Abstract
The supraspinal connectome consists of dozens of neuronal populations that project axons from the brain to the spinal cord to influence a wide range of motor, autonomic, and sensory functions. The complexity and wide distribution of supraspinal neurons present significant technical challenges, leading most spinal cord injury research to focus on a handful of major pathways such as the corticospinal, rubrospinal, and raphespinal. Much less is known about many additional populations that carry information to modulate or compensate for these main pathways, or which carry pre-autonomic and other information of high value to individuals with spinal injury. A confluence of technical developments, however, now enables a whole-connectome study of spinal cord injury. Improved viral labeling, tissue clearing, and automated registration to 3D atlases can quantify supraspinal neurons throughout the murine brain, offering a practical means to track responses to injury and treatment on an unprecedented scale. Here we discuss the need for expanded connectome-wide analyses in spinal injury research, illustrate the potential by discussing a new web-based resource for brain-wide study of supraspinal neurons, and highlight future prospects for connectome analyses.
The supraspinal connectome: an overview
A connectome refers to “the comprehensive structural description of the network of elements and connections” in a nervous system (Sporns et al., 2005). Connectomes can be described at different resolutions, ranging from microscale analyses of subcellular structures like synapses to macroscale descriptions of whole regions and tracts. Between these is the mesoscale, which maps connections at the level of individual cells. The focus of this review is the supraspinal mesoscale connectome, hereafter shortened to supraspinal connectome, which encompasses the full set of pathways that descend from brain to any level of the spinal cord, with the neurons of origin mapped at single-cell resolution. The supraspinal connectome is comprised of dozens of distinct cell populations, and our knowledge of their anatomical and functional contributions comes from efforts spanning more than a century. The size and complexity prevent a detailed description here (excellent anatomical reviews are available (ten Donkelaar, 2000; Kuypers H. and Martin, 1982; Nudo and Masterton, 1988)). Below we highlight three general features of the supraspinal connectome of high relevance to spinal injury research.
The first regards the wide distribution of the cell bodies of origin, which are dispersed through the medulla, pons, midbrain, ventral forebrain, and isocortex (Leong et al., 1984; Liang et al., 2011; Loewy, 1981; Muñoz-Castañeda et al., 2020). Importantly, supraspinal neurons are not generally well segregated, meaning they are closely adjacent or intermingled with neurons whose axons project only intracranially. These features create dual challenges for direct delivery of potential therapeutics to supraspinal neurons. First, the large tissue volume makes it impossible to reach all supraspinal cell types by intracranial injection. Second, the intermingling makes it impossible to prevent the spread of injections to off-target cells. This necessitates a “broad but specific” delivery of therapeutics, which is considered in a section below.
Second, the supraspinal connectome carries an exceptional diversity of functions, which can be broadly categorized into motor, autonomic, and sensory (Coote and Spyer, 2018; Grillner and El Manira, 2020; Lemon, 2008). Each of these can be further sub-divided. For example, motor control encompasses a wide range of commands related to locomotion, posture, balance, fine movement, and more. Descending autonomic functions include thermal regulation, micturition and bladder control, blood pressure, metabolism, and sexual function (Hou and Rabchevsky, 2014), while sensory modulation can gate pain sensation and also fine touch (Liu et al., 2018). One obvious implication of this functional diversity is that upon injury to the spinal cord, affected individuals experience a wide range of functional deficits. Starting from a historical focus on locomotion, the field has somewhat expanded its consideration to non-locomotor effects of spinal injury, spurred by influential studies that have communicated the priorities of the SCI population (Anderson, 2004). Nevertheless, the wide range of functions that are served by supraspinal connections presents an inherent challenge to the field.
This challenge is compounded by a third feature of the supraspinal connectome: descending control is distributed across multiple tracts in a redundant and/or cooperative manner. In other words, virtually all behaviors valued by individuals with SCI are controlled not by a pathway arising from a single population of neurons, but rather by the coordinated output of nuclei distributed across multiple brain regions. Importantly, in normal conditions these distributed inputs maintain an appropriate balance of excitatory and inhibitory drive. This is achieved both by distinct excitatory and inhibitory transmitters in different supraspinal neurons themselves (for example, about 40% of supraspinal brainstem neurons are Gaba- or glycinergic), and by a balance of excitation and inhibition in spinal circuits that are targeted by descending axons (Boulenguez et al., 2010; Bouvier et al., 2015; Chen et al., 2018; Holstege, 1991). Thus, to understand the functional consequences of any given injury SCI researchers must consider the specific contributions of distinct tracts, with special attention paid to the broader balance of inhibitory and excitatory drive.
Hand function, a high priority for cervically injured individuals (Anderson, 2004), serves as an illustrative example of distributed supraspinal control. In animal models, hand function is often modeled by reaching and grasping tasks. Corticospinal tract neurons have long been recognized as important for fine motor tasks and have been the focus of many restorative tests in animal models. Importantly, however, the red nucleus and reticular populations in the brainstem have also been identified as direct spinal projections that control distinct phases of reaching behavior (Esposito et al., 2014; Ruder et al., 2021). It is also important to consider the compensatory postural adjustments that support a successful reach, which may be controlled in part by reticular neurons in the pons (Takakusaki et al., 2016). Ongoing work in uninjured animals is continuing to parse the distinct contributions of various cell types, and it is likely that there are more relevant regions that await discovery.
The distributed control of reaching behavior presents core challenges for SCI research. After injury, there are likely limits to the amount of function that can be restored by repairing any single tract, even one as important as the corticospinal. If so, difficult questions arise. How plastic is each additional population, meaning to what extent can their axons sprout and form new connections, and to what extent can such remodeling compensate for the loss of the CST and one another (Raineteau et al., 2001)? Conversely, what is the minimal set of connections, and from which of the populations, that must be repaired to restore discrete arm and hand functions? Another important question is the extent to which sensorimotor adaptations, initiated early in recovery and involving diverse supraspinal centers, must be “unlearned” for long term functional gains (Lei and Perez, 2021). Progress has been made in linking lesions in discrete tracts to functional deficits (Hurd et al., 2013), but overall the field remains in the early stages of addressing these questions. Importantly, reaching and grasping behavior is just one exemplar of the general rule of multi-source control. Even autonomic functions such as the regulation of blood pressure and body temperature are influenced by multiple supraspinal centers (Dampney, 1994; Llewellyn-Smith, 2009). Thus, whether the goal is to predict the degree of functional deficit after injury, to explain the innate capacity for natural compensation and recovery, or to assess the potential benefit of pro-regenerative therapies, a broad consideration of multiple populations is essential.
The understudied supraspinal connectome: causes and consequences
To what extent has spinal injury research considered the diverse range of supraspinal populations? Figure 1 presents an analysis of the Pubmed-referenced research in which broad search terms for various injuries, axon tracts, and growth responses were combined with “spinal cord” and the names of forty known supraspinal brain regions (Fig. 1A). The results show that nearly all spinal injury studies, upwards of 95%, have focused on only a handful of major pathways, notably the corticospinal, rubrospinal, and raphespinal (Fig 1B). Reticulospinal pathways have also been studied, but, importantly, “reticular” is an umbrella term that encompasses many regionally segregated populations with both motor and pre-autonomic outputs. Searches for individual reticular populations shows that in SCI research the subpopulations are not usually distinguished (but see (Vavrek et al., 2007) for an exception).
Figure 1. The focus of spinal injury research on selected supraspinal populations.
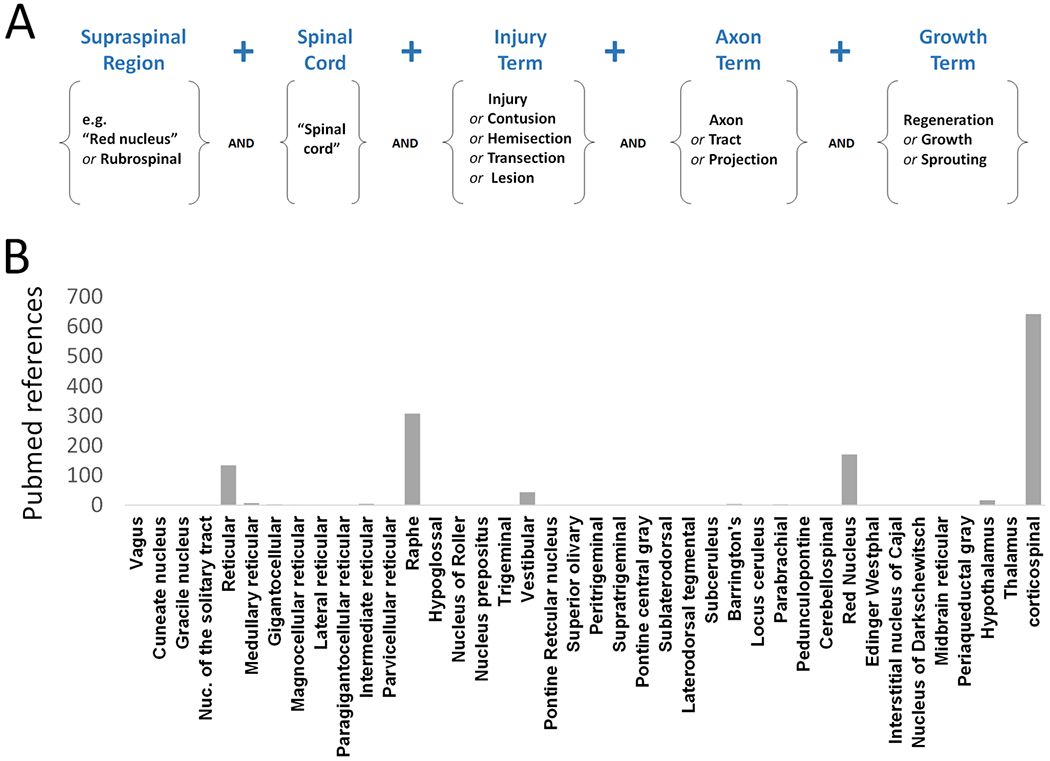
(A) A Pubmed search strategy to identify articles concerning axon growth and plasticity after spinal cord injury. (B) The number of articles returned by Pubmed when search terms were combined with 40 known supraspinal areas. Note that for each area the searches were made as inclusive as possible by using “or” functions with multiple synonymous terms.
This is not to argue that the field has ignored other parts of the supraspinal connectome entirely. For example, in some cases researchers have employed retrograde tracing and manual scoring of brain sections to survey more broadly the cell types that have extended axons into grafts of transplanted cells (Iannotti et al., 2004; Plant et al., 2003; Vavrek et al., 2007). A brain-wide analysis of synapse formation by injured axons stands out as particularly thorough in this regard (Adler et al., 2017). It is also likely that the Pubmed searches may have missed data within manuscripts that did not reach the abstract or keywords. Nevertheless, it is clear that research on axonal growth and plasticity has aimed mostly at several supraspinal populations. Many other pathways that are essential for motor, autonomic and sensory integration such as pedunculopontine, parabrachial, specific hypothalamic nuclei, or the pontine reticular formation are addressed in just a few or in no studies.
What has kept most SCI research away from large swaths of the supraspinal connectome? One challenge is essentially informational and educational. With dozens of discrete populations that are distributed and sometimes intermingled throughout the brain, considerable effort is required to master the detailed anatomical knowledge needed for their study. Information is available but is scattered widely across nuclei-specific reports that use different species and sometimes inconsistent nomenclature that makes it difficult to pinpoint exact location or compare between studies. Neuronal locations that are described in reference to (unknown) anatomical landmarks and drawn in two dimensions present a circular problem of inaccessibility for the novice attempting to create a three-dimensional understanding. Certainly, individuals and labs with strong anatomical knowledge exist in the SCI field. We would argue, however, that it is not atypical for trainees in SCI to rise with a general understanding of the major tracts but without nuanced understanding of many other supraspinal populations that carry critical inputs.
The primary limitations, however, are technical. Our collective knowledge of supraspinal circuits was gathered over more than a century of study, with early work employing techniques of orthograde degeneration and targeted electrical stimulation. Much of our understanding comes from the development of axonally transported tracers (Bentivoglio et al., 1980; Cavada et al., 1984; Köbbert et al., 2000). In rodent brains, arguably the most comprehensive picture has emerged from studies involving retrograde tracers (Leong et al., 1984; Liang et al., 2011). In these experiments retrogradely transported dyes are injected into selected levels of the spinal cord, followed by serial sectioning of the brain. Skilled anatomists then count or subsample labeled neurons projecting to various brain regions while assigning them to recognized locations throughout the brain. These efforts have been invaluable in surveying the distribution of supraspinal neurons and in providing within-animal estimates of their relative abundance (Leong et al., 1984; Liang et al., 2011). The chief limitations of this approach are the exceptional skill and training that are required and the effort and time needed to analyze even a single brain. Indeed, these brain-wide efforts have typically assessed fewer than five total animals, and have selected a single spinal level for analysis. These approaches are clearly incompatible with measuring connectome data in SCI experiments, which require large cohorts of animals across multiple treatments and time points.
The SCI field’s narrow focus on major supraspinal populations, although understandable and even justified by scarce resources, carries significant drawbacks. The first regards the ability of pre-clinical animal models to predict translational success. As noted above, individuals with SCI suffer from a wide range of motor and autonomic deficits, reflecting the diversity of disconnected populations. The preclinical focus on major motor pathways, however, means that proposed regenerative therapeutics can potentially advance to clinical trial with very limited understanding of how – or if – they may impact the regrowth of many supraspinal populations. It is unlikely that all populations respond similarly to any given treatment, or to the same dose. It should be acknowledged that substantial differences exist in the organization of descending control in rodents versus primates, for example the rodent reticulospinal tract plays a significant role in movement control but is largely vestigial in primates (Basile et al., 2020), while the corticospinal tract in primates is dramatically expanded, is more bilateral, and unlike in rodents displays direct connectivity to spinal motor neurons (Friedli et al., 2015; Lemon, 2008) . Others tracts, for example many reticulospinal spinal and micturition pathways, are likely more conserved (Fowler et al., 2008; Ruder and Arber, 2019). Overall, although clinical predictions from rodent data must always consider species differences, it is reasonable to wonder how the near absence of information about treatment effects on most supraspinal cell types may further undercut the ability of rodent studies to predict human efficacy.
Limited connectome data is also a major hurdle at the level of basic SCI research. Recent attention has focused on the so-called neuroanatomical-functional paradox; we refer readers to an outstanding recent review on the topic (Fouad et al., 2021). This paradox refers to the fact that even when every effort is made to impose highly consistent injuries, individual animals display a range of functional recovery. Moreover, detailed analysis of the spinal injury cannot fully predict or explain these subject-to-subject differences. The field is left with an uncontrolled and unexplained source of variability, both within and especially between labs, which can result in both false positive and false negative conclusions about treatment effects. This in turn likely contributes to problems with reproducibility in the field (Steward et al., 2012). As discussed by Fouad et al. the neuroanatomical-functional paradox is complex and driven by a variety of factors, but we would echo the point that among these is incomplete anatomical information.
We propose that one driver of the apparent paradox is incomplete connectome data. Descending tracts are close and intermingled in spinal white matter, so small and uncontrollable differences in injuries will inevitably damage a different set of axons in each animal. In Figure 2 we present a simple model. It starts with uninjured animals and considers all supraspinal populations that connect via axons to distal spinal cord, setting the starting number in each as 100 percent. Upon injury, each individual population will experience some loss in connection across the injury, leaving it with a “sparing score” that ranges from 100 (all neurons keep axonal connection across the lesion) to 0 (all neurons lose connection) (Figure 2B). We refer to this as a “post-injury connectome”, and we emphasize again that this connectome is likely unique to each individual due to a distinctive combination of sparing in each population. Study of the spinal lesion itself can at best approximate the post-injury connectome, because very small changes in lesion geometry could have large effects on specific axon tracts. We conjecture that functional recovery from injury may be better predicted and explained by connectome data than by lesion data per se.
Figure 2. A connectome-based approach to spinal injury research.
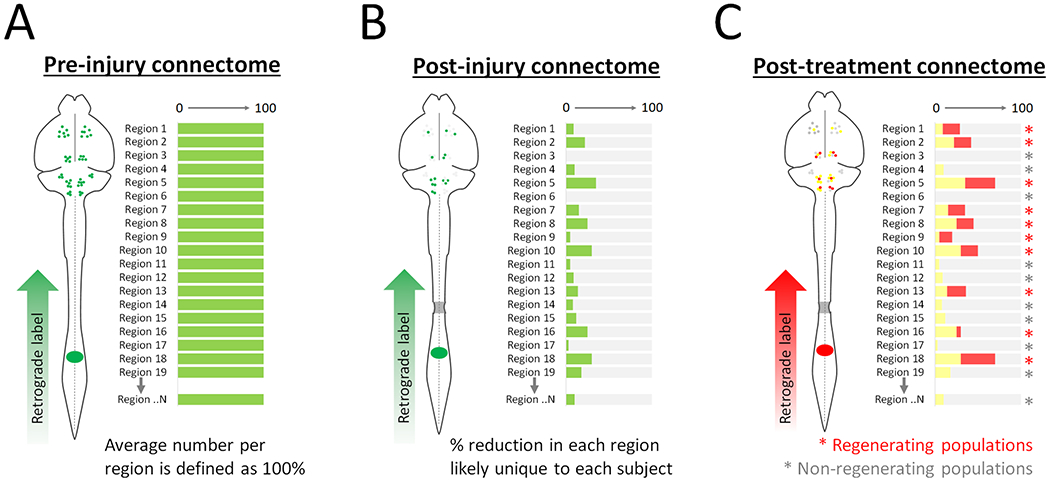
(A) Prior to injury, a retrograde label applied to the spinal cord will mark supraspinal neurons in regions throughout the brain. In each region, the number of labeled neurons establishes a pre-injury baseline of 100 percent based on average results in cohorts of uninjured animals. (B) After a partial spinal injury, retrograde label placed distal to the injury will reach only supraspinal neurons with spared axons that span the injury. The number of neurons counted in region, relative to the previously established baseline, creates a sparing index. The collection of sparing indexes across all regions establishes a unique post-injury connectome in each animal. (C) After treatment of injured animals, a second retrograde label is placed distal to the injury. Spared neurons are marked by both labels (yellow), while neurons that regrew after treatment are marked only by the second label. In this way the effect of treatment can be quantified in each population, identifying both regenerating regions (red asterisk) and non-regenerating regions (grey asterisk).
Taking the concept one step further, when regenerative treatments are applied it is likely that axon growth is evoked more successfully in some populations than others (for example the long-standing observation that brainstem neurons extend axons into peripheral nerve transplants more successfully than CST (Richardson et al., 1984)). Full information about the “post-treatment connectome”, when compared to the connectome immediately post-injury, would distinguish regenerating from non-regenerating populations (i.e. identify cells that responded to treatment by re-establishing axonal connection to the site of distal tracer), while also explaining differences in recovered functions (Figure 2C). In principle, improved connectome information could dramatically enhance understanding of experimental variability, innate plasticity, and experimental treatments.
New techniques for global study of the supraspinal connectome
Complete data on supraspinal connectomes in mouse models are now feasible in SCI experiments, owing to a confluence of technical advances. Briefly, these consist of tissue clearing and 3D imaging, improved viral methods for retrograde labeling, and methods using deep-learning algorithms for cell detection and automatic 3D registration to standardized brain atlases. Below we describe each technical component separately, and then discuss a recently developed pipeline that combines the approaches and applies them to the problem of spinal injury.
Optical clearing and 3D imaging
Methods to render fixed tissue transparent, pioneered over a century ago by Walter Spalteholz, have seen a resurgence in recent years. A variety of solutions and protocols are now available to clear brain and spinal cord tissue (Ueda et al., 2020a; Weiss et al., 2021). Our experience has been that the various 3DISCO approaches achieve the fastest and most effective clearing of neural tissue, although with the caveat that fluorescent signal is unstable and is best imaged the day of clearing (Soderblom et al., 2015; Wang et al., 2018). In parallel, the beginning of this century has seen major advances in light microscopy, particularly related to the neurosciences. One area of rapid development has been light sheet fluorescence microscopy (LSFM), which offers ever-improving speed, resolution and isotropy when imaging cleared tissue (Chakraborty et al., 2019; Glaser et al., 2020). As discussed below, the output of LSFM is a series of individual images, each corresponding to successive 2D planes through the sample, which can then be reconstructed to yield a 3D representation. These developments are enabling researchers to visualize the cellular architecture of the nervous system in 3D with unprecedented detail (Ueda et al., 2020b). For SCI research, an essential point is that these techniques enable relatively rapid imaging of an entire rodent brain, thus avoiding the laborious step of preparing and imaging serial tissue sections that previously limited whole-brain analyses. In principle, the potential speed of data acquisition dramatically increases the number of animals and opens the door to analyzing replicate animals across multiple treatments.
A new generation of neurotropic viruses and optimized fluorescent proteins
Even with optical clearing and imaging in hand, however, significant hurdles remain. First, the approach depends on relatively bright fluorescence. Traditional tracers such as biotinylated dextran amine (BDA) or the retrograde cholera toxin are localized in vesicles and tend to be dim and minimally effective for detection and counting in cleared tissue (Bray et al., 2017; Renier et al., 2014). Antibody amplification may partly resolve this problem, particularly when antibody penetration is aided by electrophoresis, pressurization, reduced antibody size, or repetitive infusion (Cai et al., 2019; Kim et al., 2015; Lai et al., 2018; Lee et al., 2014). In some cases these methods were developed for embryonic brain or thick tissue sections and their suitability for whole brains has yet to be established (see (Ueda et al., 2020b) for a comprehensive review). The fact remains that despite the proliferation of methods of clearing combined with immunohistochemistry certain hurdles remain and no clear winner has emerged that can be adopted widely.
Alternatively, viral tracing approaches now offer a solution to achieving cell detection in cleared tissue. Strictly neurotropic viruses have been used for several decades to study neural circuitry, but biosafety issues and toxicity somewhat slowed widespread within the neuroscience community. In recent years adeno-associated virus (AAV) has become the virus of choice for delivering genes of interest into the brain due to their relative biosafety and tropism to neurons. Many AAV serotypes exist, with unique capsid structures that confer distinct tropism and patterns of intraneuronal transport (Aschauer et al., 2013; Bohlen et al., 2020; Srivastava, 2016). Most serotypes are taken up predominantly by cell bodies and are most suitable for anterograde tracing approaches, although some do display at least some degree of uptake from axonal terminals and subsequent retrograde transport (Engmann et al., 2020; Hilton et al., 2016). Using directed evolution or rational design, AAV capsids can be altered to modify their tropism or mode of transport. Using these approaches the AAV2 capsid has been modified to produce new variants that are transported retrogradely and that are now widely used in neuroscience (Tervo et al., 2016; Wang et al., 2018). In the context of spinal injury this is a highly significant development. By injecting retrograde virus to the spinal cord, where descending axons converge to a relatively small tissue volume, transgenes can be readily distributed to the cell bodies of origin that are spread broadly through the brain. Moreover, because exposure to the virus is limited to the neurons with spinal projections, off-target expression is limited. Thus retrograde AAV offers an elegant solution to the problem of delivering genes to supraspinal neurons in a “broad but specific” manner.
We and others have combined retrograde viral vectors with tissue clearing and 3D imaging to detect supraspinal neurons in whole brain tissue (Asboth et al., 2018; Steward et al., 2021; Wang et al., 2018). Even in these studies, however, we noted that the success of detection is highly variable between different cell types. When using AAV2-retro expressing cytoplasmic tdTomato or GFP, we found corticospinal tract neurons to be strongly transduced and easily detected. In the brainstem, however, retrogradely labeled neurons displayed dimmer fluorescence, and suffered from optic interference by the numerous bundles of descending axons that also expressed tdTomato. To address this, we recently showed that detection of these dimmer populations was improved more than ten-fold by using nuclear-localized fluorophores rather than cytoplasmic, and by deploying latest-generation monomeric FPs including mGreenLantern and mScarlet (Campbell et al., 2020). With these optimized FP retrograde AAV vectors, spinal injection of the virus to the spinal cord enabled detection of tens of thousands of neurons distributed throughout cortex, midbrain, and brainstem (Wang et al., 2021).
Automated cell detection and registration
Finally, there is the challenge of counting retrogradely labeled nuclei and assigning them to specific brain regions. Spot detection tools are available in open-source software such as ImageJ, Napari, and commercial software (e.g. Imaris) and can assist in the quantification of cell nuclei. Determining the correct location and identity of labeled cell nuclei, however, is more difficult. Manual tracing is only practical and sufficiently precise for populations that are large and discrete, for example the corticospinal tract. Many other populations, however, are closely adjacent and cannot be easily distinguished in 3D space. The emergence of standard 3D atlases offers a solution (Niedworok et al., 2016). By mapping user-supplied images onto standard atlases with pre-defined boundaries marking the borders of discrete brain regions, labeled cells can be placed precisely. For example, a set of integrated and open-source tools, developed by the BrainGlobe initiative, enables a series of optical sections of the sort produced by LSFM to be registered to the 3D atlas utilized by the Allen Brain Atlas (Claudi et al., 2021; Niedworok et al., 2016). In a more recent development, an open-source tool called cellfinder utilizes a user-trained deep learning algorithm to identify and quantify labeled cells or cell nuclei (Tyson et al., 2020). Importantly, these registration tools are robust in the face of the tissue shrinkage or expansion that often accompanies tissue clearing. Thus, regardless of some variation in the absolute size of the input images, the software imposes conformity to a standard, idealized brain atlas (Renier et al., 2016; Tyson and Margrie, 2021). Indeed, tools now exist for the registration of brain sections prepared by standard microscopy (Tappan et al., 2019). Collectively, these and future developments might solve the problem of taking data from disparate labs and preparations and then assigning detected supraspinal neurons to their brain regions of origin in a standard and quantitative manner. In turn this raises the exciting possibility of creating common data repositories in which registered and quantified data can be shared and directly compared across the field.
An interactive atlas of the supraspinal connectome
We have recently presented in preprint form a description of a web-based resource (3Dmousebrain.com) for brain-wide assessment of supraspinal populations that project to various spinal levels, with and without graded levels of spinal injury (Wang et al., 2021). Figure 3 provides an overview of the resource. Users can select example brains of interest based on the spinal level of innervation and injury status (Fig. 3A), interact with 3D representations of supraspinal neurons overlaid on a standard atlas (Fig. 3B), view 2D sections with region boundaries (Fig. 3C), and view graphs that quantify supraspinal abundance in all identified regions (Fig. 3D). Details of this pipeline are available elsewhere (Wang et al., 2021). Here we highlight some general insights that emerged and their implications for further development of this strategy.
Figure 3. Overview of 3Dmousebrain.com, and online resource to visualize supraspinal neurons in intact and injured moue brains.
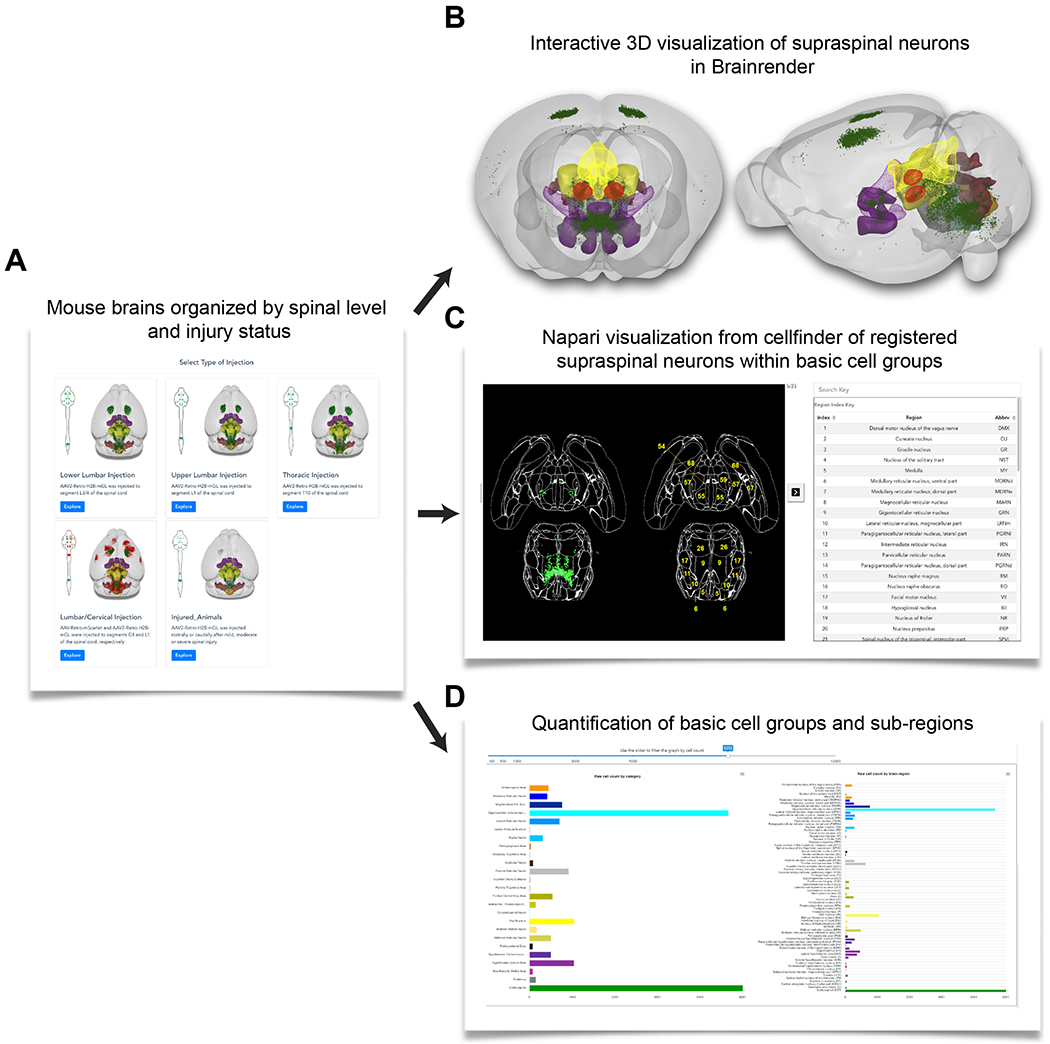
(A) An initial menu of sample types, organized by the spinal level of retrograde injection and injury status. Each type contains two or more example animals with associated data. (B) For each sample users can manipulate (zoom and rotate) a 3D representation of supraspinal neurons (green dots) overlaid on a standard atlas. (C) 2D representation of supraspinal neurons in which individuals cells are registered to defined brain regions in horizontal brain sections. (D) Graphs provide the number of supraspinal neurons that were identified in each of 69 brain regions (right), or in 25 summary regions that combine adjacent areas. The 3D representation was created by Brainrender, and the 2D and quantitative data were generated using cellfinder; both are open source tools created by the Brainglobe initiative.
Demystifying supraspinal diversity
The 3D images and associated quantification underscore the distributed nature of supraspinal neurons, which registered to a total of 69 identified regions throughout brainstem, midbrain, and forebrain. As expected, many neurons were detected in the most commonly studied populations (e.g. corticospinal, rubrospinal, raphespinal), yet more than half of the total supraspinal nuclei were located outside of these areas. The reticular formation emerged as a major and complex source of supraspinal input, with a large and centrally located gigantocellular population surrounded ventrally, laterally, and dorsally by identified subregions with abundant supraspinal neurons. The midbrain and hypothalamus also stood out as major regions of supraspinal input. Some of the outputs from the 3D imaging and registration pipeline were initially surprising, for example a direct projection from the amygdala to the cervical spinal cord. A search of the literature, however, revealed discovery of this projection in monkeys and cats more than thirty years ago and more recently in mice (Liang et al., 2011; Mizuno et al., 1985; Sandrew et al., 1986). Indeed, the same was true for virtually all of the 69 supraspinal regions detected by registration; prior work, often decades old, had already established their existence and sometimes function (one possible exception is the superior olivary complex, in which we detected sparse label but were unable to locate a prior description of descending axons). We emphasize that the essential value of the imaging and registration pipeline is not the (re)discovery of supraspinal populations, but rather the ease of visualizing and quantifying them. These accessible tools are intended to increase awareness and understanding of less-studied supraspinal populations, lowering conceptual barriers that may hinder broader consideration of these populations in the study of SCI.
Mapping cervical- and lumbar-specific connectomes
Supraspinal regions display specificity and topographic mapping with respect to their innervation of specific spinal levels. For example, lumbar-projecting CST neurons reside medially and caudally to their cervically-projecting counterparts, while lumbar-projecting rubrospinal neurons are ventral-medial to the cervical projection neurons (Flumerfelt and Gwyn, 1974; Tennant et al., 2011). For many supraspinal populations, however, the corresponding topography is obscure. Clarifying level-specific projections is important for SCI researchers for two reasons. First, it is needed when potential therapeutics or tracers are delivered by intracranial injection, as these must be matched to the level of injury (e.g. after thoracic injury it is essential to treat and trace those neurons that normally project to locations distal to the injury). Second, the pre-injury topography must be well characterized in order to benchmark its restoration upon treatment.
3D imaging and quantification provides an opportunity for within-brain comparison of supraspinal connectomes that innervate different spinal levels. This is achieved simply by delivering different fluorescent proteins in the same animal to the spinal levels of interest, for example lumbar versus cervical, and then imaging and registering the resulting retrograde label to a common 3D space. 3Dmousebrain.com provides examples of this strategy in which mice received lumbar injection of mGreenLantern and cervical injection of mScarlet, enabling direct comparison of the two projectomes. This approach revealed a number of regions that project predominantly to one of the two levels, and revealed new spatial patterns of topography. An important consideration for this approach is the degree to which tracers are taken up by fibers of passage versus synaptic terminals. In the case of AAV2-retro, our results and others indicate that fibers of passage are not transduced, as evidenced by the fact that cervical and lumbar injections in the same animal drive FP expression in almost completely distinct sets of CST neurons, meaning the cervically-injected virus largely fails to enter axons as they pass to lumbar targets (Steward et al., 2021; Wang et al., 2018, 2021). Thus brain-wide imaging and quantification can provide essential insights into the arrangement of supraspinal circuitry and establish needed benchmarks for restorative therapies for injuries at different spinal levels.
The neuroanatomical-functional paradox revisited
Can connectome data help explain variability in functional outcomes after SCI? The prospects remain speculative, but early analyses offer encouraging signs. For example, using brain-wide connectome data it is possible to analyze cohorts of animals and correlate the degree of individual recovery of locomotion after spinal injury with the residual number of spared neurons in specific brain regions. In principle such an analysis can determine broadly whether variability in the sparing within any given population of neurons explains differences in locomotor impairment between animals. This approach builds on prior work that related locomotor recovery with the size and position of damage within the spinal cord (Loy et al., 2002; Schucht et al., 2002), but adds more precise information about the number and source of axons whose axons span the injury. Indeed, in our recent analysis we found that sparing in some populations, such as the corticospinal and hypothalamic, correlated poorly with locomotor recovery while sparing in others such as the gigantocellular and pedunculopontine was more predictive (Wang et al., 2021). These results, although only correlational and based on a relatively simple motor score, show the intriguing possibility that improved supraspinal connectome data may point toward populations of relevance for specific motor tasks. An important caveat to this approach is that it assigns to each supraspinal neuron an essentially binary readout of axonal connection to target tissue but is insensitive to changes in the number of target cells. Thus, changes in the “influence” of supraspinal neurons, meaning the number of spinal neurons innervated by each, are missed. Nevertheless, full information about the distribution and number of supraspinal neurons with axons that span and injury still marks a significant advancement beyond the current state of knowledge and may help to resolve some of the lingering difficulties with interpreting variance in functional recovery.
Near-term applications and refinements
Improved connectome/function relationships
The correlation between locomotor scores and sparing in specific brain regions is an important proof-of-concept demonstration, but with key refinements much more is possible. First, it will be important in future work to utilize more sophisticated behavioral analyses, for example kinematic-based assessment of locomotion or forelimb trajectories (Becker and Person, 2019; Capogrosso et al., 2018; Engmann et al., 2020; Pocratsky et al., 2017; Whishaw, 1996). Although kinematics analysis of reaching is rare in the clinic due to the high baseline variability in human extension and grasping trajectories, in a research setting the exercise of correlating the spared neurons in defined populations with distinct aspects of rodent movement could dramatically improve understanding of how and if each can potentially contribute to recovery. Similarly, it will be important to expand functional assessment beyond motor control, for example bladder function, blood pressure regulation, and thermoregulation, in order to improve understanding of their distributed control networks after injury. The hypotheses generated by this correlative approach can lead to more direct causal links by functional tests using chemo- or optogenetic tools (Vlasov et al., 2018) that are targeted to selected populations using transgenic approaches or custom AAVs with specific enhancer/promoters (Haery et al., 2019).
The second main refinement involves the set of neurons that are analyzed. Although we have focused so far on the supraspinal connectome, the ascending sensory connectome and the propriospinal connectome are also critically important for normal function and for recovery from SCI (Bareyre et al., 2004; Benthall et al., 2017; Giovanelli Barilari and Kuypers, 1969; Shepard et al., 2021). Indeed, even changes in the vagal connectome can produce both positive and negative influences on functional outcomes of spinal injury (Ganzer et al., 2018; Holmes, 2012). A current technical limitation is that the AAV2-retro utilized is less effective in sensory neurons in the DRG and in serotonergic neurons (Tervo et al., 2016; Wang et al., 2018). Indeed, even using the improved nuclear-localized fluorophores described above, we recently confirmed with 5-HT immunohistochemistry that descending serotonergic neurons were rarely transduced. Other AAV serotypes have previously been reported to transduce sensory or serotonergic types (Leibinger et al., 2021; Soderblom et al., 2015; Yu et al., 2016) and thus a simple near-term solution would be to package nuclear fluorescent proteins in these alternative vectors and inject a mixture of serotypes to broaden the tropism. In parallel, as additional retrogradely transported AAVs and other neurotropic AAVs are developed, it will be important to systematically test their efficacy in distinct classes of ascending and descending neurons. Given the rapid development of viral technology, it is likely that targeting options for both descending and ascending connectomes will soon expand (Bedbrook et al., 2018; Haery et al., 2019).
Regarding propriospinal neurons, we showed recently that after lumbar injection of AAV2-retro several thousand retrogradely labeled nuclei were readily detected in the cervical enlargement after clearing and 3D imaging (Wang et al., 2021). An important development is the recent description of a 3D atlas of the mouse spinal cord, which raises the prospect of mapping and quantifying these propriospinal neurons to defined lamina in a process analogous to the supraspinal analysis described above (Fiederling et al., 2021).
Finally, it is important to note that after spinal injury, connectome changes occur not only in direct supraspinal projections but also intracranially as well (Fink and Cafferty, 2016). For example, spinal injury can trigger strengthening of rubro-raphe (Siegel et al., 2015) or cortico-reticular circuits (Asboth et al., 2018). The formation of such relay circuits within the brain is likely to be another critical determinant of the functional recovery in the post-injury connectome. Combining brain clearing and registration with viruses that spread trans-synaptically, particularly those that can transduce only first-order presynaptic neurons (Adler et al., 2017) , would offer a way to map these changes in relay circuitry. In other words, by using viruses with transsynaptic properties to label both direct supraspinal neurons and the neurons that supply their immediate input, it would be possible to detect the emergence of new relays after injury and to quantify more subtle changes in the number and distribution of upstream neurons that innervate the direct supraspinal circuitry. Quantifying these changes in intracranial circuitry, in combination with the quantification of direct supraspinal, propriospinal, and sensory connectomes described above, may offer unprecedented insight into distributed neural control and variable recovery from injury.
An improved preclinical pipeline
What types of pre-clinical injuries, species, and outcome measurements constitute an appropriate burden of proof for progression to clinical trial (Failli et al., 2021; Lilley et al., 2020)? These difficult questions are a source of long-standing debate in the SCI field (Kwon et al., 2011). It is interesting to note that many – perhaps most – pro-regenerative treatments have proceeded to clinical trial with only partial information about their effects on diverse supraspinal populations. As discussed above, technical limitations have focused attention on several descending populations. Interestingly, even within this limited set of cell types large differences in growth responses are observed (e.g. propriospinal and raphespinal cell types, but not corticospinal, regenerate axons into Schwann Cell grafts (Xu et al., 1997)). Thus, for any given pro-growth treatment, it is likely that different types of neurons will display a range of growth responses (Han et al., 2019). Although some experiments have attempted to survey the regrowth of axons more globally, in general the range of growth across all supraspinal cell types is unknown. This gap makes it difficult even in principle to predict or interpret the potential clinical effects. For example, without knowing how Barrington’s nucleus, hypothalamic nuclei, and pre-autonomic reticular neurons respond to a given transplant, clinical predictions of potential effects on micturition, blood pressure, and thermoregulation are difficult.
The 3D imaging and registration pipeline described above offers a practical solution. In principle, treatments can be applied to mouse models of spinal injury, followed by retrograde labeling and brain-wide quantification of supraspinal populations with connections to spinal tissue distal to the injury. By injecting one tracer immediately after injury to label spared neurons, and then injecting a second label at extended periods after treatment, it should be possible to distinguish spared neurons from neurons that initiated post-injury growth. By comparing these values to uninjured and treatment controls, it would be possible to profile the percent of neurons in each region that reconnected to distal spinal regions, thereby quantifying the regenerative response or its absence across the brain. Such a standard platform would enable quantitative comparison, population by population, between different treatment options in order to identify the most efficacious from a brain-wide perspective. Furthermore, if different treatments evoked growth responses in complementary sets of supraspinal populations, this approach would provide a rational basis for combining treatments to maximize the connectome-wide response.
Functional connectomes
Tissue clearing and quantification methods may help to answer key questions at the intersection of motor control and spinal injury research. For many types of movement, such as reaching motions (above), the full set of supraspinal neurons is yet to be determined (Ferreira-Pinto et al., 2018; Ruder and Arber, 2019). After spinal injury the situation is even more complicated. Motor maps are known to shift within supraspinal populations, and compensatory connections can form between supraspinal populations to support functional recovery (Dancause et al., 2005; Fink and Cafferty, 2016; Liao et al., 2017; Nudo, 2006; Raineteau et al., 2001; Zörner et al., 2014). Thus, the set of neurons that support a given movement is likely to shift after spinal injury. Using whole-brain clearing and imaging, there may be a practical way to quantify and map all neurons that are active during specific behaviors, both before and after injury. Most of the methods of neuronal activity are based on electrophysiology. However, in the last few decades genetic method with various reporters have been developed to make them accessible to tissue clearing.
A variety of tools now exist to fluorescently label neurons that are active during discrete time windows (DeNardo and Luo, 2017). Importantly, in several cases this is achieved using viral injections which then depend on transgenic design (DeNardo et al., 2019) or synthetic promoters (Sørensen et al., 2016) to achieve fluorophore production that is restricted to active neurons. Thus, the relevant viral vectors could be delivered in retrograde fashion to the spinal cord, and time windows for expression could be set as animals engaged in specific trained behavior such as standard locomotion, horizontal ladder crossing, pellet retrieval, or other tasks commonly used to assess recovery from spinal injury. In this way, fluorescent protein expression would occur only in neurons that met two criteria: they project axons to the spinal site of injection, and they were active during the behavior of interest. Combined with subsequent tissue clearing and brain-wide quantification, this method would provide detailed information about the set of supraspinal neurons normally active during specific behaviors. A similar approach in injured or treated animals would detail shifts in the utilization of neurons in response to injury. Multiple fluorescent proteins, some cre-dependent and others not, would allow within-animal quantification of all spared neurons and the subset active during selected tasks. In practical terms, the success of this approach will depend on expression vectors that are sufficiently bright for tissue clearing and achieving a satisfactory signal to noise ratio when comparing task-exposed animals to controls. Likely multiple tools will need to be tested to find those most suited to detect supraspinal activity (DeNardo et al., 2019; Guenthner et al., 2013). The larger point is that tissue clearing and registration is not restricted to mapping only physical connections, but can also identify subsets of connections that meet secondary criteria based on activity or molecular state. In this way connectome analyses can reveal not just number and location of neurons, but also vital information about system-level characteristics in naïve and injured animals.
The future: supraspinal transcriptomes and beyond
Connectomic analyses offer a powerful means to establish patterns and correlations; they can determine which neural populations are most affected by injury, which respond to potential pro-growth treatments, and which adjust their activity. Once established, however, these patterns will need explanation and utilization. Put another way, with connectome data in hand the dominant questions will become why and how: why do some neurons succeed in regrowing axons or displaying compensatory activity, and how can the response by non-growers be improved? These are long-standing and central questions in regenerative neuroscience, but have previously focused on disparate cell types and species (e.g. peripheral versus central neurons, zebrafish versus mammalian, etc.) (Blackmore, 2012).
As detailed above, regenerating versus non-regenerating cells can now be detected and compared at a much finer scale, both within and between distinct populations in the mammalian brain. In parallel, advances in single cell sequencing now allow the possibility of transcriptionally distinguishing supraspinal populations on the same brain-wide scale as the connectome mapping described above (Chen et al., 2019; Kebschull et al., 2016; Sun et al., 2021; Tanay and Regev, 2017). In practical terms a retrograde label would be applied to the spinal cord, and labeled cells or cell nuclei could be isolated by fluorescent activated cell sorting, INTACT, or other means (Chongtham et al., 2021; Huang et al., 2020; Muñoz-Castañeda et al., 2020; Tasic et al., 2018). Single cell analyses would then identify sets of molecularly similar cells. In uninjured animals, this would generate a molecular description of different populations of supraspinal neurons: the supraspinal transcriptomes (Fig. 4A). A similar experiment performed after injury could determine, population-by-population, the set of transcripts that change as neurons respond to injury. These post-injury transcriptomes could potentially explain differences in innate growth responses (Fig 4B).
Figure 4. An approach of combined connectome and transcriptome analyses after spinal injury.
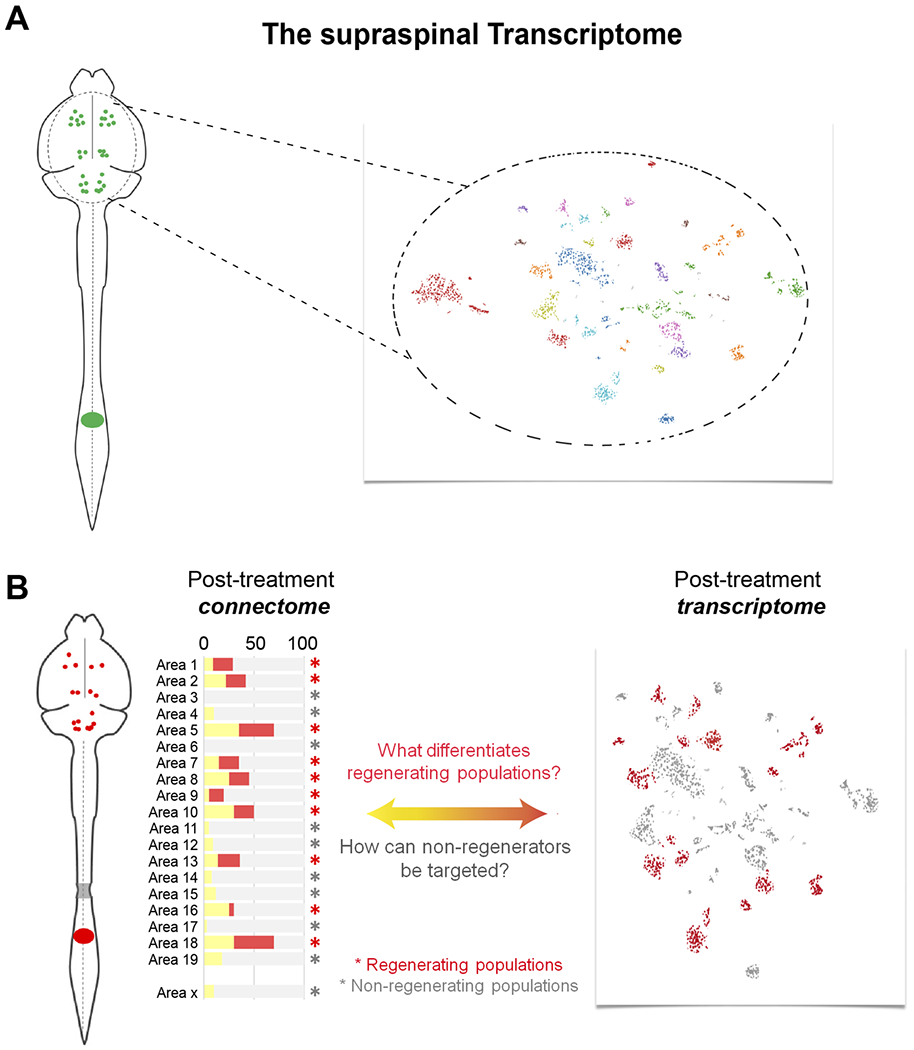
(A) Supraspinal neurons can be retrogradely labeled (left), purified based on fluorescence, and then analyzed using single cell techniques to reveal clusters of molecularly similar cells (right). This approach would molecularly define the full set of supraspinal cell types. (B) Retrograde labeling after injury and treatment can quantify the response to treatment of individual cell populations (left) and the transcriptional profile of populations as they respond to injury and treatment. Comparing the two datasets will yield insight into the basis for regeneration and non-regeneration.
After treatment, by specifically labeling neurons that successfully reconnected to a site of retrograde labeling placed distal to an injury, the transcription profiles of regenerating neurons could help explain the underlying differences and guide future efforts to coax the non-regenerators into growth. Neurons that shift their activity levels after injury could be similarly profiled. Further, the advent of spatial transcriptomics, which allows the visualization of hundreds of transcripts, provides new ways to profile neurons that regenerate axons and detect transcriptional differences from non-regenerating neighbors (Ortiz et al., 2021).
Importantly, the brain-wide connectome and transcriptome data are mutually reinforcing and could create a cycle of rapid advance. Starting from a candidate pro-repair treatment, the first step would be to use connectome data to globally assess the efficacy across all relevant populations and to detect differences in growth response between populations. With this map in hand, the corresponding transcriptome data could identify molecular correlates and generate candidate treatments to target non-regenerators. In turn, when these modified treatments are tested, brain-wide connectome data after injury could gauge the resulting changes in axon growth, or their absence, across all populations. Importantly, although it is hoped that increases in axon growth would correlate with positive improvements in function, it is also possible that ectopic connections would correlate with negative outcomes. Thus, it is conceivable that the goal will be to foster more growth from some cell types, and less from others. A back-and-forth iteration of mapping the types of supraspinal neurons that were stimulated to regenerate axons and then determining the transcriptional signatures that distinguish regenerating versus non-regenerating cells could dramatically accelerate understanding and successful manipulation of the regenerative response.
Finally, the conception that different supraspinal populations may require distinct interventions to encourage (or reign in) axon growth raised the problem of cellular targeting. As discussed above, retrograde gene delivery can achieve selective gene delivery to supraspinal neurons as a broad class, but how can treatments be delivered to only some subtypes? Recent developments at the intersection of single-cell transcriptional and epigenome profiling now offer a solution. Briefly, once the single-cell approach has identified transcripts that are specifically expressed in subsets of supraspinal neurons (e.g. “marker” genes), examination of the DNA accessibility and histone modification can identify enhancer regions that are responsible for the selective expression. These enhancers can then be deployed in gene therapy vectors to drive expression of transgenes specifically in selected populations. This strategy has been used successfully in variety of neurons (Graybuck et al., 2021; Mich et al., 2021), opening the door to more targeted therapies that address the potentially specific needs of subsets of supraspinal neurons.
Summary
Historically, technical challenges have hindered a broad consideration of diverse supraspinal cell types in pre-clinical studies of spinal cord injury. The focus on major tracts has drawbacks, including difficulties in interpreting functional outcomes and the emergence of apparent paradoxes, the understudy of autonomic functions of high importance to individuals with spinal injury, and arguably challenges to reproducibility. Now a confluence of advances in viral gene delivery, 3D imaging, and deep learning-assisted image analysis have positioned the field for rapid expansion in understanding how diverse cell types respond to injury and treatment. By capitalizing on these developments, the field has new opportunities to move towards badly needed insights and additional treatment options for the wide range of deficits that come with spinal injury.
Acknowledgements
We would like to especially thank Troy Margrie, Adam Tyson, Luigi Petrucco and other investigators at the Brainglobe project for making available and helping to solve issues that arose during the installation and use of their neuroanatomical tools. We would like to thank Yania Ondaro-Martinez at the Miami Project Viral Core and Yan Shi at the Miami Project Image Core facility.
Funding
This work was supported by NINDS R01NS107807 (MB and PT), the Bryon Riesch Paralysis Foundation (MB), The Miami Project, The Buoniconti fund and the state of Florida Red Light Camera Fund (PT).
Footnotes
Declarations of interest: none.
References
- Adler AF, Lee-Kubli C, Kumamaru H, Kadoya K, and Tuszynski MH (2017). Comprehensive Monosynaptic Rabies Virus Mapping of Host Connectivity with Neural Progenitor Grafts after Spinal Cord Injury. Stem Cell Reports 8, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KD (2004). Targeting recovery: priorities of the spinal cord-injured population. J. Neurotrauma 21, 1371–1383. [DOI] [PubMed] [Google Scholar]
- Asboth L, Friedli L, Beauparlant J, Martinez-Gonzalez C, Anil S, Rey E, Baud L, Pidpruzhnykova G, Anderson MA, Shkorbatova P, et al. (2018). Cortico-reticulo-spinal circuit reorganization enables functional recovery after severe spinal cord contusion. Nat. Neurosci 21, 576–588. [DOI] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, and Rumpel S (2013). Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre FM, Kerschensteiner M, Raineteau O, Mettenleiter TC, Weinmann O, and Schwab ME (2004). The injured spinal cord spontaneously forms a new intraspinal circuit in adult rats. Nat. Neurosci 7, 269–277. [DOI] [PubMed] [Google Scholar]
- Basile GA, Quartu M, Bertino S, Serra MP, Boi M, Bramanti A, Anastasi GP, Milardi D, and Cacciola A (2020). Red nucleus structure and function: from anatomy to clinical neurosciences. Brain Struct. Funct 2020 2261 226, 69–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MI, and Person AL (2019). Cerebellar Control of Reach Kinematics for Endpoint Precision. Neuron 103, 335–348.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook CN, Deverman BE, and Gradinaru V (2018). Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci 41, 323–348. [DOI] [PubMed] [Google Scholar]
- Benthall KN, Hough RA, and McClellan AD (2017). Descending propriospinal neurons mediate restoration of locomotor function following spinal cord injury. J. Neurophysiol 117, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentivoglio M, Kuypers HGJM, Catsman-Berrevoets CE, Loewe H, and Dann O (1980). Two new fluorescent retrograde neuronal tracers which are transported over long distances. Neurosci. Lett 18, 25–30. [DOI] [PubMed] [Google Scholar]
- Blackmore MG (2012). Molecular Control of Axon Growth: Insights from Comparative Gene Profiling and High-Throughput Screening. Int. Rev. Neurobiol 105, 39–70. [DOI] [PubMed] [Google Scholar]
- Bohlen MO, McCown TJ, Powell SK, El-Nahal HG, Daw T, Basso MA, Sommer MA, and Jude Samulski R (2020). Adeno-associated virus capsid-promoter interactions in the brain translate from rat to the nonhuman primate. Hum. Gene Ther 31, 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulenguez P, Liabeuf S, Bos R, Bras H, Jean-Xavier C, Brocard C, Stil A, Darbon P, Cattaert D, Delpire E, et al. (2010). Down-regulation of the potassium-chloride cotransporter KCC2 contributes to spasticity after spinal cord injury. Nat. Med 2010 163 16, 302–307. [DOI] [PubMed] [Google Scholar]
- Bouvier J, Caggiano V, Leiras R, Caldeira V, Bellardita C, Balueva K, Fuchs A, and Kiehn O (2015). Descending Command Neurons in the Brainstem that Halt Locomotion. Cell 163, 1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray ER, Noga M, Thakor K, Wang Y, Lemmon VP, Park KK, and Tsoulfas P (2017). 3D Visualization of Individual Regenerating Retinal Ganglion Cell Axons Reveals Surprisingly Complex Growth Paths. Eneuro 4, ENEURO.0093-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Pan C, Ghasemigharagoz A, Todorov MI, Förstera B, Zhao S, Bhatia HS, Parra-Damas A, Mrowka L, Theodorou D, et al. (2019). Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci 22, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BC, Nabel EM, Murdock MH, Lao-Peregrin C, Tsoulfas P, Blackmore MG, Lee FS, Liston C, Morishita H, and Petsko GA (2020). mGreenLantern: a bright monomeric fluorescent protein with rapid expression and cell filling properties for neuronal imaging. Proc. Natl. Acad. Sci. U. S. A 117, 30710–30721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capogrosso M, Wagner FB, Gandar J, Moraud EM, Wenger N, Milekovic T, Shkorbatova P, Pavlova N, Musienko P, Bezard E, et al. (2018). Configuration of electrical spinal cord stimulation through real-time processing of gait kinematics. Nat. Protoc 13, 2031–2061. [DOI] [PubMed] [Google Scholar]
- Cavada C, Huisman AM, and Kuypers HGJM (1984). Retrograde double labeling of neurons: the combined use of horseradish peroxidase and diamidino yellow dihydrochloride (DY·2HCl) compared with true blue and DY·2HCl in rat descending brainstem pathways. Brain Res. 308, 123–136. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Driscoll MK, Jeffery E, Murphy MM, Roudot P, Chang BJ, Vora S, Wong WM, Nielson CD, Zhang H, et al. (2019). Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Li Y, Yu B, Zhang Z, Brommer B, Williams PR, Liu Y, Hegarty SV, Zhou S, Zhu J, et al. (2018). Reactivation of Dormant Relay Pathways in Injured Spinal Cord by KCC2 Manipulations. Cell 174, 521–535.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sun YC, Zhan H, Kebschull JM, Fischer S, Matho K, Huang ZJ, Gillis J, and Zador AM (2019). High-Throughput Mapping of Long-Range Neuronal Projection Using In Situ Sequencing. Cell 179, 772–786.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongtham MC, Butto T, Mungikar K, Gerber S, and Winter J (2021). INTACT vs. FANS for Cell-Type-Specific Nuclei Sorting: A Comprehensive Qualitative and Quantitative Comparison. Int. J. Mol. Sci 22, 5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudi F, Tyson AL, Petrucco L, Margrie TW, Portugues R, and Branco T (2021). Visualizing anatomically registered data with brainrender. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote JH, and Spyer KM (2018). Central control of autonomic function. Brain Neurosci. Adv 2, 239821281881201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampney RAL (1994). Functional organization of central pathways regulating the cardiovascular system. Physiol. Rev 74, 323–364. [DOI] [PubMed] [Google Scholar]
- Dancause N, Barbay S, Frost SB, Plautz EJ, Chen D, Zoubina EV, Stowe AM, and Nudo RJ (2005). Extensive cortical rewiring after brain injury. J. Neurosci 25, 10167–10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo L, and Luo L (2017). Genetic strategies to access activated neurons. Curr. Opin. Neurobiol 45, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo LA, Liu CD, Allen WE, Adams EL, Friedmann D, Fu L, Guenthner CJ, Tessier-Lavigne M, and Luo L (2019). Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat. Neurosci 22, 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Donkelaar HJ (2000). Development and Regenerative Capacity of Descending Supraspinal Pathways in Tetrapods: A Comparative Approach (Berlin, Heidelberg: Springer Berlin Heidelberg; ). [DOI] [PubMed] [Google Scholar]
- Engmann AK, Bizzozzero F, Schneider MP, Pfyffer D, Imobersteg S, Schneider R, Hofer AS, Wieckhorst M, and Schwab ME (2020). The gigantocellular reticular nucleus plays a significant role in locomotor recovery after incomplete spinal cord injury. J. Neurosci 40, 8292–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito MS, Capelli P, and Arber S (2014). Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature 508, 351–356. [DOI] [PubMed] [Google Scholar]
- Failli V, Kleitman N, Lammertse DP, Hsieh JTC, Steeves JD, Fawcett JW, Tuszynski MH, Curt A, Fehlings MG, Guest JD, et al. (2021). Experimental Treatments for Spinal Cord Injury: What you Should Know. Top. Spinal Cord Inj. Rehabil 27, 50–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Pinto MJ, Ruder L, Capelli P, and Arber S (2018). Connecting Circuits for Supraspinal Control of Locomotion. Neuron 100, 361–374. [DOI] [PubMed] [Google Scholar]
- Fiederling F, Hammond LA, Ng D, Mason C, and Dodd J (2021). Title Tools for efficient analysis of neurons in a 3D reference atlas of whole mouse spinal cord. BioRxiv 2021.05.06.443008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink KL, and Cafferty WBJ (2016). Reorganization of Intact Descending Motor Circuits to Replace Lost Connections After Injury. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flumerfelt BA, and Gwyn DG (1974). Proceedings: The red nucleus of the rat: its organization and interconnexions. J. Anat 118, 374, 376. [PubMed] [Google Scholar]
- Fouad K, Popovich PG, Kopp MA, and Schwab JM (2021). The neuroanatomical–functional paradox in spinal cord injury. Nat. Rev. Neurol 17, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CJ, Griffiths D, and De Groat WC (2008). The neural control of micturition. Nat. Rev. Neurosci 9, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli L, Rosenzweig ES, Barraud Q, Schubert M, Dominici N, Awai L, Nielson JL, Musienko P, Nout-Lomas Y, Zhong H, et al. (2015). Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci. Transl. Med 7, 302ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzer PD, Darrow MJ, Meyers EC, Solorzano BR, Ruiz AD, Robertson NM, Adcock KS, James JT, Jeong HS, Becker AM, et al. (2018). Closed-loop neuromodulation restores network connectivity and motor control after spinal cord injury. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanelli Barilari M, and Kuypers HGJM (1969). Propriospinal fibers interconnecting the spinal enlargements in the cat. Brain Res. 14, 321–330. [DOI] [PubMed] [Google Scholar]
- Glaser A, Bishop K, Barner L, Serafin R, and Liu J (2020). A hybrid open-top light-sheet microscope for multi-scale imaging of cleared tissues. BioRxiv 2020.05.06.081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck LT, Daigle TL, Sedeño-Cortés AE, Walker M, Kalmbach B, Lenz GH, Morin E, Nguyen TN, Garren E, Bendrick JL, et al. (2021). Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron 109, 1449–1464.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S, and El Manira A (2020). Current principles of motor control, with special reference to vertebrate locomotion. Physiol. Rev 100, 271–320. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haery L, Deverman BE, Matho KS, Cetin A, Woodard K, Cepko C, Guerin KI, Rego MA, Ersing I, Bachle SM, et al. (2019). Adeno-Associated Virus Technologies and Methods for Targeted Neuronal Manipulation. Front. Neuroanat 13, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Ordaz JD, Liu NK, Richardson Z, Wu W, Xia Y, Qu W, Wang Y, Dai H, Zhang YP, et al. (2019). Descending motor circuitry required for NT-3 mediated locomotor recovery after spinal cord injury in mice. Nat. Commun 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton BJ, Anenberg E, Harrison TC, Boyd JD, Murphy TH, and Tetzlaff W (2016). Re-Establishment of Cortical Motor Output Maps and Spontaneous Functional Recovery via Spared Dorsolaterally Projecting Corticospinal Neurons after Dorsal Column Spinal Cord Injury in Adult Mice. J. Neurosci 36, 4080–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes GM (2012). Upper gastrointestinal dysmotility after spinal cord injury: is diminished vagal sensory processing one culprit? Front. Physiol 0, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege J (1991). Ultrastructural evidence for GABAergic brain stem projections to spinal motoneurons in the rat. J. Neurosci 11, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, and Rabchevsky AG (2014). Autonomic consequences of spinal cord injury. Compr. Physiol 4, 1419–1453. [DOI] [PubMed] [Google Scholar]
- Huang L, Kebschull JM, Fürth D, Musall S, Kaufman MT, Churchland AK, and Zador AM (2020). BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals. Cell 182, 177–188.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd C, Weishaupt N, and Fouad K (2013). Anatomical correlates of recovery in single pellet reaching in spinal cord injured rats. Exp. Neurol 247, 605–614. [DOI] [PubMed] [Google Scholar]
- Iannotti C, Ping Zhang Y, Shields CB, Han Y, Burke DA, and Xu XM (2004). A neuroprotective role of glial cell line-derived neurotrophic factor following moderate spinal cord contusion injury. Exp. Neurol 189, 317–332. [DOI] [PubMed] [Google Scholar]
- Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, and Zador AM (2016). High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 91, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, Ruelas L, Hubbert A, McCue M, Vassallo SL, et al. (2015). Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. U. S. A 112, E6274–E6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köbbert C, Apps R, Bechmann I, Lanciego JL, Mey J, and Thanos S (2000). Current concepts in neuroanatomical tracing. Prog. Neurobiol 62, 327–351. [DOI] [PubMed] [Google Scholar]
- Kuypers H, and Martin G (1982). Descending Pathways to the Spinal Cord. Prog. Brain Res 57. [DOI] [PubMed] [Google Scholar]
- Kwon BK, Okon EB, Tsai E, Beattie MS, Bresnahan JC, Magnuson DK, Reier PJ, McTigue DM, Popovich PG, Blight AR, et al. (2011). A grading system to evaluate objectively the strength of pre-clinical data of acute neuroprotective therapies for clinical translation in spinal cord injury. J. Neurotrauma 28, 1525–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HM, Liu AKL, Ng HHM, Goldfinger MH, Chau TW, DeFelice J, Tilley BS, Wong WM, Wu W, and Gentleman SM (2018). Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat. Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Park J-H, Seo I, Park S-H, and Kim S (2014). Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev. Biol 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, and Perez MA (2021). Cerebellar contribution to sensorimotor adaptation deficits in humans with spinal cord injury. Sci. Rep 11, 2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibinger M, Zeitler C, Gobrecht P, Andreadaki A, Gisselmann G, and Fischer D (2021). Transneuronal delivery of hyper-interleukin-6 enables functional recovery after severe spinal cord injury in mice. Nat. Commun 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon RN (2008). Descending Pathways in Motor Control. Annu. Rev. Neurosci 31, 195–218. [DOI] [PubMed] [Google Scholar]
- Leong SK, Shieh JY, and Wong WC (1984). Localizing spinal-cord-projecting neurons in adult albino rats. J. Comp. Neurol 228, 1–17. [DOI] [PubMed] [Google Scholar]
- Liang H, Paxinos G, and Watson C (2011). Projections from the brain to the spinal cord in the mouse. Brain Struct. Funct 215, 159–186. [DOI] [PubMed] [Google Scholar]
- Liao Y, Tong L, Tang L, and Wu S (2017). The role of cold-inducible RNA binding protein in cell stress response. Int. J. Cancer 141, 2164–2173. [DOI] [PubMed] [Google Scholar]
- Lilley E, Andrews MR, Bradbury EJ, Elliott H, Hawkins P, Ichiyama RM, Keeley J, Michael-Titus AT, Moon LDF, Pluchino S, et al. (2020). Refining rodent models of spinal cord injury. Exp. Neurol 328. [DOI] [PubMed] [Google Scholar]
- Liu Y, Latremoliere A, Li X, Zhang Z, Chen M, Wang X, Fang C, Zhu J, Alexandre C, Gao Z, et al. (2018). Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature 561, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn-Smith IJ (2009). Anatomy of synaptic circuits controlling the activity of sympathetic preganglionic neurons. J. Chem. Neuroanat 38, 231–239. [DOI] [PubMed] [Google Scholar]
- Loewy AD (1981). Descending pathways to sympathetic and parasympathetic preganglionic neurons. J. Auton. Nerv. Syst 3, 265–275. [DOI] [PubMed] [Google Scholar]
- Loy DN, Magnuson DSK, Ping Zhang Y, Onifer SM, Mills MD, Cao QL, Darnall JB, Fajardo LC, Burke DA, and Whittemore SR (2002). Functional redundancy of ventral spinal locomotor pathways. J. Neurosci 22, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich JK, Graybuck LT, Hess EE, Mahoney JT, Kojima Y, Ding Y, Somasundaram S, Miller JA, Kalmbach BE, Radaelli C, et al. (2021). Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 34, 108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Takahashi O, Satoda T, and Matsushima R (1985). Amygdalospinal projections in the macaque monkey. Neurosci. Lett 53, 327–330. [DOI] [PubMed] [Google Scholar]
- Muñoz-Castañeda R, Zingg B, Matho KS, Wang Q, Chen X, Foster NN, Narasimhan A, Li A, Hirokawa KE, Huo B, et al. (2020). Cellular Anatomy of the Mouse Primary Motor Cortex. BioRxiv 2020.10.02.323154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedworok CJ, Brown APY, Jorge Cardoso M, Osten P, Ourselin S, Modat M, and Margrie TW (2016). AMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nat. Commun 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudo RJ (2006). Mechanisms for recovery of motor function following cortical damage. Curr. Opin. Neurobiol 16, 638–644. [DOI] [PubMed] [Google Scholar]
- Nudo RJ, and Masterton RB (1988). Descending pathways to the spinal cord: A comparative study of 22 mammals. J. Comp. Neurol 277, 53–79. [DOI] [PubMed] [Google Scholar]
- Ortiz C, Carlén M, and Meletis K (2021). Spatial Transcriptomics: Molecular Maps of the Mammalian Brain. Annu. Rev. Neurosci 44. [DOI] [PubMed] [Google Scholar]
- Plant GW, Christensen CL, Oudega M, and Bunge MB (2003). Delayed transplantation of olfactory ensheathing glia promotes sparing/regeneration of supraspinal axons in the contused adult rat spinal cord. J. Neurotrauma 20, 1–16. [DOI] [PubMed] [Google Scholar]
- Pocratsky AM, Burke DA, Morehouse JR, Beare JE, Riegler AS, Tsoulfas P, States GJR, Whittemore SR, and Magnuson DSK (2017). Reversible silencing of lumbar spinal interneurons unmasks a task-specific network for securing hindlimb alternation. Nat. Commun 8, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineteau O, Fouad K, Noth P, Thallmair M, and Schwab ME (2001). Functional switch between motor tracts in the presence of the mAb IN-1 in the adult rat. Proc. Natl. Acad. Sci 98, 6929–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M (2014). IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. [DOI] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PM, Issa VM, and Aguayo AJ (1984). Regeneration of long spinal axons in the rat. J. Neurocytol 13, 165–182. [DOI] [PubMed] [Google Scholar]
- Ruder L, and Arber S (2019). Brainstem Circuits Controlling Action Diversification. Annu. Rev. Neurosci 42, 485–504. [DOI] [PubMed] [Google Scholar]
- Ruder L, Schina R, Kanodia H, Valencia-Garcia S, Pivetta C, and Arber S (2021). A functional map for diverse forelimb actions within brainstem circuitry. Nature 590, 445–450. [DOI] [PubMed] [Google Scholar]
- Sandrew BB, Edwards DL, Poletti CE, and Foote WE (1986). Amygdalospinal projections in the cat. Brain Res. 373, 235–239. [DOI] [PubMed] [Google Scholar]
- Schucht P, Raineteau O, Schwab ME, and Fouad K (2002). Anatomical correlates of locomotor recovery following dorsal and ventral lesions of the rat spinal cord. Exp. Neurol 176, 143–153. [DOI] [PubMed] [Google Scholar]
- Shepard CT, Pocratsky AM, Brown BL, Rijswijck M.A. Van, Zalla RM, Burke DA, Morehouse JM, Reigler AS, Whittemore SR, and Magnuson DSK (2021). Silencing long ascending propriospinal neurons after spinal cord injury improves hindlimb stepping in the adult rat. BioRxiv 2021.05.18.444653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel CS, Fink KL, Strittmatter SM, and Cafferty WBJ (2015). Plasticity of intact rubral projections mediates spontaneous recovery of function after corticospinal tract injury. J. Neurosci 35, 1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderblom C, Lee D-H, Dawood A, Carballosa M, Santamaria AJ, Benavides FD, Jergova S, Grumbles RM, Thomas CK, Park KK, et al. (2015). 3D Imaging of Axons in Transparent Spinal Cords from Rodents and Nonhuman Primates. ENeuro 2, ENEURO.0001-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AT, Cooper YA, Baratta MV, Weng FJ, Zhang Y, Ramamoorthi K, Fropf R, Laverriere E, Xue J, Young A, et al. (2016). A robust activity marking system for exploring active neuronal ensembles. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, and Kötter R (2005). The human connectome: A structural description of the human brain. PLoS Comput. Biol 1, 0245–0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A (2016). In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol 21, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Popovich PG, Dietrich WD, and Kleitman N (2012). Replication and reproducibility in spinal cord injury research. Exp. Neurol 233, 597–605. [DOI] [PubMed] [Google Scholar]
- Steward O, Yee KM, Metcalfe M, Willenberg R, Luo J, Azevedo R, Martin-Thompson JH, and Gandhi SP (2021). Rostro-Caudal Specificity of Corticospinal Tract Projections in Mice. Cereb. Cortex 31, 2322–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-C, Chen X, Fischer S, Lu S, Zhan H, Gillis J, and Zador AM (2021). Integrating barcoded neuroanatomy with spatial transcriptional profiling enables identification of gene correlates of projections. Nat. Neurosci 24, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakusaki K, Chiba R, Nozu T, and Okumura T (2016). Brainstem control of locomotion and muscle tone with special reference to the role of the mesopontine tegmentum and medullary reticulospinal systems. J. Neural Transm 123, 695–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, and Regev A (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappan SJ, Eastwood BS, O’Connor N, Wang Q, Ng L, Feng D, Hooks BM, Gerfen CR, Hof PR, Schmitz C, et al. (2019). Automatic navigation system for the mouse brain. J. Comp. Neurol 527, 2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant KA, Adkins DL, Donlan NA, Asay AL, Thomas N, Kleim JA, and Jones TA (2011). The organization of the forelimb representation of the C57BL/6 mouse motor cortex as defined by intracortical microstimulation and cytoarchitecture. Cereb. Cortex 21, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson AL, and Margrie TW (2021). Mesoscale microscopy for micromammals: image analysis tools for understanding the rodent brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson AL, Rousseau CV, Niedworok CJ, Keshavarzi S, Tsitoura C, and Margrie TW (2020). A deep learning algorithm for 3D cell detection in whole mouse brain image datasets. BioRxiv 2020.10.21.348771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Ertürk A, Chung K, Gradinaru V, Chédotal A, Tomancak P, and Keller PJ (2020a). Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci 21, 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Dodt HU, Osten P, Economo MN, Chandrashekar J, and Keller PJ (2020b). Whole-Brain Profiling of Cells and Circuits in Mammals by Tissue Clearing and Light-Sheet Microscopy. Neuron 106, 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavrek R, Pearse DD, and Fouad K (2007). Neuronal populations capable of regeneration following a combined treatment in rats with spinal cord transection. J. Neurotrauma 24, 1667–1673. [DOI] [PubMed] [Google Scholar]
- Vlasov K, Van Dort CJ, and Solt K (2018). Optogenetics and Chemogenetics. In Methods in Enzymology, (Academic Press Inc.), pp. 181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Maunze B, Wang Y, Tsoulfas P, and Blackmore MG (2018). Global connectivity and function of descending spinal input revealed by 3D microscopy and retrograde transduction. J. Neurosci 1196–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Romanski A, Mehra V, Wang Y, Campbell BC, Petsko GA, Tsoulfas P, Blackmore M, authors C, and Appel Alzheimer R (2021). Brain-wide quantification of the supraspinal connectome. BioRxiv 2021.06.10.447885. [Google Scholar]
- Weiss KR, Voigt FF, Shepherd DP, and Huisken J (2021). Tutorial: practical considerations for tissue clearing and imaging. Nat. Protoc 16, 2732–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whishaw IQ (1996). An endpoint, descriptive, and kinematic comparison of skilled reaching in mice (Mus musculus) with rats (Rattus norvegicus). Behav. Brain Res 78, 101–111. [DOI] [PubMed] [Google Scholar]
- Xu XM, Chen A, Guénard V, Kleitman N, and Bunge MB (1997). Bridging Schwann cell transplants promote axonal regeneration from both the rostral and caudal stumps of transected adult rat spinal cord. J. Neurocytol 26, 1–16. [DOI] [PubMed] [Google Scholar]
- Yu H, Fischer G, and Hogan QH (2016). AAV-mediated gene transfer to dorsal root ganglion. In Methods in Molecular Biology, (Humana Press Inc.), pp. 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörner B, Bachmann LC, Filli L, Kapitza S, Gullo M, Bolliger M, Starkey ML, Röthlisberger M, Gonzenbach RR, and Schwab ME (2014). Chasing central nervous system plasticity: The brainstem’s contribution to locomotor recovery in rats with spinal cord injury. Brain 137, 1716–1732. [DOI] [PubMed] [Google Scholar]


