Abstract
Background
Telomere maintenance is increasingly recognized as being fundamental to glioma oncogenesis with longer leukocyte telomere length (LTL) reported to increase risk of glioma. To gain further insight into the relationship between telomere genetics and risk of glioma, we conducted several complementary analyses, using genome-wide association studies data on LTL (78 592 individuals) and glioma (12 488 cases and 18 169 controls).
Methods
We performed both classical and summary Mendelian randomization (SMR), coupled with heterogeneity in dependent instruments tests, at genome-wide significant LTL loci to examine if an association was mediated by the same causal variant in glioma. To prioritize genes underscoring glioma-LTL associations, we analyzed gene expression and DNA methylation data.
Results
Genetically increased LTL was significantly associated with increased glioma risk, random-effects inverse variance weighted ORs per 1 SD unit increase in the putative risk factor (odds ratio [OR]SD) 4.79 (95% confidence interval: 2.11-10.85; P = 1.76 × 10−4). SMR confirmed the previously reported LTL associations at 3q26.2 (TERC; PSMR = 1.33 × 10−5), 5p15.33 (TERT; PSMR = 9.80 × 10−27), 10q24.33 (STN1 alias OBFC1; PSMR = 4.31 × 10−5), and 20q13.3 (STMN3/RTEL1; PSMR = 2.47 × 10−4) glioma risk loci. Our analysis implicates variation at 1q42.12 (PSMR = 1.55 × 10−2), 6p21.3 (PSMR = 9.76 × 10−3), 6p22.2 (PSMR = 5.45 × 10−3), 7q31.33 (PSMR = 6.52 × 10−3), and 11q22.3 (PSMR = 8.89 × 10−4) as risk factors for glioma risk. While complicated by patterns of linkage disequilibrium, genetic variation involving PARP1, PRRC2A, CARMIL1, POT1, and ATM-NPAT1 was implicated in the etiology of glioma.
Conclusions
These observations extend the role of telomere-related genes in the development of glioma.
Keywords: glioma, Mendelian randomization, risk factors, telomere length
Key Points.
We explore the genetic relationship between telomere maintenance and glioma development.
Our analyses implicate genetic variation at a number of novel loci as risk factors for glioma.
Our observations extend the role of telomere-related genes in glioma development.
Importance of the Study.
As yet, we have little understanding of the etiological basis of glioma. In this study, we attempt to elucidate the genetic relationship between telomere maintenance and risk of glioma. This association has been reported by previous epidemiological studies, with longer leukocyte telomere length (LTL) leading to an increased risk of glioma. We conducted complementary analyses, using two forms of Mendelian randomization and heterogeneity in dependent instruments tests, leveraging the largest glioma GWAS dataset published to date. In addition to confirming previously reported LTL associations, our study implicates variation at 1q42.12, 6p21.3, 6p22.2, 7q31.33, and 11q22.3 as risk factors for glioma risk. Although complicated patterns of linkage disequilibrium are present at these loci, we implicate genetic variation involving PARP1, PRRC2A, CARMIL1, POT1, and ATM-NPAT1 in the etiology of glioma and thereby extend the role of telomere-related genes in the development of glioma.
Gliomas account for approximately 80% of adult primary malignant brain tumors, and they are generally associated with poor clinical outcomes irrespective of treatment.1 The etiology of glioma is poorly understood, the only modifiable risk factor consistently linked to risk being exposure to ionizing radiation, which accounts for very few cases.2 Our understanding of inherited predisposition to glioma has been transformed by genome-wide association studies (GWAS), which have so far identified 25 loci influencing risk.3 Many of the risk loci exhibit subtype specificities, with a recent study proposing 2q37.3 and 7p22.3 as additional risk loci for low-grade gliomas.4Elucidating the functional basis of these risk loci is, therefore, an important step toward the development of testable hypotheses regarding the biological processes involved in the development of glioma.
Large-scale sequencing of glioma has highlighted dysregulated telomere biology as central to glioma oncogenesis, with somatic mutations in the promoter of the telomerase reverse transcriptase gene (TERTp) being common in primary glioblastoma multiforme (GBM) and grades II and III oligodendroglioma.5 Telomeres protect chromosome ends during DNA replication, shortening with each normal cell division until a critical point when apoptosis ensues. The enzyme telomerase, which consists of TERT and telomerase RNA component (TERC), is involved in the maintenance of telomere length. It is generally essential for tumors to avoid senescence and apoptosis by acquisition of telomere length maintenance strategies, often through telomerase reactivation.6 In glioma, this is typically through upregulation of telomerase by TERTp mutation or the alternative lengthening of telomeres pathway by ATRX mutation.7
Telomere length is determined by the balance of processes that shorten and lengthen the telomere resulting in variation between individuals at the same age. The relationship between telomere length and cancer risk has been widely investigated, and longer leukocyte telomere length (LTL) has been reported as a risk factor for a number of cancers, including glioma, implying a general relationship.8–12 Heritable factors contribute to the variation in telomere length and GWAS have identified multiple loci influencing LTL,13–27 with genetically predicted higher LTL reported to be associated with increased risk of glioma.28–30
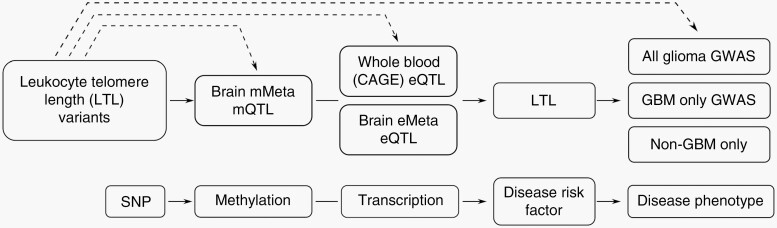
To gain further insight into the relationship between telomere genetics and risk of glioma, we conducted complementary analyses, making use of LTL GWAS data of 78 592 individuals13 and a GWAS of 12 488 glioma cases and 18 169 controls.3 Firstly, we conducted Mendelian randomization analyses to estimate the effect of LTL on glioma risk. Secondly, we performed summary Mendelian randomization (SMR) and heterogeneity in dependent instruments (HEIDI) tests at the 20 genome-wide significant LTL loci to examine if an association was mediated by the same causal variant (indicative of pleiotropy) (Figure 1). Thirdly, to prioritize genes that are most likely influenced by the genetic variation at each locus, we leveraged large-scale human genomic data integrated with tissue gene expression and DNA methylation data, coupled with knowledge-driven manual curation. Finally, to estimate the genome-wide heritability and genetic correlation between LTL and risk of glioma, we conducted a cross-trait linkage disequilibrium adjusted kinships (LDAK) analysis.
Fig. 1.
Overview of study design. Solid arrows represent a possible mechanism by which genetic variation influencing leukocyte telomere length (LTL) is associated with glioma risk. Dashed arrows correspond to summary Mendelian randomization analyses.
Materials and Methods
Ethics
Ethical approval for this study was not required as our analyses were based on summary statistics from published GWAS or the data were publicly accessible and no individual-level data were used.
Datasets
Glioma GWAS data were obtained from the most recent meta-analysis of 12 488 cases and 18 169 controls of European descent.3 Different subtypes of glioma, defined in part by malignancy grade (eg, pilocytic astrocytoma—World Health Organization [WHO] grade I, diffuse “low-grade” glioma—WHO grade II, anaplastic glioma—WHO grade III, and glioblastoma [GBM]—WHO grade IV) can be distinguished. For brevity, we considered gliomas as being either GBM (n = 6183) or non-GBM (n = 5820).
LTL GWAS data were obtained from the recent European Network for Genetic and Genomic Epidemiology (ENGAGE), European Prospective Investigation into Cancer (EPIC)-CVD, and EPIC-Interact consortium study involving 78 592 individuals of European descent.13
We explored the functional basis of associations by analyzing gene expression and methylation in tissues. Specifically, expression quantitative trait loci (e-QTL) data from peripheral whole blood (CAGE, n = 2765) and brain tissue (Brain eMeta, n = 1194) were from Lloyd-Jones et al.31 and Qi et al.,32 respectively. Brain methylation quantitative trait loci (me-QTL) data (n = 1160) were from Qi et al.32 Individual methylation probes were attributed to gene transcripts if the genomic location of the probe overlapped the start and end site of Ensembl v101 transcript locations lifted from hg38 to hg37 (for each gene, the canonical transcript was used; otherwise, the longest transcript was used).33
r 2 and D′ metrics of linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) in the European population (CEU) were obtained using LDlink.34HaploReg v4.1 was used to investigate the presence of tissue-specific “Enhancer Histone Marks” in all variants in strong LD (r2 > 0.8) with lead LTL-associated SNPs.35
Statistical Analysis
Two-sample MR was conducted using the TwoSampleMR R package.36 For each SNP, the chromosome position, the effect estimate expressed in standard deviations (SD) of the trait per-allele, and the corresponding standard error (SE) were recovered (Supplementary Table 1). SNPs were only considered as potential instruments if they were associated at P < 5 × 10-8 in a GWAS of European populations and had a minor allele frequency (MAF) > 0.01. To avoid co-linearity between SNPs, correlated SNPs were excluded (LD threshold, r2 ≥ .01). For each SNP, causal effect estimates were generated for glioma, GBM, and non-GBM tumors as ORs per 1 SD unit increase in the putative risk factor (ORSD), with 95% confidence intervals (CIs), using the Wald ratio. Causal effects were estimated using a random-effects inverse weighted variance (IVW-RE) model, which assumes that each SNP identifies a different causal effect. These causal effects are averaged to elucidate the true causal effect, due to balanced pleiotropy.36 To assess the robustness of our findings, we compared the causal estimates and associated P-values using weighted median and weighted mode-based methods (Supplementary Table 2).37,38 We examined the potential impact of outlying and pleiotropic SNPs on causal estimates using a leave-one-out strategy (Supplementary Table 3). The directional pleiotropy was assessed using MR-Egger regression (Supplementary Table 2).39 The heterogeneity (I2) within the SNP associations between LTL and each glioma dataset (all glioma, GBM, and non-GBM) was calculated from Cochran’s Q-value (Supplementary Table 4).40,41
We carried out SMR analyses, as per Zhu et al.42 To distinguish pleiotropic associations from those driven by linkage, we tested for HEIDI in each region; under the hypothesis of pleiotropy, β SMR values for SNPs in LD with the causal variant should be identical. Default settings were used adopting a window +/− 1 Mb from the SMR probe’s location. To account for multiple testing, a Benjamini-Hochberg false discovery rate (FDR) correction (FDR q ≤ 0.05) was applied. The HEIDI test result was based upon the 20 maximally associated SNPs within a region, after pruning for LD, and a P-value < .01 was considered significant. For the SMR analysis of e-QTL and me-QTL data, we limited our analysis to LTL-glioma concordant associations signals.13
The heritability of LTL and glioma and genetic correlation attributable to common variation were estimated using LDAK (v5.1), under the baseline linkage disequilibrium (BLD)-LDAK and LDAK-Thin models, respectively.43 As advocated, SNPs were filtered from respective GWAS on the basis of an INFO score > 0.9, MAF > 0.01, harmonized to HapMap3 with 1000 Genomes European MAF > 0.05, excluding indels, structural variants, and strand-ambiguous SNPs. Transformation of observed scale heritability estimates of glioma to the liability scale was carried out assuming a lifetime risk of 0.24%, 0.13%, and 0.11% for all, GBM, and non-GBM glioma, respectively.44,45P-values were estimated using a Wald Test (Supplementary Table 5).
Results
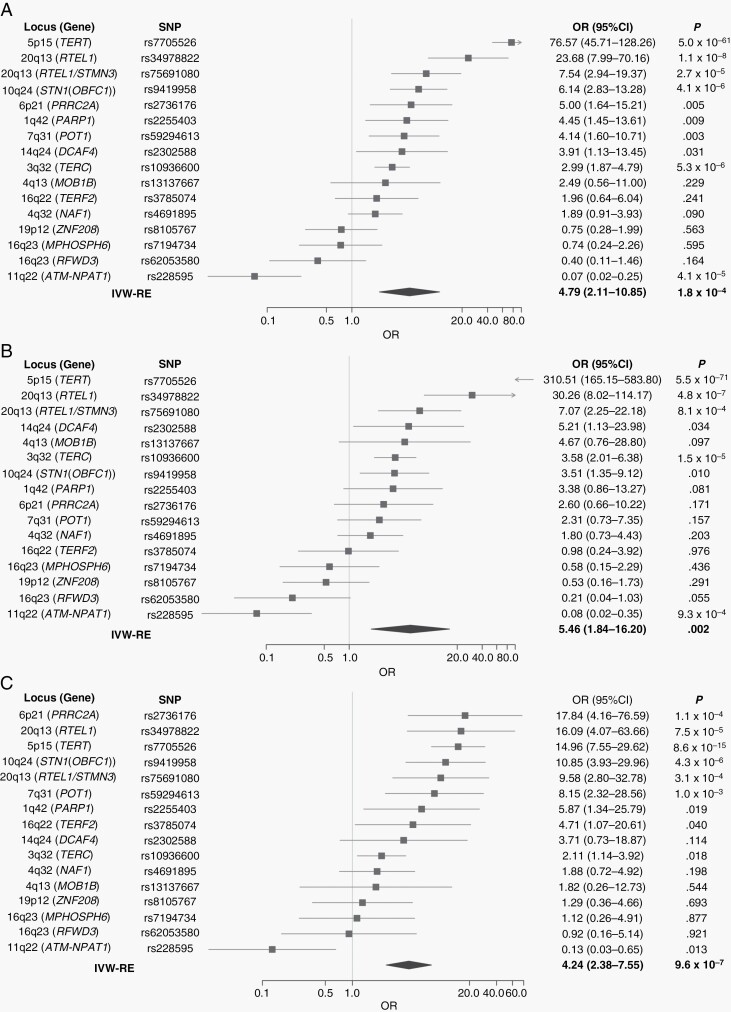
A significant relationship between genetically increased LTL and an increased risk of glioma was observed under an IVW-RE model; ORs per SD of 4.79 (95% CI: 2.11-10.85; P = 1.76 × 10−4), 5.46 (95% CI: 1.84-16.20; P = 2.23 × 10−3), and 4.24 (95% CI: 2.38-7.55, P = 9.56 × 10−7) were estimated for all glioma, GBM, and non-GBM, respectively (Figure 2A-C). These associations were robustly significant across a number of different models (Supplementary Table 2). Leave-one-out analysis results confirmed that no single variant was responsible for the strong associations (Supplementary Table 3). It was observed that the strength of these associations was primarily determined by variants at the 5p15.33 (TERT), 20q13.33 (RTEL1), 10q24.33 (STN1 alias OBFC1), and 11q22.3 (ATM) loci. A significant amount of heterogeneity existed among the SNPs used as IVs with values of Phet = 1.78 × 10−37; I2 = 93%, 2.40 × 10−45; I2 = 94% and 1.03 × 10−7; I2 = 76% for all glioma, GBM, and non-GBM, respectively (Supplementary Table 4).
Fig. 2.
Forest plot showing the effect of alleles associated with longer leukocyte telomere length on risk of (A) all glioma, (B) GBM only, and (C) non-GBM only. Diamonds represent overall causal effects estimated using random-effects inverse-variance-weighted (IVW-RE) models. Confidence intervals (CIs) are indicated by diamond width. The vertical line denotes the null value (ORSD = 1). Abbreviations: GBM, glioblastoma multiforme; ORSD, ORs per 1 SD unit increase in the putative risk factor.
Table 1 (Supplementary Table 6) presents the risk of glioma associated with each of the 20 LTL SNPs as estimated using the SMR methodology. Eleven of the 20 SNPs, mapping to 9 distinct genomic regions, showed an association with glioma risk (ie, FDR q ≤ 0.05). These included the previously documented associations at 3q26.2 (rs10936600), 5p15.33 (rs7705526 and rs2853677), 10q24.33 (rs9419958), and 20q13.33 (rs34978822 and rs75691080). All 4 of these regions contain well-recognized risk loci for glioma, the most significant glioma GWAS signals at each being provided by rs3772190 (3q26.2; P = 2.25 × 10−6), rs10069690 (5q15.33; P = 2.32 × 10−66), rs11598018 (10q24.33; P = 3.54 × 10−7), and rs2297440 (20q13.33; P = 2.53 × 10−42). Additional to these loci, novel LTL-glioma associations were observed at 1q42.12 (rs2255403; P = 1.55 × 10−2), 6p21.33 (rs2736176; P = 9.76 × 10−3), 6p22.2 (rs34991172; P = 5.45 × 10−3), 7q31.33 (rs59294613; P = 6.52 × 10−3), and 11q22.3 (rs228595; P = 8.89 × 10−4). With the exception of the 11q22.3 (rs228595) association, genetically predicted longer telomeres were associated with an increased risk of glioma. Subtype analysis was consistent with the 3q26.2 and 5p15.33 LTL-glioma associations being primarily driven by GBM (Supplementary Table 7), while the 6p21.33, 7q31.33, and 10q24.33 associations relate to non-GBM tumors (Supplementary Table 7).
Table 1.
Summary Mendelian Randomization Analysis Using LTL-Associated Variants and Glioma GWAS Data
| Locus | SNP | Proximal Gene | P Glioma | P LTL | P SMR | qSMR | P HEIDI |
|---|---|---|---|---|---|---|---|
| 5p15 | rs7705526 | TERT | 5.01 × 10−61 | 5.34 × 10−45 | 9.80 × 10−27 | 1.86 × 10−25 | 1.71 × 10−3 |
| 5p15 | rs2853677 | TERT | 1.08 × 10−28 | 3.35 × 10−31 | 9.68 × 10−16 | 9.20 × 10−15 | 3.88 × 10−8 |
| 3q26 | rs10936600 | TERC | 5.34 × 10−6 | 7.18 × 10−51 | 1.33 × 10−5 | 8.42 × 10−5 | .871 |
| 20q13 | rs34978822 | RTEL1 | 1.12 × 10−8 | 7.26 × 10−10 | 2.81 × 10−5 | 1.34 × 10−4 | 7.86 × 10−8 |
| 10q24 | rs9419958 | STN1(OBFC1) | 4.15 × 10−6 | 5.05 × 10−19 | 4.31 × 10−5 | 1.64 × 10−4 | .160 |
| 20q13 | rs75691080 | RTEL1/STMN3 | 2.67 × 10−5 | 5.99 × 10−14 | 2.47 × 10−4 | 7.82 × 10−4 | 3.21 × 10−3 |
| 11q22 | rs228595 | ATM | 4.09 × 10−5 | 1.43 × 10−8 | 8.89 × 10−4 | 2.41 × 10−3 | 2.56 × 10−3 |
| 6p22 | rs34991172 | CARMIL1 | 1.55 × 10−3 | 6.19 × 10−9 | 5.45 × 10−3 | 1.29 × 10−2 | .829 |
| 7q31 | rs59294613 | POT1 | 3.46 × 10−3 | 1.17 × 10−13 | 6.52 × 10−3 | 1.38 × 10−2 | .767 |
| 6p21 | rs2736176 | PRRC2A | 4.57 × 10−3 | 3.53 × 10−10 | 9.76 × 10−3 | 1.85 × 10−2 | .449 |
| 1q42 | rs2255403 | PARP1 | 8.92 × 10−3 | 1.72 × 10−10 | 1.55 × 10−2 | 2.68 × 10−2 | .711 |
| 14q24 | rs2302588 | DCAF4 | 3.09 × 10−2 | 1.68 × 10−8 | 4.38 × 10−2 | .069 | .409 |
| 4q32 | rs4691895 | NAF1 | .089 | 1.58 × 10−21 | .095 | .139 | .291 |
| 20q13 | rs73624724 | RTEL1/ZBTB46 | .138 | 6.33 × 10−12 | .147 | .200 | 2.35 × 10−6 |
| 16q23 | rs62053580 | RFWD3 | .164 | 4.06 × 10−8 | .178 | .225 | .139 |
| 16q22 | rs3785074 | TERF2 | .241 | 4.64 × 10−10 | .249 | .278 | .750 |
| 4q13 | rs13137667 | MOB1B | .228 | 2.43 × 10−8 | .240 | .285 | .335 |
| 16q23 | rs7194734 | MPHOSPH6 | .594 | 6.94 × 10−10 | .596 | .596 | .494 |
| 19p12 | rs8105767 | ZNF208 | .563 | 5.42 × 10−13 | .565 | .596 | .430 |
Abbreviations: GWAS, genome-wide association studies; HEIDI, heterogeneity in dependent instruments; LTL, leukocyte telomere length; SMR, summary Mendelian randomization.
P LTL published P-value.13 Complete results are detailed in Supplementary Table 6.
At the 1q42.12, 3q26.2, 6p21.33, 6p22.2, 7q31.33, and 10q24.33 loci, the association signals for LTL and glioma risk were concordant—HEIDI test results consistent with causal associations (ie, pleiotropic with both phenotypes being influenced by the same genetic variant) (Table 1; Supplementary Table 6 and Supplementary Figure 1). In contrast, HEIDI test results for 5p15.33, 11q22.3, and 20q13.33 loci were consistent with the associations being primarily driven by linkage (ie, genetic variants in LD independently influencing phenotypes) (Table 1; Supplementary Table 6 and Supplementary Figure 1).
The strongest LTL association for glioma was provided by the 5p15.33 SNPs rs7705526 and rs2853677. There is LD between these SNPs, albeit not particularly strong (r2 = .16, D′= 0.48), and conditional analysis has been reported to support 2 independent LTL association signals.13 Both associations for glioma are, however, several orders of magnitude weaker than that afforded by the GWAS glioma sentinel SNP rs10069690 (P = 5.01 × 10−61 and 1.08 × 10−28, respectively). Conditional analysis of glioma GWAS data does not support the existence of an additional association to that provided by rs10069690. Hence, the results of the HEIDI analysis are compatible with the LTL associations for glioma being a consequence of linkage at this locus, presumably reflecting the LD with the glioma risk SNP rs10069690 (LD with rs7705526: r2 = .33, D′ = 0.63, which in turn is in LD with rs2853677: r2 = .16, D′ = 0.48).
A similar scenario may also apply to the 20q13 associations for glioma with LTL SNPs rs75691080 (P = 2.67 × 10−5) and rs34978822 (P = 1.12 × 10− 8). The LTL SNPs are acting as weak proxies for the GWAS glioma risk SNP rs2297440 (P = 2.53 × 10−42). The modest association P-values may be reflective of the limited LD with rs2297440 (rs75691080, r2 = .023, D′ = 1.0 and rs34978822, r2 = .051, D′ = 1.0).
Haplotype fine-mapping, centered on each of the LTL-associated variants, revealed that 5 variants, which had shown an association with glioma risk, existed in haplotype blocks with variants associated with brain tissue-specific enhancer histone marks (Supplementary Table 8) at 10q24.33 (STN1 alias OBFC1), 20q13.33 (RTEL1), 1q42.21 (PARP1), 6p22.2 (CARMIL1), and 7q31.33 (POT1).35 This observation lends weight to the proposition that significantly concordant variants are exerting their regulatory effect on both LTL and glioma in brain tissue.
To gain insight into a possible biological basis of the relationship between these LTL loci with glioma risk, we analyzed e-QTL data from peripheral whole blood (CAGE, n = 2765),15 and brain tissue (Brain eMeta, n = 1194),31,32 as well as brain me-QTL data (n = 1160),32 applying a similar SMR-based strategy as before (Tables 2-4 and Supplementary Tables 9-11). Among the significant relationships we identified, a number were especially noteworthy as they implicate genes with strong candidacy based on proximity and plausibility a priori (Supplementary Table 12), specifically, (1) me-QTLs—5q15.33 (TERT), 6p21.3 (PRRC2A, BAG6, FLOT1), and 7q31.33 (POT1); (2) e-QTL—20q13.33 (STMN3); (3) e-QTL and me-QTL—1q42.12 (PARP1), 10q24.33 (STN1[OBFC1], ITRIP, GSTO2), and 11q22.3 (ATM/NPAT1).
Table 2.
Relationship Between Gene Expression (Whole Blood e-QTL) With Leukocyte Telomere Length (LTL) SNPsa
| LTL Locus | Proximal Gene | SNP | P LTL | P eQTL | P SMR | qSMR | P HEIDI |
|---|---|---|---|---|---|---|---|
| 6p21 | HSPA1A | rs494620 | 1.28 × 10−6 | 1.40 × 10−74 | 2.85 × 10−6 | 1.12 × 10−4 | 1.63 × 10−3 |
| 20q13 | STMN3 | rs3865523 | 3.63 × 10−7 | 2.49 × 10−34 | 2.64 × 10−6 | 1.56 × 10−4 | 7.20 × 10−5 |
| 6p21 | IER3 | rs2233980 | 6.19 × 10−7 | 2.05 × 10−60 | 1.85 × 10−6 | 2.18 × 10−4 | .030 |
| 6p21 | FLOT1 | rs3130985 | 2.67 × 10−6 | 6.93 × 10−37 | 1.07 × 10−5 | 3.15 × 10−4 | .029 |
| 10q24 | SLK | rs10883954 | 9.18 × 10−6 | 2.29 × 10−25 | 4.49 × 10−5 | 5.89 × 10−4 | .011 |
| 6p22 | BTN3A2 | rs9393710 | 2.33 × 10−5 | 9.06 × 10−237 | 2.72 × 10−5 | 6.42 × 10−4 | 7.79 × 10−3 |
| 6p21 | HLA-C | rs2523578 | 2.63 × 10−5 | 1.55 × 10−65 | 4.47 × 10−5 | 6.59 × 10−4 | .012 |
| 6p21 | HCP5 | rs2596495 | 7.39 × 10−7 | 5.73 × 10−14 | 3.56 × 10−5 | 7.00 × 10−4 | 1.62 × 10−3 |
| 11q22 | NPAT | rs228601 | 3.38 × 10−7 | 5.93 × 10−12 | 4.17 × 10−5 | 7.03 × 10−4 | .236 |
| 10q24 | SLK | rs7076157 | 1.05 × 10−5 | 8.50 × 10−22 | 6.21 × 10−5 | 7.33 × 10−4 | 1.75 × 10−3 |
| 6p21 | LY6G5C | rs1144708 | 1.86 × 10−5 | 2.25 × 10−24 | 7.91 × 10−5 | 8.49 × 10−4 | 2.01 × 10−4 |
| 20q13 | LIME1 | rs6062509 | 8.71 × 10−5 | 2.89 × 10−65 | 1.31 × 10−4 | 1.29 × 10−3 | .014 |
| 6p21 | RNF5 | rs192471087 | 6.88 × 10−5 | 2.49 × 10−37 | 1.45 × 10−4 | 1.31 × 10−3 | .338 |
| 6p22 | BTN2A1 | rs3734544 | 1.62 × 10−4 | 3.34 × 10−189 | 1.83 × 10−4 | 1.54 × 10−3 | 3.49 × 10−3 |
| 6p21 | NEU1 | rs3117573 | 9.82 × 10−6 | 1.75 × 10−8 | 5.05 × 10−4 | 3.97 × 10−3 | .473 |
| 20q13 | UCKL1 | rs111319089 | 6.79 × 10−4 | 2.53 × 10−28 | 1.16 × 10−3 | 8.58 × 10−3 | .169 |
| 6p21 | PBX2 | rs114544105 | 3.62 × 10−4 | 1.29 × 10−9 | 2.11 × 10−3 | 1.46 × 10−2 | .163 |
| 10q24 | ITPRIP | rs34970111 | 1.99 × 10−3 | 3.70 × 10−30 | 2.84 × 10−3 | 1.86 × 10−2 | .193 |
| 11q22 | ACAT1 | rs35188041 | 3.23 × 10−3 | 1.49 × 10−145 | 3.43 × 10−3 | 2.13 × 10−2 | 5.51 × 10−3 |
| 5p15 | ZDHHC11 | rs56007868 | 3.45 × 10−3 | 1.60 × 10−12 | 6.88 × 10−3 | 4.06 × 10−2 | .812 |
| 3q26 | GPR160 | rs2160901 | 7.64 × 10−3 | 1.43 × 10−119 | 8.04 × 10−3 | 4.31 × 10−2 | .584 |
| 6p22 | BTN3A2 | rs9379862 | 7.55 × 10−3 | 3.08 × 10−255 | 7.74 × 10−3 | 4.35 × 10−2 | 1.59 × 10−3 |
| 1q42 | PARP1 | rs12240196 | 7.60 × 10−3 | 2.09 × 10−54 | 8.52 × 10−3 | 4.37 × 10−2 | .127 |
| 20q13 | LKAAEAR1 | rs7271530 | 8.20 × 10−3 | 8.51 × 10−30 | 1.00 × 10−2 | 4.93 × 10−2 | .805 |
Abbreviations: e-QTL, expression quantitative trait loci; HEIDI, heterogeneity in dependent instruments; SMR, summary Mendelian randomization.
aAnalysis +/− 1 Mb around LTL SNPs. Only significant e-QTL probes (FDR q ≤ .05) are shown.
Complete results are detailed in Supplementary Table 9.
Table 4.
Relationship Between Gene Methylation (Brain me-QTL) With Leukocyte Telomere Length (LTL) SNPsa
| LTL Locus | Proximal Gene | SNP | P LTL | P meQTL | P SMR | qSMR | P HEIDI |
|---|---|---|---|---|---|---|---|
| 5p15 | TERT | rs2736100 | 2.28 × 10−41 | 1.20 × 10−33 | 2.30 × 10−19 | 1.48 × 10−16 | .106 |
| 20q13 | ZBTB46 | rs58150746 | 7.18 × 10−10 | 2.79 × 10−78 | 4.82 × 10−9 | 8.46 × 10−7 | .053 |
| 1q42 | PARP1 | rs2048425 | 1.92 × 10−10 | 8.04 × 10−25 | 6.15 × 10−8 | 6.60 × 10−6 | .153 |
| 6p21 | LY6G6C | rs707930 | 7.43 × 10−8 | 1.00 × 10−300 | 1.01 × 10−7 | 8.88 × 10−6 | .018 |
| 6p21 | PRRC2A | rs3091280 | 1.71 × 10−7 | 7.51 × 10−57 | 6.83 × 10−7 | 4.71 × 10−5 | .028 |
| 7q31 | POT1 | rs4311608 | 1.32 × 10−7 | 1.17 × 10−19 | 5.10 × 10−6 | 2.47 × 10−4 | .265 |
| 6p21 | FLOT1 | rs9262132 | 4.87 × 10−7 | 7.72 × 10−20 | 1.06 × 10−5 | 4.09 × 10−4 | .146 |
| 6p21 | DDAH2 | rs707916 | 2.62 × 10−5 | <1 × 10−307 | 2.88 × 10−5 | 7.23 × 10−4 | .012 |
| 6p21 | VARS2 | rs1264309 | 9.68 × 10−5 | 2.84 × 10−290 | 1.06 × 10−4 | 1.47 × 10−3 | .011 |
| 6p21 | HLA-C | rs2524074 | 5.47 × 10−5 | 4.22 × 10−42 | 1.10 × 10−4 | 1.48 × 10−3 | .018 |
| 11q22 | ATM | rs624888 | 5.84 × 10−7 | 1.69 × 10−9 | 1.20 × 10−4 | 1.54 × 10−3 | .258 |
| 20q13 | LIME1 | rs6011058 | 6.98 × 10−5 | 2.44 × 10−40 | 1.39 × 10−4 | 1.70 × 10−3 | .039 |
| 6p21 | HSPA1A | rs2763979 | 8.40 × 10−6 | 1.45 × 10−12 | 1.63 × 10−4 | 1.91 × 10−3 | .123 |
| 10q24 | STN1 | rs11191841 | 1.85 × 10−6 | 1.78 × 10−9 | 1.86 × 10−4 | 2.05 × 10−3 | .052 |
| 6p21 | CCHCR1 | rs7750641 | 4.98 × 10−6 | 8.85 × 10−10 | 2.51 × 10−4 | 2.47 × 10−3 | .027 |
| 6p21 | HCP5 | rs2844503 | 2.43 × 10−4 | 5.04 × 10−14 | 9.71 × 10−4 | 6.92 × 10−3 | .032 |
| 6p21 | BAG6 | rs3130047 | 3.44 × 10−4 | 1.77 × 10−13 | 1.28 × 10−3 | 8.65 × 10−3 | .051 |
| 20q13 | UCKL1 | rs7264220 | 1.29 × 10−3 | 2.43 × 10−11 | 3.74 × 10−3 | 2.24 × 10−2 | .455 |
| 10q24 | ITPRIP | rs34970111 | 1.99 × 10−3 | 1.26 × 10−10 | 5.32 × 10−3 | 3.02 × 10−2 | .075 |
| 10q24 | GSTO2 | rs276204 | 6.09 × 10−3 | <1 × 10−307 | 6.23 × 10−3 | 3.48 × 10−2 | .122 |
| 11q22 | ACAT1 | rs35219733 | 5.43 × 10−3 | 9.86 × 10−17 | 8.37 × 10−3 | 4.40 × 10−2 | .033 |
Abbreviations: HEIDI, heterogeneity in dependent instruments; me-QTL, methylation quantitative trait loci; SMR, summary Mendelian randomization.
aAnalysis shows only the most significantly associated probe results, shown by the HEIDI test to be truly causal or pleiotropic, at genes that had a significant concordant association signal in the previous analyses. Complete results are detailed in Supplementary Table 11.
Table 3.
Relationship Between Gene Expression (Brain e-QTL) With Leukocyte Telomere Length (LTL) SNPsa
| LTL Locus | Proximal Gene | SNP | P LTL | P eQTL | P SMR | qSMR | P HEIDI |
|---|---|---|---|---|---|---|---|
| 6p21 | BAG6 | rs2523593 | 4.28 × 10−7 | 2.14 × 10−287 | 5.51 × 10−7 | 5.95 × 10−5 | .011 |
| 6p21 | C4A | rs501942 | 4.90 × 10−6 | 9.49 × 10−48 | 1.31 × 10−5 | 4.72 × 10−4 | 4.65 × 10−3 |
| 6p22 | BTN3A2 | rs9366653 | 8.34 × 10−6 | <1 × 10−302 | 9.59 × 10−6 | 5.18 × 10−4 | 4.92 × 10−4 |
| 6p21 | FLOT1 | rs2596495 | 7.39 × 10−7 | 4.53 × 10−17 | 2.00 × 10−5 | 5.40 × 10−4 | .133 |
| 6p21 | CCHCR1 | rs2523593 | 4.28 × 10−7 | 1.13 × 10−10 | 6.92 × 10−5 | 1.50 × 10−3 | 2.94 × 10−3 |
| 11q22 | ATM | rs10890822 | 4.22 × 10−7 | 1.71 × 10−9 | 1.07 × 10−4 | 1.93 × 10−3 | 6.11 × 10−4 |
| 6p21 | CYP21A1P | rs6463 | 1.30 × 10−5 | 2.92 × 10−13 | 1.82 × 10−4 | 2.81 × 10−3 | .362 |
| 6p22 | BTN3A3 | rs13220495 | 2.29 × 10−5 | 5.56 × 10−13 | 2.61 × 10−4 | 3.52 × 10−3 | .079 |
| 20q13 | ZBTB46 | rs6011149 | 5.80 × 10−5 | 6.21 × 10−15 | 3.52 × 10−4 | 4.22 × 10−3 | 3.32 × 10−4 |
| 6p21 | DDAH2 | rs1144708 | 1.86 × 10−5 | 4.64 × 10−10 | 4.18 × 10−4 | 4.51 × 10−3 | .039 |
| 6p21 | VARS2 | rs2507989 | 6.69 × 10−4 | 1.26 × 10−17 | 1.57 × 10−3 | 1.54 × 10−2 | .227 |
| 3q26 | AC008040.1 | rs13315158 | 2.96 × 10−4 | 3.03 × 10−8 | 2.45 × 103 | 2.20 × 102 | .435 |
| 10q24 | STN1 | rs3850670 | 5.55 × 10−4 | 8.50 × 10−9 | 3.06 × 10−3 | 2.55 × 10−2 | .021 |
| 20q13 | STMN3 | rs6011016 | 1.40 × 10−3 | 1.74 × 10−10 | 4.28 × 10−3 | 3.30 × 10−2 | .230 |
| 10q24 | GSTO2 | rs276203 | 6.59 × 10−3 | <1 × 10−302 | 6.71 × 10−3 | 4.83 × 10−2 | .494 |
Abbreviations: e-QTL, expression quantitative trait loci; HEIDI, heterogeneity in dependent instruments; SMR, summary Mendelian randomization.
aAnalysis +/− 1 Mb around LTL SNPs. Only significant e-QTL probes (FDR q ≤ .05) are shown.
Complete results are detailed in Supplementary Table 10.
The above analyses exploring the relationship between telomere genetics and glioma risk have been restricted to examining the LTL genome-wide significant SNPs. To explore the inter-relationship between heritable variation in telomeres and glioma risk on a genome-wide basis, we conducted a cross-trait LDAK analysis. The heritability of the glioma and LTL associated with all common genetic variation was 17.2% (SD: 1.6%, P = 5.93 × 10−27) and 3.1% (SD: 1.2%, P = 9.79 × 10−3), respectively, with 0.8% (SD: 0.9%, P = .35) co-heritability (Supplementary Table 5).
Discussion
Our analysis is predicated on the assumption that any genetic variation influencing telomere length will have a generic component, and hence LTL will be correlated with telomere length in the presumptive glioma cell and/or tissue of origin. The LD patterns that are present at some loci, such as 20q13 (RTEL1), mean that 2 variants in LD with one another may exert their regulatory effect in different tissues, with the lead glioma risk SNP (rs10069690) having the strongest regulatory effect in glial cells, whereas the LTL lead SNPs (rs34978822/rs75691080) having the strongest regulatory effect in leukocytes.46 Fine-mapping of the LTL-associated variant haplotype blocks revealed that 5 LTL loci (PARP1, CARMIL1, POT1, STN1, and RTEL1) existed in haplotype blocks with variants that were known to have enhancer histone marks in brain tissue, further strengthening the assumption that this study is based on.
This study provides additional support for genetically predicted longer telomeres being the basis for glioma predisposition at a subset of the risk loci. As well as confirming a number of the previously reported LTL associations at 3q26.2, 5p15.33, 10q24.33, and 20q13.33, our analysis implicates variation at 1q42.12, 6p21.3, 6p22.2, and 7q31.33 as risk factors for glioma risk. Perhaps not surprisingly, the strongest associations observed were those mapping to the known glioma GWAS risk loci, with implicated target genes having well-established roles in telomere biology and glioma oncogenesis—5p15.33 (TERT), 3q26.2 (TERC), 10q24.33 (STN1, formerly OBFC1), and 20q13.33 (RTEL1). In addition, our analysis implicates variation at 1q42.12, 6p21.3, 6p22.2, 7q31.33, and 11q22.3 as risk factors for glioma. We and others have previously shown that glioma risk loci often exhibit associations with particular molecular subtypes, particularly based on combinations of IDH mutation (IDH1 or IDH2), 1p/19q co-deletion, and TERT promoter mutation.5,47 It has been reported that the majority of grade IV gliomas (ie, GBM tumors) contain TERT promoter mutations, and most grades II and III gliomas contain IDH mutations.5,48 Therefore, future large-scale glioma association studies stratifying by molecular subgroups are likely to provide increased insight into the nature of the relationship between glioma risk polymorphisms and telomere maintenance mechanisms. Indeed, we note that SNP associations at telomere-related loci 3q26 (TERC), 5p15.33 (TERT), and 20q13.33 (RTEL1) show a consistent direction of effect with those in our study for GBM glioma and in the analysis of Eckel-Passow et al.5 for gliomas with TERT promoter mutation.
The correlation between LTL and telomere length in brain tissue is imperfect.49 Despite this limitation, 3 of the prioritized genes mapping to the newly identified glioma risk loci on the basis of LTL association have recognized roles in regulation of telomeres—PARP1 (1q42.12), POT1 (7q31.33), and ATM (11q22.3). Telomeres function to prevent the 3′ single-stranded overhang at the end of the chromosome from being detected as a double-stranded DNA break. This is achieved through binding of the Shelterin complex (TERF1, TERF2, TERF2IP, TINF2, ACD, and POT1), which acts to block activation of DNA damage response (DDR) pathways. Shelterin also binds a number of accessory factors that facilitate processing and replication of the telomere, including the DNA helicase RTEL1. Shelterin also interacts with the CST complex that regulates telomerase access to the telomeric DNA.50 The associated loci contain one of the Shelterin components POT1, the helicase RTEL1, and the CST component STN1 (formerly OBFC1). Intriguingly, inactivating germline mutations in POT1 have been reported to be associated with familial oligodendroglioma.51 The poly(ADP-ribosyl)ation of proteins is catalyzed by PARP1 in number of cellular pathways, including DNA repair. Additionally, PARP1 regulates the binding of TERF2 to telomeric DNA through post-translational modifications.52 While the PRRC2A locus is a gene-dense region with complex LD structure BAG6, whose expression is linked to DNA damage signaling and apoptosis is a strong candidate a priori.53 Although telomere-binding proteins and structure play a role in aim to inhibiting activation of the DDR, there is paradoxical involvement of the DDR in telomere maintenance, involving prioritized genes, ATM and PARP1.54 While TERF2 inhibits ATM activation and the classical non-homologous end joining at telomeres, ATM activation is required for telomere elongation, possibly through ATM-mediated phosphorylation of TERF1.55,56
With caveats, the observed co-heritability correlation that we report is consistent with less than 10% of the heritable risk of glioma being enshrined in genetic loci influencing telomere biology (for 80% power required genetic correlation needing to be > 0.12).57 It is, however, the case that a number of the associations and the genes that they annotate support the role of genetically determined telomere dysfunction as being biologically important in glioma. Excluding TERT, there is effectively no overlap between genes related to the respective susceptibility loci and somatic mutated genes in glial tumors suggesting that they each may act at different moments in the development of the tumourigenesis. Finally, where appropriate, this analysis serves to illustrate the value of hypothesis-driven discovery as an adjunct to conventional agnostic GWAS-based approaches to risk factor identification.
Supplementary Material
Acknowledgments
We thank Drs. Codd and Langenberg for kindly making the LTL GWAS summary statistics available.
Funding
This work was supported by a grant from Cancer Research UK [grant number C1298/A8362].
Conflict of interest statement. The authors declare that they have no competing interests.
Authorship statement. C.N.S., B.K., and R.C. performed statistical analyses. C.N.S., B.K., and R.S.H. drafted the manuscript. All authors reviewed, read, and approved the final manuscript.
Collaborators
Elizabeth B. Claus,3,4 Dora Il′yasova,5–7 Joellen Schildkraut,6,7 Jill S. Barnholtz-Sloan,1 Sara H. Olson,8 Jonine L. Bernstein,8 Rose K. Lai,9 Stephen Chanock,10 Preetha Rajaraman,10 Christoffer Johansen,11,12 Robert B. Jenkins,13 Beatrice S. Melin,14 Margaret R. Wrensch,15,16 Marc Sanson,17,18 Melissa L. Bondy2
1Department of Population and Quantitative Health Sciences and the Cleveland Center for Health Outcomes Research, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; 2Section of Epidemiology and Population Sciences, Department of Medicine, Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; 3School of Public Health, Yale University, New Haven, CT 06510, USA; 4Department of Neurosurgery, Brigham and Women’s Hospital, Boston, MA 02115, USA; 5Department of Epidemiology and Biostatistics, School of Public Health, Georgia State University, Atlanta, GA 30303, USA; 6Duke Cancer Institute, Duke University Medical Center, Durham, NC 27710, USA; 7Cancer Control and Prevention Program, Department of Community and Family Medicine, Duke University Medical Center, Durham, NC 27710, USA; 8Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY 10017, USA; 9Departments of Neurology and Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA; 10Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD 20892, USA; 11Danish Cancer Society Research Center, Survivorship, Danish Cancer Society, Copenhagen 2100, Denmark; 12Oncology Clinic, Finsen Centre, Rigshospitalet, University of Copenhagen, Copenhagen 2100, Denmark; 13Department of Laboratory Medicine and Pathology, Mayo Clinic Comprehensive Cancer Center, Mayo Clinic, Rochester, MI 55905, USA; 14Department of Radiation Sciences, Umeå University, Umeå 901 87, Sweden; 15Department of Neurological Surgery, School of Medicine, University of California, San Francisco, CA 94143, USA; 16Institute of Human Genetics, University of California, San Francisco, CA 94143, USA; 17Sorbonne Université, Inserm, CNRS, UMR S 1127, Institut du Cerveau et de la Moelle épinière, ICM, F-75013, Paris; 18AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neurologie 2-Mazarin, Paris 75013, France.
Data Availability
Meta-analyzed glioma GWAS data were obtained from the study by Melin et al.,3 which is a meta-analysis of 8 independent GWAS studies (UK58, French,59 German,60 MDA,61 UCSF-SFAGS,61 GliomaScan,62 GICC,63 and UCSF/Mayo64). Genotype data from the Glioma International Case-Control Consortium Study GWAS are available from the database of Genotypes and Phenotypes (dbGaP) under accession phs001319.v1.p1. Additionally, genotypes from the GliomaScan GWAS can be accessed through dbGaP accession phs000652.v1.p1. Summary statistics from the glioma GWAS meta-analysis are available from the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under accession number EGAS00001003372. Summary statistics from leukocyte telomere length GWAS meta-analysis were provided by Dr. V. Codd.13 Summary e-QTL data were accessed from https://cnsgenomics.com/software/smr/#eQTLsummarydata. Summary me-QTL data were accessed from https://cnsgenomics.com/software/smr/#mQTLsummarydata.31,32
References
- 1. Protsenko E, Rehkopf D, Prather AA, Epel E, Lin J. Are long telomeres better than short? Relative contributions of genetically predicted telomere length to neoplastic and non-neoplastic disease risk and population health burden. PLoS One. 2020;15(10):e0240185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bondy ML, Scheurer ME, Malmer B, et al. ; Brain Tumor Epidemiology Consortium . Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113(7 Suppl):1953–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; GliomaScan Consortium . Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckel-Passow JE, Drucker KL, Kollmeyer TM, et al. Adult diffuse glioma GWAS by molecular subtype identifies variants in D2HGDH and FAM20C. Neuro Oncol. 2020;22(11):1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18(3):175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse Glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersson U, Degerman S, Dahlin AM, et al. The association between longer relative leukocyte telomere length and risk of glioma is independent of the potentially confounding factors allergy, BMI, and smoking. Cancer Causes Control. 2019;30(2):177–185. [DOI] [PubMed] [Google Scholar]
- 9. Wang S, Chen Y, Qu F, et al. Association between leukocyte telomere length and glioma risk: a case-control study. Neuro Oncol. 2014;16(4):505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Svenson U, Roos G, Wikström P. Long leukocyte telomere length in prostate cancer patients at diagnosis is associated with poor metastasis-free and cancer-specific survival. Tumour Biol. 2017;39(2):1010428317692236. [DOI] [PubMed] [Google Scholar]
- 11. Samavat H, Xun X, Jin A, Wang R, Koh WP, Yuan JM. Association between prediagnostic leukocyte telomere length and breast cancer risk: the Singapore Chinese Health Study. Breast Cancer Res. 2019;21(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luu HN, Qi M, Wang R, et al. Association between leukocyte telomere length and colorectal cancer risk in the Singapore Chinese health study. Clin Transl Gastroenterol. 2019;10(5):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li C, Stoma S, Lotta LA, et al. Genome-wide association analysis in humans links nucleotide metabolism to leukocyte telomere length. Am J Hum Genet. 2020;106(3):389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Codd V, Nelson CP, Albrecht E, et al. ; CARDIoGRAM consortium . Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45(4):422–42 7, 427e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mangino M, Richards JB, Soranzo N, et al. A genome-wide association study identifies a novel locus on chromosome 18q12.2 influencing white cell telomere length. J Med Genet. 2009;46(7):451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walsh KM, Codd V, Smirnov IV, et al. ; ENGAGE Consortium Telomere Group . Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prescott J, Kraft P, Chasman DI, et al. Genome-wide association study of relative telomere length. PLoS One. 2011;6(5):e19635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeiger AM, White MJ, Eng C, et al. Genetic determinants of telomere length in African American Youth. Sci Rep. 2018;8(1):13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dorajoo R, Chang X, Gurung RL, et al. Loci for human leukocyte telomere length in the Singaporean Chinese population and trans-ethnic genetic studies. Nat Commun. 2019;10(1):2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Codd V, Mangino M, van der Harst P, et al. ; Wellcome Trust Case Control Consortium . Common variants near TERC are associated with mean telomere length. Nat Genet. 2010;42(3):197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delgado DA, Zhang C, Chen LS, et al. Genome-wide association study of telomere length among South Asians identifies a second RTEL1 association signal. J Med Genet. 2018;55(1):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu J, Chen M, Shete S, et al. A genome-wide association study identifies a locus on chromosome 14q21 as a predictor of leukocyte telomere length and as a marker of susceptibility for bladder cancer. Cancer Prev Res (Phila). 2011;4(4):514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JH, Cheng R, Honig LS, et al. Genome wide association and linkage analyses identified three loci-4q25, 17q23.2, and 10q11.21-associated with variation in leukocyte telomere length: the Long Life Family Study. Front Genet. 2013;4:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levy D, Neuhausen SL, Hunt SC, et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci U S A. 2010;107(20):9293–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Cao L, Li Z, et al. A genome-wide association study identifies a locus on TERT for mean telomere length in Han Chinese. PLoS One. 2014;9(1):e85043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mangino M, Christiansen L, Stone R, et al. DCAF4, a novel gene associated with leucocyte telomere length. J Med Genet. 2015;52(3):157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mangino M, Hwang SJ, Spector TD, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012;21(24):5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walsh KM, Codd V, Rice T, et al. ; ENGAGE Consortium Telomere Group . Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6(40):42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saunders CN, Cornish AJ, Kinnersley B, et al. Searching for causal relationships of glioma: a phenome-wide Mendelian randomisation study. Br J Cancer. 2021;124(2):447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang C, Ostrom QT, Semmes EC, et al. ; Glioma International Case-Control Study (GICC) . Genetic predisposition to longer telomere length and risk of childhood, adolescent and adult-onset ependymoma. Acta Neuropathol Commun. 2020;8(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lloyd-Jones LR, Holloway A, McRae A, et al. The genetic architecture of gene expression in peripheral blood. Am J Hum Genet. 2017;100(2):228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi T, Wu Y, Zeng J, et al. ; eQTLGen Consortium . Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9(1):2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 42. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 43. Speed D, Holmes J, Balding DJ. Evaluating and improving heritability models using summary statistics. Nat Genet. 2020;52(4):458–462. [DOI] [PubMed] [Google Scholar]
- 44. Surveillance Epidemiology and End Results (SEER) Program. DevCan database: “SEER 18 Incidence and Mortality, with Kaposi Sarcoma and Mesothelioma”. Produced by National Cancer Institute, DCCPS, Surveillace Research Program, Surveillance Systems Branch. 2014. www.cdc.gov/nchs. Accessed September 2019. [Google Scholar]
- 45. Kinnersley B, Mitchell JS, Gousias K, et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep. 2015;5:17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walsh KM, Wiencke JK, Lachance DH, et al. Telomere maintenance and the etiology of adult glioma. Neuro Oncol. 2015;17(11):1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Labreche K, Kinnersley B, Berzero G, et al. Diffuse gliomas classified by 1p/19q co-deletion, TERT promoter and IDH mutation status are associated with specific genetic risk loci. Acta Neuropathol. 2018;135(5):743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arita H, Yamasaki K, Matsushita Y, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Demanelis K, Jasmine F, Chen LS, et al. Determinants of telomere length across human tissues. Science (80-). 2020;369(6509). doi: 10.1126/SCIENCE.AAZ6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giraud-Panis MJ, Teixeira MT, Géli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39(5):665–676. [DOI] [PubMed] [Google Scholar]
- 51. Bainbridge MN, Armstrong GN, Gramatges MM, et al. ; Gliogene Consortium . Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst. 2015;107(1):384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gomez M, Wu J, Schreiber V, et al. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol Biol Cell. 2006;17(4):1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krenciute G, Liu S, Yucer N, et al. Nuclear BAG6-UBL4A-GET4 complex mediates DNA damage signaling and cell death. J Biol Chem. 2013;288(28):20547–20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harvey A, Mielke N, Grimstead JW, et al. PARP1 is required for preserving telomeric integrity but is dispensable for A-NHEJ. Oncotarget. 2018;9(78):34821–34837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007;14(9):832–840. [DOI] [PubMed] [Google Scholar]
- 56. McKerlie M, Lin S, Zhu XD. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res. 2012;40(9):3975–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Visscher PM, Hemani G, Vinkhuyzen AA, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10(4):e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cardis E, Richardson L, Deltour I, et al. The INTERPHONE study: design, epidemiological methods, and description of the study population. Eur J Epidemiol. 2007;22(9):647–664. [DOI] [PubMed] [Google Scholar]
- 59. Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20(14):2897–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kinnersley B, Labussière M, Holroyd A, et al. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat Commun. 2015;6:8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rajaraman P, Melin BS, Wang Z, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131(12):1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Amirian ES, Armstrong GN, Zhou R, et al. The Glioma International Case-Control Study: a report from the Genetic Epidemiology of Glioma International Consortium. Am J Epidemiol. 2016;183(2):85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Meta-analyzed glioma GWAS data were obtained from the study by Melin et al.,3 which is a meta-analysis of 8 independent GWAS studies (UK58, French,59 German,60 MDA,61 UCSF-SFAGS,61 GliomaScan,62 GICC,63 and UCSF/Mayo64). Genotype data from the Glioma International Case-Control Consortium Study GWAS are available from the database of Genotypes and Phenotypes (dbGaP) under accession phs001319.v1.p1. Additionally, genotypes from the GliomaScan GWAS can be accessed through dbGaP accession phs000652.v1.p1. Summary statistics from the glioma GWAS meta-analysis are available from the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under accession number EGAS00001003372. Summary statistics from leukocyte telomere length GWAS meta-analysis were provided by Dr. V. Codd.13 Summary e-QTL data were accessed from https://cnsgenomics.com/software/smr/#eQTLsummarydata. Summary me-QTL data were accessed from https://cnsgenomics.com/software/smr/#mQTLsummarydata.31,32