrix remodeling is restored when catabolic tissue destruction by aberrant matrix metalloproteinase release is resolved. 8 The decreased expression of these two genes in joints treated with integrin α10 selected MSCs may be indicative of earlier recovery from the posttraumatic pro-inflammatory and catabolic state.
In the original study, the synovial membrane 6 months postinjury showed a considerable mononuclear cellular infiltrate in the treated joints compared with the control, an unexpected finding that initiated this a posteriori investigation. 1 It has been previously shown that MSCs are detected within injured tissues only during the first 3 to 8 days after administration, thereafter they undergo transdifferentiation or they disappear.9,10 In mice, following intravenous injection, MSCs are found in the lungs where they recruit macrophages and are subsequently phagocytosed within 72 hours, inducing a regulatory macrophage phenotype. 11 The aforementioned studies have reported that autologous, allogeneic, and xenogeneic MSCs are capable of triggering macrophage recruitment and phagocytosis of MCSs, subsequently inducing macrophage polarization to an immunosuppressive phenotype. These events could account for the findings in our study.
The use of the NanoString nCounter platform allowed us to gain valuable insight into long-term changes in gene expression occurring within the synovial membrane following intra-articular traumatic injury and injection of integrin α10-selected MSCs. While methods such as real-time quantitative polymerase chain reaction (RT-qPCR), microarrays and RNA-seq can also be used to analyze RNA gene expression, they require higher quantity and quality of RNA than NanoString. 12 Archived FFPE tissue samples contain RNA that can be valuable for use in retrospective clinical studies. 13 However, the process of formaldehyde fixation, embedding in warm paraffin, and room temperature storage, collectively result in reduced quality and yield of extracted RNA. 14 The NanoString nCounter platform is sensitive, specific, reproducible, and does not require the same complex manipulation stages as platforms like RNA-seq. 12 Additionally, the nCounter platform requires no amplification, as it utilizes a dual-probe system that uses a combination of target-specific capture probe and color-coded reporter probe to directly quantify RNA sequences. The probes recognize a 100-base target region, and therefore, RNA degradation does not typically affect quality of the data which is important for studies involving FFPE samples.
In an ideal study design, tissues are a priori preserved for RNA isolation using snap freezing in liquid nitrogen. 15 However, for a posteriori analysis of musculoskeletal tissues preserved using FFPE, NanoString is a suitable platform to analyze up to 800 genes from clinical samples. NanoString exhibits multiple advantages over other RNA expression methods, including RT-qPCR, microarray, and RNA-seq because of its documented sensitivity, specificity, and robustness. Use of the NanoString nCounter platform can aid in answering mechanistic questions and connect the dots when analyzing histological changes seen in archived clinical samples.
Footnotes
Acknowledgments and Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by Xintela AB, who also supplied the integrin α10 selected cells. Xintela AB had no input into study design, data analysis, or manuscript preparation.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: All procedures performed in accordance with the PHS Policy on Humane Care and Use of Laboratory Animals, federal and state regulations, and were approved by the Cornell University Institutional Animal Care and Use Committee.
ORCID iDs: Lisa A. Fortier  https://orcid.org/0000-0003-1072-5059
https://orcid.org/0000-0003-1072-5059
Liliya Becktell  https://orcid.org/0000-0001-8547-1204
https://orcid.org/0000-0001-8547-1204
Marta Cercone  https://orcid.org/0000-0002-8922-7927
https://orcid.org/0000-0002-8922-7927
References
- 1. Delco ML, Goodale M, Talts JF, Pownder SL, Koff MF, Miller AD, et al. Integrin α10β1-selected mesenchymal stem cells mitigate the progression of osteoarthritis in an equine talar impact model. Am J Sports Med. 2020;48(3):612-23. doi: 10.1177/0363546519899087 [DOI] [PubMed] [Google Scholar]
- 2. Aswad M, Assi S, Schif-Zuck S, Ariel A. CCL5 promotes resolution-phase macrophage reprogramming in concert with the atypical chemokine receptor D6 and apoptotic polymorphonuclear cells. J Immunol. 2017;199(4):1393-404. doi: 10.4049/jimmunol.1502542 [DOI] [PubMed] [Google Scholar]
- 3. Mathiessen A, Conaghan PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res Ther. 2017;19(1):18. doi: 10.1186/s13075-017-1229-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haseeb A, Haqqi TM. Immunopathogenesis of osteoarthritis. Clin Immunol. 2013;146(3):185-96. doi: 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Córdova LA, Loi F, Lin T, Gibson E, Pajarinen Nabeshima A, et al. CCL2, CCL5 and IGF-1 participate in the immunomodulation of osteogenesis during M1/M2 transition in vitro. J Biomed Mater Res Part A. 2018;105(11):3069-76. doi: 10.1002/jbm.a.36166.CCL2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage M1-M2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rooney T, Roux-Lombard P, Veale DJ, FitzGerald O, Dayer JM, Bresnihan B. Synovial tissue and serum biomarkers of disease activity, therapeutic response and radiographic progression: analysis of a proof-of-concept randomised clinical trial of cytokine blockade. Ann Rheum Dis. 2010;69(4):706-14. doi: 10.1136/ard.2009.108324 [DOI] [PubMed] [Google Scholar]
- 8. Galboiz Y, Shapiro S, Lahat N, Miller A. Modulation of monocytes matrix metalloproteinase-2, MT1-MMP and TIMP-2 by interferon-γ and -β: implications to multiple sclerosis. J Neuroimmunol. 2002;131(1-2):191-200. doi: 10.1016/S0165-5728(02)00266-7 [DOI] [PubMed] [Google Scholar]
- 9. Eggenhofer E, Luk F, Dahlke MH, Hoogduijn MJ. The life and fate of mesenchymal stem cells. Front Immunol. 2014;5:148. doi: 10.3389/fimmu.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrera MB, Bussolati B, Bruno S, Morando L, Mauriello-Romanazzi G, Sanavio F, et al. Exogenous mesenchymal stem cells localize to the kidney by means of CD44 following acute tubular injury. Kidney Int. 2007;72(4):430-41. doi: 10.1038/sj.ki.5002334 [DOI] [PubMed] [Google Scholar]
- 11. de Witte SFH, Luk F, Sierra Parraga JM, Gargesha M, Merino A, Korevaar SS, et al. Immunomodulation by therapeutic mesenchymal stromal cells (MSC) is triggered through phagocytosis of MSC by monocytic cells. Stem Cells. 2018; 36(4):602-15. doi: 10.1002/stem.2779 [DOI] [PubMed] [Google Scholar]
- 12. Tsang HF, Xue VW, Koh SP, Chiu YM, Ng LPW, Wong SCC. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Expert Rev Mol Diagn. 2017;17(1):95-103. doi: 10.1080/14737159.2017.1268533 [DOI] [PubMed] [Google Scholar]
- 13. Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, et al. Evaluating robustness and sensitivity of the nanostring technologies ncounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75(13):2587-93. doi: 10.1158/0008-5472.CAN-15-0262 [DOI] [PubMed] [Google Scholar]
- 14. Evers DL, He J, Kim YH, Mason JT, O’Leary TJ. Paraffin embedding contributes to RNA aggregation, reduced RNA yield, and low RNA quality. J Mol Diagn. 2011;13(6):687-94. doi: 10.1016/j.jmoldx.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salehi Z, Najafi M. RNA preservation and stabilization. Biochem Physiol. 2014;3(1):126. doi: 10.4172/2168-9652.1000126 [DOI] [Google Scholar]


