Abstract
Aims
Visceral adipose tissue inflammation is a fundamental mechanism of insulin resistance in obesity and type 2 diabetes. Translocation of intestinal bacteria has been suggested as a driving factor for the inflammation. However, although bacterial DNA was detected in visceral adipose tissue of humans with obesity, it is unclear to what extent this is contamination or whether the gut microbiota is causally involved. Effects of fecal microbiota transplantation (FMT) on bacterial translocation and visceral adipose tissue inflammation in individuals with obesity and insulin resistance were assessed.
Material and Methods
Eight individuals with clinically severe obesity (body mass index [BMI] >35 kg/m2) and metabolic syndrome received lean donor FMT 4 weeks prior to elective bariatric surgery. The participants were age‐, sex‐, and BMI‐matched to 16 controls that underwent no fecal transplantation. Visceral adipose tissue was collected during surgery. Bacterial translocation was assessed by 16S rRNA gene sequencing of adipose tissue and feces. Pro‐inflammatory cytokine expression and histopathological analyses of visceral adipose tissue were performed to assess inflammation.
Results
Fecal microbiota transplantation significantly altered gut microbiota composition. Visceral adipose tissue contained a very low quantity of bacterial DNA in both groups. No difference in visceral bacterial DNA content between groups was observed. Also, visceral expression of pro‐inflammatory cytokines and macrophage infiltration did not differ between groups. No correlation between inflammatory tone and bacterial translocation was observed.
Conclusions
Visceral bacterial DNA content and level of inflammation were not altered upon FMT. Thus, bacterial translocation may not be the main driver of visceral adipose tissue inflammation in obesity.
Keywords: bacterial translocation, fecal microbiota transplantation, gut microbiota, visceral adipose tissue inflammation
1. INTRODUCTION
An extensive body of research has shown that obesity and insulin resistance are characterized by a state of chronic low‐grade inflammation, originating from visceral adipose tissue. 1 , 2 , 3 , 4 , 5 , 6 Visceral adipose tissue secretes many cytokines, termed adipocytokines, involved in inflammatory processes. For example, in both diet‐ and genetically‐induced obesity, expression of pro‐inflammatory cytokines such as tumor necrosis factor (TNF)‐α, interleukin (IL)‐1β and IL‐6 is increased in adipose tissue. 6 , 7 , 8 Moreover, in participants with obesity, IL‐1β expression was found to be 100–1000 times higher in visceral adipose tissue compared to subcutaneous adipose tissue and liver. 7 These pro‐inflammatory cytokines directly interfere with insulin signaling through insulin receptor substrate‐1 in muscle tissue. 9
The mechanistic link between high‐fat diet and visceral adipose tissue inflammation remains to be determined. In this regard, altered intestinal microbiota has been suggested as a contributing factor to chronic low‐grade inflammation in obesity. The approximately 1013 bacteria in the gut live in close interaction with the host immune system and play a large role in nutrient digestion and energy handling. 10 Moreover, translocation of bacteria or their metabolites into the blood or adipose tissue may result in a systemic inflammatory response. For example, using fluorescently labeled Escherichia coli, Amar et al. showed high‐fat diet‐induced translocation of live bacteria into blood and mesenteric visceral adipose tissue, resulting in visceral adipose tissue inflammation and insulin resistance in mice. 11 Bacterial translocation was dependent on microbial pattern recognition receptors cluster of differentiation (CD)14 and Nod1 and could be reversed by manipulation of the gut microbiota, resulting in improvement of the animals' inflammatory and metabolic status. 14
The role of bacterial translocation into visceral adipose in humans is less clear. Pyrosequencing of mesenteric visceral adipose of humans with obesity revealed DNA of several bacterial species to be present. 12 However, whether this represents active translocation or merely contamination remains unclear. This study thus aimed to assess the effect of fecal microbiota transplantation (FMT) on bacterial translocation in visceral adipose tissue inflammation in individuals with clinically severe obesity and insulin resistance. It was hypothesized that FMT results in altered bacterial translocation and visceral adipose tissue inflammation compared to no FMT.
2. MATERIALS AND METHODS
2.1. Patient selection
Individuals with clinically severe obesity (body mass index [BMI] ≥35 kg/m2) who were on the waiting list for Roux‐en‐Y gastric bypass (RYGB) bariatric surgery in the Slotervaart Hospital in Amsterdam were recruited via the outpatient clinic and screened for the characteristics of metabolic syndrome (≥3/5 criteria: waist circumference >102 cm for men or >88 cm for women; blood pressure ≥130 mmHg systolic or ≥85 mmHg diastolic or treatment for hypertension; fasting plasma glucose ≥5.6 mmol/L; high‐density lipoprotein cholesterol (HDL‐C) <1.03 mmol/L for men or <1.29 mmol/L for women; fasting triglycerides ≥1.7 mmol/L 13 ). Participants who had a medical history of recent weight loss, a cardiovascular event, use of any medication known to influence gut microbiota composition, compromised immunity, a diagnosis of diabetes, and participants who actively smoked were excluded from the study. After including participants for lean donor allogenic FMT, controls who received no FMT were matched from the BARIA cohort, an ongoing prospective cohort study of participants undergoing bariatric surgery, based on age, sex and BMI, of whom visceral adipose tissue was sampled during surgery. For FMTs, lean healthy fecal donors with BMI 18.5–25 kg/m2 and no medication use were recruited via local advertisement and screened to exclude the risk of transmission of infectious disease, as described below.
Written informed consent was obtained from all participants. The study was approved by the local Institutional Review Board of the Amsterdam UMC, location AMC in Amsterdam, the Netherlands and conducted in accordance with the Declaration of Helsinki (version 2013). This study was registered as NTR5141 in the Dutch Trial Registry (https://www.trialregister.nl/trial/4894).
2.2. Study design
Participants visited the hospital after an overnight fast of at least 10 h. After measurement of height, weight, waist and hip circumference, and blood pressure; blood was drawn for screening. Participants who were included in the FMT arm of the study returned 4 weeks prior to planned RYGB after an overnight fast of at least 10 h, for collection of a blood sample, morning fecal sample, and duodenal mucosal tissue samples collected during gastro‐duodenoscopy and FMT as described below. Collection of these samples was repeated the day before the surgery. In participants who were included as matched controls, a fasting blood sample and morning fecal sample were collected on the day of the surgery. During surgery, the gastro‐intestinal surgeon collected mesenteric visceral adipose tissue samples from the epiploic appendages of the transverse colon. Tissue was either snap frozen in liquid nitrogen and stored at −80°C until further analysis or fixed in formaldehyde for histopathological analysis. Sterilized equipment was used to handle all samples. Participants were asked to keep 3‐day online food diaries in the days preceding all study visits except for the screening to ensure a stable diet.
2.3. Fecal microbiota transplantation
Fecal microbiota transplantation was performed as previously described. 14 In short, during gastro‐duodenoscopy, duodenal biopsies were collected and immediately snap frozen in liquid nitrogen and stored at −80°C until further analysis. A nasoduodenal probe was then inserted, of which correct positioning was confirmed by an abdominal X‐ray. Bowel lavage was then performed by infusing 2–4 L of Klean‐Prep (a laxative consisting of dissolved macrogol/electrolytes) through the probe at a rate of 1 L/h, until stools were clear. Participants were matched to one of three donors using computerized randomization. A donor fecal sample of at least 50 g was used within 6 h of production. The sample was processed by homogenizing with 500 ml 0.9% sterile saline and filtering through a sieve. The solution was stored in a sterile glass 500 ml bottle. The homogenized 500 ml fecal suspension was then administered through the nasoduodenal tube. To exclude the risk of transfer of pathogens, the lean healthy donors were screened for the presence of infectious diseases using questionnaires and fecal and blood tests for known pathogens. 15
2.4. Biochemical analyses
After an overnight fast, blood was collected in sterile Vacutainer® tubes containing heparin, Ethylenediaminetetraacetic acid, or spray‐coated silica and a polymer gel for serum separation (Beckton Dickinson), centrifuged at 1550 g (15 min, 4°C), and plasma and serum were stored at −80°C until further analyses. Fasting plasma glucose was determined with a commercial assay on the Cobas 8000 c702 analyzer (Roche). Plasma insulin was determined using the ADVIA Centaur XP Immunoassay System (Siemens), according to the manufacturer's protocol. Plasma total cholesterol, HDL‐C, and triglycerides were determined using commercial assays (Diasys and WAKO) on the Selectra® (Sopachem) according to the manufacturer's instructions.
2.5. Bacterial translocation
Bacterial translocation was assessed by 16S sequencing of feces, small intestinal biopsies, visceral adipose tissue, as well as plasma samples. DNA was extracted using a repeated bead beating protocol. 16 For tissue DNA extraction, as a prestep proteinase K was added, and samples were shaken at 56°C for 60 min. DNA was then purified using a Maxwell RSC Whole Blood DNA Kit. 16S rRNA gene amplicons were generated using a single step polymerase chain reaction (PCR) protocol targeting the V3‐V4 region. 17 Polymerase chain reaction products were purified using Ampure XP beads, and purified products were equimolar pooled. The libraries were sequenced using a MiSeq platform using V3 chemistry with 2 × 251 cycles.
Forward and reverse reads were truncated to 240 and 210 bases, respectively and merged using USEARCH. 18 Merged reads that did not pass the Illumina chastity filter, had an expected error rate higher than 2, or were shorter than 380 bases were filtered. Amplified Sequence Variants (ASVs) were inferred for each sample individually with a minimum abundance of four reads. Unfiltered reads were then mapped against the collective ASV set to determine abundances. Taxonomy was assigned using the RDP classifier 19 and SILVA 20 16S ribosomal database V132. Contaminating ASVs were removed after identification using decontam (https://www.biorxiv.org/content/10.1101/221499v2) and manual curation. Non‐contaminant ASVs present in both fecal and visceral adipose tissue samples from the same individual were marked as possible translocations. Raw sequence data was submitted to ENA repository under study PRJEB35645. Microbiota data was further analyzed and visualized using phyloseq, 21 vegan, 22 and picante. 23
2.6. Visceral adipose tissue cytokine expression
Visceral adipose tissue samples were collected by a gastro‐intestinal surgeon during RYGB at the start of the surgery from the epiploic appendages of the transverse colon using sterile laparoscopic scissors and graspers and immediately frozen in liquid nitrogen and stored at −80°C until further analysis, or fixed in formaldehyde for histopathological analysis (see below). RNA was isolated from adipose tissue biopsies using TriPure Isolation Reagent (Roche) according to the manufacturer's instructions. Complementary DNA (cDNA) was prepared using the SensiFAST cDNA Synthesis Kit (Bioline), and mRNA expression was measured using the SensiFAST SYBR No‐ROX Kit (Bioline). The primers for that were used are shown in Supplementary Table S1. Expression levels were normalized to 36B4 and expressed as arbitrary units (AU).
2.7. Macrophage infiltration in visceral adipose tissue
After fixation in formaldehyde of at least 18 h, the visceral adipose tissue samples were embedded in paraffin and stored at room temperature until further analysis. The samples were cut into sections of 5 μm, dewaxed in xylene (3 × 10 min) and rehydrated in four steps (100%, 100%, 96%, and 70% ethanol for 1 min each, respectively) followed by ddH2O. The assay involved 20 min of heat‐induced antigen retrieval using 10 mmol/L sodium citrate buffer (pH 6.0) and endogenous peroxidase quenching with a 1% peroxide block in methanol. Next, samples were incubated with Ultravision protein block (Fisher Scientific) at room temperature (RT) for 10 min and then incubated with CD68‐antibody PGM1 (DAKO, Jena, Germany) overnight at 4°C. Samples were incubated with BrightVision Poly‐HRP goat anti‐mouse IgG, HRP labeled (Immunologic) for 30 min at RT. Next, samples were visualized using a Perma RED kit (Diagnostic BioSystems) per instructions of the manufacturer for 4 min. Finally, samples were counterstained with hematoxylin for 1 min, after which a cover slip with VectaMount Mounting Medium (Vector Laboratories) was placed over the tissue. The areas of most abundant CD68 positivity were photographed at ×20 magnification. Macrophages in these photos were manually counted. The average of 10 photographs for each sample that contained most macrophages was used in the analyses. Second, the amount of crown‐like structures (CLSs) was assessed at ×10 magnification and normalized for total sample area. All assessments were done in blinded fashion.
2.8. Statistical analyses
Data were assessed for normality with the Shapiro–Wilk test and by visually inspecting histograms. Effects FMT compared to controls were assessed using unpaired t‐tests for normal continuous variables, the Kolmogorov–Smirnov Z test for non‐normal continuous variables and Fisher's exact test for proportions. Effects of FMT within groups were assessed using paired t‐tests for normal continuous variables and Wilcoxon signed rank tests for other variables. Shift in taxonomic microbiome composition after FMT was tested using adonis with appropriate repeated measure permutation design. Bivariate correlations were assessed using the Spearman's correlation test. Statistical analyses were performed using SPSS Statistics software, version 24 (IBM). Data are provided as mean with standard deviation or median with interquartile range (IQR). p‐values <0.05 were considered statistically significant.
3. RESULTS
Eight participants received donor FMT prior to RYGB surgery. The participants were age‐, sex‐, and BMI‐matched to controls who did not receive FMT in a 1:2 fashion. Baseline characteristics are shown in Table 1. The individuals included in the study did not receive any particular diet before FMT or before surgery. There were no adverse events in any of the participants.
TABLE 1.
Baseline characteristics
| Characteristics | FMT (n = 8) | Controls (n = 16) | p |
|---|---|---|---|
| Male gender, n (%) | 4 (50) | 8 (50) | 1.000 |
| Age, years | 42.3 (10.9) | 44.3 (9.0) | 0.658 |
| BMI, kg/m2 | 39.8 (3.1) | 40.7 (2.5) | 0.453 |
| Waist circumference, cm | 124.3 (8.2) | 127.4 (12.7) | 0.534 |
| Systolic blood pressure, mmHg | 137.0 (11.7) | 136.4 (12.5) | 0.907 |
| Diastolic blood pressure, mmHg | 87.1 (5.7) | 83.9 (11.3) | 0.464 |
| HDL‐cholesterol, mmol/L a | 1.19 [1.08–1.48] | 1.01 [0.78–1.18] | 0.058 |
| Triglycerides, mmol/L a | 0.96 [0.57–1.59] | 1.89 [1.25–2.75] | 0.126 |
| CRP, mg/L a | 2.6 [2.1–6.6] | 5.2 [2.9–7.8] | 0.233 |
| Fasting glucose, mmol/L | 6.0 (0.8) | 5.8 (0.6) | 0.479 |
| Fasting insulin, pmol/L | 141.3 (37.1) | 135.7 (50.6) | 0.789 |
| HOMA‐IR | 5.5 (2.0) | 5.0 (1.9) | 0.574 |
Note: p values were calculated using unpaired t‐tests and Kolmogorov–Smirnov Z tests for normally and non‐normally distributed data, respectively. Data are mean (SD) unless otherwise specified.
Abbreviations: BMI, body mass index; CRP, C‐reactive protein; FMT, fecal microbiota transplantation; HDL, high‐density lipoprotein; HOMA‐IR, homeostasis model assessment of insulin resistance; IQR, interquartile range; n, number of patients; SD, standard deviation.
Data are median (IQR).
3.1. Effect of FMT versus no FMT on bacterial translocation
At 4 weeks after FMT, both fecal (MANOVA p = 0.015) and small intestinal (MANOVA p = 0.006) microbiota composition were significantly altered (Figure 1). As previously described, there was a very low abundance of 16S rRNA gene in visceral adipose tissue in both groups. While sequencing did show a bacterial signal in visceral adipose tissue, most of the detected ASVs (median 98%, IQR 95%–99%) could be marked as kit contaminants. After removing the contaminants, most samples had some remaining ASVs that could indicate true presence of bacterial DNA. The possible translocations, quantified as ASVs present in both fecal and visceral adipose tissue samples from the same individual, did not differ between treatment groups (Wilcoxon p = 0.086, Figure 2). Also, in overnight fasted plasma samples, obtained reads were minimal (median 180 reads) and not significantly different between groups. Direct comparisons of visceral adipose tissue 16S sequencing data between groups could not be made due to technical confounders.
FIGURE 1.
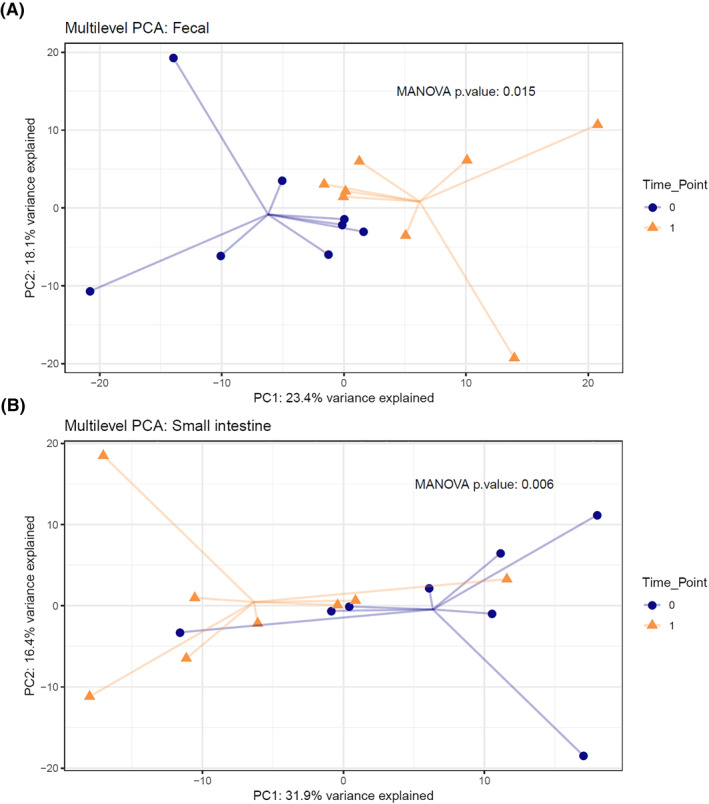
Multilevel principal component analysis (PCA) plot of (A) fecal and (B) small intestinal microbiota composition before (time point 0) and after (time point 1) fecal microbiota transplantation
FIGURE 2.
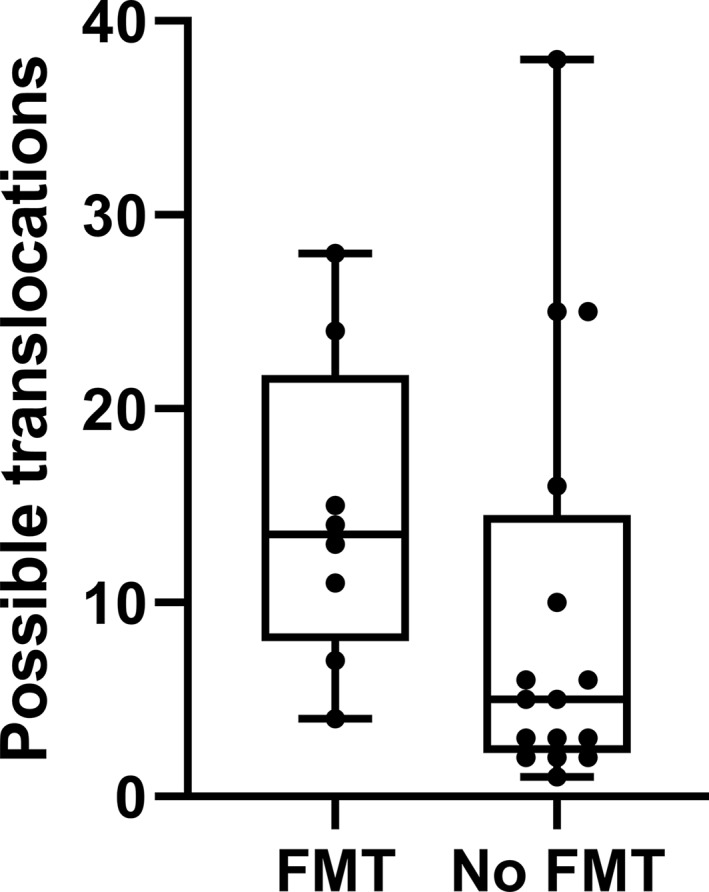
Boxplot showing possible translocations, defined as Amplified Sequence Variants present in both visceral adipose tissue and feces within one participant. Boxes show median, interquartile range, and range. FMT, fecal microbiota transplantation
3.2. Effect of donor FMT on visceral adipose tissue inflammation
Visceral adipose tissue gene expression (quantitative polymerase chain reaction) of the adipocytokines IL‐1β, IL‐6, IL‐10, and TNF‐α did not significantly differ between the FMT group and control group. Also, there was no significant difference in visceral adipose tissue expression of monocyte chemoattractant protein (MCP)‐1, CD11b, CD68, adiponectin, and leptin between groups (Table 2). Only nuclear factor kappa‐light‐chain‐enhancer of activated B cells (NFκB) expression was significantly increased upon donor FMT compared to controls (Figure 3). There was no correlation between fecal microbiota composition and expression of pro‐inflammatory cytokines in visceral adipose tissue (data not shown).
TABLE 2.
Effects of donor FMT on inflammatory gene expression in visceral adipose tissue
| Marker | FMT group | Control group | p |
|---|---|---|---|
| MCP‐1 | 0.63 [0.42–0.84] | 0.61 [0.37–0.84] | 0.804 |
| CD11b | 1.82 [1.06–2.77] | 0.85 [0.49–1.63] | 0.068 |
| CD68 | 0.51 [0.37–0.78] | 0.40 [0.36–0.48] | 0.809 |
| Adiponectin | 0.24 [0.22–0.52] | 0.37 [0.21–0.45] | 0.441 |
| Leptin | 0.53 [0.27–0.88] | 0.26 [0.15–0.40] | 0.139 |
Note: Expression of inflammatory genes in visceral adipose tissue, measured using qPCR, in the donor FMT group (n = 8) versus matched controls (n = 16). Expression was normalized for expression of a housekeeping gene (36B4) and expressed as arbitrary units. p values were calculated using Kolmogorov–Smirnov Z tests. Data are median (IQR).
Abbreviations: CD, cluster of differentiation; FMT, fecal microbiota transplantation; IQR, interquartile range; MCP, monocyte chemoattractant protein; qPCR, quantitative polymerase chain reaction.
FIGURE 3.
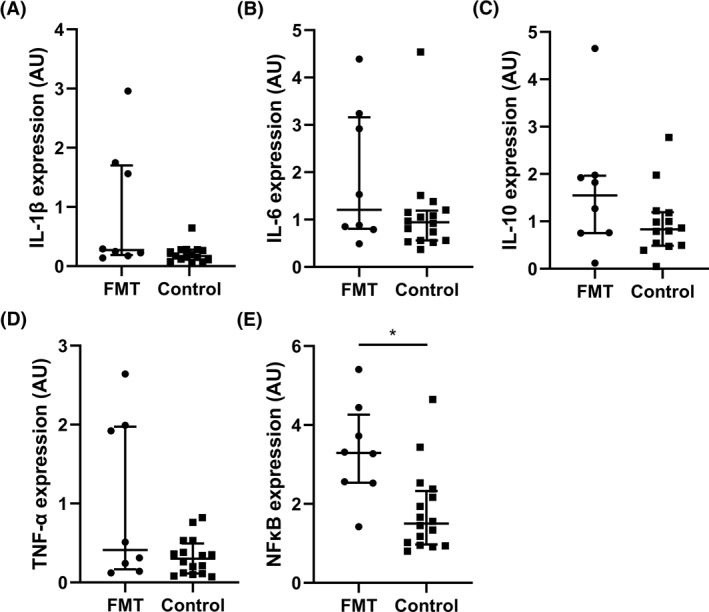
Expression of (A) interleukin (IL)‐1β, (B) IL‐6, (C) IL‐10, (D) tumor necrosis factor (TNF)‐α, and (E) nuclear factor kappa‐light‐chain‐enhancer of activated B cells (NFκB) in mesenteric visceral adipose tissue after fecal microbiota transplantation (FMT) versus controls, measured using quantitative polymerase chain reaction (qPCR). Expression was normalized for expression of a housekeeping gene (36B4) and expressed as arbitrary units (AU). Lines show median and interquartile range
In line, histopathological analysis of visceral adipose tissue showed no significant difference in CLSs (donor FMT group (median [IQR]), 0.59 [0.43–1.05] CLS/cm2 versus control group, 0.51 [0.28–1.19] CLS/cm2; p = 0.641; Figure 4a) or macrophage infiltration (donor FMT group (median [IQR]), 10.30 [7.13–12.40] macrophages versus control group, 8.10 [5.38–12.28] macrophages; p = 0.571; Figure 4b). Examples of histopathological adipose tissue samples are shown in Supplementary Figures S1 and S2.
FIGURE 4.
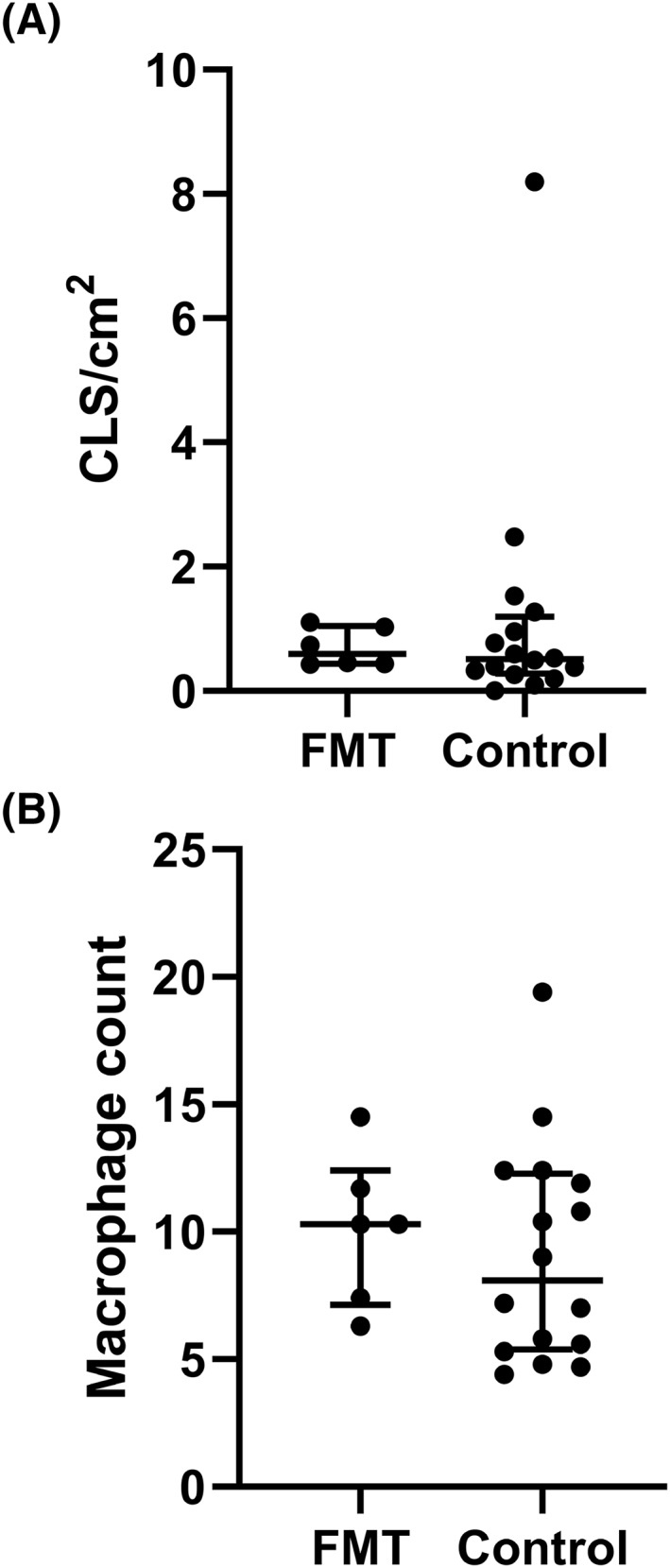
(A) Amount of crown‐like structures (CLSs) and (B) macrophages in mesenteric visceral adipose tissue after fecal microbiota transplantation (FMT) versus controls in histological samples. Macrophages counts and CLSs were assessed by CD68 staining. Lines show median and interquartile range
4. DISCUSSION
Recently, lean donor FMT studies in humans have shown a clinically relevant effect on glucose metabolism, specifically insulin sensitivity. 14 , 24 , 25 , 26 As mesenteric visceral adipose tissue inflammation is an important driver of insulin resistance in obesity, in this study the effect of lean donor FMT on visceral adipose tissue was evaluated. This was the first study in which the effect of lean donor FMT on bacterial translocation and visceral adipose tissue inflammation in humans with obesity was assessed. Although (beyond level of contamination) small amounts of bacterial DNA in visceral adipose tissue that could indicate bacterial translocation were found, there was no difference in translocation between the group that received donor FMT and the matched controls. Moreover, no significant difference in level of visceral adipose tissue inflammation was observed. Finally, there was no correlation between presence of bacterial DNA and inflammatory markers in visceral adipose tissue. Taken together, these results do not provide evidence in support of the hypothesis of bacterial translocation as driver of visceral adipose tissue inflammation.
Translocation of bacteria or bacteria‐derived metabolites from the gut and subsequent mesenteric visceral adipose tissue inflammation in obesity has been postulated to occur in when the gut barrier function is impaired. 5 Indeed, in animal models, obesity is associated with increased intestinal permeability and translocation of gut‐derived lipopolysaccharide (LPS). 27 , 28 In turn, this has been shown to drive chronic low‐grade inflammation and subsequent insulin resistance, a mechanism that has been dubbed metabolic endotoxemia. 28 In turn, inflamed visceral adipocytes may promote the expression of tight junction proteins in intestinal epithelial cells, improving gut barrier function, through secretion of adipocytokines. 29 For example, in a diet‐induced mouse model of obesity, a high‐fat diet resulted in increased gut permeability and macrophage infiltration in visceral adipose tissue, in conjunction with increased IL‐6. 5 Moreover, gut‐derived LPS increased the number of CLSs in white adipose tissue and shifted the adipose tissue macrophages towards a pro‐inflammatory phenotype. 30
Bacterial translocation in humans has mainly been investigated in the setting of inflammatory bowel disease, which is associated with compromised gut barrier function. 31 Using immunofluorescence, Enterococcus faecalis was detected in mesenteric adipose tissue of Crohn's disease patients undergoing surgery for complications. 31 Moreover, ex vivo infection with E. faecalis resulted in proliferation of preadipocytes and adipocytes, which suggests that adipose tissue expansion may be a result of bacterial translocation. 31 One study found that in patients who received surgery for colorectal neoplasia, culturing of mesenteric adipose tissue resulted in bacterial growth in 27% in those with inflammatory bowel disease. However, this was not statistically significantly different from positive culture rates in controls without inflammatory bowel disease. 32
Studies of bacterial translocation into visceral adipose tissue in humans with obesity are scarce. 11 , 33 , 34 Previously, omental adipose tissue of 14 participants with obesity with and without diabetes and five normal‐weight participants was obtained during abdominal surgery. 35 16S rRNA gene sequencing of isolated adipocytes revealed that the majority of reads belonged to Clostridium histolyticum species, of which an enzyme that was used in adipocyte isolation was purified. 5%–10% of reads were unclassified, and less than 1.5% of reads were classified as “other.” In whole adipose tissue, qRT‐PCR of the 16s rRNA gene resulted in only femtograms of bacterial DNA, which was comparable to the no‐template control amount. Cultured specimens also remained negative. 35 Although these findings are interesting, contamination of used reagents has always been an issue when studying these relatively small amounts of bacterial DNA compared to human genomic DNA in human tissue. 36 , 37 , 38
Indeed, this study further underscores the technical difficulty in assessing bacterial translocation based on presence of 16S rRNA gene in tissues with low biomass. As there is a relatively high risk of contamination of samples during sampling and during each step of DNA extraction and sequencing, the utmost care to keep samples sterile was taken. Moreover, all positive signal of known contaminants was removed. Furthermore, all materials used during the sequencing process were sequenced, and positive signal that matched both the materials and the tissues was also removed. In order to further avoid improper labeling of bacterial DNA as translocation from the gut, translocation was defined as ASV reads that were both present in visceral adipose tissue and fecal samples. However, it is important to note that this is not a standard way of assessing translocation. The majority of reads (median 98%) of bacterial DNA in adipose tissue were found to be contaminant‐derived. Thus, it was difficult to interpret the relatively low signal of bacterial DNA which did not fit the criteria for contaminants. Of note, some reads in visceral adipose tissue that were not clear contaminants were nevertheless not marked as translocation, since these reads were not found in feces and were thus not gut inhabitants in the individuals in this study. These strict precautions were used in order to avoid falsely labeling reads as translocation. However, this approach may have resulted in an underestimation of bacterial translocation into visceral adipose tissue.
In the plasma samples, there was not enough signal to discern true translocation from contamination. In several studies, bacterial DNA was found in plasma of healthy participants. 39 , 40 , 41 , 42 , 43 One study even reported positive blood cultures in otherwise healthy blood donors, suggesting the presence of live bacteria in the blood stream. 41 Moreover, in a large prospective population study, plasma 16S rRNA gene sequences were found to be markers of cardiovascular disease risk. 44 Nevertheless, intervention studies showing possible causality in humans so far were lacking.
Importantly, mesenteric visceral adipose tissue inflammation was also assessed using inflammatory gene expression and histology. A difference in inflammatory tone between the FMT group and the controls was not observed. Interestingly, visceral adipose tissue expression of NFκB, a central regulator of inflammatory pathways, was increased in the FMT group compared to the group that did not receive FMT. Nevertheless, other inflammatory parameters were not different between groups. Moreover, the lack of correlation between bacterial signature and inflammation might be in further support of the hypothesis that bacterial translocation may not be the main driver of visceral adipose tissue inflammation. Importantly, although this study focused on bacterial translocation, the gut microbiota may impact visceral adipose tissue inflammation through other mechanisms, such as production of secondary bile acids, short‐chain fatty acids, or trimethylamine‐N‐oxide. 45
This study had several limitations. As the participants were scheduled for bariatric surgery, mesenteric visceral adipose tissue could be sampled, which is the compartment that is closest to the gut. In a study comparing bacterial DNA levels in different adipose tissue compartments, concentrations were the highest in mesenteric adipose tissue, followed by omental adipose tissue, and lowest in subcutaneous adipose tissue. 46 However, as participants only underwent one surgery, it was not possible to compare samples at different time points (e.g., before and after intervention) within participants. Second, as the control group was derived from a cohort study, these participants did not receive autologous fecal transplantation. However, in previous studies no major microbiota and metabolic effects upon autologous FMT were observed, 14 , 26 and the current study design did allow us to match the groups for age, sex, and BMI. Finally, the sample size was relatively small. However, as in other FMT studies, 14 , 24 , 26 a significant effect of fecal transplantation on gut microbiota composition after 4 weeks was observed, and FMT was the principal component in the multilevel principal component analysis. Thus, the intervention had significant impact and changes in translocation of gut bacteria into visceral adipose tissue should have been detected, had it occurred. Nevertheless, it cannot be excluded that long‐term changes in gut microbiota composition do have an effect on bacterial translocation. Future studies might be aimed at investigating effects of long‐term gut microbiota manipulation on bacterial translocation within individuals.
In conclusion, this study shows that bacterial translocation and mesenteric visceral adipose tissue inflammation were not altered after FMT in participants with obesity. This suggests that bacterial translocation may not be the main driver of visceral adipose tissue inflammation in obesity. This study further highlights technical issues with 16S rRNA gene sequencing of tissues with very low biomass.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Guido J. Bakker, Albert K. Groen, Hilde Herrema, and Max Nieuwdorp conceived and designed the study and analyzed the data. Guido J. Bakker, Abraham S. Meijnikman, Ömrüm Aydin, Thomas C.C. Boerlage, L. Maurits de Brauw, Arnold W. van de Laar, and Victor E. Gerdes carried out the experiments and collected the data. Mark Davids, and Daniël H. van Raalte analyzed the data. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Supporting information
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4
ACKNOWLEDGMENTS
Guido J. Bakker was supported by an ICAR grant 2016. Max Nieuwdorp is supported by a ZONMW‐VIDI grant 2013 [016.146.327], ICaR‐VU talent grant and a Dutch Heart Foundation CVON IN CONTROL Young Talent Grant 2013. Daniël H. van Raalte is supported by a Junior Fellowship of the Dutch Diabetes Foundation [2015.81.1840] and by a Marie Skłodowska‐Curie Actions Global Fellowship [708193]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Bakker GJ, Meijnikman AS, Scheithauer TP, et al. Fecal microbiota transplantation does not alter bacterial translocation and visceral adipose tissue inflammation in individuals with obesity. Obes Sci Pract. 2022;8(1):56‐65. 10.1002/osp4.545
REFERENCES
- 1. Weisberg SP, McCann D, Desai M, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity‐related insulin resistance. J Clin Invest. 2003;112(12):1821‐1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith JD, Borel AL, Nazare JA, et al. Visceral adipose tissue indicates the severity of cardiometabolic risk in patients with and without type 2 diabetes: results from the INSPIRE ME IAA study. J Clin Endocrinol Metab. 2012;97(5):1517‐1525. [DOI] [PubMed] [Google Scholar]
- 5. Lam YY, Ha CW, Campbell CR, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet‐induced obese mice. PLoS One. 2012;7(3):e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontana L, Eagon JC, Trujillo ME, et al. Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes. 2007;56(4):1010‐1013. [DOI] [PubMed] [Google Scholar]
- 7. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor‐alpha: direct role in obesity‐linked insulin resistance. Science. 1993;259(5091):87‐91. [DOI] [PubMed] [Google Scholar]
- 8. Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hotamisligil GS, Peraldi P, Budavari A, et al. IRS‐1‐mediated inhibition of insulin receptor tyrosine kinase activity in TNF‐alpha‐ and obesity‐induced insulin resistance. Science. 1996;271(5249):665‐670. [DOI] [PubMed] [Google Scholar]
- 10. Aydin O, Nieuwdorp M, Gerdes V. The gut microbiome as a target for the treatment of type 2 diabetes. Curr Diabetes Rep. 2018;18(8):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amar J, Chabo C, Waget A, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3(9):559‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Udayappan SD, Kovatcheva‐Datchary P, Bakker GJ, et al. Intestinal Ralstonia pickettii augments glucose intolerance in obesity. PLoS One. 2017;12(11):e0181693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alberti KG, Eckel RH, Grundy SM, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640‐1645. [DOI] [PubMed] [Google Scholar]
- 14. Kootte RS, Levin E, Salojarvi J, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611‐619.e6. [DOI] [PubMed] [Google Scholar]
- 15. Cammarota G, Ianiro G, Tilg H, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66(4):569‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costea PI, Zeller G, Sunagawa S, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35(11):1069‐1076. [DOI] [PubMed] [Google Scholar]
- 17. Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112‐5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460‐2461. [DOI] [PubMed] [Google Scholar]
- 19. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261‐5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res. 2013;41(Database issue):D590‐D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oksanen J, Guillaume Blanchet F, Kindt R, et al. Vegan: Community Ecology Package. R Package Version 2.2‐1; 2015.
- 23. Kembel SW, Cowan PD, Helmus MR, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26(11):1463‐1464. [DOI] [PubMed] [Google Scholar]
- 24. de Groot P, Scheithauer T, Bakker GJ, et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. 2019;69(3):502‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Clercq NC, Frissen MN, Davids M, Groen AK, Nieuwdorp M. Weight gain after fecal microbiota transplantation in a patient with recurrent underweight following clinical recovery from anorexia nervosa. Psychother Psychosom. 2019;88(1):58‐60. [DOI] [PubMed] [Google Scholar]
- 26. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913‐916.e7. [DOI] [PubMed] [Google Scholar]
- 27. Cani PD, Delzenne NM. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr Opin Pharmacol. 2009;9(6):737‐743. [DOI] [PubMed] [Google Scholar]
- 28. Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470‐1481. [DOI] [PubMed] [Google Scholar]
- 29. Barrenetxe J, Villaro AC, Guembe L, et al. Distribution of the long leptin receptor isoform in brush border, basolateral membrane, and cytoplasm of enterocytes. Gut. 2002;50(6):797‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caesar R, Reigstad CS, Backhed HK, et al. Gut‐derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61(12):1701‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zulian A, Cancello R, Ruocco C, et al. Differences in visceral fat and fat bacterial colonization between ulcerative colitis and Crohn's disease. An in vivo and in vitro study. PLoS One. 2013;8(10):e78495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peyrin‐Biroulet L, Gonzalez F, Dubuquoy L, et al. Mesenteric fat as a source of C reactive protein and as a target for bacterial translocation in Crohn's disease. Gut. 2012;61(1):78‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manfredo Vieira S, Hiltensperger M, Kumar V, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359(6380):1156‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kell DB, Pretorius E. On the translocation of bacteria and their lipopolysaccharides between blood and peripheral locations in chronic, inflammatory diseases: the central roles of LPS and LPS‐induced cell death. Integr Biol (Camb). 2015;7(11):1339‐1377. [DOI] [PubMed] [Google Scholar]
- 35. Zulian A, Cancello R, Cesana E, et al. Adipose tissue microbiota in humans: an open issue. Int J Obes (Lond). 2016;40(11):1643‐1648. [DOI] [PubMed] [Google Scholar]
- 36. Theis KR, Romero R, Winters AD, et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real‐time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol. 2019;220(3):267.e1–267.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stinson LF, Keelan JA, Payne MS. Identification and removal of contaminating microbial DNA from PCR reagents: impact on low‐biomass microbiome analyses. Lett Appl Microbiol. 2019;68(1):2‐8. [DOI] [PubMed] [Google Scholar]
- 39. Kowarsky M, Camunas‐Soler J, Kertesz M, et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell‐free DNA. Proc Natl Acad Sci U. S. A. 2017;114(36):9623‐9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paisse S, Valle C, Servant F, et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56(5):1138‐1147. [DOI] [PubMed] [Google Scholar]
- 41. Gosiewski T, Ludwig‐Galezowska AH, Huminska K, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next‐generation sequencing method–the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36(2):329‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev. 2015;39(4):567‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Damgaard C, Magnussen K, Enevold C, et al. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One. 2015;10(3):e0120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amar J, Lange C, Payros G, et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PLoS One. 2013;8(1):e54461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716‐1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Troseid M, Nestvold TK, Rudi K, Thoresen H, Nielsen EW, Lappegard KT. Plasma lipopolysaccharide is closely associated with glycemic control and abdominal obesity: evidence from bariatric surgery. Diabetes Care. 2013;36(11):3627‐3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4


