Abstract
Shiga toxin 2 (Stx2) has been reported as the main Shiga toxin associated with human disease. In addition, the Stx2 toxin type can have a profound impact on the degree of tissue damage in animal models. We have characterized the stx2 subtype of 168 Shiga toxin-producing Escherichia coli (STEC) isolates of which 146 were derived from ovine sources (principally feces and meat) and 22 were isolated from humans. The ovine STEC isolates were of serotypes that have been shown to occur commonly in the gastrointestinal tract of healthy sheep. The major stx2 subtype in the ovine isolates was shown to be stx2d-Ount (119 of 146 [81.5%]) and was predominantly associated with serotypes O75:H−/H8/H40, O91:H−, O123:H−, O128:H2, and OR:H2. However, 17 of 18 (94.4%) ovine isolates of serotype O5:H− possessed a stx2d-O111/OX3a subtype. Furthermore, STEC isolates of serotypes commonly found in sheep and recovered from both clinical and nonclinical human infections also contained a stx2d (stx2d-Ount/O111/OX3a) subtype. These studies suggest that a specific stx2 subtype(s) associates with serotype and may have important epidemiological implications for tracing sources of E. coli during outbreaks of STEC-associated diseases in humans.
Multiple virulence factors contribute to the pathogenicity of Shiga toxin-producing Escherichia coli (STEC). Although Shiga toxins are the primary virulence factor, the ability to produce intimin (encoded by eaeA) and the possession of a plasmid encoding enterohemolysin (ehxA) are also important (2, 5, 11, 14). Shiga toxins comprise two immunologically non-cross-reactive groups designated Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2). Stx1 is virtually identical to the Shiga toxin of Shigella dysenteriae (21). Stx2 is considered to be the most important virulence factor associated with human disease (5, 22). In addition, Stx2 is about 400-fold more toxic to mice than Stx1 and has also been shown to induce fetoplacental resorption, intrauterine hematoma, fibrin deposition, and neutrophil infiltration when injected intravenously into mice on day 5 of pregnancy (32, 34). Unlike for stx1, considerable sequence variation among stx2 genes has been reported (12, 26, 30, 33). More importantly, differences in the degree of pathogenicity of STEC serotypes have been associated with variations in the stx2 subtype (13, 16, 17).
At least 10 stx2 gene variants have been described (10, 12, 19, 24, 25, 26, 29, 30, 33). The most prevalent Stx2 variants are stx2c, stx2d, and stx2e (26, 30, 33). stxc was isolated from E. coli O157:H− strain E32511 and is closely related to stx2 and stx2vha (30). The stx2d cluster as defined by Pierard et al. (26) comprises stx2d-O111 (24), stx2d-OX3a (25), and stx2d-Ount variants, and these subtypes were identified in non-O157 STEC strains isolated from humans and meat (26, 27). However, stx2d-positive STEC strains are not observed in the most virulent serogroups for humans, including O157, O26, O103, O111, and O145 and have been reported to be less frequently associated with diarrhea and hemolytic uremic syndrome (HUS) (26). stx2e is predominantly associated with edema disease in swine (33) and is rarely recovered from humans.
The importance of characterizing Stx2 types has been recently highlighted by the observation that mouse or human colonic mucin (18) can activate some Stx2 toxins. The Vero cell cytotoxicity of intestinal mucus-treated Stx2vha/b was reported to increase 35- to 350-fold compared to non-mucin-treated Stx2vha/b. Mucin activation provides an explanation for the observation that STEC strains expressing Stx2vh are highly virulent (50% lethal dose of <10 CFU) when fed to streptomycin-treated CD-1 mice compared to STEC strains expressing Stx2c (50% lethal dose of 1010 CFU) (16, 17).
Recent studies of sheep in eastern Australia have demonstrated that the predominant STEC serotypes containing accessory virulence factors (enterohemolysin and/or intimin) are O5:H−, O75:H8, O91:H−, O123:H−, and O128:H2 (6a), and several of these serotypes have been occasionally isolated from clinically affected patients. More than 60 different serotypes of STEC have been isolated from humans with clinical infections (1). Many STEC isolates of ovine origin contain stx2 and express toxin (6a). However, only a few reports have examined stx2 subtypes among STEC isolates recovered from ruminant sources, particularly sheep. The aims of this study were (i) to determine the stx2 subtype(s) of STEC isolates derived from ovine sources and (ii) to determine the stx2 subtypes among human STEC isolates that possess a serotype commonly associated with sheep, with the purpose of determining if sheep represent a source of STEC for human infections.
MATERIALS AND METHODS
STEC isolates.
One hundred sixty-eight STEC isolates were used in this study (Table 1). The Elizabeth Macarthur Agricultural Institute (New South Wales, Australia) provided 124 isolates, which were isolated using methods described by Djordjevic et al. (6a). Of these, 121 were isolated from healthy sheep and 3 were isolated from diagnostic submissions in which STEC was not necessarily implicated as the cause of the disease. Thirty-four isolates were obtained from the Victorian Infectious Diseases Laboratory (Melbourne, Australia). These consisted of 11 isolates of human origin, 9 isolates from lamb meat, 2 isolates from sheep feces, 1 isolate from a meat sausage, and 10 isolates from lamb carcasses. Andre Burnens from the National Reference Laboratory for Foodborne Diseases (Bern, Switzerland) provided 10 human isolates from patients with diarrhea or HUS (6, 7). The Swiss isolates possessed serotypes not commonly found in STEC isolates recovered from ovine sources and were included in this study for comparative purposes only. The Swiss isolates were serotyped by Kim Ziebel and Roger Johnson from the Guelph Laboratory, Health Canada, Guelph, Ontario, Canada.
TABLE 1.
Virulence factor profiles and stx2 subtypes of ovine and human STEC isolates
| Serotype | Sourcea | Total no. of isolates | Virulence Profile
|
No. of isolates containing stx2 variant:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | ehxA | eaeA | stx2 | stx2d-Ount | stx2d-OX3a/O111 | stx2vha | stx2vhb | |||
| O5:H− | Sheep feces, NSW (E) | 17 | + | + | + | − | 17 | ||||
| O5:H− | Sheep feces, NSW (E) | 1 | − | + | + | − | 1 | ||||
| O5:H− | Human, Australia, HUS (V) | 1 | + | + | + | − | 1 | ||||
| O8:H14 | Human, HUS (S) | 1 | − | + | − | − | 1 | ||||
| O7:H− | Human, HUS (S) | 1 | − | + | + | + | 1 | ||||
| O15:H−b | Human, HUS (S) | 1 | − | + | + | + | |||||
| O26:H− | Human, HUS (S) | 2 | − | + | − | + | 2 | ||||
| O75:H− | Sheep feces, NSW (E) | 1 | + | + | + | − | 1 | ||||
| O75:H8 | Sheep feces, NSW (E) | 16 | + | + | + | − | 16 | ||||
| O75:H40 | Sheep feces, NSW (E) | 1 | + | + | + | − | 1 | ||||
| O91:H− | Sheep feces, NSW (E) | 36 | + | + | + | − | 34 | 2 | |||
| O91:H− | Human, Australia, Symptomless (V) | 1 | + | + | − | − | 1 | ||||
| O91:H− | Human, Australia, Diarrhea (V) | 1 | + | + | − | − | 1 | ||||
| O91:H− | Lamb carcasses, Queensland (V) | 9 | + | + | + | − | 8 | 1 | |||
| O91:H− | Lamb carcasses, Queensland (V) | 1 | − | + | − | − | 1 | ||||
| O91:H− | Lamb meat, New Zealand (V) | 6 | + | + | + | − | 6 | ||||
| O91:H− | Lamb meat, New Zealand (V) | 3 | + | + | − | − | 3 | ||||
| O91:H− | Sheep feces, New Zealand (V) | 1 | + | + | + | − | 1 | ||||
| O91:H− | Sheep feces, USA (V) | 1 | + | + | + | − | 1 | ||||
| O91:H− | Meat sausage, Australia (V) | 1 | + | + | − | − | 1 | ||||
| O91:H2 | Sheep feces, NSW (E) | 1 | + | + | + | − | 1 | ||||
| O91:H10 | Human, Australia, Diarrhea (V) | 1 | − | + | − | − | 1 | ||||
| O91:H21c | Human, New Zealand, Diarrhea (V) | 1 | − | + | + | − | 1 | 1 | |||
| O103:H38 | Sheep feces, NSW (E) | 1 | + | + | + | − | 1 | ||||
| O121:H19 | Human, HUS (S) | 1 | − | + | + | + | 1 | ||||
| O121:H19 | Human, Diarrhea (S) | 1 | − | + | − | + | 1 | ||||
| O123:H− | Sheep feces, NSW (E) | 22 | + | + | + | − | 21 | 1 | |||
| O123:H− | Sheep feces, Diagnostic, NSW (E) | 3 | + | + | + | − | 3 | ||||
| O123:H− | Human, Australia, Diarrhea (V) | 1 | + | + | + | − | 1 | ||||
| O128:H2 | Sheep feces, NSW (E) | 12 | + | + | + | − | 12 | ||||
| O128:H2 | Human, Australia, Diarrhea (V) | 3 | + | + | + | − | 3 | ||||
| O128:H2 | Human, New Zealand, Diarrhea (V) | 1 | + | + | + | − | 1 | ||||
| O128:H2 | Human, Australia, Diarrhea (V) | 1 | + | + | − | − | 1 | ||||
| O128:H2 | Human, New Zealand, Diarrhea (V) | 1 | + | + | − | − | 1 | ||||
| O128:H− | Sheep feces, NSW (E) | 2 | + | + | + | − | 2 | ||||
| O145:H− | Human, HUS and diarrhea (S) | 2 | − | + | − | + | 1 | 1 | |||
| O153:H− | Sheep feces, NSW (E) | 2 | + | + | + | − | 2 | ||||
| O153:H25 | Sheep feces, NSW (E) | 1 | + | + | + | − | 1 | ||||
| O157:H− | Sheep feces, NSW (E) | 2 | + | + | + | + | 2 | ||||
| O157:H− | Sheep feces, NSW (E) | 1 | − | + | + | + | 1 | ||||
| O157:H21 | Sheep feces, NSW (E) | 1 | + | + | + | + | 1 | ||||
| OR:H2 | Sheep feces, NSW (E) | 4 | + | + | + | − | 4 | ||||
| OX3:H8 | Human, HUS (S) | 1 | + | + | − | − | 1 | ||||
E, isolates obtained from Elizabeth Macarthur Agricultural Institute, New South Wales, Australia; V, isolates obtained from Victorian Infectious Diseases Laboratory, Victoria, Australia; S, isolates obtained from National Reference Laboratory for Foodborne Diseases, Bern, Switzerland.
stx2 untypeable.
Strain with two different stx2 subtypes.
Multiplex PCR analysis of STEC isolates.
All isolates were prepared and subjected to multiplex PCR for the detection of STEC virulence factors stx1, stx2, ehxA, and eaeA as described by Paton and Paton (23), with the following modification. For DNA preparation, Instagene matrix (Bio-Rad, Richmond, Calif.) was used as described by Fagan et al. (8). Amplified DNA fragments were resolved by gel electrophoresis (28) using 2% (wt/vol) agarose. Gels were stained with ethidium bromide, visualized with UV illumination, and imaged using a GelDoc 1000 image analysis station (Bio-Rad).
stx2 subtyping.
Ovine and human STEC isolates (Table 1) containing stx2 were subjected to stx2 subtyping as described by Pierard et al. (26) and Bastian et al. (3). The primer sequences used are listed in Table 2. For this report stx2d (stx2d-Ount, stx2d-O111, and stx2d-OX3a) is defined as a nucleotide sequence variant of stx2 as described by Peirard et al. (26) and does not refer to the mucin-inducible Stx2d toxin subtype (encoded by stx2vha and stx2vhb) as defined by Melton-Celsa et al. (17).
TABLE 2.
Primers used to amplify stx2
| Primer | Sequence (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|
| Primers for typing stx2 | |||
| VT2-e | AATACATTATGGGAAAGTAATA | 348 | 26 |
| VT2-f | TAAACTGCACTTCAGCAAAT | ||
| LinF | GAACGAAATAATTTATATGT | 900a | 15 |
| LinR | TTTGATTGTTACAGTCAT | ||
| Primers for sequencing stx2 | |||
| Stx2F | TATCTGCGCCGGGTCT | 1,280 | This study |
| Stx2R | CAAAKCCKGARCCTGA | ||
| Paton F | GGCACTGTCTGAAACTGCTCC | 256 | 26 |
| Paton R | TCGCCAGTTATCTGACATTCTG | ||
| Gannon F | CCATGACAACGGACAGCAGTT | 779 | 9 |
| Gannon R | CCTGTCAACTGAGCACTTTG |
The size of the Lin amplicon varies by a few nucleotides depending on the variant.
stx2 amplified with VT2-e and VT2-f primers (Table 2) was subjected to restriction endonuclease digestion with HaeIII and PvuII as described by Pierard et al. (26). The PCR product obtained with the LinF and LinR primers (Table 2) was digested with HincII and AccI as described by Bastian et al. (3). PCR products (10 μl) were incubated with 5 U of appropriate enzyme in the buffer provided by the manufacturer. Restriction fragments were separated by agarose gel electrophoresis. stx2 subtypes were identified based on their restriction profiles (Table 3).
TABLE 3.
Sizes of restriction fragments used for restriction fragment length polymorphism analysis of stx2
| Primers used to amplify fragment | Restriction enzyme | Expected fragment size(s) (bp) for:
|
Reference | ||||
|---|---|---|---|---|---|---|---|
| stx2 | stx2vha | stx2vhb | stx2d-Ount | stx2d-OX3a/O111 | |||
| VT2-e, VT2-f | HaeIII | 348 | 216, 132 | 216, 132 | 216, 132 | 167, 132, 49a | 26 |
| PvuII | 323, 25a | 323, 25a | 250, 73, 25a | 200, 120, 28a | 200, 120, 28a | ||
| LinF, LinR | HincII | 556, 263, 62,a 25a | 556, 322, 25a | 556, 347 | 881, 25a | 881, 25a | 3 |
| AccI | 554, 352 | 551, 352 | 551, 352 | 906 | 554, 352 | ||
Fragment was too small to visualize under the electrophoresis conditions used.
stx2 DNA sequence analysis.
An O91:H− isolate (isolate 122-A1) was chosen as a source of stx2 for sequencing studies for the following reasons. Firstly, O91:H− is the most common ovine STEC serotype recovered from Australian sheep. Secondly, the stx2d restriction fragment length polymorphism profile indicated that it possessed a stx2d-Ount subtype, which is the most common stx2 subtype observed among STEC isolates from the feces of Australian sheep. The A and B subunits of stx2 from isolate 122-A1 were amplified using oligonucleotide primers Stx2F and Stx2R (Table 2). PCRs were carried out in a 50-μl total volume containing 5 μl of nucleic acid (extracted with Instagene matrix) from the isolate, 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 2 mM MgCl2, 10 pmol of each primer, 200 μM (each) deoxynucleotide triphosphates, and 1 U of Taq DNA polymerase. After an initial denaturing step of 5 min at 95°C, the samples were subjected to 35 cycles of denaturation (95°C, 30 s), annealing (60°C, 45 s), and extension (72°C, 90 s), followed by a single final extension step of 5 min at 72°C. PCR products were analyzed by agarose gel electrophoresis and purified using a QIAquick DNA purification kit (Qiagen, Hilden, Germany). Primers used for sequencing are listed in Table 2. DNA sequence reactions were performed using the Big Dye terminator cycle sequencing ready reaction DNA sequencing kit and electrophoresed on an ABI prism 377 DNA sequencer (Perkin-Elmer, Santa Clara, Calif.). Compilation and analysis of DNA sequence data were performed using Auto Assembler software (Perkin-Elmer). Nucleotide and amino acid homology analysis was performed using the Blast program located on the Australian National Genomic Information Service website (http://www.angis.org.au).
Nucleotide sequence accession number.
The sequence of stx2 from the ovine O91:H− isolate (122-A1) has been submitted to the GenBank database under the accession no. AF298816.
RESULTS
Detection of STEC virulence factors using multiplex PCR.
All 146 ovine STEC isolates contained stx2. Of these, 143 (97.9%) contained stx1 and stx2, 139 (95.2%) contained stx1, stx2, and ehxA, and 3 (2%) contained all four virulence factors. All 22 human STEC isolates contained stx2. Eleven (50%) of these contained stx1 and stx2, three (13.6%) contained stx2, ehxA, and eaeA, and none contained all four virulence factors. The virulence factor profiles for all isolates are presented in Table 1.
Subtyping of stx2.
The most common stx2 subtype observed among STEC isolates from sheep was stx2d-Ount (Fig. 1; Table 3). Specifically, 55 of 58 (94.8%) O91:H−, 16 of 16 (100%) O75:H8, 24 of 25 (96%) O123:H−, 12 of 12 (100%) O128:H2, and 4 of 4 (100%) OR:H2 STEC isolates from sheep contained stx2d-Ount. Seventeen of 18 (94.4%) O5:H−, 3 of 58 (5.1%) O91:H−, and 1 of 25 (4%) O123:H− STEC isolates from sheep were found to contain either stx2d-O111 or stx2d-OX3a. These latter two stx2 variants were not differentiated due to their high nucleotide sequence homology (99%). Of the 10 human isolates with serotypes commonly isolated from sheep, 9 (90%) also contained stx2d-Ount. The human O5:H− isolate contained stx2d-O111 and/or stx2d-OX3a. The four ovine O157:H−/H21 isolates possessed stx2vha. Other human isolates with serotypes not commonly found in sheep contained either stx2 or stx2vhb variants (Fig. 2; Table 3). One strain of serotype O91:H21 from a human source contained stx2 in combination with stx2vhb. The stx2 variant in the human strain with serotype O15:H− was untypable.
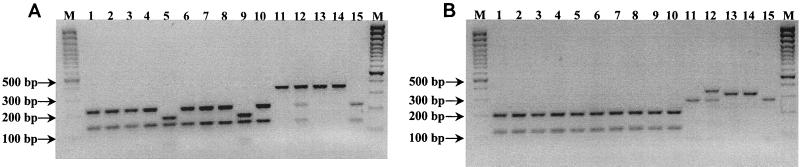
FIG. 1.
HaeIII (A) and PvuII (B) digests of PCR products obtained with VT2-e and VT2-f primers. Lanes: M, 100-bp Plus marker; 1, O91:H− (ovine); 2, O123:H− (ovine); 3, O128:H2 (ovine); 4, O75:H8 (ovine); 5, O5:H− (ovine); 6, O91:H− (human); 7, O123:H− (human); 8, O128:H2 (human); 9, O5:H− (human); 10, OX3:H8 (human); 11, O91:H10 (human); 12, O91:H21 (human); 13, O121:H19 (human); 14, O145:H− (human); 15, O8:H14 (human).
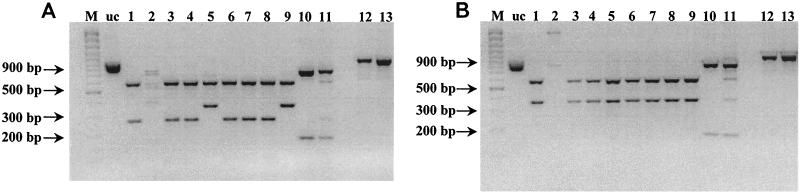
FIG. 2.
HincII (A) and AccI (B) digests of PCR products obtained with LinF and LinR primers. Lanes: M, 100-bp Plus marker; uc, undigested PCR product; 1, O121:H19 (human); 2, O15:H− (human); 3, O145:H− (human); 4, O7:H− (human); 5, O145:H− (human); 6, O26:H− (human); 7, O121:H19 (human); 8, O26:H− (human); 9, O8:H14 (human); 10, O26:H11 (negative control: human isolate with stx1 only); 11, O111:H8 (positive stx2 control; human isolate with both stx1 and stx2); 12, O91:H− (ovine); 13, O5:H− (ovine).
stx2 sequence analysis.
DNA sequence analysis of stx2 from the O91:H− (122-A1) isolate showed 99% homology with stx2d-Ount (GenBank accession no. AF043627). The stx2 DNA sequence was also highly homologous (97%) to stx2d-OX3a (accession no. X65949) and stx2d-O111 (accession no. L11078). These stx2 variants are grouped together as stx2d as described by Pierard et al. (26).
DISCUSSION
Although STEC may contain at least four well-characterized virulence factors (Stx1, Stx2, intimin, and enterohemolysin), Stx2 is considered the most important factor affecting human health (5, 22, 34). In this study the stx2 subtypes of 146 STEC isolates from sheep and 22 human isolates were determined. stx2d variants were most predominant among ovine isolates (141 of 146 [96.6%]). Of these, 119 were stx2d-Ount positive, which was found in association with serotypes O75:H−/H8/H40, O91:H−, O123:H−, O128:H2/H−, OR:H2, and O153:H25/H−. stx2d-O111/OX3a subtypes were found in the remaining 22 ovine isolates, of serotypes O5:H−, O91:H−, O91:H2, and O123:H−. The four ovine isolates of serotype O157:H−/H21 possessed a stx2vha subtype, and the single ovine O5:H− isolate possessed a stx2 subtype.
Of the 22 human STEC isolates, 10 possessed serotypes commonly associated with STEC derived from ovine feces (6a). Nine STEC isolates (six of serotype O128:H2, two of O91:H−, and one of O123:H−) were recovered from seven patients with diarrhea and from two asymptomatic carriers and possessed the stx2d-Ount subtype. Furthermore, isolates OX3:H8 (Switzerland) and O5:H− (Australia) were each recovered from HUS patients and possessed the stx2d-Ount subtype and the stx2d-OX3a and/or stx2d-O111 subtypes, respectively. The O5:H− isolate from the HUS patient is genetically indistinguishable from several epidemiologically unrelated O5:H− isolates recovered from sheep by pulsed-field gel electrophoresis (31). Collectively, these observations suggest that this isolate had an ovine origin.
Twelve human isolates were of serotypes not commonly associated with sheep (O7:H−, O8:H14, O15:H−, O26:H−, O91:H10, O91:H21, O121:H19, O145:H−, and OX3:H8). These were recovered from patients with symptoms ranging from diarrhea to HUS and also included an isolate from a symptomless carrier. All of these isolates possessed stx2 and stx2vhb subtypes, and one isolate (O91:H21) contained two subtypes of stx2 and stx2vhb. However, it is important to emphasize that none of the human isolates from Switzerland possessed a serotype representative of the vast majority of isolates recovered from ovine sources. These data are consistent with studies by Pierard et al. (26) showing that STEC strains normally associated with human disease (serogroups O157, O111, O26, O103, and O145) do not possess a stx2d subtype and that stx2d-positive isolates are less frequently associated with HUS. These and previous studies reinforce the hypothesis that certain serotypes of STEC seem to be associated with their animal host species (4, 6a, 20). Studies in our laboratories demonstrate that STEC isolates recovered from ovine sources possess serotypes rarely observed among STEC isolates recovered from bovine sources (Hornitzky et al., unpublished results). Furthermore, we very rarely observe stx2d subtypes among STEC isolates recovered from bovine sources in Australia (Brett et al., unpublished results). Collectively, these results are consistent with the observation that different stx2 subtypes associate with certain serotypes and these data have significant ramifications in epidemiological studies of STEC infections. These observations also suggest that lamboid phages carrying different stx2 subtypes lysogenize distinct E. coli populations, which may be determined by their serotype.
Vero cell assays of ovine isolates possessing stx2d subtypes are generally toxigenic, with titers down to 10−7 (6a). We did not determine the contribution of Stx1 toxin (which is present in almost all sheep isolates used in this study) to Vero cell toxicity. However, Paton et al. (24, 25) reported a low cytoxicity to Vero cells for the two stx2d variants (stx2d-O111 and stx2d-OX3a), as did Pierard et al. (26) for the single isolate tested in that study. Pierard et al. (26) suggested that Stx2d-producing strains may be a marker for less-pathogenic STEC, since they often failed to possess associated virulence factors. We did not observe the eaeA gene among any of the ovine STEC isolates that possessed stx2d in this study. This result is consistent with the observations of Pierard et al. (26), who failed to observe eaeA among 65 isolates displaying stx2d variant genes. However, in contrast to the findings of Pierard et al. (26), 141 of 146 isolates recovered from ovine sources possessed the ehxA gene. These data suggest that further studies need to be carried out to determine the pathogenicity of ovine STEC to humans.
ACKNOWLEDGMENTS
V.R. is a recipient of an Overseas Postgraduate Research Scholarship and a University of Wollongong Postgraduate Award. This work was supported by funds from Meat and Livestock, Australia.
We thank Jody Wilton and Wendy Forbes for technical assistance with sequencing and Kim Ziebel and Roger Johnson for serotyping the isolates from Switzerland.
REFERENCES
- 1.Acheson D W K. How does Escherichia coli O157:H7 testing in meat compare with what we are seeing clinically? J Food Prot. 2000;63:819–821. doi: 10.4315/0362-028x-63.6.819. [DOI] [PubMed] [Google Scholar]
- 2.Asakura H, Makino S, Shirahata T, Tsukamato T, Kurazono H, Ikeda T, Takeshi K. Detection and long term existence of Shiga toxin (stx)-producing Escherichia coli in sheep. Microbiol Immunol. 1998;42:683–688. doi: 10.1111/j.1348-0421.1998.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 3.Bastian S N, Carle T, Grimont F. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res Microbiol. 1998;149:457–472. doi: 10.1016/s0923-2508(98)80001-6. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Geier D, Steinruck H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerlin P, McEwen S A, Boerlin-Petzold F, Wilson J B, Johnson R P, Gyles C L. Association between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnens A P, Boss P, Orskov F, Orskov I, Schaad U B, Mueller F, Heinzle R, Nicolet J. Occurrence and phenotypic properties of verotoxin producing Escherichia coli in sporadic cases of gastroenteritis. Eur J Clin Microbiol. 1992;11:631–634. doi: 10.1007/BF01961673. [DOI] [PubMed] [Google Scholar]
- 6a.Djordjevic, S. P., M. A. Hornitzky, G. Bailey, P. Gill, B. Vanselow, K. Walker, and K. A. Bettelheim. 2001. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian slaughter-age sheep. J. Clin. Microbiol. 39:•••–•••. [DOI] [PMC free article] [PubMed]
- 7.Essers B, Burnens A P, Lanfranchini F M, Somaruga S G E, von Vigier R O, Schaad U B, Aebi C, Bianchetti M G. Acute community-acquired diarrhea requiring hospital admission in Swiss children. Clin Infect Dis. 2000;30:192–196. doi: 10.1086/313901. [DOI] [PubMed] [Google Scholar]
- 8.Fagan P K, Hornitzky M A, Bettelheim K A, Djordjevic S P. Detection of Shiga-like toxin (Stx1 and Stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65:868–872. doi: 10.1128/aem.65.2.868-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gannon V P J, King R K, Kim J K, Golsteyn Thomas E J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gannon V P J, Teerling C, Masri S A, Gyles C L. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J Gen Microbiol. 1990;136:1125–1135. doi: 10.1099/00221287-136-6-1125. [DOI] [PubMed] [Google Scholar]
- 11.Gyles C, Johnson R, Gao A, Ziebell K, Pierard D, Aleksic S, Boerlin P. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of Shiga-like toxin-producing E. coli of human and bovine origins. Appl Environ Microbiol. 1998;64:4134–4141. doi: 10.1128/aem.64.11.4134-4141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 13.Kokai-Kun J F, Melton-Celsa A R, O'Brien A D. Elastase in intestinal mucus enhances the cytotoxicity of Shiga toxin type 2d. J Biol Chem. 2000;275:3713–3721. doi: 10.1074/jbc.275.5.3713. [DOI] [PubMed] [Google Scholar]
- 14.Lehmacher A, Meier H, Aleksic S, Bockemühl J. Detection of hemolysin variants of Shiga toxin-producing Escherichia coli by PCR and culture on vancomycin-cefixime-cefsulodin blood agar. Appl Environ Microbiol. 1998;64:2449–2453. doi: 10.1128/aem.64.7.2449-2453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S, Yamasaki S, Kurazono H, Ohmura M, Karasawa T, Inoue T, Sakamoto S, Suganami T, Takeoka T, Taniguchi Y, Takeda Y. Cloning and sequencing of two new verotoxin 2 variant genes of Escherichia coli isolated from cases of human and bovine diarrhea. Microbiol Immunol. 1993;37:451–459. doi: 10.1111/j.1348-0421.1993.tb03236.x. [DOI] [PubMed] [Google Scholar]
- 16.Lindgren S W, Melton A R, O'Brien A D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect Immun. 1993;61:3832–3842. doi: 10.1128/iai.61.9.3832-3842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melton-Celsa A R, Rogers J E, Schmitt C K, Darnell S C, O'Brien A D. Virulence of Shiga toxin-producing Escherichia coli (STEC) in orally-infected mice correlates with the type of toxin produced by the infecting strain. Jpn J Med Sci Biol. 1998;51:108–114. doi: 10.7883/yoken1952.51.supplement1_s108. [DOI] [PubMed] [Google Scholar]
- 18.Melton-Celsa A R, Darnell S C, O'Brien A D. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer T, Karch H, Hacker J, Bocklage H, Heesemann J. Cloning and sequencing of a Shiga-like toxin II-related gene from Escherichia coli O157:H7 strain 7279. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;276:176–188. doi: 10.1016/s0934-8840(11)80004-6. [DOI] [PubMed] [Google Scholar]
- 20.Montenegro M A, Bülte M, Trumpf T, Aleksić S, Reuter G, Bulling E, Helmuth R. Detection and characterization of fecal verotoxin-producing Escherichia coli from healthy cattle. J Clin Microbiol. 1990;28:1417–1421. doi: 10.1128/jcm.28.6.1417-1421.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien A D, LaVeck G D, Thompson M R, Formal S B. Productions of Shigella dysenteriae 1-like cytotoxin by Escherichia coli. J Infect Dis. 1982;146:763–769. doi: 10.1093/infdis/146.6.763. [DOI] [PubMed] [Google Scholar]
- 22.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–999. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton A W, Paton J C, Goldwater P N, Heuzenroeder M W, Manning P A. Sequence of a variant Shiga-like toxin type I operon of Escherichia coli O111:H−. Gene. 1993;129:87–92. doi: 10.1016/0378-1119(93)90700-d. [DOI] [PubMed] [Google Scholar]
- 25.Paton A W, Paton J C, Heuzenroeder M W, Goldwater P N, Manning P A. Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of sudden infant death syndrome. Microb Pathog. 1992;13:225–236. doi: 10.1016/0882-4010(92)90023-h. [DOI] [PubMed] [Google Scholar]
- 26.Pierard D, Muyldermans G, Moriau L, Stevens D, Lauwers S. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J Clin Microbiol. 1998;36:3317–3322. doi: 10.1128/jcm.36.11.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierard D, Stevens D, Moriau L, Lior H, Lauwers S. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin Microbiol Infect. 1997;3:531–540. doi: 10.1111/j.1469-0691.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler L H, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. J Clin Microbiol. 2000;66:1205–1208. doi: 10.1128/aem.66.3.1205-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt C K, McKee M L, O'Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr M, Bennett-Wood V, Bigham A K, de Koning-Ward T F, Bordun A M, Lightfoot D, Bettelheim K A, Jones C L, Robins-Browne R M. Hemolytic-uremic syndrome following urinary tract infection with enterohemorrhagic Escherichia coli: case report and review. Clin Infect Dis. 1998;27:310–315. doi: 10.1086/514656. [DOI] [PubMed] [Google Scholar]
- 32.Tesh V L, Burris J A, Owens J W, Gordon V M, Wadolkowski E A, O'Brien A D, Samuel J E. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect Immun. 1993;61:3392–3402. doi: 10.1128/iai.61.8.3392-3402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein D L, Jackson M P, Samuel J E, Holmes R K, O'Brien A D. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimura K, Fujii J, Tanimoto A, Yutsudo T, Kashimura M, Yoshida S. Effects of Shiga toxin 2 on lethality, fetuses, delivery, and puerperal behavior in pregnant mice. Infect Immun. 2000;68:2254–2258. doi: 10.1128/iai.68.4.2254-2258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]