Abstract
Objective
Osteoarthritis (OA) is a degenerative joint disease characterized by deterioration of articular cartilage functions. Previous studies have confirmed the role of circular RNAs (circRNAs) in OA, but the role of mechanical stress–related circRNA (circRNA-MSR) in OA is unknown.
Design
The human chondrocytes C28/I2 were cultured and treated with lipopolysaccharide (LPS) to establish the OA model. The mRNA and protein levels were measured by qRT-PCR or Western blot. Cell viability was analyzed by MTT assay. Flow cytometry was carried out to detect cell apoptosis. The levels of TNF-α, IL-1β, and IL-6 were determined by enzyme-linked immunosorbent assay (ELISA). Pull-down assay was conducted to measure circRNA-MSR-related miRNA. Dual-luciferase reporter gene detection was performed to detect the target relationships between miR-643 and circRNA-MSR or Mitogen-activated protein kinase kinase 6 (MAP2K6). The RNA–fluorescence in situ hybridization (RNA-FISH) assay was conducted to verify the localization of circRNA-MSR and miR-643.
Results
The expressions of circRNA-MSR were upregulated in LPS stimulated C28/I2 cells. Knockdown of circRNA-MSR can inhibit LPS-induced apoptosis, inflammatory response, and extracellular matrix (ECM) degradation, and promote cell C28/I2 cells proliferation. Moreover, circRNA-MSR directly targeted miR-643. RNA-FISH exhibited that circRNA-MSR may act as a competing endogenous RNA (ceRNA) of miR-643. Over-expression of miR-643 could alleviate LPS-induced C28/I2 chondrocyte injury and promote cell proliferation. Besides, miR-643 directly bound to MAP2K6 mRNA. MiR-643 inhibition or MAP2K6 overexpression can reverse the role of circRNA-MSR knockdown on LPS-treated chondrocytes.
Conclusion
circRNA-MSR can upregulate MAP2K6 by targeting miR-643, thereby inhibiting cell proliferation and promoting apoptosis of C28/I2 cells.
Keywords: osteoarthritis, circRNA-MSR, miR-643, MAP2K6
Introduction
Osteoarthritis (OA) is a kind of degenerative disease that is characterized by structural damage and functional failure of articular cartilage. The etiology of OA is complex, including inflammation, oxidative stress, cartilage degradation, and so on. 1 The progression of OA presents irreversibility, mainly showing swelling, pain, stiffness, and even limited movement of the joint, which greatly affects people’s quality of life. 2 Chondrocytes are the only resident cells in the extracellular matrix (ECM) and are essential to maintain the dynamic balance between anabolism and catabolism in the ECM.3,4 The degree of chondrocyte apoptosis in OA patients is closely associated with the degradation of articular cartilage, and the incidence of chondrocyte apoptosis is high in the severely degenerated cartilage matrix. 4 The activation of macrophages and synovial lining cells can lead to the release of inflammatory cytokines, including inflammatory cytokines tumor necrosis factor (TNF)-α, interleukin (IL)-1, and oncostatin M (OSM) to stimulate cartilage degradation. 5 Therefore, it has become the focus of clinical research to find the regulation mechanism of chondrocyte injury.
Circular RNAs (circRNAs) are newly found endogenous noncoding RNAs, which develop covalent closed loops and serve as miRNA sponges that regulate gene expression through binding to targeted miRNAs and further affect the pathophysiological process of multiple diseases, such as atherosclerosis and nervous system disorders.6,7 Chondrocyte ECM-related circRNA (circRNA-CER) regulates the expression of matrix metalloproteinase 13 (MMP13) by serving as a competing endogenous RNA (ceRNA) during OA, and participating in the degradation process of chondrocyte ECM.7,8 As reported in previous study, circRNA_Atp9b acts as a ceRNA that binds to miR-138-5p and regulates OA progression by mediating ECM catabolism and inflammation of chondrocytes. 9 The previous study identified that the expression of circRNA-MSR increased in chondrocytes under mechanical stress. 10 Moreover, circRNAs-MSR knockdown could inhibit the expression of TNF-α and promote ECM formation. 10 However, there are few reports on the molecular mechanism of circRNA-MSR on OA process, and it is of value for further research.
MiRNAs are noncoding RNAs that are 17- to 25-nucleotide long. MiRNA can regulate genes expression by directly binding to the 3′-untranslated region (3′-UTR) of the target mRNAs. Interestingly, miR-643 was reported to be involved in tumor and inflammatory disease.11,12 A study by Wang et al. 12 has identified that miR-643 can act as a tumor suppressor in osteosarcoma by directly targeting zinc finger E-box-binding homeobox 1 (ZEB1). Zhao et al. 11 showed that miR-643 can attenuate lipopolysaccharide (LPS)-induced activation of nuclear factor-κB (NF-κB) pathway and inflammatory factors secretion by targeting tumor necrosis factor receptor-associated factor 6 (TRAF6) during endometritis progression. Based on Circular RNA Interactome prediction databases, miR-643 is identified as a target of circRNA-MSR. However, the interaction between circRNA-MSR and miR-643 in chondrocytes as well as the underlying mechanism is largely unknown.
Mitogen-activated protein kinase kinase 6 (MAP2K6) is an upstream kinase of the p38/MAPK signal pathway, which can be activated under the status of stress and inflammation. 13 MAP2K6 plays a key role in multiple physiological and pathological processes, such as cell cycle regulation, cell growth, development, division, apoptosis, and inflammatory reactions. 13 Previous study has demonstrated that the expression of MAP2K6 is upregulated in OA patients. 14 Nevertheless, the role of MAP2K6 in the progression of OA remains unknown.
In this study, we have focused on a signaling cascade as reported in the cartoon (see Supplemental Material) and our data evidenced that circRNA-MSR can up-regulate MAP2K6 expression by targeting miR-643, thereby inhibiting chondrocyte proliferation and promoting cells apoptosis. These data might expand our insight to understand the molecular mechanism of circRNA-MSR in OA progression.
Materials and Methods
Cell Culture and Treatment
The human chondrocytes C28/I2 were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in complete Roswell Parker Memorial Institute (RPMI)-1640 medium (Gibco-BRL, USA) containing 10% fetal bovine serum (FBS, Gibco-BRL) at 37 °C with 5% CO2. LPS was utilized to stimulate C28/I2 cells to establish the model of OA in vitro. 15 In this study, to construct the OA model, cells were treated by 0, 1, 5, and 10 µg/mL LPS (Sigma, USA), respectively for 12 hours.
Cell Transfection
The circRNA-MSR or MAP2K6 overexpressed vectors (circRNA-MSR or MAP2K6) were generated by inserting the corresponding full-length sequences into pcDNA3.1 vector (ThermoFisher Scientific, USA), with pcDNA3.1 empty vector (vector) as the negative control. The si-circRNA-MSR, miR-643 mimic and miR-643 inhibitor, as well as their corresponding negative controls (si-NC, oe-NC, inhibitor NC) were designed and provided by GenePharma (Shanghai, China). For cell transfection, C28/I2 cells were adjusted the density at 60% and inoculated on 6-well plate, then cell transfection was performed by Lipofectamine 3000 reagent (Invitrogen, USA) for 48 hours according to the manufacturer’s instructions.
RNA Extraction and Quantitative RT-PCR
Total RNA in LPS-treated C28/I2 cells was extracted using Trizol reagent (Invitrogen, USA) as per the manufacturer’s instructions. The OD value was measured to determine the concentration and purity of RNA. Total RNA was reverse transcribed into cDNAs using the high-capacity cDNA reverse transcription kit (Applied Biosystems, USA) according to the protocols provided by the manufacturer. Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay was conducted using the TaqManTM MicroRNA Assay Quantitative PCR Kit (4427975, Applied Biosystems) and SYBR Green real-time PCR Master mix (Toyobo, Japan) on ABI PRISM 7300 Sequence Detection system (Applied Biosystems, CA, USA). GAPDH and U6 were used as the endogenous references. The specific primer sequences used in this work were listed as follows:
circRNA-MSR (F): 5′-TCAAGTTCGCTGACAGGTCC-3′
circRNA-MSR (R): 5′-CGGCTCAGCTCTTCTCTAGC-3′
miR-643 (F): 5′-GCCCACTTGTATGCTAGCTC-3′
miR-643 (R): 5′-GTGCAGGGTCCGAGGT-3′
MAP2K6 (F): 5′-TCAATGCTCTCGGTCAAGTG-3′
MAP2K6 (R): 5′-GTGATGCCCAGACTCCAAAT-3’
GAPDH (F): 5′-CTGACTTCAACAGCGACACC-3′
GAPDH (R): 5′-GTGGTCCAGGGGTCTTACTC-3′
U6 (F): 5′-CTCGCTTCGGCAGCACA-3′
U6 (R): 5′-AACGCTTCACGAATTTGCGT-3′
Total Protein Extraction and Western Blot Assay
C28/I2 cells were lysed using the RIPA reagent (Beyotime, China) and the protein levels were quantified by the BCA protein assay kit (Bio-Rad Laboratories, USA). The equal amount of total protein for each sample was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidine fluoride (PVDF) membranes (Invitrogen, USA). Then the proteins were blocked with 5% skimmed milk diluted in Tris-buffered saline (TBS) containing 0.1% of Tween 20 (TBST buffer). The protein specific primary antibodies including anti-MMP13 (1:2000 dilution, Abcam, UK), anti-aggrecan (1:1000 dilution, Abcam, UK), anti-collagen II (1:3000 dilution, Abcam, UK), anti-Bax (1:1000 dilution, Cell Signaling Technology, USA), anti-Bcl-2 (1:1000 dilution, Cell Signaling Technology, USA), anti-cleaved-caspase-3 (1:500 dilution, Abcam, UK) and anti-GAPDH antibody (1:1000 dilution, Bioss, China) were added and incubated overnight at 4 °C. The secondary horseradish peroxidase conjugated goat anti-rabbit IgG antibodies (1:5000 dilution, Beyotime Biotechnology, China) were incubated for 1 hour at room temperature. Subsequently, the membranes were rinsed with the TBST buffer 3 times for 10 minutes, respectively. The target proteins were visualized using enhanced chemiluminescence reagent (Beyotime, China). The relative band density of each protein was analyzed using the ImageJ software.
MTT Assay
Cell viability was measured by MTT assay using the MTT kit (Sigma, USA) in line with the manufacturer’s protocol. C28/I2 cells were inoculated at 1 × 104 cells per well into a 96-well plate, and then treated by 0, 1, 5, and 10 µg/mL LPS, respectively. The cells were received until reaching 80% confluence. 10 μL of MTT reagent (0.5 mg/mL) were added to each well and incubated for 4 hours. Then 100 μL of dimethyl sulfoxide (DMSO; Sigma, USA) was added to each well and the cells were incubated for 1 hour for dissolution. The absorbance at 490 nm was detected and analyzed using a microplate reader (Bio-Rad, USA). The cell viability was determined by normalizing to the corresponding control group.
Apoptosis Detection by Annexin V-FITC/PI With Flow Cytometry
Cell apoptosis of C28/I2 cells was determined by flow cytometry using annexin V-FITC/PI staining kit (Invitrogen, USA). The C28/I2 cells were trypsinized, rinsed with cold phosphate buffered saline (PBS) buffer and stained with annexin V-FITC/PI solution diluted in 1 × binding buffer. Then flow cytometry detection was conducted using the FACScan flow cytometer (Beckman Coulter, USA). The cell apoptosis rate was analyzed by FlowJo software (version 10, TreeStar, USA) and calculated by the percentage of cells with annexin V-FITC-positive and PI-positive/negative.
Detection of Inflammatory Factors by ELISA
C28/I2 cells were inoculated at 4 × 104 cells per well into a 24-well plate, and then treated by 5 µg/mL LPS. The supernatant of C28/I2 cells was collected and centrifuged at 2000 × g for 20 minutes at 4 °C. After dilution, the levels of TNF-α, IL-1β, and IL-6 in cell supernatant were quantitatively detected using the specific ELISA kits (ThermoFisher Scientific, USA) following the instrument. The optical density (OD) value at 450 nm was recorded by Flexstation 3 microplate reader (Molecular Devices, USA). The standard curve was created and analyzed by CurveExpert 1.3 software (Cabit Information Technology, China) program and normalized to cell protein concentrations.
Pull-Down Assay
The pull-down assay was performed to determine circRNA-MSR-associated miR-643 through specific biotin-labeled circRNA-MSR probe. A total of 1 × 107 C28/I2 cells were harvested, lysed, and sonicated. The circRNA-MSR probe was incubated with C-1 magnetic beads (Life Technologies, USA) for 2 hours at 25 °C. Cell lysate was incubated with circRNA-MSR probe or oligo probe overnight at 4 °C. After rinsing with washing buffer, the RNA mixture was eluted and extracted with RNeasy Mini Kit (Qiagen, Germany) for qRT-PCR assay.
Dual-Luciferase Reporter Gene Detection
The fragment of wide-type (WT-circRNA-MSR) or mutant-type (MUT-circRNA-MSR) CircRNA-MSR involving the possible binding site of miR-643 was cloned and inserted into pMIR-REPORT plasmid (Obio Technology, China). Meanwhile, wild-type or mutant MAP2K6 fragment was constructed and inserted downstream of the luciferase reporter gene of the pMIR-REPORT plasmid. C28/I2 cells were co-transfected with miR-643 mimics and the luciferase reporter plasmid using Lipofectamine 3000 (Invitrogen, USA). After 48 hours of transfection, the relative ratio of Rluc/Luc was analyzed using the Dual-Luciferase reporter system (Promega, USA) as per the instructions.
RNA-FISH Assay
The RNA–fluorescence in situ hybridization (RNA-FISH) assay was conducted to verify the localization of circRNA-MSR and miR-643 in C28/I2 cells in accordance with the manufacturer’s protocol (Sino Biological Inc., China). After the fixed slides were dehydrated, the probes point to circRNA-MSR and miR-643 were added to the slides and pre-denatured at 78 °C for 5 minutes. Subsequently, hybridization was performed overnight at 42 °C. Cell nucleus was counterstained with 4,6-diamidino-2-phenylindole (DAPI, Sigma, USA). Images were tested with the Zeiss LSM 700 Meta confocal microscope (Jena, Germany).
Statistical Analysis
All the tests in this study were repeated at least 3 times with similar results. The measurement data were represented by means ± standard deviations (SD). The pairwise comparison was conducted by using student t test. The results of the Student t test are shown in the Figure 3C and D . For multigroup comparison, nonparametric analysis of variance Friedman test was applied for paired data to firstly compare multiple groups and then the Dunn’s post hoc test was used to find pairwise differences among 2 groups. P < 0.05 represented the difference was statistically significant.
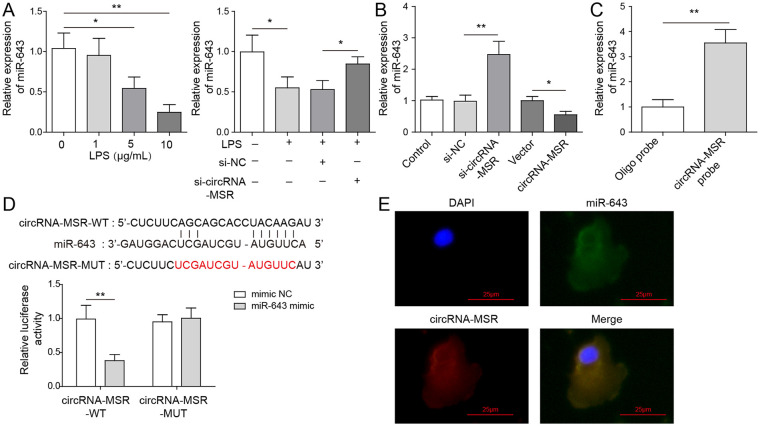
Figure 3.
MiR-643 is proved to be the molecular target of circRNA-MSR. (A, B) The expression of miR-643 in C28/I2 cells was measured by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay. (C) Pull-down assay was conducted to measure circRNA-MSR-related miRNA through a specific biotin-labeled circRNA-MSR probe. (D) The interaction of circRNA-MSR and miR-643 was detected by dual-luciferase reporter detection. (E) The RNA–fluorescence in situ hybridization (RNA-FISH) assay was conducted to verify the localization of circRNA-MSR and miR-643. The measurement data were represented by mean ± standard deviation. Scale bar = 25 μm. *P < 0.05, **P < 0.01. All the above assay were executed independently 3 times. The results of the analysis shown in C and D was using the Student t test. Multiple comparisons were using nonparametric analysis of variance.
Results
The Expression of circRNA-MSR Is Upregulated in C28/I2 Cells With LPS Treatment
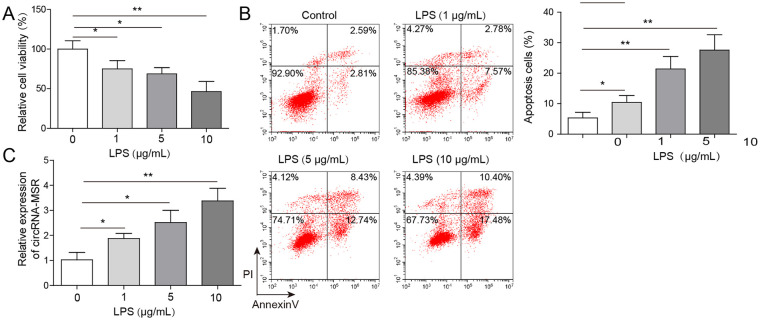
The cell viability of C28/I2 cells was determined by MTT assay. As shown in Figure 1A , the relative cell viability of C28/I2 cells after LPS treatment was remarkably decreased, and in a dose-dependent manner. Flow cytometry was used to quantify LPS-induced cell apoptosis, and the results showed that cell apoptosis was increased following exposure to LPS with dose-dependence ( Fig. 1B ). qRT-PCR assay showed that the expression levels of circRNA-MSR in LPS stimulated C28/I2 cells were upregulated and was dose-dependent ( Fig. 1C ).
Figure 1.
CircRNA-MSR level was increased in C28/I2 cells with lipopolysaccharide (LPS) stimulation. (A) Cell viability of C28/I2 cells after LPS induction was measured by MTT assay. (B) Cell apoptosis of C28/I2 cells induced by LPS was determined by flow cytometry. (C) The expression of circRNA-MSR in C28/I2 cells with LPS treatment was measured by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay. The measurement data were expressed in term of mean ± standard deviation. *P < 0.05, **P < 0.01. All the above assays were executed for three times. Multiple comparisons were using nonparametric analysis of variance.
Knockdown of circRNA-MSR Can Inhibit LPS-Induced Cell Apoptosis and Promote Proliferation of C28/I2 Cells
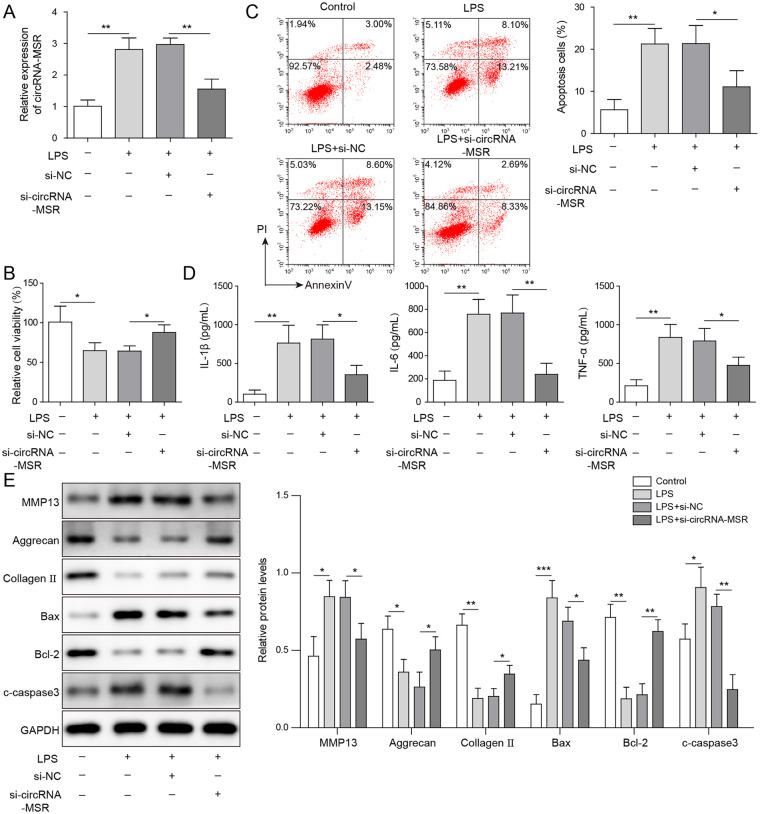
To explore the effect of circRNA-MSR on LPS-induced cell apoptosis, C28/I2 cells were transfected with siRNAs targeting circRNA-MSR (si-circRNA-MSR), and the correlative tests were carried out. As shown in Figure 2A , qRT-PCR results indicated that the level of circRNA-MSR was upregulated in C28/I2 cells induced by LPS and significantly decreased after circRNA-MSR knockdown. Subsequently, MTT assay indicated that the cell activity of LPS-induced C28/I2 cells was remarkably decreased, while that was increased after circRNA-MSR knockdown ( Fig. 2B ). Moreover, the apoptosis rate of C28/I2 cells induced by LPS was dramatically increased while it is decreased after knockdown of circRNA-MSR ( Fig. 2C ). Besides, it was found that cellular inflammatory cytokines TNF-α, IL-1β, and IL-6 levels were increased in LPS-induced C28/I2 cells and remarkably decreased after circRNA-MSR knockdown ( Fig. 2D ). The protein expressions of pro-apoptotic factors Bax and cleaved-caspase 3, anti-apoptotic factor Bcl-2, ECM degradation markers aggrecan, collagen II, and MMP13 were detected by Western blot assay. As shown in Figure 2E , Bax, cleaved-caspase-3, and MMP13 expression increased and the expression of Bcl-2, aggrecan, and collagen II decreased in C28/I2 cells after LPS stimulation, while knockdown of circRNA-MSR decreased the expression level of Bax, cleaved-caspase 3, and MMP13 and promoted the expression of Bcl-2, aggrecan, and collagen II. These results indicated that knockdown of circRNA-MSR could alleviate cell apoptosis and ECM degradation caused by LPS.
Figure 2.
CircRNA-MSR knockdown suppressed lipopolysaccharide (LPS)-induced apoptosis of C28/I2 cells and enhanced cell viability. (A) The expression of circRNA-MSR in C28/I2 cells was measured by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay. (B) Cell viability of C28/I2 cells was measured by MTT assay. (C) Cell apoptosis of C28/I2 cells was determined by flow cytometry. (D) The tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6 levels were detected by enzyme-linked immunosorbent assay (ELISA). (E) The expression levels of Bax, cleaved-caspase-3, matrix metalloproteinase-13 (MMP13), Bcl-2, aggrecan, and Collagen II were measured by Western blot in C28/I2 cells. The measurement data were represented by mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. All the above assay were executed independently 3 times. Multiple comparisons were using nonparametric analysis of variance.
miR-643 Is the Molecular Target of circRNA-MSR
To further analyze the mechanism of circRNA-MSR on LPS-induced C28/I2 cell apoptosis, its downstream target genes were investigated. As shown in Figure 3A , LPS treatment of C28/I2 cells could reduce the level of miR-643 in a dose-dependent manner. In addition, knockdown of circRNA-MSR could resist the inhibition of LPS on miR-643. Meanwhile, overexpression of circRNA-MSR could reduce the expression of miR-643, while knockdown of circRNA-MSR could elevate the level of miR-643 ( Fig. 3B ). Pull-down assay was conducted to measure circRNA-MSR-related miRNA through a specific biotin-labeled circRNA-MSR probe. As shown in Figure 3C , miR-643 could be enriched on the biotin-labeled circRNA-MSR probe in C28/I2 cells. Dual-luciferase reporter gene assay indicated that fluorescence intensity of the group co-transfected with WT-circRNA-MSR and miR-643 mimic was reduced compared with that of the group co-transfected with WT-circRNA-MSR and mimic NC, and the fluorescence intensity of MUT-circRNA-MSR group was basically unchanged ( Fig. 3D ). Furthermore, RNA-FISH assay was performed in C28/I2 cells to measure the immune-localization, and the results showed that circRNA-MSR and miR-643 co-localized in cytoplasm ( Fig. 3E ).
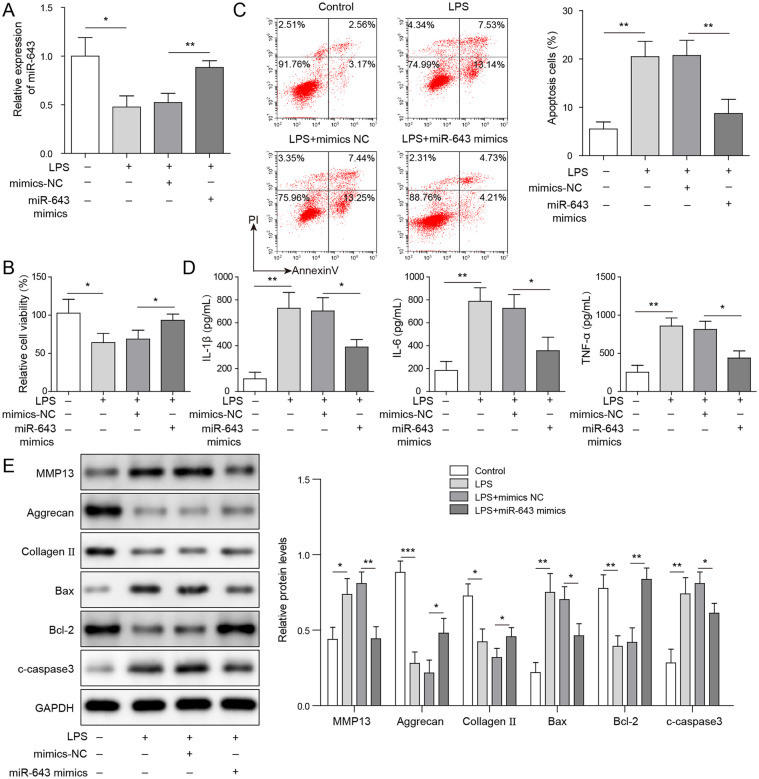
Overexpression of miR-643 Inhibits LPS-Induced C28/I2 Chondrocytes Apoptosis and Promotes Cell Proliferation
To confirm the role of miR-643, C28/I2 cells were transfected with miR-643 mimic and stimulated by LPS. The results of qRT-PCR assay showed that miR-643 levels decreased in C28/I2 cells after LPS induction and increased after miR-643 overexpression ( Fig. 4A ). As shown in Figure 4B , cell viability was dramatically decreased in C28/I2 cells with LPS treatment, while miR-643 mimic transfection could alleviate LPS-induced decrease of cell viability. Moreover, the apoptosis of C28/I2 cells induced by LPS was remarkably increased while it is decreased after overexpression of miR-643 ( Fig. 4C ). Cellular inflammatory cytokines (TNF-α, IL-1β, and IL-6) levels were detected. The results showed that TNF-α, IL-1β, and IL-6 levels in LPS-induced C28/I2 cells was obviously increased, and that in C28/I2 cells with miR-643 mimic transfection was decreased significantly ( Fig. 4D ). Besides, it was found that LPS stimulation resulted in increased the expression of Bax, cleaved caspase-3, and MMP13 and decreased the expression of Bcl-2, aggrecan, and collagen II in C28/I2 cells, while overexpression of miR-643 could reduce the expression levels of Bax, cleaved caspase-3, and MMP13, and promote expression of Bcl-2, aggrecan, and Collagen II ( Fig. 4E ). The results indicated that overexpression of miR-643 could alleviate the apoptosis and ECM degradation caused by LPS.
Figure 4.
Overexpression of miR-643 alleviates chondrocyte injury caused by lipopolysaccharide (LPS). (A) The expression of miR-643 in C28/I2 cells was measured by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay. (B) Cell viability of C28/I2 cells was measured by MTT assay. (C) Cell apoptosis of C28/I2 cells was determined by flow cytometry. (D) The tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6 levels were detected by enzyme-linked immunosorbent assay (ELISA). (E) The protein expressions of Bax, cleaved-caspase-3, matrix metalloproteinase-13 (MMP13), Bcl-2, aggrecan, and collagen II were measured by Western blot in C28/I2 cells. The measurement data were represented by mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. All the above assays were executed independently 3 times. Multiple comparisons were using nonparametric analysis of variance.
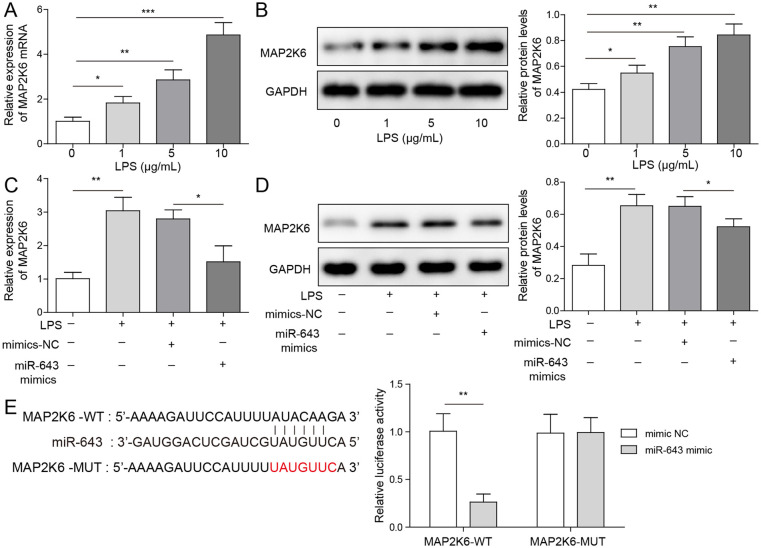
miR-643 Can Target MAP2K6 and Inhibit Its Expression
To further explore the mechanism of miR-643 on LPS-induced C28/I2 cell injury, its downstream target genes were investigated. As shown in Figure 5A and B , LPS treatment could increase MAP2K6 mRNA and protein level in C28/I2 cells and showed a dose-dependent trend. Overexpression of miR-643 alleviates the increased MAP2K6 mRNA and protein expression induced by LPS stimulation ( Fig. 5C and D ). Furthermore, dual-luciferase reporter gene experiments showed that compared with the group co-transfected with mimic NC and WT-MAP2K6, the fluorescence intensity of WT-MAP2K6 and miR-643 mimic co-transfection group decreased markedly, while the fluorescence intensity of the MUT-MAP2K6 group was basically unchanged ( Fig. 5E ). Collectively, it was suggested that miR-643 can directly target MAP2K6 and inhibit its expression.
Figure 5.
MiR-643 can target MAP2K6 and inhibit its expression. (A, B) The mRNA and protein expressions of MAP2K6 in C28/I2 cells were detected by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay and Western blot. (C, D) The mRNA and protein expressions of MAP2K6 in C28/I2 cells with miR-643 mimic transfection was measured. (E) The interaction between miR-643 and MAP2K6 was detected by dual-luciferase reporter detection. The measurement data were represented by mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. All the above assays were executed independently 3 times. Multiple comparisons were using nonparametric analysis of variance.
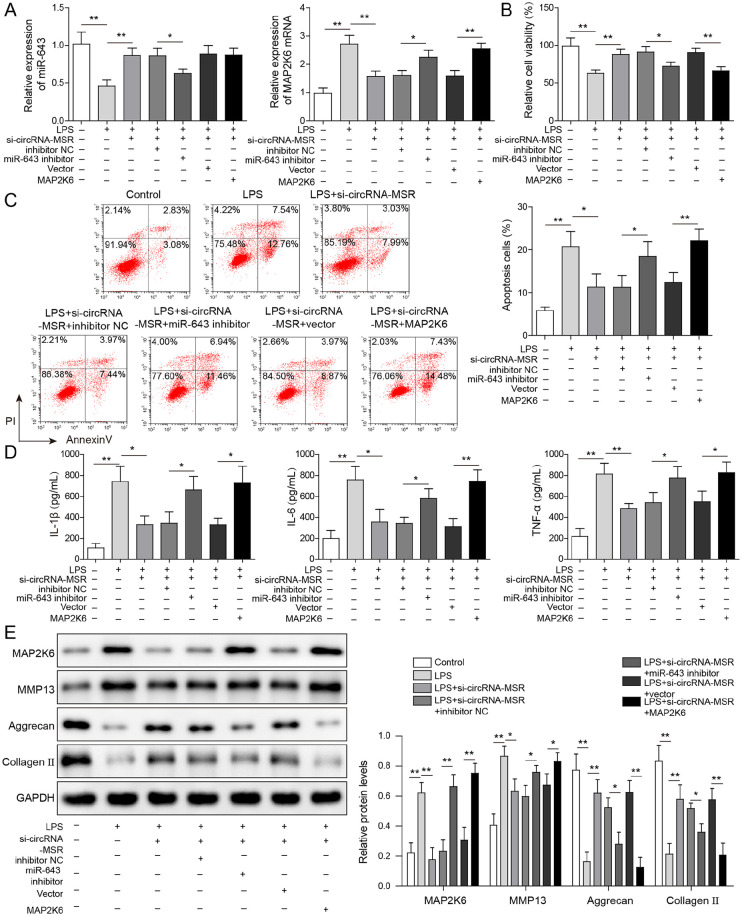
circRNA-MSR Inhibited Proliferation and Promoted Apoptosis of C28/I2 Cells through miR-643/MAP2K6 Signaling Pathway
As shown in Figure 6A , compared with the control group, miR-643 level was decreased and MAP2K6 mRNA level was increased in the LPS-treated group. Compared with LPS treatment group, miR-643 level was upregulated and MAP2K6 mRNA level was downregulated in C28/I2 cells with circRNA-MSR knockdown. Moreover, compared with the LPS + si-circRNA-MSR + inhibitor NC group, the miR-643 level in the LPS + si-circRNA-MSR + miR-643 inhibitor group decreased markedly, while the MAP2K6 mRNA level increased obviously. Transfection of MAP2K6 overexpression vector can increase MAP2K6 mRNA level but has no effect on miR-643 level. The results of MTT and flow cytometry assay indicated that compared with the control group, LPS treatment group showed decreased cell viability and increased apoptosis, while compared with the LPS-treated group, the cell viability was elevated, and apoptosis was reduced after circRNA-MSR knockdown ( Fig. 6B and C ). Besides, Inhibition of miR-643 or overexpression of MAP2K6 can resist the effect of circRNA-MSR knockdown on LPS-treated C28/I2 cells, resulting in decreased cell viability and increased apoptosis of C28/I2 cells. These results suggested that circRNA-MSR inhibits proliferation and promotes apoptosis of C28/I2 cells via miR-643/MAP2K6 pathway. The results of ELISA showed that compared with the control group, the secretion of inflammatory cytokines (IL-1β, IL-6, TNF-α) increased in the LPS-treated group, while the secretion of IL-1β, IL-6, and TNF-α was decreased after knockdown of circRNA-MSR compared with the LPS-treated group ( Fig. 6D ). Meanwhile, inhibition of miR-643 or overexpression of MAP2K6 resists the effect of circRNA-MSR knockdown and increased the secretion of inflammatory factors ( Fig. 6D ). In addition, compared with the control group, the expressions of MAP2K6 and MMP13 protein in the LPS treatment group were increased, while the expressions of aggrecan and collagen II were decreased ( Fig. 6E ). After transfection with si-circRNA-MSR, the expressions of MAP2K6 and MMP13 were reduced, and aggrecan and collagen II expressions were enhanced. Inhibition of miR-643 or overexpression of MAP2K6 can resist the effect of circRNA-MSR knockdown ( Fig. 6E ). The above data implicated that circRNA-MSR promotes inflammatory factors secretion of chondrocytes and ECM degradation through miR-643/MAP2K6 pathway.
Figure 6.
circRNA-MSR promotes apoptosis, inflammatory factors secretion and extracellular matrix degradation of chondrocytes through miR-643/MAP2K6 pathway. (A) The mRNA expression of MAP2K6 and miR-643 in C28/I2 cells were detected by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) assay. (B) Cell viability of C28/I2 cells was measured by MTT assay. (C) Cell apoptosis of C28/I2 cells was determined by flow cytometry. (D) The tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6 levels were detected by enzyme-linked immunosorbent assay (ELISA). (E) The protein expressions of Bax, cleaved caspase-3, MMP13, Bcl-2, aggrecan, and collagen II were measured by Western blot. The measurement data were represented by mean ± standard deviation. *P < 0.05, **P < 0.01. All the above assays were executed independently 3 times. Multiple comparisons were using nonparametric analysis of variance.
Discussion
OA has been shown to be a kind of degenerative disease which is characterized by deterioration of articular cartilage functions and osteophyte formation. 16 It was reported that circRNA-MSR (hsa_circ_0005567) was increased in cartilage tissue of OA patients. 10 Knockdown of circRNA-MSR inhibits inflammatory cytokine TNF-α expression and promotes the degradation of ECM. 10 However, there are few reports on the molecular mechanism of circRNA-MSR regulating OA process, so it is of great value for further research. Human normal chondrocytes C28/I2 cells stimulated by LPS were served as the model of OA. 15 It was proved here that the expression levels of circRNA-MSR in LPS stimulated C28/I2 cells were upregulated and was dose-dependent, indicating that circRNA-MSR was likely to be involved in the disease progression of OA. Knockdown of circRNA-MSR can inhibit LPS-induced apoptosis, inflammatory cytokine secretion and ECM degradation, and promote proliferation of C28/I2 cells. These results elucidate that circRNA-MSR plays a vital role in the process of OA and may be a target of OA treatment.
circRNAs has been proved to modulate gene expression at the transcriptional level or post-transcriptional level. 17 In this study, it was found that circRNA-MSR was co-located with miR-643 in the cytoplasm, suggesting that circRNA-MSR might function as a miRNA sponge. The Circular RNA Interactome prediction website indicated that circRNA-MSR could target miR-643. Pull-down assay and dual-luciferase reporter gene detection conducted here validated the interacting relationship between miR-643 and circRNA-MSR. All these results suggest that circRNA-MSR directly target miR-643 by acting as a sponge, and miR-643 may also play a regulatory role in OA progression.
Up to now, more and more miRNAs have been found to have potential implications of cartilage homeostasis and OA progression. 18 For example, miR-145 negatively regulate TNF-α-modulated signaling cascade and induction of cartilage matrix degradation in OA process by restraining the MKK4-JNK/p38-c-Jun/ATF2 axis. 19 miR-146a could promote cell proliferation and suppress apoptosis of chondrocytes in OA through directly targeting TRAF6 to repress its expression and inhibiting the activation of NF-κB signal axis. 20 As demonstrated by Zhao et al., 11 miR-643 can inhibit endometritis caused by LPS by targeting TRAF6. Besides, the downregulation of TRAF6 inhibits IL-1β-induced chondrocyte apoptosis. 21 Therefore, it is speculated that miR-643 may alleviate the OA process. Here, our findings first imply that miR-643 exhibited anti-inflammatory and anti-apoptotic effects on C28/I2 cells with LPS treatment. In mechanism, we demonstrated that miR-643 could directly target MAP2K6. While it has been proved that stress and inflammation can induce MAP2K6 activation. 22 Tang et al. 23 demonstrated that circ_016719 mediated the expression of MAP2K6 markedly contributed to neuron cell apoptosis induced by ischemia/reperfusion. Based on these studies, we speculated that MAP2K6 may play a regulatory role by participating in the proliferation and apoptosis of chondrocytes during OA process. As demonstrated in this study, miR-643 could directly target MAP2K6, and inhibition of miR-643 or overexpression of MAP2K6 can resist the effects of circRNA-MSR knockdown on LPS-treated C28/I2 cells. In the current work, it was showed that circRNA-MSR could upregulate the expression of MAP2K6 by targeting miR-643, thereby inhibiting chondrocyte proliferation and promoting apoptosis, inflammatory factors secretion, and ECM degradation.
Taken together, our study demonstrated that circRNA-MSR expression level was increased in LPS stimulated C28/I2 cells. CircRNA-MSR down-regulation alleviated LPS induced C28/I2 cells apoptosis, inflammatory factors secretion and extracellular matrix degradation and promoted cell proliferation. Mechanically, circRNA-MSR might act as a sponge for miR-643 to regulate MAP2K6 signaling pathway, which provided a novel target for the cascades in LPS-induced chondrocyte injury. These findings suggested that circRNA-MSR/miR-643/MAP2K6 axis could be used as diagnostic biomarkers in OA treatment. However, chondrocyte injury is one of the factors that contribute to OA. Thus, the regulatory mechanism of circRNA-MSR in synovium tissue or other cell types in OA still need exploration.
Supplemental Material
Supplemental material, sj-tif-1-car-10.1177_19476035211044826 for CircRNA-MSR Regulates LPS-Induced C28/I2 Chondrocyte Injury through miR-643/MAP2K6 Signaling Pathway by Zhen Jia and Qing-Jun Wei in CARTILAGE
Footnotes
Supplementary material for this article is available on the Cartilage website at http://cart.sagepub.com/supplemental.
Acknowledgments and Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iD: Qing-Jun Wei  https://orcid.org/0000-0002-3458-9736
https://orcid.org/0000-0002-3458-9736
References
- 1. Browne JL, Hooshmand S, Elam M, Fe Resin R, Arjmandi B. Relationship between inflammation, oxidative stress, and oxidative damage with severity of knee osteoarthritis (OA). Phytomed Int J Phytother Phytopharmacol. 2012;20:470-80. [Google Scholar]
- 2. Xu Q, Chen B, Wang Y, Wang X, Han D, Ding D, et al. The effectiveness of manual therapy for relieving pain, stiffness and dysfunction in knee osteoarthritis: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2015;23:A387. [PubMed] [Google Scholar]
- 3. Kim H, Kang D, Cho Y, Kim JH. Epigenetic regulation of chondrocyte catabolism and anabolism in osteoarthritis. Mol Cells. 2015;38:677-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas CM, Fuller CJ, Whittles CE, Sharif M. Chondrocyte death by apoptosis is associated with cartilage matrix degradation. Osteoarthritis Cartilage. 2007;15:27-34. [DOI] [PubMed] [Google Scholar]
- 5. Liu Q, Zhao J, Tan R, Zhou H, Lin Z, Zheng M, et al. Parthenolide inhibits pro-inflammatory cytokine production and exhibits protective effects on progression of collagen-induced arthritis in a rat model. Scand J Rheumatol. 2015;44:182-91. [DOI] [PubMed] [Google Scholar]
- 6. Zhang P, Ming Y, Ye Q, Niu Y. Comprehensive circRNA expression profile during ischemic postconditioning attenuating hepatic ischemia/reperfusion injury. Sci Rep. 2019;9:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 “Sponge” in human cartilage degradation. Sci Rep. 2016;6:22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang M, Sampson ER, Jin H, Jia L, Ke QH, Im HJ, et al. MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res Ther. 2013;15:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou BZ, Du D, Huang GX, Chen A, Zhu L. Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene. 2018;646:203-9. [DOI] [PubMed] [Google Scholar]
- 10. Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y, et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao R, Wang J, Zhang X, Chen Y. MiR3 inhibits lipopolysaccharide induced endometritis progression by targeting TRAF6. Cell Biol Int. 2020;44:1059-67. [DOI] [PubMed] [Google Scholar]
- 12. Wang H, Xing D, Ren D, Feng W, Chen Y, Zhao Z, et al. MicroRNA643 regulates the expression of ZEB1 and inhibits tumorigenesis in osteosarcoma. Mol Med Rep. 2017;16:5157-64. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Li Z, Li N, Shen L. MAP2K6 is associated with radiation resistance and adverse prognosis for locally advanced nasopharyngeal carcinoma patients. Cancer Manag Res. 2018;10:6905-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiang W, Li Y, Zhang Z, Fang Y, Li X, Sun Y, et al. Bioinformatics analysis of gene expression profiles of osteoarthritis. Acta Histochem. 2015;117:40-6. [DOI] [PubMed] [Google Scholar]
- 15. Luo X, Wang J, Wei X, Wang S, Wang A. Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sci. 2019;240:117019. [DOI] [PubMed] [Google Scholar]
- 16. Huang J, Ballou LR, Hasty KA. Cyclic equibiaxial tensile strain induces both anabolic and catabolic responses in articular chondrocytes. Gene. 2007;404:101-9. [DOI] [PubMed] [Google Scholar]
- 17. Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiroshi A. Current status and strategy of microRNA research for cartilage development and osteoarthritis pathogenesis. J Bone Metab. 2016;23:121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu G, Zhao X, Wang C, Geng Y, Zhao J, Xu J, et al. MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 2017;8:e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhong JH, Li J, Liu CF, Liu N, Bian RX, Zhao SM, et al. Effects of microRNA-146a on the proliferation and apoptosis of human osteoarthritis chondrocytes by targeting TRAF6 through the NF-κB signalling pathway. Biosci Rep. 2017;37:BSR20160578. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Zhou Q, Xiao Z, Zhou R, Zhou Y, Qian Q. Ubiquitin-specific protease 3 targets TRAF6 for deubiquitination and suppresses IL-1β induced chondrocyte apoptosis. Biochem Biophys Res Commun. 2019;514:482-9. [DOI] [PubMed] [Google Scholar]
- 22. Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689-737. [DOI] [PubMed] [Google Scholar]
- 23. Tang C, Ou J, Kou L, Deng J, Lou S. Circ_016719 plays a critical role in neuron cell apoptosis induced by I/R via targeting miR-29c/Map2k6. Mol Cell Probes. 2020;49:101478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-tif-1-car-10.1177_19476035211044826 for CircRNA-MSR Regulates LPS-Induced C28/I2 Chondrocyte Injury through miR-643/MAP2K6 Signaling Pathway by Zhen Jia and Qing-Jun Wei in CARTILAGE