Abstract
Objective
The knowledge about functions of caspases, usually associated with cell death and inflammation, keeps expanding also regarding cartilage. Active caspases are present in the growth plate, and caspase inhibition in limb-derived chondroblasts altered the expression of osteogenesis-related genes. Caspase inhibitors were reported to reduce the severity of cartilage lesions in osteoarthritis (OA), and caspase-3 might represent a promising biomarker for OA prognosis. The objective of this investigation was to decipher the transcriptomic regulation of caspase inhibition in chondrogenic cells.
Design
Limb-derived chondroblasts were cultured in the presence of 2 different inhibitors: Z-VAD-FMK (FMK) and Q-VD-OPH (OPH). A whole transcriptome RNA sequencing was performed as the key analysis.
Results
The analysis revealed a statistically significant increase in the expression of 252 genes in the FMK samples and 163 genes in the OPH samples compared with controls. Conversely, there was a significant decrease in the expression of 290 genes in the FMK group and 188 in the OPH group. Among the top up- and downregulated genes (more than 10 times changed), almost half of them were associated with OA. Both inhibitors displayed the highest upregulation of the inflammatory chemokine Ccl5, the most downregulated gene was the one for mannose receptors Mrc1.
Conclusions
The obtained datasets pointed to a significant impact of caspase inhibition on the expression of several chondro-/osteogenesis-related markers in an in vitro model of endochondral ossification. Notably, the list of these genes included some encoding for factors associated with cartilage/bone pathologies such as OA.
Keywords: caspase inhibitors, chondroblasts, osteoarthritis, RNA sequencing
Introduction
Functions of caspases are traditionally linked to inflammation and cell death. Knowledge about these processes and the underlying mechanisms progressed during the last decade. 1 This trend applies to cartilage as well. Chondrocyte death by apoptosis and associations with cartilage matrix degradation were reported during cartilage development and homeostasis. 2 Notably, strong correlations exist between chondrocyte apoptosis and pathogenesis, including osteoarthritis (OA). 3 Recently, also other types of cell death such as autophagy were investigated in connection with OA. 4 Notably, the biomarkers of chondrocyte apoptosis and autophagy in OA include cysteine proteases (caspases). 5
Due to the key role of caspases in cell death and inflammation, their pathways have been investigated particularly in pathological processes, and also with respect to therapeutic approaches.6,7 Broad-spectrum caspase inhibitors, small cell-permeable molecules which bind caspase active site and irreversibly block caspase activity, belong to the attractive targets. 6 Caspase inhibitors contain carboxyterminal group: the classical Z-VAD-FMK (FMK) caspase inhibitor includes fluoromethyl ketone, whereas the more effective “new generation” Q-VD-OPH (OPH) inhibitor includes O-phenoxy group.8,9
The therapeutic effects of a broad spectrum of caspase inhibitors were examined in experimentally induced OA in rabbits. In this context, injections of general caspase inhibitor reduced the severity of cartilage lesions in the knee joint. 10 Beneficial effects of intraarticular caspase inhibition therapy were also observed in the case of experimental osteochondral injury. 11 Inhibition of chondrocyte cell death was further performed using the general inhibitor in porcine cartilage explants, 12 or in the case of induced non-apoptotic cell death in the pre-chondrogenic ATDC5 cell line. 13 More recently, the effects of caspase inhibition on cell survival rate were examined in rabbit articular chondrocytes 14 and TMJ chondrocytes. 15 General caspase inhibitor was tested also in engineered cartilage analogues to reduce cell death 16 related to posttraumatic OA. Caspase inhibitors were shown to reduce the severity of cartilage lesions in experimental OA 10 ; however, the specific modifications of gene expression were not investigated.
As apparent from the literature, general caspase inhibitors have been used to prevent cell death and thus to protect chondrocytes and cartilage. However, there is a lack of evidence about their effects beyond apoptosis 1 despite the first reports are emerging.17-19
Therefore, the objective of this investigation was to decipher the transcriptomic regulation of caspase inhibition in chondrogenic cells. General caspase inhibition followed by whole-genome RNA sequencing was performed to test this hypothesis. The effects of Z-VAD-FMK, as the most commonly used broad-spectrum inhibitor, and Q-VD-OPH, as a “new generation” inhibitor, 7 were investigated in parallel.
Material and Methods
Micromass Cultures
Cells for micromass cultures were obtained from mouse forelimbs at embryonic day (E) 12. Fresh post mortem limbs were collected in PSA buffer (Puck’s Saline A liquid) and cut into pieces by a needle. Pieces of tissue were incubated for 1 to 2 hours at 37 °C with Dispase I (Gibco, final activity 1 U/mL) solved in PSA buffer with 10% fetal bovine serum (FBS; Sigma-Aldrich). Dispase I was inactivated by adding culture medium, cells were passed through a 40-µm cell strainer (Corning), centrifuged (100 g/5 minutes), washed with fresh medium, and counted. Subsequently, cell suspension at concentration of 2 × 107/mL were spotted in 10 µL drops on the culture plate and left in 5% CO2/37 °C to adhere. After 60 minutes, spots were covered by a fresh culture medium. Culture medium which supports chondrogenic differentiation was composed of DMEM (Sigma-Aldrich) and Nutrient Mixture F12 (Sigma-Aldrich) in proportion 2:3, 10% FBS (Sigma Aldrich), penicillin/streptomycin (Sigma-Aldrich, final concentration 100 U/mL and 100 µg/mL), L-glutamine (Sigma-Aldrich, final concentration 2 mM), β-glycerol phosphate (Sigma-Aldrich, final concentration 10 mM), and ascorbic acid (Sigma-Aldrich, final concentration 50 µg/mL). Cells were cultured without treatment overnight to adhere to the surface. For caspase inhibition, pharmacological inhibitors Z-VAD-FMK (FMK001, R&D Systems) and Q-VD-OPH (OPH001, R&D Systems) were applied to the micromass cultures at a concentration of 100 µM, according to the manufacturer’s recommendation and previous studies. 18 In the controls, DMSO (dimethyl sulfoxide) as an inhibitor vehicle was added and the control micromass cultures were run in parallel. The medium with caspase inhibitors or DMSO was changed every second day up to 7 days of cultivation. The experiments were performed in 3 biological replicates.
RNA Isolation and RNA Sequencing
The cultured cells were harvested using 350 µL RLT lysis buffer (Qiagen) with β-mercaptoethanol (Sigma-Aldrich). RNeasy Mini Kit (Qiagen) with included DNase I treatment was used for RNA isolation. Isolated RNA of triplicates of samples treated with FMK, OPH, and DMSO was sent to Lexogen GmbH, Campus Vienna Biocenter 5, Wien, for QuantSeq analysis, where the samples were processed and sequenced according to the company's standard operating process. Briefly, the library for sequencing was constructed using QuantSeq 3′mRNA-Seq Library Prep kit FWD for Illumina (Lexogen) and Lexogen i5 6 nt Unique Dual Indexing Add-on kit (Lexogen). Single read RNA-Seq (1 × 75) was conducted using an Illumina NextSeq 500 system (Illumina).
Analysis of RNA-Seq Data
Data analysis including demultiplexing, read quality control, trimming and filtering procedures, mapping and read counting, and differential expression analysis was performed by Lexogen GmbH using their standard pipeline for QuantSeq-FWD data. Briefly, quality control of reads was performed using FastQC, 20 RSeQC, 21 and MultiQC 22 packages. Samples were trimmed using cutadapt tool 23 and aligned to the reference genome using STAR aligner. 24 R package DeSeq2 25 was used for exploration and differential expression of the count data.
Enrichment Analysis
Significantly differentially expressed genes (Foldchange > 2, Padj < 0.01) were analyzed by MouseMine warehouse. 26 The pathway enrichments of Reactome 27 were visualized using Cytoscape and Cytoscape 28 plugins ClueGO 29 and CluePedia. 30 Differentially expressed genes were further analyzed by Singular Enrichment Analysis (SEA) for GO terms enrichment by using agriGO. 31
Results
Transcriptomic Analysis of Caspase-Inhibited Primary Chondroblasts
Mouse limb–derived primary chondroblasts were separately treated with 2 different caspase inhibitors (FMK and OPH) and compared with control (DMSO). A total of 81,455,442 high-quality reads were obtained for 9 samples with an average of 9,050,605 reads for each sample ( Table 1 ). The reads of each sample were mapped to the Mus musculus genome sequence when 78% of the reads were mapped to the genome sequence. A total of 13,633 genes from 52,550 annotated genes were identified; among these, 542 genes were significantly differentially expressed (Foldchange > 2, Padj < 0.01) in FMK samples and 351 in OPH samples compared with control ( Fig. 1 , Supplementary file 1). RNA sequencing analysis of transcriptome revealed a statistically significant increase in expression of 252 genes in FMK samples and 163 genes in OPH samples ( Fig. 1 , Supplementary File 1). Statistically significant downregulated were 290 genes in FMK samples and 188 genes in OPH samples. Focused on the genes that were common between the 2 inhibitors, 130 were upregulated and 153 were downregulated ( Fig. 1 ). The genes upregulated only in FMK samples were the most enriched for processes such as “response to stimulus,” “response to stress,” “response to organic substance,” whereas genes upregulated in OPH samples were the most enriched for processes such as “apoptotic process” or “response to stress.” The downregulated genes were the most enriched for processes like “cell surface receptor signaling pathway,” “cellular response to stimulus,” “cell communication,” and only in FMK samples (Supplementary File 2).
Table 1.
Number of Mapped Reads for Each Sample.
| Replicate | Number of Input Reads | Uniquely Mapped Reads Number | Uniquely Mapped Reads % |
|---|---|---|---|
| DMSO1 | 8,939,952 | 6,928,158 | 77.5 |
| DMSO2 | 8,510,216 | 6,628,394 | 77.89 |
| DMSO3 | 9,406,483 | 7,237,556 | 76.94 |
| FMK1 | 9,242,993 | 7,151,392 | 77.37 |
| FMK2 | 9,687,655 | 7,515,678 | 77.58 |
| FMK3 | 7,886,898 | 6,092,014 | 77.24 |
| OPH1 | 9,591,813 | 7,505,620 | 78.25 |
| OPH2 | 9,554,312 | 7,451,843 | 77.99 |
| OPH3 | 8,635,120 | 6,720,930 | 77.83 |
Figure 1.
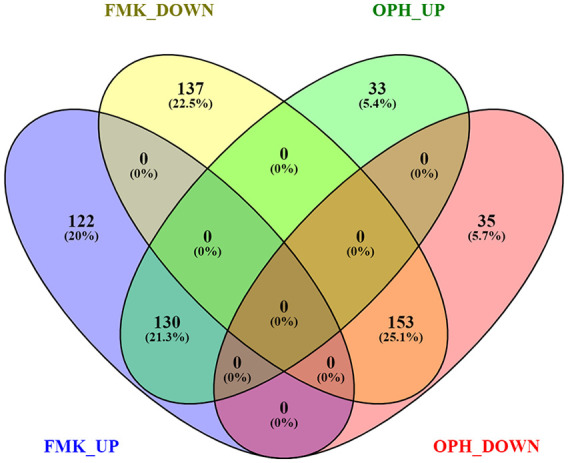
Overlap between genes differentially regulated after FMK and OPH inhibitor treatment. FMK UP = genes upregulated after FMK inhibitor; FMK DOWN = genes downregulated after FMK inhibitor; OPH UP = genes upregulated after OPH inhibitor; OPH DOWN = genes downregulated after OPH inhibitor. ≥2-fold difference and P < 0.01 were used as cutoffs. Visualization was done using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
The top 20 upregulated and downregulated genes for each inhibitor are shown in Table 2 . The majority of these genes were related to OA. After FMK as well as OPH inhibition the top (over 50 times) upregulation was applied for the pro-inflammatory cytokine Ccl5. To the others highly expressed genes connected with OA, Cxcl10, Ifi44, Isg15, Irf7, Mx1, and Cxcl9 were upregulated in the top 20 for both inhibitors. The most downregulated OA-related genes were Laptm5 in FMK inhibitor and Mrc1 in OPH inhibitor. Upregulated genes were the most enriched for GO components like “cell,” “cell part,” “intracellular,” or “nucleus.” Downregulated genes were connected with terms such as “plasma membrane,” “cell periphery,” “plasma membrane part,” or “membrane” (Supplementary File 2).
Table 2.
Top 20 Significantly Upregulated and Downregulated Genes in FMK and OPH Samples.
| FMK-UP |
OPH-UP |
FMK-DOWN |
OPH-DOWN |
||||
|---|---|---|---|---|---|---|---|
| Gene | Fold Change | Gene | Fold Change | Gene | Fold Change | Gene | Fold Change |
| Ccl5 | 87.96 | Ccl5 | 51.90 | Tyrobp | −50.06 | Mrc1 | −10.53 |
| Plac8 | 62.56 | Plac8 | 34.23 | Laptm5 | −37.85 | Adgre1 | −10.04 |
| Cxcl10 | 43.30 | Cxcl10 | 32.77 | Adgre1 | −37.47 | Tyrobp | −8.47 |
| Ifi44 | 34.85 | Ifi44 | 29.71 | Lcp1 | −34.18 | Trem2 | −8.08 |
| Usp18 | 30.79 | Usp18 | 27.17 | Msr1 | −33.00 | Cyth4 | −7.27 |
| Isg15 | 28.24 | Cxcl9 | 24.57 | Csf1r | −32.16 | Cd68 | −7.07 |
| Irf7 | 27.35 | Gbp3 | 24.56 | Nckap1 | −26.79 | Slc37a2 | −7.03 |
| Gbp3 | 25.68 | Ifit1 | 22.56 | Fyb | −24.34 | Mmp9 | −7.01 |
| Ifit1 | 23.57 | Isg15 | 22.18 | Trem2 | −23.95 | Hpgds | −6.98 |
| Ifih1 | 23.37 | Irf7 | 21.56 | Hpgds | −21.50 | Ms4a7 | −6.61 |
| Mx1 | 21.92 | H2-Q4 | 20.57 | Ms4a7 | −21.48 | Slc7a8 | −6.50 |
| Cxcl9 | 21.39 | Ifih1 | 19.64 | Ctss | −19.24 | Csf1r | −6.37 |
| H2-Q4 | 19.70 | Mx1 | 18.25 | Clec4a2 | −18.09 | Fermt3 | −6.34 |
| Oas1b | 19.63 | Oas1b | 18.21 | Spi1 | −16.90 | Fyb | −6.14 |
| Rsad2 | 18.34 | Rsad2 | 17.16 | Ptpn6 | −16.71 | Fcgr2b | −6.12 |
| Zbp1 | 17.19 | Oasl2 | 14.61 | C1qa | −16.46 | Vav1 | −6.12 |
| Herc6 | 17.11 | Herc6 | 13.98 | Fermt3 | −15.18 | Lcp1 | −6.03 |
| Gbp2 | 15.83 | Ecel1 | 13.11 | Myo1f | −14.72 | Pf4 | −5.96 |
| Oasl2 | 15.60 | Gbp2 | 12.95 | Coro1a | −14.37 | Pik3cg | −5.94 |
| Gbp9 | 15.01 | Tap1 | 12.34 | Igsf6 | −13.87 | Nckap1l | −5.79 |
FMK UP = genes upregulated after FMK inhibitor; FMK DOWN = genes downregulated after FMK inhibitor; OPH UP = genes upregulated after OPH inhibitor; OPH DOWN = genes downregulated after OPH inhibitor.
Bold—affected by both inhibitors (FMK and OPH) and being related to OA. Fold changes are included in the right columns. The complete list of upregulated and downregulated genes including fold changes is in Supplementary File 1.
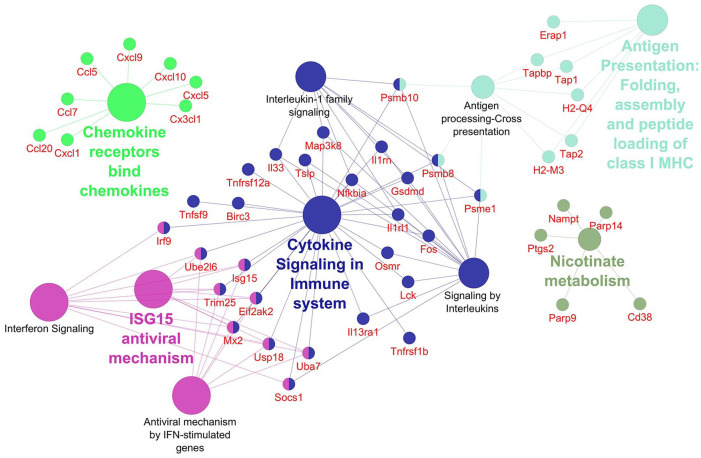
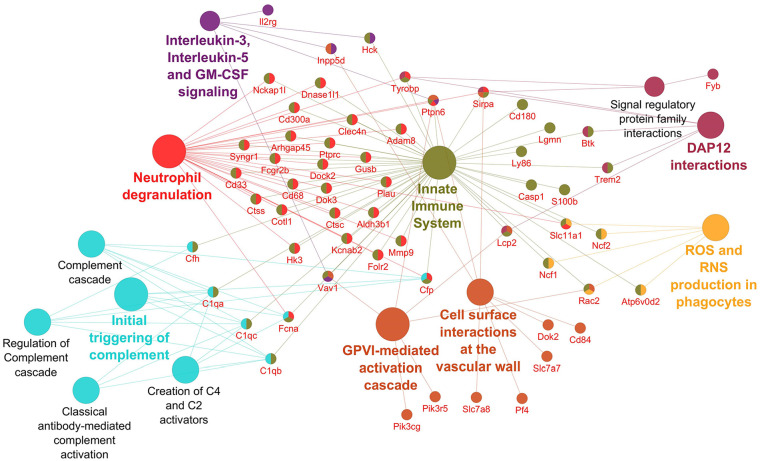
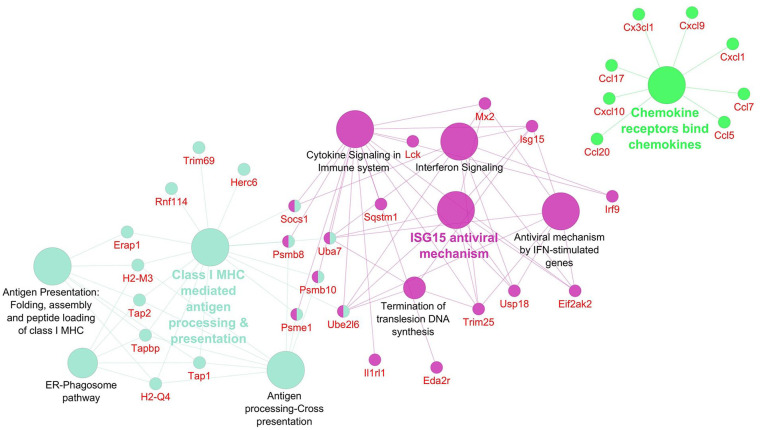
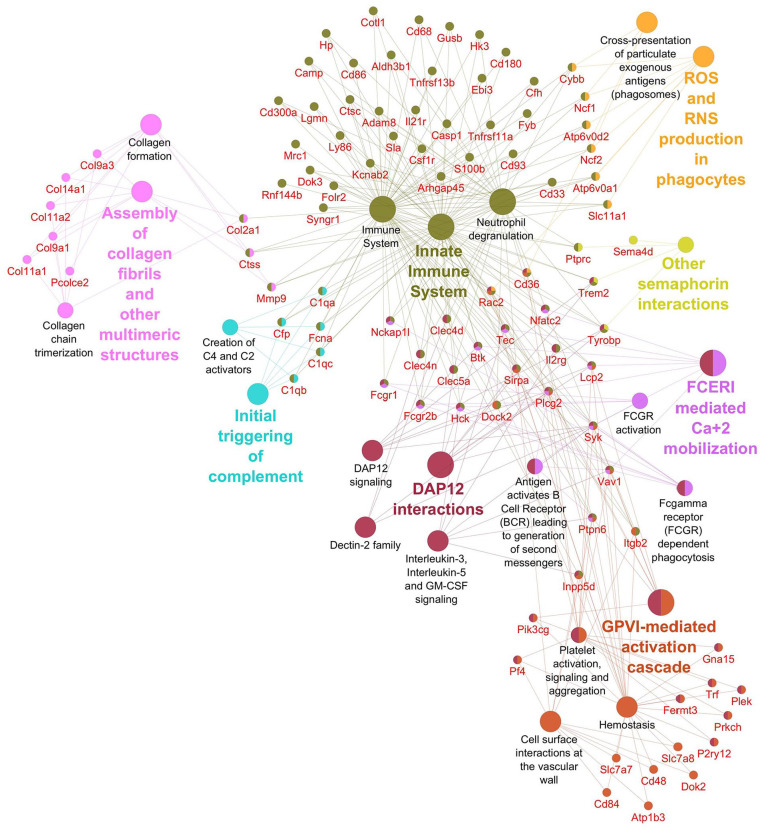
Networks displaying impacted reactome pathways for both general caspase inhibitors compared with control are shown in Figures 2 to 5 where the impact on the immune system is emphasized. In the case of upregulated genes in the FMK ( Fig. 2 ) and OPH ( Fig. 3 ) samples, the affected reactome pathways included the cell-cell communications based on cytokine signaling mediated by interleukins (e.g., IL3, IL5) as well as interferons (IFN). The alterations in interferon-signaling were linked also with ubiquitin-like proteins and the corresponding mechanisms including damage/repair responses. The impact on the innate immune system was apparent also in the spectrum of downregulated genes associated with triggering of the complement system and activating signals in natural killer cells as well as neutrophil degranulation as shown for FMK ( Fig. 4 ) and OPH ( Fig. 5 ) groups.
Figure 2.
Pathways significantly enriched with upregulated transcripts after inhibition using FMK inhibitor. Significantly enriched Reactome pathways 27 (P < 0.01) and related significantly upregulated transcripts (Foldchange > 2, Padj < 0.01) after caspase inhibition using FMK inhibitor. The network was created using Cytoscape 28 and Cytoscape plugins ClueGO 29 and CluePedia. 30
Figure 5.
Pathways significantly enriched with downregulated transcripts after inhibition using OPH inhibitor. Significantly enriched Reactome pathways 27 (P < 0.01) and related significantly downregulated transcripts (Foldchange > 2, Padj < 0.01) after caspase inhibition using OPH inhibitor. The network was created using Cytoscape 28 and Cytoscape plugins ClueGO 29 and CluePedia. 30
Figure 3.
Pathways significantly enriched with upregulated transcripts after inhibition using OPH inhibitor. Significantly enriched Reactome pathways 27 (P < 0.01) and related significantly upregulated transcripts (Foldchange > 2, Padj < 0.01) after caspase inhibition using OPH inhibitor. The network was created using Cytoscape 28 and Cytoscape plugins ClueGO 29 and CluePedia. 30
Figure 4.
Pathways significantly enriched with downregulated transcripts after inhibition using FMK inhibitor. Significantly enriched Reactome pathways 27 (P < 0.01) and related significantly downregulated transcripts (Foldchange > 2, Padj < 0.01) after caspase inhibition using FMK inhibitor. The network was created using Cytoscape 28 and Cytoscape plugins ClueGO 29 and CluePedia. 30
Taken together, both inhibitors caused massive changes in homeostasis and the immune system factors. Moreover, OPH inhibitor affects the cell cycle and adaptive immune system, especially upregulates transcripts related to DNA damage checkpoints and antigen processing and presentation, respectively. FMK inhibitor additionally caused the downregulation of transcripts associated with extracellular matrix organization, in particular collagen formation and degradation.
Datasets generated and analyzed during the current study are available in the GEO repository under the GEO accession number GSE164835.
Discussion
In this investigation, the impact of caspase inhibition on the transcriptome profile of primary chondrogenic cultures was evaluated. Micromass system is a primary cell culture providing 3D microenvironment to follow chondrogenesis in vitro and as such is the most commonly used model.32,33
Two different general caspase inhibitors, Z-VAD-FMK (FMK) and Q-VD-OPH (OPH), were used to achieve caspase inhibition in the cultures. The main reason for using these 2 most common commercially available general caspase inhibitors comes from the fact that they do not display the same efficiency in inhibiting individual caspases. Moreover, in the case of FMK, other cysteine proteases (cathepsins) may be affected along with caspases. 7 The results from both types of inhibitions are provided in parallel among the data presented here, but only those OA-related molecules being significantly impacted by both inhibitors are discussed. Notably, almost half of them are somehow associated also with osteoarthritis.
The top upregulated gene for both inhibitors (FMK and OPH) was Ccl5. Ccls have the ability to induce the release of enzymes associated with cartilage damage and immune response.34,35 Elevated levels of Ccl5 have been identified in leukocytes and synovial tissue of patients with rheumatoid arthritis, reactive arthritis, as well as osteoarthritis.36,37 In human synovial fibroblasts, Ccl5 was demonstrated to promote IL6 production,38,39 a pro-inflammatory cytokine with a confirmed clinical diagnosis of OA. 40 The upregulation of Ccl5 after caspase inhibition is in agreement with complementary results from the RNA-Seq analysis, where other Ccls (namely, Ccl2, Ccl7, Ccl20) were also upregulated. Ccl2 was shown to mediate monocyte recruitment followed by inflammation and cartilage destruction. 39 Ccl7 has been detected in synovial tissue of OA patients. 36 Regarding Ccl20, its concentration within the synovial fluid correlates with the severity of OA. 41 Notably, intraarticular application of caspase inhibitors resulted in a significant reduction of cartilage degradation 10 supporting interplay of pro- and anti-osteoarthritic factors in vivo.
Among the top 20 genes upregulated by caspase inhibitors, there were several interferon-related molecules, such as Ifi44, Isg15, Irf7, Ifit1, and Mx1. Ifi44 (interferon-induced protein 44) gene encodes a protein able to aggregate to form microtubular structures. A study focused on the phenotype of osteoarthritis suggested that Ifi44 was downregulated in damaged cartilage. 42 Isg15 (interferon-stimulated gene 15) acts both as an extracellular cytokine and intracellularly, as a protein modifier. Recently, Isg15 as well as Ifit1 (interferon-induced protein with tetratricopeptide repeats 1) were identified as hub nodes in protein-protein interactions behind the effects of 1,25(OH)2D3 on primary cultures of chondrocytes from patients with osteoarthritis. 43 Irf7 (interferon regulatory factor 7) was associated with several disorders44,45 and was investigated also in synoviocyte innate responses. 46 Additionally, Irf7 was among differentially expressed genes in a gene expression profiling study related to osteoarthritis. 47
Mrc1 (mannose-receptor-C-type1), an anti-inflammatory marker, was the top downregulated gene in OPH and among the top 5 in FMK treated samples. MRC-mediating signaling is involved in collagen reorganization and lysosomal degradation of the cartilage. 48 MRC expressing macrophages have recently been considered as a potential target for cartilage regeneration. 49 Additionally, there is a significant association of Mrc1 with COMP (cartilage oligomeric matrix protein) being a biomarker of OA. 50 Recently, osteoarthritis-associated basic calcium phosphate (BCP) crystals have been shown to downregulate the expression of Mrc1. 51 Notably, treatment with BCP upregulated mRNA levels of Cxcl9 and Cxcl10. This association is in agreement with our observation when Mrc1 downregulation was accompanied by a significant upregulation of Cxcl10 (within top 3) and Cxcl9 in caspase inhibited samples.
The top 20 genes downregulated by caspase inhibition included also other genes being associated with cartilage homeostasis and OA disorders. Laptm5 (lysosomal-associated protein transmembrane 5) was earlier reported as one of the molecules affected in methylation analyses in patients with OA52,53 and in a microarray study related to the severity of OA. 54 Trem2 (triggering receptor expressed on myeloid cells) was investigated with respect to systemic inflammation related also to osteoarthritis. 55 Csf1r (colony-stimulating factor 1 receptor) was earlier examined in osteoarthritis patients in relation to the modulation of inflammatory reaction. 56 Csf1r was recently shown to positively correlate with Cd68 expression. 57 This is in agreement with our observation; Cd68 expression also decreased in the inhibited samples. Notably, a decrease in synovial CD68-positive macrophages represents one of the biomarkers of the effectiveness of arthritis treatment. 58
Mmp9 (matrix metalloproteinase 9), also appearing as a potential biomarker, was included in the top 10 downregulated genes after caspase inhibition. High levels of Mmp9 were detected in OA patients and might thus take part in the cartilage destruction in OA.59,60 Additionally, miR-targeted Mmp9 caused a reduction of articular cartilage degradation. 61
The analysis of interactions among impacted genes/proteins pointed to the major effects of caspase inhibition within the immune system components. The effect was associated not only with cytokine signaling but also with the regulation of IFN-γ signaling and neutrophils. The role of the immune system in the progression of osteoarthritis 62 has also been investigated in the case of other types of arthritis, such as rheumatoid arthritis. 63 IFN-γ plays role in IL-1β driven matrix metalloproteinases synthesis resulting in the downregulation of MMP-1 and MMP-3 production in vitro. 64 Besides this, IFN-γ was found to induce a dose-dependent decrease in cell proliferation and proteoglycan synthesis in cultured human chondrocytes. 65 Recently, the interest in the immunogenic properties of chondrocytes was stimulated by their possible implication in chondrocyte transplantations for therapeutic purposes. 66 The other components of the reactome impacted by both inhibitors included molecules involved in signal transduction, metabolism, and transport of small molecules. MAPK signalization, upregulated in both groups, was described as regulating the degradation of cartilage ECM. 67 Furthermore, PECAM-1 also decreased in both investigated groups. PECAM-1 inhibition was suggested for clinical treatment of rheumatoid arthritis. 68 The direct impact on cartilage differentiation can be connected also to a decrease of SNARE disinhibition participating in lysosomal degradation of ECM. v-SNARE proteins have potential roles in lysosomal exocytosis in hypertrophic chondrocytes. 69
The results from the present research provide a comprehensive view of the impact of general caspase inhibition on the transcriptional profile in primary chondrogenic cells. As a consequence of caspases inhibitions, a large number of genes were up- or downregulated. Additionally, impacts of the 2 most common caspase inhibitors (FMK and OPH) were compared and the overlapping spectrum of genes, as well as differences, were illustrated.
Although in vitro approaches were applied and a classical model of chondrogenesis was challenged, the results also indicated possible fragile points in chondrocyte homeostasis impacted by caspase inhibition. The obtained data thus offer a solid background for further extrapolations and complementary investigations toward chondrocyte differentiation, maintenance, and dysregulations. With this respect, the evaluation emphasized expression modifications of genes associated with osteoarthritis.
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Supplemental material, sj-xlsx-1-car-10.1177_19476035211044823 for General Caspase Inhibition in Primary Chondrogenic Cultures Impacts Their Transcription Profile Including Osteoarthritis-Related Factors by Barbora Vesela, Martina Zapletalova, Eva Svandova, Alice Ramesova, Jaroslav Doubek, Hervé Lesot and Eva Matalova in CARTILAGE
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Supplemental material, sj-xlsx-2-car-10.1177_19476035211044823 for General Caspase Inhibition in Primary Chondrogenic Cultures Impacts Their Transcription Profile Including Osteoarthritis-Related Factors by Barbora Vesela, Martina Zapletalova, Eva Svandova, Alice Ramesova, Jaroslav Doubek, Hervé Lesot and Eva Matalova in CARTILAGE
Footnotes
Supplementary material for this article is available on the Cartilage website at https://journals.sagepub.com/home/car.
Acknowledgments and Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Supported by the ITA Grant Agency of the University of Veterinary and Pharmaceutical Sciences Brno (FVL/Matalová/ITA2019; https://www.vfu.cz/cz/ita-vfu-brno-2019).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval was not sought for the present study because the mouse samples were obtained post mortem in agreement with the recent legislative in the Czech Republic, law 359/2012 Sb., where according paragraph 3, part t) there is no specific requirement for approvals in case when organ/tissue samples are collected post mortem.
Informed Consent: Informed consent was not sought for the present study because only animal samples were used.
ORCID iD: Barbora Vesela  https://orcid.org/0000-0001-8585-2082
https://orcid.org/0000-0001-8585-2082
References
- 1. Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22(4):526-39. doi: 10.1038/cdd.2014.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomas CM, Fuller CJ, Whittles CE, Sharif M. Chondrocyte death by apoptosis is associated with cartilage matrix degradation. Osteoarthritis Cartilage. 2007;15(1):27-34. doi: 10.1016/j.joca.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 3. Hwang H, Kim H. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 2015;16(11):26035-54. doi: 10.3390/ijms161125943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. An S, Hu H, Li Y, Hu Y. Pyroptosis plays a role in osteoarthritis. Aging Dis. 2020;11(5):1146-57. doi: 10.14336/AD.2019.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Musumeci G, Castrogiovanni P, Trovato F, Weinberg AM, Al-Wasiyah MK, Alqahtani MH, et al. Biomarkers of chondrocyte apoptosis and autophagy in osteoarthritis. Int J Mol Sci. 2015;16(9):20560-75. doi: 10.3390/ijms160920560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudelova J, Fleischmannova J, Adamova E, Matalova E. Pharmacological caspase inhibitors: research towards therapeutic perspectives. J Physiol Pharmacol. 2015;66(4):473-82. doi: 10.1179/tex.1992.23.1.97 [DOI] [PubMed] [Google Scholar]
- 7. Chauvier D, Ankri S, Charriaut-Marlangue C, Casimir R, Jacotot E. Broad-spectrum caspase inhibitors: from myth to reality? Cell Death Differ. 2007;14(2_suppl):387-91. doi: 10.1038/sj.cdd.4402044 [DOI] [PubMed] [Google Scholar]
- 8. Caserta TM, Smith AN, Gultice AD, Reedy MA, Brown TL. Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis. 2003;8(4):345-52. doi: 10.1023/A:1024116916932 [DOI] [PubMed] [Google Scholar]
- 9. Van Noorden CJF. The history of Z-VAD-FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem. 2001;103(3):241-51. doi: 10.1078/0065-1281-00601 [DOI] [PubMed] [Google Scholar]
- 10. D’Lima D, Hermida J, Hashimoto S, Colwell C, Lotz M. Caspase inhibitors reduce severity of cartilage lesions in experimental osteoarthritis. Arthritis Rheum. 2006;54(6):1814-21. doi: 10.1002/art.21874 [DOI] [PubMed] [Google Scholar]
- 11. Dang AC, Warren AP, Kim HT. Beneficial effects of intra-articular caspase inhibition therapy following osteochondral injury. Osteoarthritis Cartilage. 2006;14(6):526-32. doi: 10.1016/j.joca.2005.12.010 [DOI] [PubMed] [Google Scholar]
- 12. Otsuki S, Brinson DC, Creighton L, Kinoshita M, Sah RL, D’Lima D, et al. The effect of glycosaminoglycan loss on chondrocyte viability: a study on porcine cartilage explants. Arthritis Rheum. 2008;58(4):1076-85. doi: 10.1002/art.23381 [DOI] [PubMed] [Google Scholar]
- 13. Morimoto R, Obinata A. Overexpression of hematopoietically expressed homeoprotein induces nonapoptotic cell death in mouse prechondrogenic ATDC5 cells. Biol Pharm Bull. 2011;34(10):1589-95. doi: 10.1248/bpb.34.1589 [DOI] [PubMed] [Google Scholar]
- 14. Maeda T, Toyoda F, Imai S, Tanigawa H, Kumagai K, Matsuura H, et al. Lidocaine induces ROCK-dependent membrane blebbing and subsequent cell death in rabbit articular chondrocytes. J Orthop Res. 2016;34(5):754-62. doi: 10.1002/jor.23092 [DOI] [PubMed] [Google Scholar]
- 15. Zhou S, Xie Y, Li W, Huang J, Wang Z, Tang J, et al. Conditional deletion of Fgfr3 in chondrocytes leads to osteoarthritis-like defects in temporomandibular joint of adult mice. Sci Rep. 2016;6(1):24039. doi: 10.1038/srep24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohanraj B, Meloni GR, Mauck RL, Dodge GR. A high-throughput model of post-traumatic osteoarthritis using engineered cartilage tissue analogs. Osteoarthritis Cartilage. 2014;22(9):1282-90. doi: 10.1016/j.joca.2014.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janečková E, Bíliková P, Matalová E. Osteogenic potential of caspases related to endochondral ossification. J Histochem Cytochem. 2018;66(1):47-58. doi: 10.1369/0022155417739283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vesela B, Svandova E, Ramesova A, Kratochvilova A, Tucker AS, Matalova E. Caspase inhibition affects the expression of autophagy-related molecules in chondrocytes. Cartilage. Epub 2020 Jul 4. doi: 10.1177/1947603520938444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolberg-Stolberg J, Sambale M, Hansen U, Raschke ASM, Bertrand J, Pap T, et al. Cartilage trauma induces necroptotic chondrocyte death and expulsion of cellular contents. Int J Mol Sci. 2020;21(12):4204. doi: 10.3390/ijms21124204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wingett SW, Andrews S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184-5. doi: 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 22. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047-8. doi: 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10-2. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 24. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motenko H, Neuhauser SB, O’Keefe M, Richardson JE. MouseMine: a new data warehouse for MGI. Mamm Genome. 2015;26(7-8):325-30. doi: 10.1007/s00335-015-9573-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabergat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2019;48(D1):D498-D503. doi: 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091-3. doi: 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661-3. doi: 10.1093/bioinformatics/btt019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. AgriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45(W1):W122-W129. doi: 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mello MA, Tuan RS. High density micromass cultures of embryonic limb bud mesenchymal cells: an in vitro model of endochondral skeletal development. Vitr Cell Dev Biol Anim. 1999;35(5):262-9. doi: 10.1007/s11626-999-0070-0 [DOI] [PubMed] [Google Scholar]
- 33. Butterfield NC, Qian C, Logan MPO. Pitx1 determines characteristic hindlimb morphologies in cartilage micromass culture. PLoS One. 2017;12(7):e0180453. doi: 10.1371/journal.pone.0180453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yoshida S, Arakawa F, Higuchi F, Ishibashi Y, Goto M, Sugita Y, et al. Gene expression analysis of rheumatoid arthritis synovial lining regions by cDNA microarray combined with laser microdissection: up-regulation of inflammation-associated STAT1, IRF1, CXCL9, CXCL10, and CCL5. Scand J Rheumatol. 2012;41(3):170-9. doi: 10.3109/03009742.2011.623137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agere SA, Akhtar N, Watson JM, Ahmed S. RANTES/CCL5 induces collagen degradation by activating MMP-1 and MMP-13 expression in human rheumatoid arthritis synovial fibroblasts. Front Immunol. 2017;8:1341. doi: 10.3389/fimmu.2017.01341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haringman JJ, Smeets TJM, Reinders-Blankert P, Tak PP. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann Rheum Dis. 2006;65(3):294-300. doi: 10.1136/ard.2005.037176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao Z, Liu Z, Wang R, Zheng Y, Li H, Yang L. Galectin-3 is a potential mediator for atherosclerosis. J Immunol Res. 2020;2020:5284728. doi: 10.1155/2020/5284728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang CH, Hsu CJ, Fong YC. The CCL5/CCR5 axis promotes interleukin-6 production in human synovial fibroblasts. Arthritis Rheum. 2010;62(12):3615-24. doi: 10.1002/art.27755 [DOI] [PubMed] [Google Scholar]
- 39. Raghu H, Lepus CM, Wang Q, Wong HH, Lingampalli N, Oliviero F, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis. 2017;76(5):914-22. doi: 10.1136/annrheumdis-2016-210426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akeson G, Malemud CJ. A role for soluble IL-6 receptor in osteoarthritis. J Funct Morphol Kinesiol. 2017;2(3):27. doi: 10.3390/jfmk2030027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guan J, Li Y, Ding LB, Liu GY, Zheng XF, Xue W, et al. Relationship between serum and synovial fluid CCL20 concentrations with disease severity in primary knee osteoarthritis. J Musculoskelet Neuronal Interact. 2019;19(3):326-32. [PMC free article] [PubMed] [Google Scholar]
- 42. Snelling S, Rout R, Davidson R, Clark I, Carr A, Hulley PA, et al. A gene expression study of normal and damaged cartilage in anteromedial gonarthrosis, a phenotype of osteoarthritis. Osteoarthritis Cartilage. 2014;22(2_suppl):334-43. doi: 10.1016/j.joca.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang G, Gu M, Xu Y, Wu Z. A comprehensive analysis on the effects of 1,25(OH)2D3 on primary chondrocytes cultured from patients with osteoarthritis. Gene. 2020;730:144322. doi: 10.1016/j.gene.2019.144322 [DOI] [PubMed] [Google Scholar]
- 44. Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12(6):399-414. doi: 10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Antonczyk A, Krist B, Sajek M, Michalska A, Piaszyk-Borychoswska A, Plens-Galaska M, et al. Direct inhibition of IRF-dependent transcriptional regulatory mechanisms associated with disease. Front Immunol. 2019;10:1176. doi: 10.3389/fimmu.2019.01176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sweeney SE, Kimbler TB, Firestein GS. Synoviocyte innate immune responses: II. Pivotal role of IFN regulatory factor 3. J Immunol. 2010;184(12):7162-8. doi: 10.4049/jimmunol.0903944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi T, Shen X, Gao G. Gene expression profiles of peripheral blood monocytes in osteoarthritis and analysis of differentially expressed genes. Biomed Res Int. 2019;2019:4291689. doi: 10.1155/2019/4291689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martinez-Pomares L, Wienke D, Stillion R, McKenzie EJ, Arnold JN, Harris J, et al. Carbohydrate-independent recognition of collagens by the macrophage mannose receptor. Eur J Immunol. 2006;36(5):1074-82. doi: 10.1002/eji.200535685 [DOI] [PubMed] [Google Scholar]
- 49. Fernandes TL, Gomoll AH, Lattermann C, Hernandez AJ, Bueno DF, Amano MT. Macrophage: a potential target on cartilage regeneration. Front Immunol. 2020;11:111. doi: 10.3389/fimmu.2020.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramos YFM, Metrustry S, Arden N, Bay-Jensen AC, Beekman M, de Crean AJM, et al. Meta-analysis identifies loci affecting levels of the potential osteoarthritis biomarkers sCOMP and uCTX-II with genome wide significance. J Med Genet. 2014;51(9):596-604. doi: 10.1136/jmedgenet-2014-102478 [DOI] [PubMed] [Google Scholar]
- 51. Mahon OR, Kelly DJ, McCarthy GM, Dunne A. Osteoarthritis-associated basic calcium phosphate crystals alter immune cell metabolism and promote M1 macrophage polarization. Osteoarthritis Cartilage. 2020;28(5):603-12. doi: 10.1016/j.joca.2019.10.010 [DOI] [PubMed] [Google Scholar]
- 52. Alvarez-Garcia O, Fisch KM, Wineinger NE, Akagi R, Saito M, Sasho T, et al. Increased DNA methylation and reduced expression of transcription factors in human osteoarthritis cartilage. Arthritis Rheumatol. 2016;68(8):1876-86. doi: 10.1002/art.39643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fernández-Tajes J, Soto-Hermida A, Vázquez-Mosquera ME, Cortés-Pereira E, Mosquera A, Fernández-Moreno M, et al. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann Rheum Dis. 2014;73(4):668-77. doi: 10.1136/annrheumdis-2012-202783 [DOI] [PubMed] [Google Scholar]
- 54. Rai MF, Sandell LJ, Barrack TN, Cai L, Tycksen ED, Tang SY, et al. A microarray study of articular cartilage in relation to obesity and severity of knee osteoarthritis. Cartilage. 2020;11(4):458-72. doi: 10.1177/1947603518796122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fahrenhold M, Rakic S, Classey J, Brayne C, Ince PG, Nicoll JAR, et al. TREM2 expression in the human brain: a marker of monocyte recruitment? Brain Pathol. 2018;28(5):595-602. doi: 10.1111/bpa.12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garcia S, Hartkamp LM, Malvar-Fernandez B, van Es IE, Lin H, Wong J, et al. Colony-stimulating factor (CSF) 1 receptor blockade reduces inflammation in human and murine models of rheumatoid arthritis. Arthritis Res Ther. 2016;18(1):75. doi: 10.1186/s13075-016-0973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Holmgaard RB, Zamarin D, Lesokhin A, Merghoub T, Wolchok JD. Targeting myeloid-derived suppressor cells with colony stimulating factor-1 receptor blockade can reverse immune resistance to immunotherapy in indoleamine 2,3-dioxygenase-expressing tumors. EBioMedicine. 2016;6:50-8. doi: 10.1016/j.ebiom.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hamilton JA, Tak PP. The dynamics of macrophage lineage populations in inflammatory and autoimmune diseases. Arthritis Rheum. 2009;60(5):1210-21. doi: 10.1002/art.24505 [DOI] [PubMed] [Google Scholar]
- 59. Lipari L, Gerbino A. Expression of gelatinases (MMP-2, MMP-9) in human articular cartilage. Int J Immunopathol Pharmacol. 2013;26(3):817-23. doi: 10.1177/039463201302600331 [DOI] [PubMed] [Google Scholar]
- 60. Chen C, Xu G, Sun Y, Cui Z. Transcriptome sequencing reveals dynamic changes in matrix metalloproteinases in facet joint osteoarthritis. Exp Ther Med. 2020:2475-82. doi: 10.3892/etm.2020.8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tian F, Wang J, Zhang Z, Yang J. MiR-107 modulates chondrocyte proliferation, apoptosis, and extracellular matrix synthesis by targeting PTEN. Int J Clin Exp Pathol. 2019;12(2_suppl):488-97. [PMC free article] [PubMed] [Google Scholar]
- 62. Woodell-May JE, Sommerfeld SD. Role of Inflammation and the immune system in the progression of osteoarthritis. J Orthop Res. 2020;38(2_suppl):253-7. doi: 10.1002/jor.24457 [DOI] [PubMed] [Google Scholar]
- 63. Yap HY, Tee S, Wong M, Chow SK, Peh SC, Teow SY. Pathogenic role of immune cells in rheumatoid arthritis: implications in clinical treatment and biomarker development. Cells. 2018;7(10):161. doi: 10.3390/cells7100161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Page CE, Smale S, Carty SM, Amos N, Lauder SN, Goodfellow RM, et al. Interferon-γ inhibits interleukin-1β-induced matrix metalloproteinase production by synovial fibroblasts and protects articular cartilage in early arthritis. Arthritis Res Ther. 2010;12(2_suppl):R49. doi: 10.1186/ar2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Verbruggen G, Malfait AM, Veys EM, Gyselbrecht L, Lambert J, Almqvist KF. Influence of interferon-gamma on isolated chondrocytes from human articular cartilage. Dose dependent inhibition of cell proliferation and proteoglycan synthesis. J Rheumatol. 1993;20(6):1020-6. [PubMed] [Google Scholar]
- 66. Osiecka-Iwan A, Hyc A, Radomska-Lesniewska DM, Rymarczyk A, Skopinski P. Antigenic and immunogenic properties of chondrocytes. Implications for chondrocyte therapeutic transplantation and pathogenesis of inflammatory and degenerative joint diseases. Cent Eur J Immunol. 2018; 43(2_suppl):209-19. doi: 10.5114/ceji.2018.77392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sondergaard BC, Schultz N, Madsen SH, Bay-Jensen AC, Kassem M, Karsdal MA. MAPKs are essential upstream signaling pathways in proteolytic cartilage degradation—divergence in pathways leading to aggrecanase and MMP-mediated articular cartilage degradation. Osteoarthritis Cartilage. 2010;18(3):279-88. doi: 10.1016/j.joca.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 68. Ishikawa J, Okada Y, Bird IN, Jasani B, Spragg JH, Yamada T. Use of anti-platelet-endothelial cell adhesion molecule-1 antibody in the control of disease progression in established collagen-induced arthritis in DBA/1J mice. Jpn J Pharmacol. 2002;88(3):332-40. doi: 10.1254/jjp.88.332 [DOI] [PubMed] [Google Scholar]
- 69. Bastow ER, Last K, Golub S, Stow JL, Stanley AC, Fosang AJ. Evidence for lysosomal exocytosis and release of aggrecan-degrading hydrolases from hypertrophic chondrocytes, in vitro and in vivo. Biol Open. 2012;1(4):318-28. doi: 10.1242/bio.2012547 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Supplemental material, sj-xlsx-1-car-10.1177_19476035211044823 for General Caspase Inhibition in Primary Chondrogenic Cultures Impacts Their Transcription Profile Including Osteoarthritis-Related Factors by Barbora Vesela, Martina Zapletalova, Eva Svandova, Alice Ramesova, Jaroslav Doubek, Hervé Lesot and Eva Matalova in CARTILAGE
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Supplemental material, sj-xlsx-2-car-10.1177_19476035211044823 for General Caspase Inhibition in Primary Chondrogenic Cultures Impacts Their Transcription Profile Including Osteoarthritis-Related Factors by Barbora Vesela, Martina Zapletalova, Eva Svandova, Alice Ramesova, Jaroslav Doubek, Hervé Lesot and Eva Matalova in CARTILAGE