1. Introduction
The genetic engineering of domestic animals has increased significantly in recent years, particularly with the advent of CRISPR-Cas9 gene editing tchnology. The utility of genome editing technology provides a unique opportunity for musculoskeletal investigators to consider the examination of rare phenotypes in domestic animals or perhaps more so, to develop domestic animal models of the bone disorders in which the bone remodeling process more closely resembles that observed in humans. However, the generation of large animal models requires specialized animal husbandry facilities and demands improved analytical capabilities. Accurately assessing the molecular and cellular bone phenotype of a particular genotype is not a simple task and requires the characterization of large bone biopsy specimens, the development of ex vivo culture systems from different species as well as the availability of non-invasive imaging for larger animals. Once characterized, a specific phenotype will contribute to various purposes resulting in new mechanistic insights bone remodeling, specific details of muscle-bone-tendon interactions and perhaps also provide an opportunity for improved treatment and/or therapeutic interventions that are mechanism-based. In this perspective, we interrogate the modeling of bone phenotypes in domestic animals and evaluate the role of these species in the growth of the musculoskeletal field, considering the high current preference for rodent models. Whatever the eventual outcome, it is clear that recent biotechnology developments in gene editing and the annotation of more genomes will undoubtedly bring significant changes to the way musculoskeletal phenotypes are developed, studied and evaluated.
Historically, genetically modified mice, in particular mice in which a specific gene has been inactivated (knockouts), have served as a cornerstone for models of animal and human disease and as the ultimate test for the determination of gene function. However, mice are not completely representative of human physiology, metabolism, genetics, lifespan, or size and many times engineered mice do not exhibit the same phenotype or reveal the same gene function(s) observed in humans. Large animal models of disease often offer distinct advantages because they are in many ways more representative of human physiology and represent alternative solutions to issues with genetic testing of gene action. Genetic engineering of livestock is not a new concept but has become increasingly more efficient and thus more cost effective of late, making the utilization of these valuable large animal research resources available to scientists across multiple disciplines (Table 1). It is important to note that many other livestock species disease models exist but limited space prevented the inclusion of an exhaustive list
Table 1.
Gene Edited Livestock Biomedical Models.
| Human Disease Modeled | Species | Gene target/modification | Reference |
|---|---|---|---|
| CysticFibrosis | Domestic pig | CFTR-null | Rogers 2008 [87] |
| Domestic pig | CFTR-null | Klymiuk 2012 [88] | |
| Domestic pig | CFTR-F508del | Rogers 2008 [89] | |
| Sheep | CFTR-null | Fan 2018 [90] | |
| Diabetes | Domestic pig | HNF-1α | Umeyama 2009 [91] |
| Domestic pig | GIPR | Renner 2010 [92] | |
| Cardiovascular disease | Yucatan miniature pig | LDL Receptor null | Davis 2014 [93] |
| Yucatan miniature pig | PCSK9 | Al-Mashhadi 2013 [94] | |
| Yucatan miniature pig | SCN5A | Park 2015 [95] | |
| Huntington’s disease | Sheep | HTT-CAG73 human transgene | Jacobsen 2010 [96] |
| Liběchov miniature pig | HTT-CAG145 human transgene | Baxa 2013 [97] | |
| Parkinson’s disease | Domestic pig | PARK7-null | Yao 2014 [98] |
| Banna and Bama miniature pig | PARK2- and PINK1-null | Zhou 2015 [99] | |
| Alzheimer’s disease | Göttingen miniature pig | APP695sw human transgene | Kragh 2009 [100] |
| Göttingen miniature pig | PSEN1M1461 | Jakobsen 2013 [101] | |
| Familial Hypertrophic Cardiomyopathy | German Landrace pigs | MYH7 | Montag 2018 [102] |
| DuchenneMuscularDystrophy | Domestic pig | DMDex52del | Klymiuk 2013 [103] |
| Amyotrophic Lateral Sclerosis | Tibetan miniature pig | SOD1 human transgene | Yang 2014 [104] |
| Hemophilia A | Domestic pig | FVIII-null | Kashiwakura [41] 2012 |
| Hypophosphatasia | Sheep | TNSALP | Williams 2017 [45] |
| NeuronalCeroidLipofuscinoses | Sheep | PPT1 | Eaton 2019 [105] |
| Hutchinson-GilfordProgeria syndrome | Yucatan miniature pig | LMNA | Dorado 2019 [106] |
Cystic fibrosis transmembrane conductance regulator (CFTR); Hepatocyte nuclear factor 1alpha (HNF-1α); Gastric Inhibitory Polypeptide Receptor (GIPR); Low-density lipoprotein (LDL) receptor; Proprotein convertase subtilisin/kexin type 9 (PCSK9); α subunit of the major cardiac sodium channel Na(V)1.5 (SCN5A); CAG repeats in the huntingtin-encoding sequence HTT (HTT-CAG); Parkinsonism Associated Deglycase 7 (PARK7); Parkin RBR E3 ubiquitin protein ligase (PARK2); PTEN Induced Kinase 1 (PINK1); Swedish-type mutated APP695 (APP695sw); Alzheimer’s disease-causing gene (PSEN1M146I); Gene encoding a myosin heavy chain beta (MYH7); X-linked dystrophin (DMD) gene lacking exon 52 (DMDex52del); Superoxide dismutase 1 (SOD1); Factor VIII (FVIII); Tissue non-specific alkaline phosphatase (TNSALP); Palmitoyl-protein thioesterase 1 (PPT1); Laminin A (LMNA).
The first genetically modified livestock were produced in the 1980s, with the primary focus being agricultural applications, reviewed by Pursel [1]. These initial experiments involved genetic engineering of sheep and pigs to improve production traits such as feed efficiency and meat quality [2–4]. Results of these experiments verified the usefulness of genetically engineered livestock and laid the groundwork for future research. These important studies ultimately resulted in the production of a large number of different genetically engineered animals, such as pigs resistant to various viral diseases, pigs that produce phytase to facilitate the efficient digestion of phosphorous to decrease pollution (Enviropig ™), hornless cattle, cattle with increased muscle development, goats that produce milk with a longer shelf life, as well as chickens resistant to avian influenza [5–11].
While the animals listed above were produced primarily to benefit production agriculture, i.e. to be used as food, other genetically engineered livestock were developed with the goal of producing therapeutics, pharmaceuticals and supplements that could be used to treat human and/or animal disease. ATryn® (antithrombin) was the first approved recombinant therapeutic which is an anticoagulant produced in the milk of transgenic goats [12]. More recently, Pharming’s Ruconest®, a C1-estrase inhibitor used for the treatment of hereditary angioedema, provided a second complete case study for the development of drugs from transgenic rabbits [13]. Many other livestock species (cows, sheep, goats, pigs and rabbits) have been engineered to produce therapeutic proteins in milk. These include alpha antitrypsin to treat cystic fibrosis, lactoferrin, used to treat stomach and intestinal ulcers, diarrhea and hepatitis C, albumin to treat burn patients, and recombinant human butyrylcholinesterase to treat oganophosphate poisoning, just to name a few. Transgenic cows which produce human antibodies in their blood have also been produced and promise to provide therapeutic approaches to treat a wide variety of human diseases [7,14–20].
Livestock production and utilization as models for biomedical research have become increasing available due to improved technical efficiency. The broad utility of livestock transgenics was dramatically altered after the nuclei of somatic cells from an adult mammal were used to create “Dolly” [21] and shortly thereafter Polly [22]. Genetic engineering has proven extremely useful in enabling animals to produce novel therapeutic proteins [23], and this rapidly evolved to include models of human disease. These pioneering days of genetic engineering, driven almost entirely by insertion of large gene constructs into the animal genome (transgenics), have been superseded with recent advances in the field [24,25]. The new technologies do not solely involve transgenesis and in fact allows for the generation of targeted approaches to genetically engineer animals via gene deletion or by the specific manipulation of sequences within endogenous genes. Given the large and almost daily expansion of sequenced genomes, there is now unprecedented access to detailed sequence information, including control regions, coding regions, and known allelic variants in all the major livestock species as well as the specific gene editing technology needed to modify gene function [26].
The new technologies of gene editing have been added to the molecular toolbox for genetic manipulation of various organisms. Gene editing involves the utilization of a number of DNA modifying enzymes such as zinc-finger proteins (ZFP) [27], transcription activator-like effector nuclease (TALENS) [28] or Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) [29]. All of these operate in basically the same manner by binding to specific genomic locations and inducing DNA strand breaks. ZFPs and TALENS rely on engineered protein domains to recognize DNA, which is then cut by fused FokI nucleases whereas the CRISPR system uses Watson-Crick base pairing with single-stranded RNA to recognize a target sequence that is then cut by a Cas nuclease. Once this occurs, natural DNA repair mechanisms take over and in many cases the repair event results in non-homologous end joining (NHEJ) which causes a mutation at the specific cut site. When a mutation occurs, the target gene is modified and in the case of inactivation, unable to function. Alternatively, the system can be used in conjunction with the addition of a new sequence designed for targeted insertion into the cut site via homology directed repair (HDR). Gene editing is applicable to many different organisms including mice and livestock and has proven to be remarkably efficient [30–36].
Of all the gene editing platforms, application of the CRISPR-Cas9 system has become the method of choice for establishing animal models of disease [35,37]. Indeed, CRISPR-Cas9 is now widely accepted as a simple and versatile RNA-directed system for genome editing in a wide range of different organisms and cell types, including bacteria, mice, rat, zebrafish, human cells, and a variety of livestock species [35,37]. Yet to date, the genetically engineered phenotypes generated in domestic animals have almost entirely been for improved production traits [38–40], novel/therapeutic protein expression [23], or for models of human disease [41–43] but with little specific focus on the musculoskeletal system.
These advances in genome engineering along with a desire to utilize improved models of human disease has led to significant interest in developing gene edited large animal models. Given these broad genetic capabilities are now widely available, the almost singular focus of the bone field on murine models of bone disease is in urgent need of revision. With the increasing availability of reliable genomic sequence information for domestic animals (goats, sheep, pigs and cattle) and the ubiquitous gene manipulation tools, increasing numbers of genetically engineered livestock models, such as sheep, are appearing and being utilized in biomedical research [44,45] (Table 1). This perspective discusses the potential use of genetically engineered domestic animals in bone biology to highlight the beneficial characteristics of large animal models of human disease that complement the widespread utility of available rodent models.
2. Genetic engineered models of bone disease
It is abundantly clear that murine studies have significantly contributed to our understanding of human physiology [46]. In terms of the utility of genetic manipulations in the study of the musculoskeletal system, the utility of the murine genome for manipulation has provided substantial insights into the development of the skeleton as well as a wide variety of disease states [47]. Indeed, much of what we understand about the differentiation of the osteoblast, osteocyte and osteoclast has come from the evaluation of mutant mice. From the pioneering studies that identified c-src and c-fos as critical regulators of osteoclastogenesis [48,49] and Runx2 in osteoblasts [50] as well as the importance of the PTH/PTHrP receptor [51], the contributions of genetically manipulated mice to biomedical research knowledge of bone development has been enormous. Similarly, the utility of mice in the establishment of specific gene function has had major impact on all of science, including the musculoskeletal field. Indeed, the availability of embryonic stem (ES) cells and the relative ease and low cost of manipulating the murine genome with the extensive murine genomic sequence availability compared with other species, virtually all but excluded the use of other species.
The significant cost and time advantages of mice kept attention focused on this species as the model of choice for human disease, even considering the fact that mouse models have some major limitations and do not consistently replicate the human phenotype. Their substantial differences compared with humans in body and organ size, lifespan and inbreeding result in pronounced metabolic, physiological and behavioral differences [44]. While rodents have proven excellent for elucidating gene function and pathways in the mammalian setting (e.g.: Wnt signaling pathway [52]; Runx2 halpoinsufficiency [53]), they have been somewhat less successful in recapitulating many human disease phenotypes or for predicting drug or treatment efficacies [54]. There are many reasons for this that are attributable, in large part, to species-specific differences in anatomy, physiology, metabolic rates, genetics, lifespan, and size. It is also possible that an expanded view to better exploit rodent models by changing the housing environment, diet or microbiome may provide better insight into human physiology. However, even in scenarios where rodent models do manifest the key phenotypic aspects of the human disease being modeled, they frequently pose pre-clinical limitations for purposes such as imaging, drug development, and in many cases surgical intervention [54].
It is also apparent that mice frequently respond to experimental interventions in ways that differ markedly from human responses. For example, the oncology drug endostatin is a potential anticancer therapy that has effectively treated/cured cancer in mice, but has seen limited or minimal efficacy in humans [55,56]. In certain scenarios murine models may reproduce only some of the human disease phenotype, may be more severely affected than human cases, or may have no clinical phenotype at all [57]. In addition, with specific regard to the skeleton there are several examples where human genotype and gene function are not accurately replicated in mice. For example, follicle stimulating hormone (FSH) has been shown to be a potent activator of osteoclast activity in mice [58], yet FSH was not found to regulate bone resorption in postmenopausal women [59]. Similarly, the tooth phenotype of patients with odontohypophosphatasia is not recapitulated in murine models with a modest alkaline phosphatase deficiency [60]. These mutant mice produce only mildly affected periodontal structures because they lack the characteristic tooth deciduous loss of humans [60].
Wild-type pigs, goats, and sheep have been used for decades in biomedical research due to similarities to humans with regard to physiology and anatomy [61] (Table 1). Indeed, numerous sheep (and goat) models have been studied in the orthopedic and osteoporosis research setting [62–67]. The size of these species makes them particularly useful for numerous applications, including clinical imaging, medical implants and/or device design (e.g. plates, screws, external fixators and pacemakers), development of improved surgical procedures, and perhaps most importantly the ability to collect samples in a longitudinal manner, as in human patients. In addition, naturally occurring mutations in domestic species have been widely studied as disease models, including Gaucher’s disease [68], hemophilia [41] and atherosclerosis [69], with few known bone disease-specific mutations investigated [70]. Therefore large animal models that more closely mimic human condition are valuable.
The development of any gene edited disease model requires significant commitment of time, resources, and facilities, and deserves detailed planning. Certainly, there are important cost considerations when considering a rodent or large animal disease model. Currently, the costs to generate gene edited mice or livestock are not dissimilar (at our institution ~$8k for a mouse line and ~$2500/large animal). Costs diverge with animal per diem, and human resources to care for animals are vastly different. With appropriate pasture housing sheep per diem (for example) can be as low as $1/day. However, animal care costs are comparatively greater since it requires significantly more human resources to care for and manage a 50 head herd, versus a 500+ mouse colony.
Recently, the ability to engineer the genome of livestock animals has substantially broadened their utility and enabled new studies into disease mechanisms and therapeutic interventions that were not previously possible [42,43,45] (Fig. 1). Thus, we propose that studies modeling human bone disease using strategically chosen domestic species have the potential to complement murine-driven research and will yield additional mechanistic insights of relevance to musculoskeletal disease in both veterinary and human medicine.
Fig. 1.
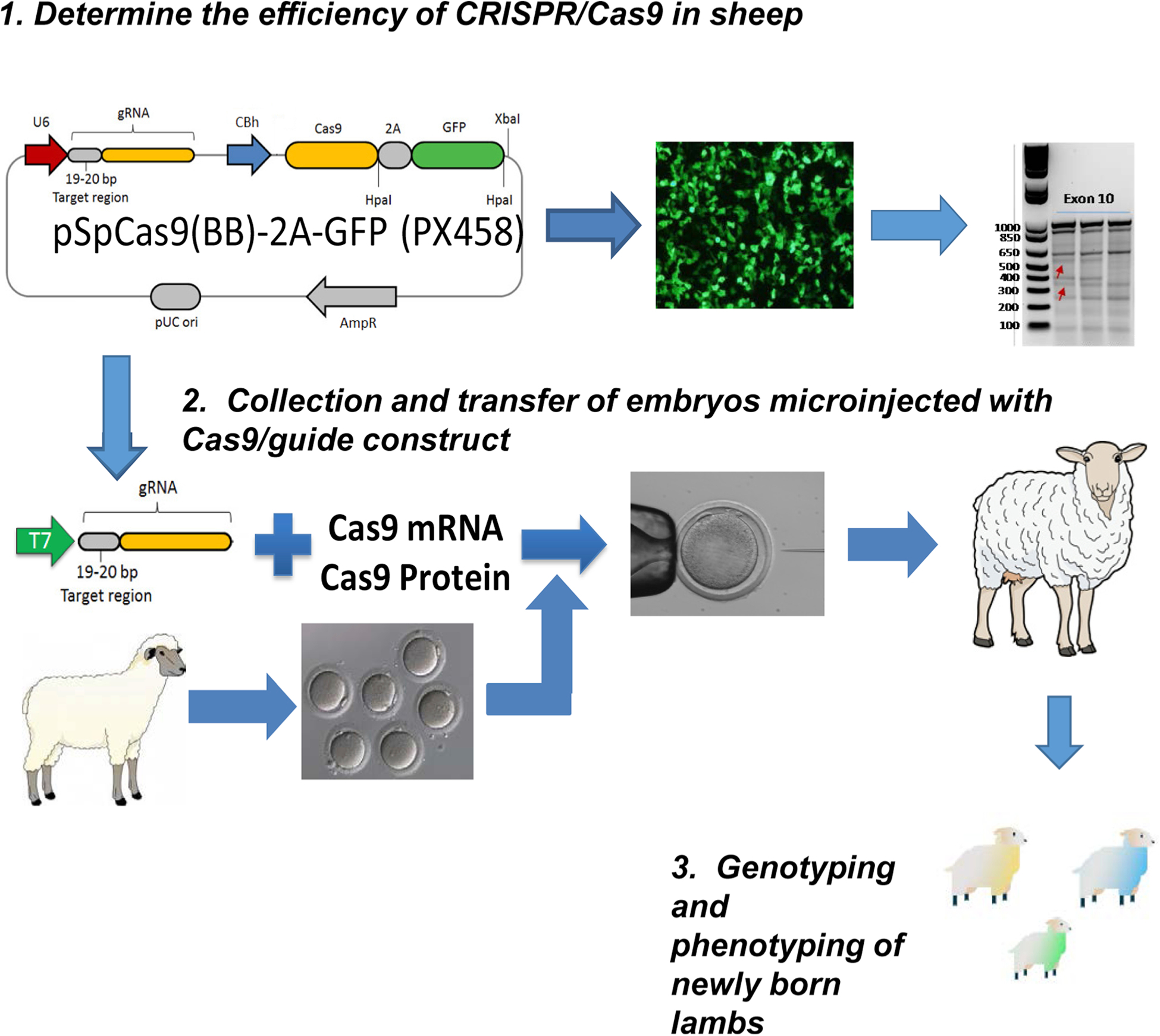
Workflow for the Generation of a Large Animal Model of HPP in Sheep via CRISPR Cas9. 1.) As in all gene editing efforts, the efficiency of the selected CRISPR/Cas9 reagents must be determined. In this case, efficiency is determined using primary cultured sheep fibroblasts with Cas9 enzymatic activity determined by T7 nuclease assay of isolated DNA. Arrows indicate enzymatic activity generating the appropriate DNA fragments in the Exon 10 gel. 2.) For subsequent embryo transfers, females are super ovulated and zygotes collected for in vitro microinjection of Cas9 reagents and the injected embryos transferred to appropriately prepare recipient ewes. 3.) Approximately 145 days after implantation, lambs are born that are subsequently genotyped by DNA sequencing and the targeted phenotype assessed.
3. Bone remodeling and the utility of studying in domestic animals
When considering the decision to embark on the development of a large animal model of bone disease, as with any genetic approach, for practical reasons it is critical to be sure that the disease to be modeled is appropriate for that species. To be clear, the many in vitro bone cell types (osteoclasts, osteoblasts and osteocytes) and assays that the field currently enjoys are extremely informative, helping to answer important questions at the mechanistic level [71]. However, they are not able to answer all concerns, nor do these assays address the critically important physiologic interactions between various organ systems or the critical structural and biomechanical issues of the skeleton. Thus, appropriate animal models are essential if a disease or condition is to be understood and appropriate treatment strategies devised. In this light, how can we design a truly representative animal model of any specific disease? [71]. Indeed, investigators must recognize that all animal models can, by their very nature, only model certain aspects of human disease, thus animal model selection becomes paramount.
For example, when planning to study the effect of overexpression of RANKL on osteoclastogenesis, the use of the mouse model is entirely appropriate. However, if the research question involves tooth development, or the regulation of bone remodeling or perhaps some combination of both, such as in hypophosphatasia (HPP), then the use of other model species must be considered. In the case of HPP (and other rare bone diseases), available murine models have limited utility for the evaluation of tooth loss and none for the examination of Haversian bone remodeling, so studies in the mouse may be less informative. In particular, if murine models do not model the bone pathology seen in patients, researchers are now equipped with the tools to make different animal model decisions.
Among the large animal models that have been utilized for many aspects of musculoskeletal research – e.g. pigs, goats and dogs – sheep have proven especially cost-effective and valuable in orthopedic research [72–74]. Due to their size, large animals are particularly convenient for studying orthopedic implants that are comparable to those used in humans [75,76]. In addition, the size of sheep allows detailed investigation of bone composition and structure of appropriately remodeling cortical and trabecular bone and even the study of the healing of metaphyseal bone, a major site for osteoporotic fractures in humans [77].
Haversian bone remodeling, the fundamental process underlying human bone turnover, is essentially absent in rodent models and many genetically-modified rodent models do not replicate overt bone phenotypes, which is likely due to many factors, including size, short life spans and the different aging processes of small animals as well as mouse housing and environmental conditions. In contrast, the bone structure of adult sheep is comparable to that of humans [71]. In particular, both the trabecular and cortical bone of sheep is characterized by abundant Haversian systems (Fig. 2), with bone remodeling performed by the basic bone multicellular units (BMUs) [78] described by Parfitt in human bone [79]. Similar to other large animals, the cortical bone of young sheep is plexiform [80–82] (Fig. 2) but becomes truly cortical bone with time. This structure, characterized by a surprising combination of woven and lamellar bone, allows the animal to grow rapidly, while maintaining the optimal mechanical properties necessary for locomotion and survival [82].
Fig. 2.
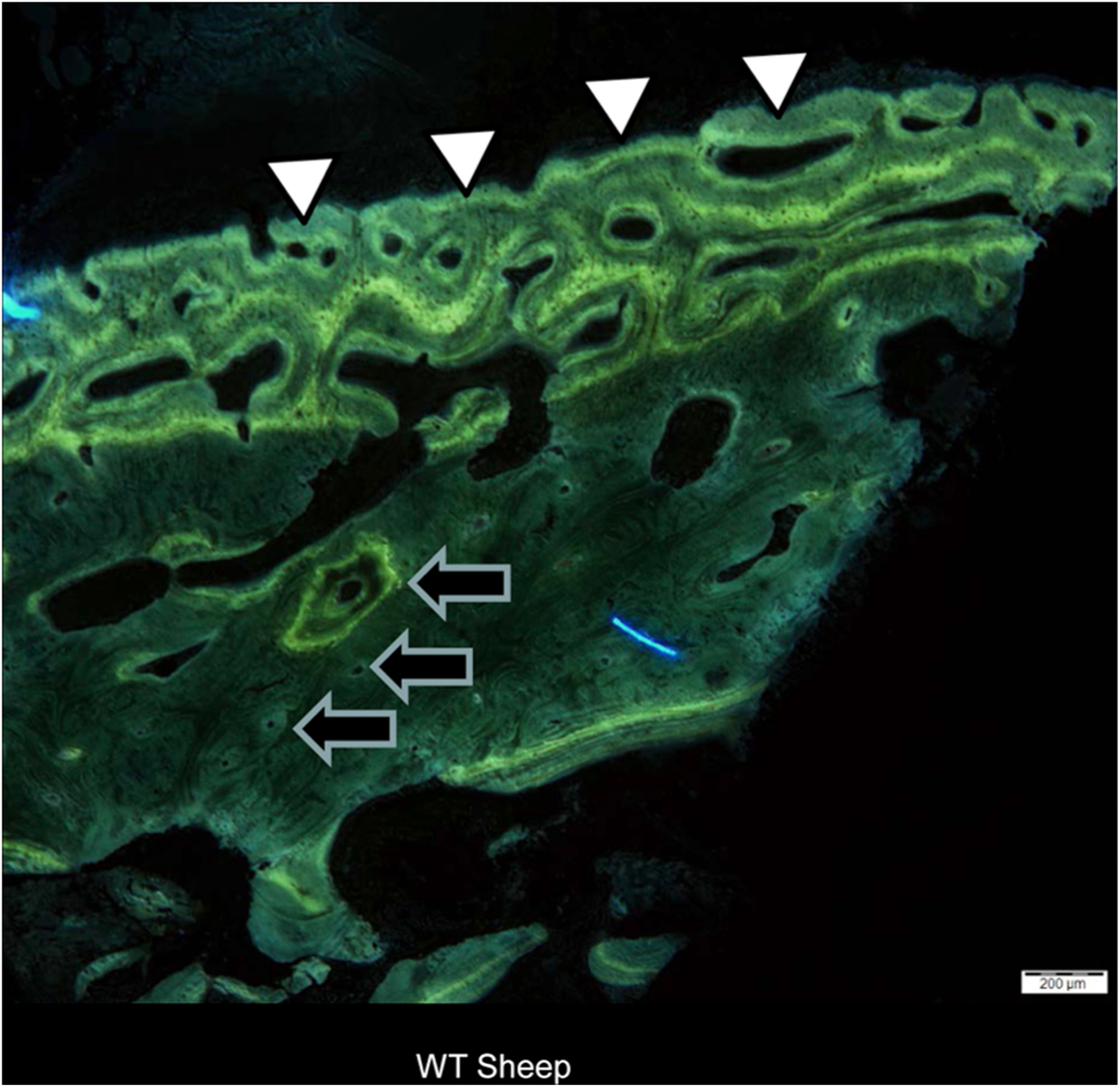
Sheep bone biopsy (6 months). White arrowheads show the appearance of plexiform bone in the developing cortex. Black arrows identify some of the numerous Haversian systems visible in sheep bone. Bar = 200um.
In general, bone formation is characterized by the rapid deposition of less oriented primary bone, followed by a slower formation of secondary (lamellar) bone [83]. Although there are functional similarities between plexiform and lamellar bone, the deposition and organization of plexiform bone shows distinct differences [81]. Older sheep (~1 year) show bone remodeling with well-developed Haversian systems [80,84], with the remodeling of all primary osteonal bone in the sheep consistently observed around 7–9 years [72,82]. Similarly, consider the remarkable differences in bone development between rodents and primates. Murine bone turnover requires around 28 days, whereas the same bone turnover cycle requires more than 150 days in larger species, including humans [85] and sheep. In addition, biochemical markers of bone turnover such as alkaline phosphatase, osteocalcin or collagen crosslinks routinely used to monitor bone turnover in mice and humans are also measurable in sheep [78,86], adding to their utility in studying the skeleton. On the other hand, sheep are not appropriate for studies dealing with orally administered drugs or oral PK/PD absorption studies because of the complex four-compartment stomach characteristic of ruminant herbivores [73].
Another important advantage of the sheep is the ability to perform repeated histomorphometric and biochemical analyses, given the significant bone marrow, blood and urine samples, as well as iliac crest (and other) bone biopsies [82] that can be routinely obtained. Accurately assessing the molecular and cellular bone phenotype of a particular genotype is not a simple task and requires the characterization of bone biopsy specimens akin to those obtained from humans, the development of ex vivo bone marrow culture systems from different species, as well as the availability of non-invasive imaging for larger animals.
The generation of large animal models does require specialized animal husbandry facilities and demands improved analytical capabilities, yet these are widely available at most schools of veterinary medicine. However, while schools of veterinary medicine do have husbandry and analytical facilities for large animals, the costs associated with using those resources can be prohibitive. In addition, many institutions lack the infrastructure to support large NIH-focused biomedical research programs, particularly focused on musculoskeletal diseases, although this is not the case at our institution. It is also critical to remember that societal and ethical implications of working with livestock species are low compared to other large animal models. In the case of sheep (and other ruminants) that are herd animals, stress can be significantly reduced by group housing (on pasture is preferable) and by avoiding daily treatment regimens. These parameters are also critical in considering the welfare and utility of other larger domestic species as bone disease models. However, choosing the right model should not only depend on the impact of gene editing on predicted bone phenotype or the assessment of gene function but must consider animal welfare. When modeling any human disease, it is important to consider which species will best represent the specific human disorder as well as recognize any potential shortcomings. There is no such thing as an ideal animal model so understanding the compromises before beginning is critical.
4. Conclusions and future directions
Other than concerns regarding the cost of large animal models, perhaps the biggest hurdle facing the development and utility of domestic animal bone disease models is simply their acceptance. From the research community and institutions as well as funding agencies more accustomed to the ease of use of rodent models, there are still questions of relevance and need. Even if the large animal disease model represents a more appropriate model of human bone disease, it is often met with significant resistance. Indeed, there are greater costs and efforts required to utilize and maintain domestic species models, although the cost to generate them is not significantly different from current gene edited mouse costs. Perhaps more detailed knowledge can be acquired by better exploiting rodent models (e.g.: changing the immediate environment, the individual microbiome) in addition to the effort to create new larger animal models. Indeed, revising the current research paradigm beyond rodents to include these additional species into the bone researchers’ armamentarium will require significant efforts from those of us able to generate the models that need to be made, and a commitment to not simply develop models that can be made.
In many cases the lack of an appropriate murine model with manifestations similar to those typically found in humans continues to hamper understanding of disease pathogenesis and/or basic physiology. Scenarios such as the bone and muscle phenotypes of human HPP and human cystic fibrosis that are not well modeled in mice are well phenocopied in sheep and pigs respectively. We propose that a strategic focus on developing these large animal models of specific human bone disorders will address relevant questions that are not answered or not possible to answer in existing rodent models. Such an effort will serve to bridge researchers with human bone disease expertise and veterinary researchers with substantial gene editing knowledge, to develop a clear plan for appropriate model design, characterization, and subsequent complete and detailed validation.
While the cost of maintaining a specific bone disease model in a domestic species is relatively high, it is clear that the costs of not having the appropriate animal model will be even higher. Has our understanding of the details of skeletal development and homeostasis been impaired by over-reliance on rodent skeletal models? What, if any, consequences have there been for the development of new treatments for rare and other bone diseases by the almost singular focus on rodents? Whatever the answers to these and other important questions may be, it is apparent that the development of additional and informative bone disease models will enhance our understanding of the skeleton. Indeed, what additional insight can large animal models provide to improve human health? We believe these models hold great promise to address important physiologic questions surrounding bone remodeling, osteocyte function, new disease mechanisms, improved predictive treatment efficacies, and improved drug and device designs. With these aspirational goals, the future for the development of large animal models in bone research seems extremely bright, and we are eager to observe and contribute to the transformation of the field.
Acknowledgements
This paper is dedicated to the memory of Dr. Marco G. Cecchini who was always considering new ways to improve our understanding and advance human health. His wit, intellect and guidance are sorely missed and the bone world diminished by his absence. Our efforts in this area are supported by NIH R21-DE028076(to DG) and by Texas A& M University College of Veterinary Medicine and Biomedical Sciences.
References
- [1].Pursel VG, Rexroad CE Jr., Status of research with transgenic farm animals, J. Anim. Sci 71 (Suppl. 3) (1993) 10–19. [DOI] [PubMed] [Google Scholar]
- [2].Pursel VG, et al. , Growth and tissue accretion rates of swine expressing an insulin-like growth factor I transgene, Anim. Biotechnol 15 (2004) 33–45. [DOI] [PubMed] [Google Scholar]
- [3].Rexroad CE Jr., Production of sheep transgenic for growth hormone genes, Biotechnology 16 (1991) 259–263. [PubMed] [Google Scholar]
- [4].Wall RJ, et al. , Synthesis and secretion of the mouse whey acidic protein in transgenic sheep, Transgenic Res. 5 (1996) 67–72. [DOI] [PubMed] [Google Scholar]
- [5].Carlson DF, et al. , Production of hornless dairy cattle from genome-edited cell lines, Nat. Biotechnol 34 (2016) 479–481. [DOI] [PubMed] [Google Scholar]
- [6].Lyall J, et al. , Suppression of avian influenza transmission in genetically modified chickens, Science 331 (2011) 223–226. [DOI] [PubMed] [Google Scholar]
- [7].Maga EA, et al. , Production and processing of milk from transgenic goats expressing human lysozyme in the mammary gland, J. Dairy Sci 89 (2006) 518–524. [DOI] [PubMed] [Google Scholar]
- [8].Meidinger RG, et al. , Digestive utilization of phosphorus from plant-based diets in the Cassie line of transgenic Yorkshire pigs that secrete phytase in the saliva, J. Anim. Sci 91 (2013) 1307–1320. [DOI] [PubMed] [Google Scholar]
- [9].Prather RS, et al. , Knockout of maternal CD163 protects fetuses from infection with porcine reproductive and respiratory syndrome virus (PRRSV), Sci. Rep 7 (2017) 13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wells KD, Prather RS, Genome-editing technologies to improve research, reproduction, and production in pigs, Mol. Reprod. Dev 84 (2017) 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cornetta K, et al. , Transgenic sheep generated by lentiviral vectors: safety and integration analysis of surrogates and their offspring, Transgenic Res. 22 (2013) 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Avidan MS, et al. , Recombinant human antithrombin III restores heparin responsiveness and decreases activation of coagulation in heparin-resistant patients during cardiopulmonary bypass, J. Thorac. Cardiovasc. Surg 130 (2005) 107–113. [DOI] [PubMed] [Google Scholar]
- [13].van Veen HA, et al. , Characterization of recombinant human C1 inhibitor secreted in milk of transgenic rabbits, J. Biotechnol 162 (2012) 319–326. [DOI] [PubMed] [Google Scholar]
- [14].Kuroiwa Y, et al. , Cloned transchromosomic calves producing human immunoglobulin, Nat. Biotechnol 20 (2002) 889–894. [DOI] [PubMed] [Google Scholar]
- [15].Koles K, et al. , N- and O-glycans of recombinant human C1 inhibitor expressed in the milk of transgenic rabbits, Glycobiology 14 (2004) 51–64. [DOI] [PubMed] [Google Scholar]
- [16].Houdebine LM, Production of pharmaceutical proteins by transgenic animals, Comp. Immunol. Microbiol. Infect. Dis 32 (2009) 107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wright G, et al. , High level expression of active human alpha-1-antitrypsin in the milk of transgenic sheep, Biotechnology (N Y) 9 (1991) 830–834. [DOI] [PubMed] [Google Scholar]
- [18].Zhou Q, et al. , Effect of genetic background on glycosylation heterogeneity in human antithrombin produced in the mammary gland of transgenic goats, J. Biotechnol 117 (2005) 57–72. [DOI] [PubMed] [Google Scholar]
- [19].Kling J, First US approval for a transgenic animal drug, Nat. Biotechnol 27 (2009) 302–304. [DOI] [PubMed] [Google Scholar]
- [20].Huang YJ, et al. , Recombinant human butyrylcholinesterase from milk of transgenic animals to protect against organophosphate poisoning, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 13603–13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH, Viable offspring derived from fetal and adult mammalian cells, Nature 385 (1997) 810–813. [DOI] [PubMed] [Google Scholar]
- [22].Schnieke AE, et al. , Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts, Science 278 (1997) 2130–2133. [DOI] [PubMed] [Google Scholar]
- [23].Ma T, et al. , An AANAT/ASMT transgenic animal model constructed with CRISPR/Cas9 system serving as the mammary gland bioreactor to produce melatonin-enriched milk in sheep, J. Pineal Res 63 (2017). [DOI] [PubMed] [Google Scholar]
- [24].Vajta G, Gjerris M, Science and technology of farm animal cloning: state of the art, Anim. Reprod. Sci 92 (2006) 211–230. [DOI] [PubMed] [Google Scholar]
- [25].Williams BO, Warman ML, CRISPR/CAS9 technologies, J. Bone Miner. Res 32 (2017) 883–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Murray JD, Maga EA, Genetically engineered livestock for agriculture: a generation after the first transgenic animal research conference, Transgenic Res. 25 (2016) 321–327. [DOI] [PubMed] [Google Scholar]
- [27].Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, Genome editing with engineered zinc finger nucleases, Nat. Rev. Genet 11 (2010) 636–646. [DOI] [PubMed] [Google Scholar]
- [28].Joung JK, Sander JD, TALENs: a widely applicable technology for targeted genome editing, Nat. Rev. Mol. Cell Biol 14 (2013) 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chu VT, et al. , Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells, Nat. Biotechnol 33 (2015) 543–548. [DOI] [PubMed] [Google Scholar]
- [30].Hammond A, et al. , A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae, Nat. Biotechnol 34 (2016) 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sinkins SP, Gould F, Gene drive systems for insect disease vectors, Nat. Rev. Genet 7 (2006) 427–435. [DOI] [PubMed] [Google Scholar]
- [32].Ni W, et al. , Efficient gene knockout in goats using CRISPR/Cas9 system, PLoS One 9 (2014) e106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gantz VM, et al. , Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi, Proc. Natl. Acad. Sci. U. S. A 112 (2015) E6736–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dow LE, et al. , Inducible in vivo genome editing with CRISPR-Cas9, Nat. Biotechnol 33 (2015) 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hsu PD, Lander ES, Zhang F, Development and applications of CRISPR-Cas9 for genome engineering, Cell 157 (2014) 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bortesi L, Fischer R, The CRISPR/Cas9 system for plant genome editing and beyond, Biotechnol. Adv 33 (2015) 41–52. [DOI] [PubMed] [Google Scholar]
- [37].Mali P, Esvelt KM, Church GM, Cas9 as a versatile tool for engineering biology, Nat. Methods 10 (2013) 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bevacqua RJ, et al. , Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system, Theriogenology 86 (1886–1896) (2016) e1881. [DOI] [PubMed] [Google Scholar]
- [39].Proudfoot C, et al. , Genome edited sheep and cattle, Transgenic Res. 24 (2015) 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Niu Y, et al. , Biallelic beta-carotene oxygenase 2 knockout results in yellow fat in sheep via CRISPR/Cas9, Anim. Genet 48 (2017) 242–244. [DOI] [PubMed] [Google Scholar]
- [41].Kashiwakura Y, et al. , Porcine model of hemophilia A, PLoS One 7 (2012) e49450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Petters RM, et al. , Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa, Nat. Biotechnol 15 (1997) 965–970. [DOI] [PubMed] [Google Scholar]
- [43].Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E, Genetically engineered pig models for human diseases, Annu. Rev. Anim. Biosci 1 (2013) 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Polejaeva IA, Rutigliano HM, Wells KD, Livestock in biomedical research: history, current status and future prospective, Reprod. Fertil. Dev 28 (2016) 112–124. [DOI] [PubMed] [Google Scholar]
- [45].Williams DK, et al. , Genetic engineering a large animal model of human hypophosphatasia in sheep, Sci. Rep 8 (2018) 16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Perlman RL, Mouse models of human disease: an evolutionary perspective, Evol. Med. Public Health 170–176 (2016) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rosen CJ, Beamer WG, Donahue LR, Defining the genetics of osteoporosis: using the mouse to understand man, Osteoporos. Int 12 (2001) 803–810. [DOI] [PubMed] [Google Scholar]
- [48].Soriano P, Montgomery C, Geske R, Bradley A, Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice, Cell 64 (1991) 693–702. [DOI] [PubMed] [Google Scholar]
- [49].Wang ZQ, et al. , Bone and haematopoietic defects in mice lacking c-fos, Nature 360 (1992) 741–745. [DOI] [PubMed] [Google Scholar]
- [50].Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G, Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation, Cell 89 (1997) 747–754. [DOI] [PubMed] [Google Scholar]
- [51].Lanske B, et al. , PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth, Science 273 (1996) 663–666. [DOI] [PubMed] [Google Scholar]
- [52].Niziolek PJ, et al. , High-bone-mass-producing mutations in the Wnt signaling pathway result in distinct skeletal phenotypes, Bone 49 (2011) 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Smith N, et al. , Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development, J. Cell. Physiol 203 (2005) 133–143. [DOI] [PubMed] [Google Scholar]
- [54].Vandamme TF, Use of rodents as models of human diseases, J. Pharm. Bioallied Sci 6 (2014) 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Whitworth A, Endostatin: are we waiting for Godot? J. Natl. Cancer Inst 98 (2006) 731–733. [DOI] [PubMed] [Google Scholar]
- [56].Kerbel RS, What is the optimal rodent model for anti-tumor drug testing? Cancer Metastasis Rev. 17 (1999) 301–304. [DOI] [PubMed] [Google Scholar]
- [57].Elsea SH, Lucas RE, The mousetrap: what we can learn when the mouse model does not mimic the human disease, ILAR J. 43 (2002) 66–79. [DOI] [PubMed] [Google Scholar]
- [58].Sun L, et al. , FSH directly regulates bone mass, Cell 125 (2006) 247–260. [DOI] [PubMed] [Google Scholar]
- [59].Drake MT, McCready LK, Hoey KA, Atkinson EJ, Khosla S, Effects of suppression of follicle-stimulating hormone secretion on bone resorption markers in postmenopausal women, J. Clin. Endocrinol. Metab 95 (2010) 5063–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Foster BL, et al. , Periodontal defects in the A116T knock-in murine model of Odontohypophosphatasia, J. Dent. Res 94 (2015) 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rogers CS, Genetically engineered livestock for biomedical models, Transgenic Res. 25 (2016) 345–359. [DOI] [PubMed] [Google Scholar]
- [62].Eckhoff DG, Turner AS, Aberman HM, Effect of age on bone formation around orthopaedic implants, Clin. Orthop. Relat. Res (1995) 253–260. [PubMed] [Google Scholar]
- [63].James AW, et al. , NELL-1 in the treatment of osteoporotic bone loss, Nat. Commun 6 (2015) 7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chavassieux P, et al. , Dose effects on ewe bone remodeling of short-term sodium fluoride administration–a histomorphometric and biochemical study, Bone 12 (1991) 421–427. [DOI] [PubMed] [Google Scholar]
- [65].Delmas PD, et al. , The anabolic effect of human PTH (1–34) on bone formation is blunted when bone resorption is inhibited by the bisphosphonate tiludronate–is activated resorption a prerequisite for the in vivo effect of PTH on formation in a remodeling system? Bone 16 (1995) 603–610. [DOI] [PubMed] [Google Scholar]
- [66].Dias IR, et al. , Preclinical and translational studies in small ruminants (sheep and goat) as models for osteoporosis research, Curr. Osteoporos. Rep 16 (2018) 182–197. [DOI] [PubMed] [Google Scholar]
- [67].Turner AS, Experiences with sheep as an animal model for shoulder surgery: strengths and shortcomings, J. Shoulder Elbow Surg 16 (2007) S158–163. [DOI] [PubMed] [Google Scholar]
- [68].Zhou H, et al. , A nucleotide substitution in exon 8 of the glucosylceramidase beta gene is associated with Gaucher disease in sheep, Anim. Genet 48 (2017) 733–734. [DOI] [PubMed] [Google Scholar]
- [69].Prescott MF, McBride CH, Hasler-Rapacz J, Von Linden J, Rapacz J, Development of complex atherosclerotic lesions in pigs with inherited hyper-LDL cholesterolemia bearing mutant alleles for apolipoprotein B, Am. J. Pathol 139 (1991) 139–147. [PMC free article] [PubMed] [Google Scholar]
- [70].Halper J, Connective tissue disorders in domestic animals, Adv. Exp. Med. Biol 802 (2014) 231–240. [DOI] [PubMed] [Google Scholar]
- [71].Oheim R, Amling M, Ignatius A, Pogoda P, Large animal model for osteoporosis in humans: the ewe, Eur. Cell. Mater 24 (2012) 372–385. [DOI] [PubMed] [Google Scholar]
- [72].Pearce AI, Richards RG, Milz S, Schneider E, Pearce SG, Animal models for implant biomaterial research in bone: a review, Eur. Cell. Mater 13 (2007) 1–10. [DOI] [PubMed] [Google Scholar]
- [73].Reinwald S, Burr D, Review of nonprimate, large animal models for osteoporosis research, J. Bone Miner. Res 23 (2008) 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ferguson JC, et al. , A large animal model for standardized testing of bone regeneration strategies, BMC Vet. Res 14 (2018) 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Borsari V, et al. , Osteointegration of titanium and hydroxyapatite rough surfaces in healthy and compromised cortical and trabecular bone: in vivo comparative study on young, aged, and estrogen-deficient sheep, J. Orthop. Res 25 (2007) 1250–1260. [DOI] [PubMed] [Google Scholar]
- [76].Rocca M, Fini M, Giavaresi G, Aldini NN, Giardino R, Osteointegration of hydroxyapatite-coated and uncoated titanium screws in long-term ovariectomized sheep, Biomaterials 23 (2002) 1017–1023. [DOI] [PubMed] [Google Scholar]
- [77].Riggs BL, Melton LJ 3rd, Involutional osteoporosis, N. Engl. J. Med 314 (1986) 1676–1686. [DOI] [PubMed] [Google Scholar]
- [78].Arens D, et al. , Seasonal changes in bone metabolism in sheep, Vet. J 174 (2007) 585–591. [DOI] [PubMed] [Google Scholar]
- [79].Parfitt AM, The cellular basis of bone remodeling: the quantum concept reexamined in light of recent advances in the cell biology of bone, Calcif. Tissue Int 36 (Suppl. 1) (1984) S37–45. [DOI] [PubMed] [Google Scholar]
- [80].Hornby SB, Ford SL, Mase CA, Evans GP, Skeletal changes in the ovar-iectomised ewe and subsequent response to treatment with 17 beta oestradiol, Bone 17 (1995) 389S–394S. [DOI] [PubMed] [Google Scholar]
- [81].Newman E, Turner AS, Wark JD, The potential of sheep for the study of osteopenia: current status and comparison with other animal models, Bone 16 (1995) 277S–284S. [DOI] [PubMed] [Google Scholar]
- [82].Turner AS, The sheep as a model for osteoporosis in humans, Vet. J 163 (2002) 232–239. [DOI] [PubMed] [Google Scholar]
- [83].Currey JD, Dean MN, Shahar R, Revisiting the links between bone remodelling and osteocytes: insights from across phyla, Biol. Rev. Camb. Philos. Soc 92 (2017) 1702–1719. [DOI] [PubMed] [Google Scholar]
- [84].Mori R, et al. , Preliminary study of histological comparison on the growth patterns of long-bone cortex in young calf, pig, and sheep, J. Vet. Med. Sci 67 (2005) 1223–1229. [DOI] [PubMed] [Google Scholar]
- [85].Recker RR, et al. , Static and tetracycline-based bone histomorphometric data from 34 normal postmenopausal females, J. Bone Miner. Res 3 (1988) 133–144. [DOI] [PubMed] [Google Scholar]
- [86].Cabrera D, et al. , Glucocorticoids affect bone mineral density and bone remodelling in OVX sheep: a pilot study, Bone Rep. 9 (2018) 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rogers CS, et al. , Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs, Science 321 (2008) 1837–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Klymiuk N, et al. , Sequential targeting of CFTR by BAC vectors generates a novel pig model of cystic fibrosis, J. Mol. Med. (Berl.) 90 (2012) 597–608. [DOI] [PubMed] [Google Scholar]
- [89].Rogers CS, et al. , Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer, J. Clin. Invest 118 (2008) 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Fan Z, et al. , A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene, JCI Insight 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Umeyama K, et al. , Dominant-negative mutant hepatocyte nuclear factor 1alpha induces diabetes in transgenic-cloned pigs, Transgenic Res. 18 (2009) 697–706. [DOI] [PubMed] [Google Scholar]
- [92].Renner S, et al. , Glucose intolerance and reduced proliferation of pancreatic beta-cells in transgenic pigs with impaired glucose-dependent insulinotropic polypeptide function, Diabetes 59 (2010) 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Davis BT, et al. , Targeted disruption of LDLR causes hypercholesterolemia and atherosclerosis in Yucatan miniature pigs, PLoS One 9 (2014) e93457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Al-Mashhadi RH, et al. , Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant, Sci. Transl. Med 5 (2013) 166ra161. [DOI] [PubMed] [Google Scholar]
- [95].Park DS, et al. , Genetically engineered SCN5A mutant pig hearts exhibit conduction defects and arrhythmias, J. Clin. Invest 125 (2015) 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Jacobsen JC, et al. , An ovine transgenic Huntington’s disease model, Hum. Mol. Genet 19 (2010) 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Baxa M, et al. , A transgenic minipig model of Huntington’s Disease, J. Huntingtons Dis 2 (2013) 47–68. [DOI] [PubMed] [Google Scholar]
- [98].Yao J, et al. , Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs, Sci. Rep 4 (2014) 6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhou X, et al. , Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer, Cell. Mol. Life Sci 72 (2015) 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kragh PM, et al. , Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer’s disease-causing dominant mutation APPsw, Transgenic Res. 18 (2009) 545–558. [DOI] [PubMed] [Google Scholar]
- [101].Jakobsen JE, et al. , Generation of minipigs with targeted transgene insertion by recombinase-mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT), Transgenic Res. 22 (2013) 709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Montag J, et al. , Successful knock-in of Hypertrophic Cardiomyopathy-mutation R723G into the MYH7 gene mimics HCM pathology in pigs, Sci. Rep 8 (2018) 4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Klymiuk N, et al. , Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle, Hum. Mol. Genet 22 (2013) 4368–4382. [DOI] [PubMed] [Google Scholar]
- [104].Yang H, et al. , Species-dependent neuropathology in transgenic SOD1 pigs, Cell Res. 24 (2014) 464–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Eaton SL, et al. , CRISPR/Cas9 mediated generation of an ovine model for infantile neuronal ceroid lipofuscinosis (CLN1 disease), Sci. Rep 9 (2019) 9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Dorado B, et al. , Generation and characterization of a novel knockin minipig model of Hutchinson-Gilford progeria syndrome, Cell Discov. 5 (2019) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]


