Abstract
Objective. Caspases, cysteine proteases traditionally associated with apoptosis and inflammation, have recently been identified as important regulators of autophagy and reported within the growth plate, a cartilaginous part of the developing bone. The aim of this research was to identify novel autophagy-related molecules affected by inhibition of pro-apoptotic caspases in chondrocytes. Design. Chondrocyte micromasses derived from mouse limb buds were treated with pharmacological inhibitors of caspases. Autophagy-related gene expression was examined and possible novel molecules were confirmed by real-time polymerase chain reaction and immunocytofluorescence. Individual caspases inhibitors were used to identify the effect of specific caspases. Results. Chondrogenesis accompanied by caspase activation and autophagy progression was confirmed in micromass cultures. Expression of several autophagy-associated genes was significantly altered in the caspases inhibitors treated groups with the most prominent decrease for Pik3cg and increase of Tnfsf10. The results showed the specific pro-apoptotic caspases that play a role in these effects. Importantly, use of caspase inhibitors mimicked changes triggered by an autophagy stimulator, rapamycin, linking loss of caspase activity to an increase in autophagy. Conclusion. Caspase inhibition significantly affects regulation of autophagy-related genes in chondrocytes cultures. Detected markers are of importance in diagnostics and thus the data presented here open new perspectives in the field of cartilage development and degradation.
Keywords: autophagy, caspase, gene expression, inhibition, limb
Introduction
Autophagy is involved in degradation of cytosol components and organelles 1 and thus represents an essential mechanism for either cell survival or death associated with tissue/organ development and maintenance.2,3 Autophagy is currently a hot topic in research promoted by the Nobel Prize award in physiology and medicine 2016, 4 which allows for a wide range of clinical applications in this field. In bone development (osteogenesis), autophagy is induced during differentiation of osteoblasts and participates in bone formation and homeostasis.5,6 In keeping with these functions, impaired regulation of autophagy is associated with several bone and cartilage disorders such as osteoporosis and osteoarthritis. 7 Recently, autophagic molecules are in focus as targets of osteoarthritic treatment. 8
In long bones, arising by endochondral ossification, a transient growth plate composed of chondroblasts/chondrocytes and their extracellular matrix allows for elongation of the bone. 9 Growth plate maturating chondrocytes display an autophagic phenotype. 10 Autophagy in these cells was recently shown to regulate secretion of collagen type II, 11 the major component of cartilage extracellular matrix. Since cartilage is mostly an avascular structure, cartilage cells grow in an under nourished, hypoxic environment. The nutrition of cells thus has to be ensured by alternative mechanism, such as autophagy, which has been proposed to allow hypertrophic cartilage cells to survive. 12 Autophagy also serves as a protective mechanism in adult cartilage 13 and is likely to precede the programmed cell death of chondrocytes.14,15 Beclin-1, one of the main autophagy markers, maintains chondrocyte survival, and its absence results in enhanced chondrocyte death. 16 Cleaved Beclin-1 loses its autophagic activity and can induce apoptosis, 17 and vice versa, overexpressed Beclin1 can inhibit apoptosis. 18
Programmed cell death (apoptosis) is mediated by caspases, a group of cysteine proteases. Two distinct pathways of caspase activation, intrinsic (mitochondrial) and extrinsic (receptor mediated), are evident. These pathways are initiated by apical caspases (-8, -9) and executed by a trio of death caspases (-3, -6, -7). 17 Notably, activation of pro-apoptotic caspases was recently demonstrated to occur also in non-apoptotic growth plate chondrocytes, 19 and in developing Meckel’s cartilage. 20 In keeping with this non-apoptotic role, caspase inhibition in vitro changed expression of osteogenic markers in developing chondrocytes.19,21 This is in agreement with caspases having novel non-apoptotic roles in addition to their role in cell death. 22 Along with engagement in apoptosis, cell cycle regulation, and cell differentiation, there is increasing evidence that pro-apoptotic caspases participate in autophagy and in autophagy-apoptosis cross-talk.18,23 Among pro-apoptotic caspases, the central caspase-3 was demonstrated as an inhibitor of autophagy in most studies. 24 The other 2 members of the “death trio,” caspase-6 and caspase-7, are also associated with a negative impact on autophagy.25,26
Application of autophagy on cartilage treatment is conditioned by understanding of autophagic processes. In the presented investigation, we aimed to define the effect of caspases inhibition to autophagy in chondrocytes. Therefore, chondrocytes were exposed to pharmacological caspase inhibitors, unique research tool, 27 to follow whether this affected the regulation of autophagy-related genes and to identify which pro-apoptotic caspase was engaged.
Material and Methods
Tissue Preparation and Immunohistochemistry
Paraffin-embedded specimens of mouse fore limbs (CD1 strain) were prepared 28 and processed to achieve 5-µm-thick histological sections. Primary antibodies (anti-Beclin, ab62557, Abcam, dilution 1:300, anti-LC3b cs3868, Cell Signaling, dilution 1:100) were applied overnight following antigen retrieval. The chromogenic reaction was mediated by a peroxidase-conjugated streptavidin biotin system (Vectastain) and substrate diaminobenzidine (Dako) to visualize positive cells as brown. Slides were counterstained by hematoxylin to distinguish the cell nuclei. In the case of immunofluorescence, Alexa Fluor 488 (Thermo Fisher Scientific) was applied as a secondary antibody. Nuclei were counterstained by ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific).
Micromass Cultures
Micromass cultures were utilized as a common in vitro method to investigate chondrogenesis. 29 Cells were obtained from mouse fore limbs at E12 by a standard protocol 30 and left until the following morning in order to attach to the surface. For caspase inhibition, pharmacological inhibitors: Pan Caspase Inhibitor Z-VAD-FMK, Caspase-2 Inhibitor Z-VDVAD-FMK, Caspase-3 Inhibitor Z-DEVD-FMK, Caspase-6 Inhibitor Z-VEID-FMK, Caspase-8 Inhibitor Z-IETD-FMK, Caspase-9 Inhibitor Z-LEHD-FMK, Pan Caspase OPH Inhibitor Q-VD (FMK001, FMK003, FMK004, FMK006, FMK007, FMK008, OPH001, R&D Systems) were applied to the micromass cultures at a concentration of 100 µM, according to the manufacturer’s recommendation and previous studies.21,31 In the controls, dimethyl sulfoxide (DMSO), the inhibitor vehicle, was added and the control micromass cultures were run in parallel. Autophagy was induced by addition of 500 nM rapamycin (R8781, Merck) to the micromass cultures. The medium, including DMSO, rapamycin, and caspase inhibitor, was changed every second day. 30 The experiments were performed in at least 3 replicates. The research was performed under the project GACR 19-12023S approved and supervised by the Czech Sciences Foundation including all ethics issues.
RNA Isolation, PCR Array, and Real-Time PCR
The cultured cells were harvested into 350 µL RLT lysis buffer (Qiagen, Valencia, CA) with β-ME (Sigma-Aldrich). RNeasy Mini Kit (Qiagen) was used for RNA isolation, Super Script VILO (Invitrogen) for cDNA transcription. PCR Arrays were applied to analyze 84 genes connected with autophagy and autophagy-apoptosis cross-talk (PAMM-084Z, Qiagen). Real-time polymerase chain reaction (PCR) was performed for expression analysis of Pik3cg (Mm00445038_m1, Thermo Fisher Scientific), Tnfsf10 (Mm01283606_m1, Thermo Fisher Scientific), and Ctss (Mm01255859_m1, Thermo Fisher Scientific). Reaction mixture contained the one-step master mix gb Ideal PCR Master Mix (Generi Biotech). Expression levels were calculated normalized against the level of actin (Mouse Actb, Mm02619580_g1, Thermo Fischer Scientific).
Immunocytofluorescence and TUNEL Assay
For immunocytofluorescence, micromass cultures were grown on cultivation glass and fixed by 4% PFA. Alcian blue staining was used for visualization of chondrogenic differentiation. Primary antibodies (Beclin, ab62557, Abcam; LC3b cs3868, Cell Signaling, Sox9, HPA001758, Sigma Aldrich), Pik3cg (MA5-26087 Thermo Fisher), Cleaved caspase-2 (PA5-39872, Thermo Fisher Scientific), Cleaved caspase-3, 9664, Cleaved caspase-6, 9761, Cleaved caspase-7, 9491, Cleaved caspase-8, 8592, and Cleaved caspase-9, 9509, all from Cell Signaling, were applied overnight/4°C. Alexa Fluor 488 or 568 (A11034, A10037, Thermo Fischer) was diluted 1:200 and then applied for 40 minutes at room temperature. Cytoskeleton was visualized by ActinGreen 488 ReadyProbes Reagent (Thermo Fischer); nuclei were detected by ProLong Gold Antifade reagent with DAPI (Thermo Fischer). For detection of apoptosis, ApopTag Fluorescein in Situ Apoptosis Detection Kit (S7110, Merck) was used. Externalization of phosphatidylserine was detected by Annexin V-FITC Apoptosis Staining/Detection Kit (ab14085, Abcam).
Statistical Analysis
PCR array data were statistically evaluated using the Qiagen Data Analysis Center (available online). The significance was determined as P < 0.05 and the threshold was established as −2/+2 fold-regulation. Three biological samples were used for all tested groups. Expression levels from real-time PCR were calculated by ΔΔCT method. Results were analyzed using 2-tailed t test or, in the case of specific caspase inhibitors, using ANOVA with Dunnett’s post hoc test.
Results
Beclin-1 and LC3b Are Expressed in the Prenatal Growth Plate
To evaluate autophagy during chondrogenesis in relation to limb development in vivo, immunohistochemistry was applied on histological sections of the mouse fore limb at different stages of development from embryonic (E) 12 (the stage corresponding to that used for micromass preparation) to E18 (when all zones of the growth plate are clearly detectable).
At E12 ( Fig. 1A ), Beclin-1 ( Fig. 1B ) was weakly, and LC3B ( Fig. 1C ) robustly, expressed in the differentiating chondrocytes. At E15, proliferation and hypertrophy of chondrocytes allowed for growth of the bone anlage, with the bone collar starting to form in the medial part of the growth plate ( Fig. 1D ). Clear positivity for Beclin-1 was observed in the resting ( Fig. 1E1 ), proliferating ( Fig. 1E2 ), and particularly pre-/hypertrophic chondrocytes ( Fig. 1E3 ). Weaker signal of LC3B was detected in the resting ( Fig. 1F1 ) and proliferating ( Fig. 1F2 ) zones, with positive cells more apparent in pre-/hypertrophic chondrocytes ( Fig. 1F3 ). At E18, when all zones of the growth plate are clearly established ( Fig. 1G ), both Beclin-1 and LC3B were clearly apparent in cells of the developing cartilage ( Fig. 1H , I , J , K ). Although Beclin-1 was detected in all cartilaginous zones with similar abundancy ( Fig. 1L1-3 ), LC3B was observed at higher levels in the proliferating ( Fig. 1M2 ) and pre-hypertrophic ( Fig. 1M3 ) zones, while the resting zone showed weak LC3B signal ( Fig. 1M1 ). Both markers were apparent at high levels in the zone of ossification ( Fig. 1L4 , 1M4 ).
Figure 1.
Expression of of Beclin-1 and LC3B in developing prenatal growth plate. Morphological appearance (stained by hematoxylin/eosin/Alcian blue) of forelimb at developmental stage E12 (A), E15 (D), E18 (G). Boxes in histology images represent areas shown in B, C, E, F, L, and M. Immunohistochemical detection of Beclin-1 (B) and LC3B (C) at E12. Detection of Beclin-1 and LC3B in resting (zone 1 in D) (E1 and F1), proliferating (zone 2 in D) (E2 and F2), and pre-/hypertrophic (zone 3 in D) (E3 and F3) cartilage at E15. Detail of cellular localization of Beclin-1 (H) and LC3B (I) in hypertrophic cartilage at E18 (green), nuclei (DAPI) in blue. Overall view of the expression of Becn1 and LC3B in mouse growth plate at E18 (J, K). Detection of Beclin-1 and LC3B in resting (zone 1 in G) (L1 and M1), proliferating (zone 2 in G) (L2 and M2), pre-/hypertrophic (zone 3 in G) (L3 and M3) and ossification (zone 4 in G) (L4 and M4) cartilage at E18. Positive cells are brown; nuclei are counterstained by hematoxylin (blue). RC (resting chondrocytes), PC (proliferating chondrocytes), PHC (pre-hypertrophic chondrocytes), HC (hypertrophic chondrocytes), OC (ossification center). Scale bar = 100 µm (A, D, G, J, K), 25 µm (B, C, E, F, L, M). Shown are typical results from 3 replicates.
Autophagy-Related Molecules Are Expressed During Micromass Progression
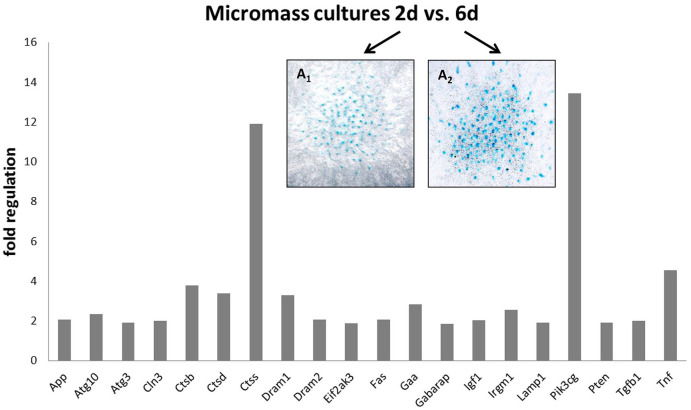
A well-established in vitro system of micromass cultures 32 was used to follow spontaneous expression of autophagic molecules during cultivation and to allow for targeted modulation of active caspases. Expression of autophagy related genes was followed during the course of standard micromass cultivation. By comparison of day 2 and day 6 ( Fig. 2A1 , A2 ), expression of 20 autophagy-related genes was found significantly increased with the most prominent change shown for cathepsin S (Ctss) and Pik3cg, the catalytic subunit of phosphoinositide-3-kinase ( Fig. 2A ). This observation may testify about spontaneous occurrence of autophagy during culture of chondrocytes in micromass.
Figure 2.
PCR array analysis of autophagy-related gene expression during micromass cultivation from 2 to 6 days (P < .05). Micromass cultures stained by Alcian blue binding to factors of extracellular matrix of cartilage (e.g., mucopolysaccharides) after 2 (A1) and after 6 days of cultivation (A2).
Pro-Apoptotic Caspases Are Activated in Cartilage Nodules in Both Apoptotic and Non-Apoptotic Patterns of Expression
To evaluate the relationship between chondrogenesis, autophagy, and caspase activation, micromass cultures were examined after 6 days of cultivation. By this time point, micromass cultures formed complex structure and formed nodules as documented by visualization of actin filaments ( Fig. 3A ). Alcian blue staining, which dyes cartilage mucopolysaccharides, confirmed the presence of cartilaginous nodules ( Fig. 3B ). Nodules were positive for Sox9, a known regulator of the chondrocyte lineage ( Fig. 3B ). 33 Apoptosis, as indicated by TUNEL staining and the presence of apoptotic bodies, was visible scattered throughout the cultures but not associated with nodules ( Fig. 3D ). The autophagy marker Beclin1 was detected in some of micromass cells, including in nodules and also was markedly positive in some cells scattered in the whole micromass ( Fig. 3E ). Interestingly, Beclin-1 positive scattered cells did not overlap with TUNEL positive cells, but the pattern was instead comparable to phosphatidylserine externalization typical for apoptosis (Supplement 1). LC3B, in contrast to Beclin-1, was detected predominantly in the nodules ( Fig. 3F ). To assess the role of apoptotic caspases during autophagy, activation of cleaved enzymes was detected. Caspase-2 activity was present in several apoptotic bodies but particularly in non-apoptotic cells within the nodules ( Fig. 3G ). Activated caspase-3 and -9 were detected mainly in apoptotic cells ( Fig. 3H , L ), with a weaker signal in the nodules. Activated caspases-6, -7, and -8 were observed in several apoptotic bodies and clearly in the cartilaginous nodules ( Fig. 3I , J , K ). Generally, cartilaginous nodules expressed autophagic markers and were positive for apoptotic caspases but also were negative for TUNEL.
Figure 3.
Detection of apoptosis, chondrogenic differentiation, autophagy, and caspase activation in micromass cultures. Alcian blue staining of micromass cultures (A), immunofluorescent detection detection of Sox9 (B), Beclin-1 (E), LC3B (F). Immunofluorescent detection cleaved caspases, caspase-2 (G), caspase-3 (H), caspase-6 (I), caspase-7 (J), caspase-8 (K), caspase-9 (L). Actin filaments visualized by actin green (C), apoptosis detected by TUNEL assay (D). Positive fluorescent signal in green, nuclei in blue (DAPI). Scale bar = 100 µm. Results shown are typical from 3 replicates. Arrows point to apoptotic cells.
Caspase Inhibition Impacts on Autophagy in Micromass Cultures
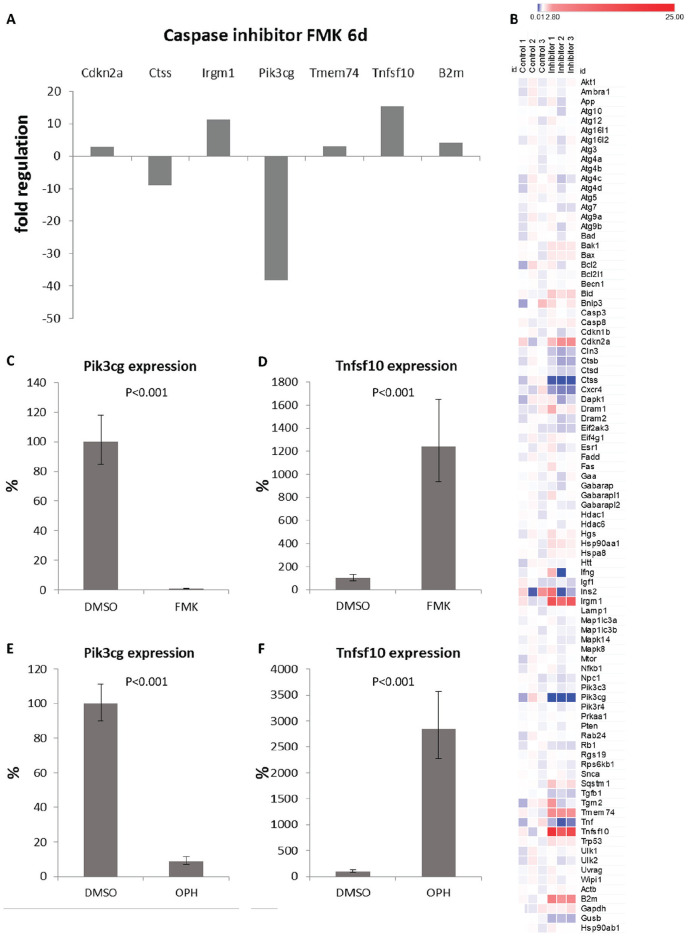
To test the role of caspases in regulating autophagy, all caspases were inhibited using a general FMK caspase inhibitor. Compared to control cultures, 6 out of 84 tested genes displayed significant alterations after 6 days of cultivation with the general caspase inhibitor. The most striking change (decrease of 38 times) was shown for Pik3cg. The other affected genes included increased expression of Cdkn2a, Irgm1, Tmem74, Tnfsf10, and decreased expression of Ctss ( Fig. 4A ). In addition, one of the housekeeping genes, beta-2 microglobulin, was increased in the treated cells. The overall results from the PCR array were visualized in a heatmap ( Fig. 4B ). To confirm the results from the PCR array, we detected expression of Pik3cg and Tnfsf10 by real-time PCR. Pik3cg decreased to 1% (P < 0.001) and Tnfsf10 increased to 1240% (P < 0.001; Fig. 4C , D ) compared to controls. The findings were further verified by use of a second general caspase inhibitor, Q-VD-OPh. This inhibitor compared to Z-VAD-FMK does not interact with other cysteine proteases such as cathepsins B, H, and L.34,35 Addition of Q-VD-OPh produced the same effect as Z-VAD, confirming the specificity of the inhibitors used. In this case expression of Pik3cg decreased to 9% (P < 0.001) and Tnfsf10 increased almost to 3000% (P < 0.001) compared to control cultures after 6 days of inhibition ( Fig. 4E , F ).
Figure 4.
PCR arrays evaluation (A) of changes in autophagy-related gene expression after 6 days of caspases inhibition (P < 0.05). Heatmap presentation (B) of the different gene expression between control and test groups. Colors represent fold change in the gene expression as compared to the mean expression in the control groups. Expression of Pik3cg and Tnfsf10 in micromass cultures detected by real-time PCR after 6 days of caspase inhibition by FMK inhibitor (C, D) and OPH inhibitor (E, F). Results are presented as means ± standard deviations.
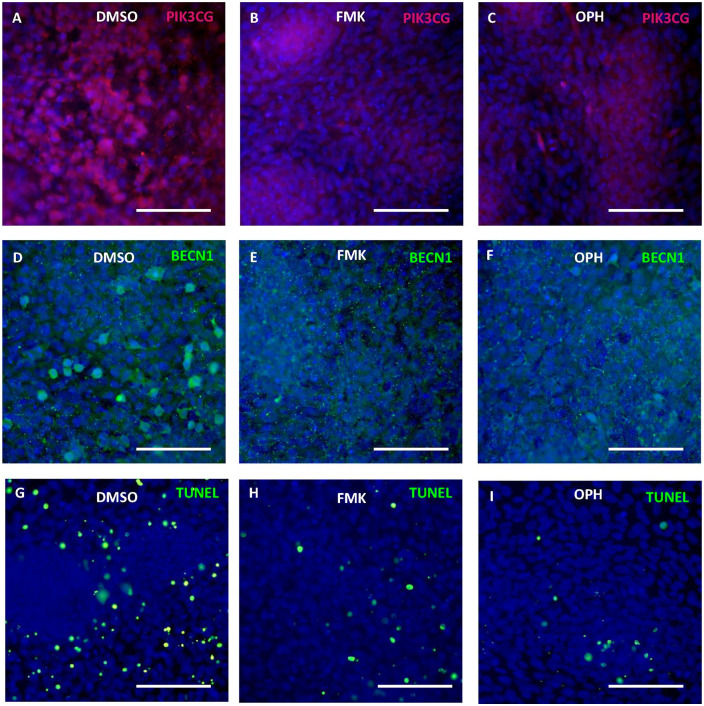
The decrease of Pik3cg in micromass cultures was visible at the protein level after both types of general caspase inhibitors ( Fig. 5A-C ). The scattered highly expressing Beclin-1 positive cells disappeared after caspase inhibition ( Fig. 5D-F ). As expected, TUNEL positive cells observed in control micromass cultures markedly decreased after caspase inhibition ( Fig. 5G-I ).
Figure 5.
Modulation of apoptosis and autophagy through caspase inhibition. Immunofluorescent detection of Pik3cg and Beclin-1 and apoptosis in micromass cultures after 6 days of cultivation with FMK inhibitor (B, E, H), OPH inhibitor (C, F, I), and control group with DMSO (A, D, G). Positive fluorescent signal in red (A-C) and in green (D-I), nuclei in blue (DAPI). Scale bar = 100 µm. Results shown are typical from 3 replicates.
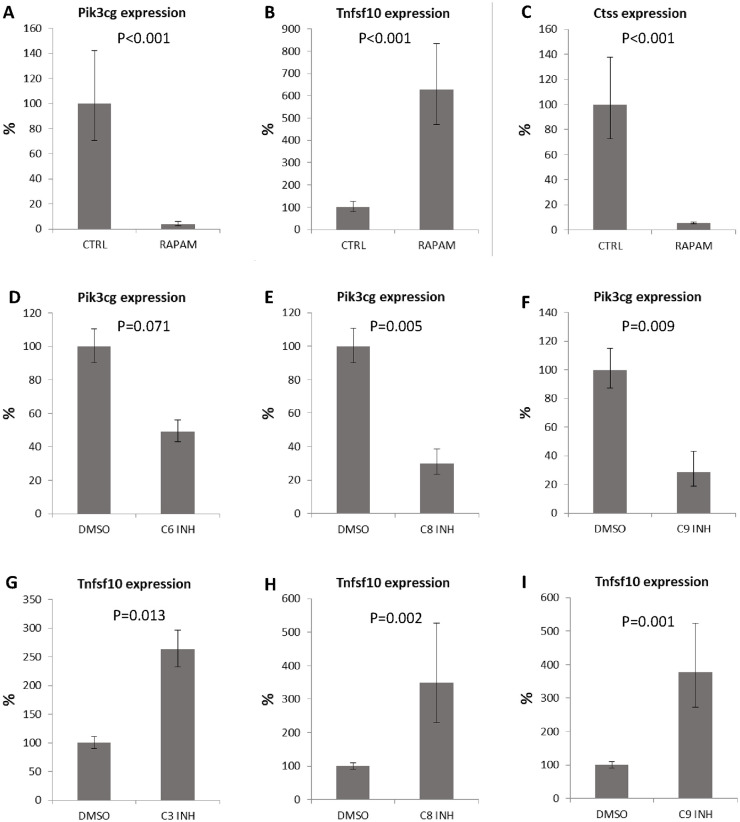
To confirm that the changes in expression detected after caspase inhibition were connected with autophagy, we induced autophagy by addition of rapamycin. After 6 days of cultivation, expression of Pik3cg was significantly decreased to 6% (P < 0.001), expression of Tnfsf10 increased to 620% (P < 0.001), and expression of Ctss decreased to 5% (P < 0.001) ( Fig. 6A , B , C ). Thus, use of a caspase inhibitor and use of an autophagy stimulator showed similar effect on the cartilage cells.
Figure 6.
Expression of autophagy related genes after rapamycin treatment and individual caspase inhibition. Expression of Pik3cg (A), Tnfsf10 (B), and Ctss (C) in micromass cultures after 6 days of cultivation with rapamycin. Pik3cg (D-F) and Tnfsf10 (G-I) expression in micromass cultures after inhibition of individual caspases. Results are presented as means ± standard deviations.
Individual Pro-Apoptotic Caspase Inhibition Impacts on the Expression of Pik3cg and Tnfsf10
To assess which caspase might be responsible for the most prominent expression changes (such as in Pik3cg and Tnfsf10), caspases-2, -3/7, -6, -8, and -9 were inhibited individually. Unfortunately, due to overlapping substrates, separate pharmacological inhibition of caspase-3 and caspase-7 is not possible. Expression of Pik3cg showed decreasing trend after inhibition of caspase-6 ( Fig. 6D ), and was significantly reduced with inhibition of caspase-8 ( Fig. 6E ) and caspase-9 ( Fig. 6F ). Tnfsf10 expression significantly increased after inhibition of caspase-3 ( Fig. 6G ), caspase-8 ( Fig. 6H ), and caspase-9 ( Fig. 6I ). Interestingly, despite its prominent activation in cartilage nodules, caspase-2 inhibition has no significant effect on these autophagy-associated genes.
Discussion
Apoptosis and autophagy are both engaged in physiologic cell removal, tightly regulated by mutual molecular crosstalk. 36 Novel functions of pro-apoptotic caspases in autophagy have been discussed recently. 23 Since apoptosis and autophagy are associated with long bone growth plate, and caspases are activated in several zones within the growth plate, the presented investigation focused on chondrocytes.
In vivo, autophagy progression is accompanied by the expression of several genes whose specific ablation can cause serious defects in a range of critical developmental steps. 37 Beclin-1 belongs to major markers of autophagy and knockout mice die at E8.5.36,37 We detected Beclin-1 in the prenatal growth plate prominently present in the hypertrophic chondrocytes and in the osteogenic zone. These data correspond with the pattern of Beclin-1 in the postnatal growth plate. 38 In the Beclin-1 positive areas, the expression of pro-apoptotic caspases has been previously described in non-apoptotic cells within the prenatal growth plate. 19 These data might point to caspase-mediated cleavage of Beclin-1, as previously demonstrated for caspase-3. 2 Caspases may thus work as a regulatory switch between autophagy, apoptosis and chondrocyte survival. 23
To follow the expression of autophagy related-genes specifically in chondrocytes, an in vitro micromass culture system was applied. The most dramatic changes observed during the culture period were for the expression of Pik3cg and Ctss. Pik3cg is an intracellular signal transducer enzyme involved in many cellular functions including regulation of autophagy. 39 Ctss is a molecule which has been be connected with apoptosis 40 as well as autophagy. 41
To investigate the effect of caspases on the expression of autophagy genes in chondrocytes, pharmacological inhibitors of caspase-activity were utilized in the micromass cultures. The most significant decrease was detected in expression of Pik3cg and the most apparent increase applied to Tnfsf10. These two markers were further investigated in detail by using inhibitors specific to individual caspases.
For Pik3cg, specific caspase inhibition identified engagement of caspase-8, -9, and probably caspase-6 but not caspase-2. Notably, apoptotic caspases, except caspase-2, are able to cleave Beclin-1 and thus these inhibitors have the potential to decrease autophagy. 23 Here we show that similarly, caspases were also able to regulate levels of Pik3cg expression, an interaction not previously reported. Pik3cg affects autophagy in a negative way by modulation of Akt signaling followed by mTOR activation. 39 This pathway is associated with growth factor mediated processes 42 and FGF signaling was recently suggested as a regulator of bone growth through autophagy. 11
Caspase-8 is one of the key apoptotic components shown to be involved in autophagy 43 and is present also in non-apoptotic chondrocytes, 20 including the growth plate. 19 Inhibition of caspase-8 in micromass culture caused a 3-fold increase in Tnfsf10 (Trail) expression. Trail promotes autophagy by induction of autophagosome formation. 44 Moreover, Trail-mediated signaling can activate caspase-8. The regulation of autophagy by caspase-8 was proposed via its substrates including Atg3, Atg5, and Beclin-1.26,45,46
A similar impact on Tnfsf10 expression was achieved in the case of inhibition of the other apical pro-apoptotic caspase, caspase-9. As for caspase-8, caspase-9, regulation of autophagy is thought to occur via Atg factors (5,7) and Beclin-1.47-49 Compared to caspase-8 and -9, inhibition of caspase-3/7 had a slightly lower effect while there was no significant difference in controls and cultures treated with caspase-2 or caspase-6 inhibitors. Among the executive caspases in autophagy, caspase-6, unlike caspase-3 and -7, can, along with ATGs and Beclin-1, cleave p62, which links the autophagy pathway and the ubiqutin–proteasome system. 50 The effect of pro-apoptotic caspases on expression of Tnfsf10 is here presented for the first time and an understanding of the mechanism may be of high importance also for TRAIL/caspase-based therapies. 51
Among upregulated genes after pan caspase inhibition, significant differences in expression after caspase inhibition were, along with Tnfsf10, observed in the case of Cdkn2a, Irgm1, and Tmem74. Cdkn2a/p16 prohibits progression of cells from G1 phase to S phase and plays a role in the induction of autophagy.52,53 The mechanism of Irgm1 action is not completely known, but Irgm was reported to interact with Ulk1 and Beclin 1 to govern their co-assembly and thus can play a direct role in the core autophagy machinery. 54 Tmem74 has been demonstrated to co-localize with apoptosis-related protein Bik in subcellular organelles and inhibit Bik-induced apoptosis. 55
Notably, there was an increased expression of B2m, beta-2-microglobulin, one of the housekeeping genes. Despite the fact that this gene is not considered in the panel of autophagy-related molecules, anti-B2m antibodies as apoptosis enhancers were recently demonstrated to inhibit autophagy. 56 Additionally, B2m may contribute to pathogenesis of osteoarthritis. 57
Knowledge about mechanisms of caspases engagement in autophagy is not yet complete. Z-FMK pan caspase inhibitors were earlier reported as modulators of autophagy 58 but the mechanism was explained particularly by inhibition of Beclin-1 cleavage and keeping retention of its autophagic properties. 24 Beclin-1 cooperates with Pik3 molecules in the formation of autophagosomes. 39 During chondrocytic cultivation, mRNA expression of Beclin-1 was kept at high levels, which was maintained even after inhibition of caspases ( Table 1 ). In addition to autophagy, Beclin-1 plays an important role during engulfment of apoptotic cells, since the process was abolished in Beclin-1 knockout cells. 59 This may explain why some Beclin-1-positive cells clearly apparent in the control cultures are almost not visible after caspase inhibition. This Beclin-1-positive cell population displays phosphatidylserine translocation rather than TUNEL positivity, indicating that they are probably at the beginning of the apoptotic process. Caspase inhibition therefore resulted in a significant reduction of apoptosis and thus possibly disruption of Beclin-1 participation during apoptotic cell engulfment.
Table 1.
Autophagy-Related Genes Expressed at High Levels (Same Range as Housekeeping Genes of the Panel) After 6 Days of Cultivation of Micromass Cultures.
| Akt1 | Casp3 | Hgs | Pik3c3 |
| App | Cdkn1b | Hsp90aa1 | Prkaa1 |
| Atg3 | Ctsb | Hspa8 | Pten |
| Atg4b | Ctsd | Igf1 | Rab24 |
| Atg5 | Dram2 | Lamp1 | Rgs19 |
| Atg9a | Eif2ak3 | Map1lc3a | Rps6kb1 |
| Bad | Eif4g1 | Map1lc3b | Sqstm1 |
| Bak1 | Gaa | Mapk14 | Tgfb1 |
| Bax | Gabarap | Mapk8 | Trp53 |
| Bcl2l1 | Gabarapl1 | Mtor | Ulk1 |
| Becn1 | Gabarapl2 | Nfkb1 | Ulk2 |
| Bnip3 | Hdac1 | Npc1 | Wipi1 |
The findings are in agreement with the fact that the decision point regulated by Beclin-1 happens at the protein level as mRNA of Becn1 was constantly high. In addition, the impact of caspase inhibition on the transcriptome must be mediated by protein changes.
Importantly, the major changes in gene expression triggered by inhibition of caspases were mimicked by stimulation of autophagy using rapamycin. Inhibition of caspases in chondrocytes therefore leads to stimulation of autophagy. Increased levels of Trail after caspase inhibition and after rapamycin treatment also supports a hypothesis that caspases can regulate autophagy through expression of Trail.
In the current investigation, novel pieces of the network puzzle connecting apoptosis and autophagy by caspases were revealed. Although autophagy was already described in cartilage, this is the first time when caspases were identified as regulators of autophagy related genes in chondrocytes as a part of physiological cartilage development. Furthermore, several novel caspase downstream factors were identified here. The results were confirmed by two different types of general caspase inhibitors (FMK and OPH) and compared to changes after rapamycin treatment. Separate inhibition of specific caspases allowed for identification of individual pro-apoptotic caspases responsible for the major effects of the inhibition. Notably, several genes whose expression was modulated by caspase inhibition are of importance in diagnostics and clinically oriented studies focused on osteoarthritis. 60 With regard to the critical function of autophagy for metabolism of chondrocytes, caspases might be potentially engaged in repair of the chondrocyte damage or slowing down of their aging. Thus, these data open new possibilities for cartilage reparation.
Supplemental Material
Supplemental material, 19-5-2020_Supplementary_figure_1 for Caspase Inhibition Affects the Expression of Autophagy-Related Molecules in Chondrocytes by Barbora Vesela, Eva Svandova, Alice Ramesova, Adela Kratochvilova, Abigail S. Tucker and Eva Matalova in CARTILAGE
Footnotes
Supplementary material for this article is available on the Cartilage website at https://journals.sagepub.com/home/nnrb.
Author Contributions: EM designed the study. BV planned experiments. BV, AR, ES, and AK performed experiments. ES and AT analyzed the data. BV, EM, and AT wrote the paper.
Acknowledgment and Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research was granted by the Czech Science Foundation, Project GA ČR 19-12023S. The CR-UK cooperation was supported by the Inter-COST project LTC18 (“Caspases as Novel Regulators in Osteogenic Cellular Networks,” running under the COST Action CA15214), the Inter-Excellence Programme provided by the Ministry of Education of the Czech Republic (www.msmt.cz).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Approval: Ethical approval was not sought for the present study. The project GACR 19-12023S was approved and supervised by the Czech Sciences Foundation including all ethics issues.
Animal Welfare: The present study followed international, national, and/or institutional guidelines for humane animal treatment and complied with relevant legislation.
ORCID iD: Barbora Vesela  https://orcid.org/0000-0001-8585-2082
https://orcid.org/0000-0001-8585-2082
References
- 1. Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535-41. [DOI] [PubMed] [Google Scholar]
- 2. Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB, 3rd. Autophagy: regulation and role in development. Autophagy. 2013;9:951-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vijayakumar K, Cho GW. Autophagy: an evolutionarily conserved process in the maintenance of stem cells and aging. Cell Biochem Funct. 2019;37:452-8. [DOI] [PubMed] [Google Scholar]
- 4. Rubinsztein DC, Frake RA. Yoshinori Ohsumi’s Nobel Prize for mechanisms of autophagy: from basic yeast biology to therapeutic potential. J R Coll Physicians Edinb. 2016;46:228-33. [DOI] [PubMed] [Google Scholar]
- 5. DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, Topi M, et al. Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy. 2014;10:1965-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shapiro IM, Layfield R, Lotz M, Settembre C, Whitehouse C. Boning up on autophagy: the role of autophagy in skeletal biology. Autophagy. 2014;10:7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeon H, Im G. Autophagy in osteoarthritis. Connect Tissue Res. 2017;58:497-508. [DOI] [PubMed] [Google Scholar]
- 9. Berendsen AD, Olsen BR. Bone development. Bone. 2015; 80:14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srinivas V, Bohensky J, Shapiro IM. Autophagy: a new phase in the maturation of growth plate chondrocytes is regulated by HIF, mTOR and AMP Kinase. Cells Tissues Organs. 2009;189:88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, Montefusco S, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272-5. [DOI] [PubMed] [Google Scholar]
- 12. Srinivas V, Shapiro IM. Chondrocytes embedded in the epiphyseal growth plates of long bones undergo autophagy prior to the induction of osteogenesis. Autophagy. 2006;2:215-6. [DOI] [PubMed] [Google Scholar]
- 13. Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15: 2865-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang RT, Zhang C, Liu Y, Zhou HH, Li ZB. Autophagy prior to chondrocyte cell death during the degeneration of Meckel’s cartilage. Anat Rec (Hoboken). 2012;295:734-41. [DOI] [PubMed] [Google Scholar]
- 16. Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 Regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207-14. [DOI] [PubMed] [Google Scholar]
- 17. McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song S, Tan J, Miao Y, Li M, Zhang Q. Crosstalk of autophagy and apoptosis: involvement of the dual role of autophagy under ER stress. J Cell Physiol. 2017;232:2977-84. [DOI] [PubMed] [Google Scholar]
- 19. Janečková E, Bíliková P, Matalová E. Osteogenic potential of caspases related to endochondral ossification. J Histochem Cytochem. 2018;66:47-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bíliková P, Švandová E, Veselá B, Doubek J, Poliard A, Matalová E. Coupling activation of pro-apoptotic caspases with autophagy in the Meckel’s cartilage. Physiol Res. 2019; 68:135-40. [DOI] [PubMed] [Google Scholar]
- 21. Adamova E, Janeckova E, Kleparnik K, Matalova E. Caspases and osteogenic markers—in vitro screening of inhibition impact. In Vitro Cell Dev Biol Anim. 2016;52:144-8. [DOI] [PubMed] [Google Scholar]
- 22. Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsapras P, Nezis IP. Caspase involvement in autophagy. Cell Death Differ. 2017;24:1369-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274: 95-100. [DOI] [PubMed] [Google Scholar]
- 26. Wirawan E, Walle EL, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kudelova J, Fleischmannova J, Adamova E, Matalova E. Pharmacological caspase inhibitors: research towards therapeutic perspectives. J Physiol Pharmacol. 2015;66:473-82. [PubMed] [Google Scholar]
- 28. Svandova E, Lesot H, Berghe TV, Tucker AS, Sharpe PT, Vandenabeele P, et al. Non-apoptotic functions of caspase-7 during osteogenesis. Cell Death Dis. 2014;5:e1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mello MA, Tuan RS. High density micromass cultures of embryonic limb bud mesenchymal cells: an in vitro model of endochondral skeletal development. Vitr Cell Dev Biol Anim. 1999;35:262-9. [DOI] [PubMed] [Google Scholar]
- 30. Chlastakova I, Liskova M, Kudelova J, Dubska L, Kleparnik K, Matalova E. Dynamics of caspase-3 activation and inhibition in embryonic micromasses evaluated by a photon-counting chemiluminescence approach. Vitr Cell Dev Biol Anim. 2012;48:545-9. [DOI] [PubMed] [Google Scholar]
- 31. Svandova E, Vesela B, Tucker AS, Matalova E. Activation of pro-apoptotic caspases in non-apoptotic cells during odontogenesis and related osteogenesis. Front Physiol. 2018;9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Underhill TM, Dranse HJ, Hoffman LM. Analysis of chondrogenesis using micromass cultures of limb mesenchyme. Methods Mol Biol. 2014;1130:251-65. [DOI] [PubMed] [Google Scholar]
- 33. Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85-9. [DOI] [PubMed] [Google Scholar]
- 34. Schotte P, Declercq W, Van Huffel S, Vandenabeele P, Beyaert R. Non-specific effects of methyl ketone peptide inhibitors of caspases. FEBS Lett. 1999;442:117-21. [DOI] [PubMed] [Google Scholar]
- 35. Keoni CL, Brown TL. Inhibition of apoptosis and efficacy of pan caspase inhibitor, Q-VD-OPh, in models of human disease. J Cell Death. 2015;8:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bohensky J, Leshinsky S, Srinivas V, Shapiro IM. Chondrocyte autophagy is stimulated by HIF-1 dependent AMPK activation and mTOR suppression. Pediatr Nephrol. 2010;25:633-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X, Long YC, Shen HM. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy. 2015;11:1711-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X, Xiong L, Yu G, Li D, Peng T, Luo D, et al. Cathepsin S silencing induces apoptosis of human hepatocellular carcinoma cells. Am J Transl Res. 2015;7:100-10. [PMC free article] [PubMed] [Google Scholar]
- 41. Huang CC, Lee CC, Lin HH, Chen MC, Lin CC, Chang YJ, et al. Autophagy-regulated ROS from xanthine oxidase acts as an early effector for triggering late mitochondria-dependent apoptosis in cathepsin S-targeted tumor cells. PLoS One. 2015;10:e0128045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ryter SW, Mizumura K, Choi AMK. The impact of autophagy on cell death modalities. Int J Cell Biol. 2014;2014:502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Y, Zhou X, Qiao J, Bao A. Autophagy is a regulator of TRAIL-induced apoptosis in NSCLC A549 cells. J Cell Commun Signal. 2017;11:219-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17:810-20. [DOI] [PubMed] [Google Scholar]
- 46. Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124-32. [DOI] [PubMed] [Google Scholar]
- 47. Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong HS, Choi HY, Lee ER, Kim JH, Jeon K, Lee HJ, et al. Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochim Biophys Acta. 2011;1813:80-90. [DOI] [PubMed] [Google Scholar]
- 49. You M, Savaraj N, Kuo MT, Wangpaichitr M, Varona-Santos J, Wu C, et al. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol Cell Biochem. 2013;374:181-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61:84-97. [DOI] [PubMed] [Google Scholar]
- 51. Mohamed MS, Bishr MK, Almutairi FM, Ali AG. Inhibitors of apoptosis: clinical implications in cancer. Apoptosis. 2017;22:1487-509. [DOI] [PubMed] [Google Scholar]
- 52. Budina-Kolomets A, Hontz RD, Pimkina J, Murphy ME. A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction. Autophagy. 2013;9:1553-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun Y, Li Q, Zhang J, Chen Z, He Q, Liu X, et al. Autophagy regulatory molecule, TMEM74, interacts with BIK and inhibits BIK-induced apoptosis. Cell Signal. 2017;36:34-41. [DOI] [PubMed] [Google Scholar]
- 56. Zhang M, He J, Liu Z, Lu Y, Zheng Y, Li H, et al. Anti-β2-microglobulin monoclonal antibodies overcome bortezomib resistance in multiple myeloma by inhibiting autophagy. Oncotarget. 2015;6:8567-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang H, Liew CC, Marshall KW. Microarray analysis reveals the involvement of beta-2 microglobulin (B2M) in human osteoarthritis. Osteoarthritis Cartilage. 2002;10: 950-60. [DOI] [PubMed] [Google Scholar]
- 58. Herzog C, Yang C, Holmes A, Kaushal GP. zVAD-fmk prevents cisplatin-induced cleavage of autophagy proteins but impairs autophagic flux and worsens renal function. Am J Physiol Renal Physiol. 2012;303:F1239-F1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Konishi A, Arakawa S, Yue Z, Shimizu S. Involvement of beclin 1 in engulfment of apoptotic cells. J Biol Chem. 2012;287:13919-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang B, Yu H, Li Y, Zhang W, Liu X. Upregulation of long noncoding TNFSF10 contributes to osteoarthritis progression through the miR-376-3p/FGFR1 axis. J Cell Biochem. 2019;120:19610-20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, 19-5-2020_Supplementary_figure_1 for Caspase Inhibition Affects the Expression of Autophagy-Related Molecules in Chondrocytes by Barbora Vesela, Eva Svandova, Alice Ramesova, Adela Kratochvilova, Abigail S. Tucker and Eva Matalova in CARTILAGE