Abstract
A successful swimming performance is a multi-factorial accomplishment, resulting from a complex interaction of physical, biomechanical, physiological and psychological factors, all of which are strongly affected by the special medium of water as well as by genetic factors. The nature of competitive swimming is unique, as most of the competitive events last less than four minutes. Yet training regimens have an endurance nature (many hours and many kilometres of swimming every day), which makes it impossible to classify swimming by definitions of aerobic-type or anaerobic-type events, as in track and field sports. Therefore, genetic variants associated with swimming performance are not necessarily related to metabolic pathways, but rather to blood lactate transport (MCT1), muscle functioning (IGF1 axis), muscle damage (IL6) and others. The current paper reviews the main findings on the leading 12 genetic polymorphisms (located in the ACE, ACTN3, AMPD1, BDKRB2, IGF1, IL6, MCT1, MSTN, NOS3, PPARA, PPARGC1A, and VEGFR2 genes) related to swimming performance, while taking into consideration the unique environment of this sport.
Keywords: Swimming, Sport, Environment, Performance, Genetic polymorphism
INTRODUCTION
Swimming is one of the most popular modes of physical activity around the world, both in recreational and in sport environments [1, 2]. Furthermore, competitive swimmers continue to break world records more frequently and with greater margins than in most other sports [3, 4]. However, despite its popularity and success, swimming has received less attention in the scientific literature compared with running or cycling (464 scientific swimming papers were published on PubMed in 2015–2020 compared to 1666 papers on running). One main reason for this may be the difficulty of collecting physiological measurements during swimming. In addition, the physical properties of water, including its density, pressure, thermal capacity and conductivity, elicit distinct physiological effects on swimmers, and represent significant challenges to investigators specializing in this unique medium.
Specifically, the horizontal body position alters the gravitational effects on circulation [5]. Although cardiac output remains unchanged, stroke volume increases and heart rate decreases compared to running for a given oxygen uptake level [6–8]. Local factors, such as peripheral circulation, capillary density, perfusion pressure and metabolic capacity of active muscles, are important determinants of the power production capacity and emphasize the role of swimming-specific training movements [9]. Breathing in swimming is restricted by stroke mechanics and the aquatic environment. The limitations on the respiratory, central circulatory or peripheral/muscular capacities for oxygen transport and utilization usually induce a reduction of about 10% in maximal oxygen uptake compared to running [10]. Thermoregulatory demands do not compete with metabolic demands during high-intensity swimming at temperatures that normally exist during training and competition [11].
Research on competitive swimming has given special emphasis to energetic and biomechanical assessments [12, 13]. It is well established that the energy expended to transport the body over a given distance increases with speed, both in water [14, 15] and in land activities such as running [16] or cycling [17]. However, at any given speed, the cost of energy is lower on land than in water [18]. This is because of the need to overcome larger resistance forces in the water (hydrodynamic resistance) than on land (aerodynamic resistance) [19]. In addition, there is a lower propelling efficiency in the water compared to on land (e.g., a lower capability to exert useful forces in water than on land) [18]. This also explains the reasons why there are differences in the energy cost of the different strokes (the energy expenditure for a given swimming speed is lowest in the front crawl, followed by the backstroke, butterfly and breaststroke) or in swimmers with different technical skills [20, 21]. This is also why the swimmers’ skill in reducing water resistance, as well as in applying propulsive forces effectively, may be more important in dictating the physiological and energetic demands of swimming than a simple kinematic analysis of race duration [22, 23].
As can be learned from the literature, performance in swimming is strongly linked to energetic variables, as these are dependent on the swimmers’ biomechanical profile and on motor strategies adopted by the swimmers [24]. Consistent with this is the strong correlation that was found between the 100 m and 2000 m swim times of elite male swimmers specializing in 100 to 800 m distances [25]. Such a correlation may indicate that factors such as the swimmers’ technique, body size (particularly limb length) and swimming motor strategies are major determinants of the swimmers’ level [26, 27]. It is therefore likely that these variables, at least partially, mask metabolic differences between swimmers of different specialties, enabling technically skilled and tall swimmers to excel at all swimming events [28, 29]. This assumption can be supported by past records of top world-class swimmers, such as Ian Thorpe (world record holder in the 200, 400 and 800 m) and Grant Hackett (world record holder in the 400, 800 and 1500 m). While strong relationships between sprint and long-distance results are characteristic for swimming, they are very uncommon in running events.
Performance in competitive swimming is the result of a complex interaction of physical, biomechanical, physiological and psychological factors, all of which are strongly affected by the special medium of water [26,28,30]. It should be noted, however, that most competitive events in swimming last less than four minutes. Yet, by tradition, elite swimmers spend many hours and many kilometres of swimming in training every day [31]. It is doubtful whether the high volume of work among these swimmers is really necessary to improve swimming records. Although research on swimming has given emphasis to biomechanical and energetics assessment to understand the special needs of swimming, in the last few years special attention has been given to studying the genetic characteristics of swimmers.
The primary aim of the current review is to outline the main genetic predictors of competitive swimming performance. This research work was designed as an overview of the scientific literature. General recommendations to improve the quality of narrative reviews were taken into account for better structuring this research [32,33]. Scientific databases such as Medline, ScienceDirect, SPORTDiscus and Google Scholar were searched for articles on the topic with relevant keywords, including “genetic*, *polymorphism*, *swimming*.
RESULTS
Identification of genes that promote athletic excellence is challenging, mainly because the contribution of each possible gene to the overall heritability is small [31, 34]. Thus, the potential use of single nucleotide polymorphism (SNP) as a tool to assist in the prediction of future athletic success is still theoretical.
Efforts to use genetics for sports selection and prediction of sports excellence are particularly relevant in young ages [35]. Children are encouraged to participate in sports at a level consistent with their abilities [36, 37]. Thus, directing children to exercise at or above their limits, or to specialize in a single sport before adolescence, is discouraged [38]. Despite this, an increasing number of children do specialize in a sport such as swimming at an early age, and compete at an “elite” level [39, 40]. Moreover, some Olympic sports have selection processes that attempt to identify future champions and initiate specialized training, even before elementary school [41]. In addition, media coverage of sports often focuses on very talented but very young competitors [42]. Even more complex is a situation where sports-talented pre-pubertal children often excel in aerobic, anaerobic, individual, as well as team sports [43]. Thus, it seems that a key factor for competitive sports success is identification of a sport event that best matches the athlete’s ability at an appropriate age [36, 44, 45]. Genetic polymorphism may be used as an additional scientific tool to assist athletes and coaches in sport selection [35]. Yet, it should be noted that although a favourable genetic profile is necessary for athletic excellence, other factors including advanced training methods, equipment and facilities; adequate nutrition; psychological health; high motivation; and familial support are also crucial for top-level excellence in sports. While sports genetics research has focused mainly on individual sports, the majority of these studies examined runners and cyclists, and studies on the genetic characteristics of swimming have emerged only in recent years with the work of Woods et al. in 2001 [46].
DISCUSSION
ACE related genes
The renin-angiotensin system (RAS) plays a key role in human circulatory homeostasis. One of the main components of the RAS is angiotensin-converting enzyme (ACE), which catalyses production of angiotensin II (ANG II) from angiotensin I (ANG I), consequently increasing blood pressure Furthermore, ACE is an essential part of the kallikrein-kinin system (KKS), where it degrades kinins into inactive fragments, thus reducing blood pressure [47, 48].
ACE
The human angiotensin converting enzyme gene (ACE) is located on chromosome 17 in position 17q23.3[49]. The enzyme coded by this gene converts angiotensin I to II and is a key element in the renin angiotensin system (RAS), a system responsible for the regulation of blood pressure. The most widely studied ACE polymorphism is the restriction fragment length polymorphism consisting of the insertion (I) or deletion (D) of a 287 base pair repeat sequence in intron 16 (rs4340). The I allele is associated with lower ACE activity in both serum and tissue compared with the D allele [50].
D allele carriers have shown greater strength gains and muscle volume after isometric strength training in quadriceps muscles [51]. The I allele was associated with endurance performance [51], greater improvements in medium duration aerobic performance [52] as well as with an increase in the proportion of free fibres (type I muscle fibres) [53]. Nevertheless, some studies do not confirm these observations [54].
Some studies [55–57] but not all [58] have shown that the I allele is more prevalent among long distance swimmers compared to controls. However, I allele prevalence was similar among short- and long-distance swimmers [56] indicating that this polymorphism cannot distinguish between the two specialties.
BDKRB2
Bradykinin (BK) is a vasodilator, released from kininogens [59, 60]. BK is involved in various biological processes and its action is mainly mediated by one of its two receptors, the bradykinin 2 receptor (BDKRB2) [61]. The receptors are located on the plasma membrane of skeletal muscle cells and the vascular endothelium [48]. The activation of BDKRB2 results in increased skeletal muscle glucose uptake during physical activity, increase of muscle blood flow, and as a result higher endurance performance [47].
BDKRB2 is encoded by the BDKRB2 gene, located on chromosome 14q32 and expressed in most human tissues. An insertion/deletion polymorphism of 9 base pairs (bp) (-9/+9, rs5810761) in exon 1 is the most frequently investigated polymorphism in the context of relationships between genotypes and athletic status [47]. The -9bp allele is associated with higher gene transcriptional activity, higher mRNA expression, and increased receptor activity [62, 63]. As a result, the -9 allele may be connected with higher skeletal muscle metabolic efficiency and endurance performance [47]. Indeed, The BDKRB2 -9 allele was found to be associated with endurance performance [47, 48, 64–66]. Specifically, The BDKRB2 -9 allele was over-represented in long-distance (800–1500 m) swimmers compared to controls (72.2 vs 45.9%; P = 0.032) [64]. The +9 allele was over-represented in short-distance (50–100 m) swimmers compared to controls (67.9 vs 54.1%; P = 0.044)[64], and the +9/+9 BDKRB2 genotype was associated with a greater improvement in swimming performance in response to training [68]. However, in another study, Grenda et. al [67] statistically significant differences in BDKRB2 genotype and allele frequencies between long-distance swimmers and the total group of swimmers or controls, meaning that the BDKRB2 -9/+9 polymorphism was not associated with swimming performance at short, middle or long distance, regardless of gender among Polish swimmers.
AMPD1
Adenosine monophosphate deaminase (AMPD) is a very important regulator of muscle energy metabolism during exercise [68–70]. AMPD plays a pivotal role in ATP production through converting AMP to inosine monophosphate (IMP) [68–72] as well as in glycolytic pathway regulation [68, 71, 72]. AMPD expression in skeletal muscle is dependent on muscle fibre composition [68, 70–72]. A decrease in AMPD activity was reported concurrent with an increase in the proportion of fast-twitch (type II) fibres affecting, therefore, intense anaerobic physical activity [73–75].
The gene encoding the isoform (AMPD1) is located on chromosome 1 (1p13). AMPD1 is mainly expressed in fast-twitch (type II) muscle fibres. Differential AMPD1 gene expression may contribute to quantitative variations in enzyme activity across muscle groups with different types of fibres [71, 72, 75]. The nonsense mutation c.34C > T (rs17602729) in exon 2 of the AMPD1 gene converts glutamine codon (CAA) into the premature stop codon (TAA), which results in the early interruption of protein synthesis and appears to be the main cause of AMPD deficiency [76].
TT carriers have extremely low skeletal muscle AMPD activity compared to CT and CC carriers [71, 75]. T allele prevalence is lower among elite endurance athletes compared to controls [70, 77, 78]. A significant deficiency of the T allele was noted among short-distance swimmers compared to controls [79] suggesting, that short-distance swimming requires endurance abilities.
NOS3
Nitric oxide (NO) is a gaseous free radical synthesized from arginine by the nitric oxide synthase (NOS) enzymes. Both neuronal NOS (nNOS, NOS1) and endothelial NOS (eNOS, NOS3) are constitutively expressed, while inducible NOS (iNOS, NOS2) is not expressed under normal circumstance, but may be induced under stress conditions [80]. NO is involved in various aspects of skeletal muscle structure and function [81], all crucial for aerobic and anaerobic performance [82, 83]. Two NOS isoforms were identified in skeletal muscles – nNOS and eNOS. nNOS is the primary skeletal muscle isoform, whereas eNOS is expressed mainly in endothelial cells and contributes mostly to vascular tone control. Endothelial nitric oxide synthase (eNOS or NOS3) is encoded by NOS3 gene that is located on chromosome 7 (7q36) [84, 85]. Several investigations have demonstrated that NOS3 polymorphic variants can lead to altered transcription and/or processing rates of eNOS, and as a result interfere with normal enzyme function [86]. NOS3 gene – polymorphisms are associated with several health/fitness, exercise and training response phenotypes [87]. The -786T/C (rs2070744) and G894T(rs1799983) variants of NOS3 have been associated with endurance performance among elite endurance athletes [66, 88], power athletes [89, 90] and soccer players [91], as well as with the differentiation of elite power from endurance athletes [92]. Recently it was found that the -786T allele and the G-T haplotype of the -786T/C and G894T polymorphisms may be beneficial for long-distance swimming [93].
PPAR genes
Peroxisome proliferator activated receptor-alpha (PPAR-a) and peroxisome proliferator activated receptor gamma coactivator 1 alpha (PPARGC1A) play an important role in muscle fibre type conversion [94, 95]. PPARs are transcriptional factors which, through stimulation, affect glucose and fat metabolism [96], inflammatory processes [97], division and differentiation of some cell types [98] and energy homeostasis [99, 100].
The PPARA gene is located on chromosome 22 (22q12–q13.1 [101]. The most frequently analysed genetic variant in this gene is a polymorphism located in intron 7 (G/C, rs4253778). The PPARA gene is activated under conditions of energy deprivation, promoting uptake, utilization, and catabolism of fatty acids [102]. There is evidence that this gene is involved in the immune responses of the human body to endurance training, as it enables activation of the FAO mitochondrial pathway [97]. The PPARA gene is expressed at high levels in tissues that catabolize fatty acids, such as the liver, skeletal muscle, and heart [103]. There are findings indicating that PPARA gene G/C polymorphism is associated with swimming performance. The G allele was over-represented in LDS compared to controls (95.8 vs 83.6%; P = 0.023), while the C allele was over-represented in SDS compared to controls (33.8 vs 16.4%; P = 0.0002) [104]. A meta-analysis on 760 endurance athletes, including swimmers, and 1792 controls, found higher frequency of the GG genotype and G allele among athletes compared to controls [105]. No association was found between PPARA G/C polymorphism and performance among power-type Ukrainian athletes, including short-distance swimmers [106]
PPARGC1A has been suggested to affect athletic performance because of its role in a wide variety of biological responses [107, 108]. It encodes peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α), a transcriptional coactivator of the peroxisome proliferator-activated receptor (PPAR) family. PGC1α regulates the expression of several key genes involved in glucose and fatty acid oxidation [109, 110]. It is also a key stimulator of mitochondrial biogenesis by activating transcription of the nuclear respiratory factors NRF1 and NRF2, inducing expression of mitochondrial transcription factor A (TFAM)[111]. PGC1α is also important for skeletal muscle fibre conversion. Over-expression of PPARGC1A leads to the conversion of fast-twitch type IIb muscle fibres to type IIa and slow-twitch type I fibres [112]. Furthermore, PPARGC1A expression correlates with both short-term exercise and endurance training in rodents and humans [113–115].
The PPARGC1A gene is located on chromosome 4 (4p15.2). The Gly482Ser (rs8192678) gene polymorphism is the most frequently analysed. This polymorphism was reported to be associated with type 2 diabetes, obesity and elevated blood pressure [116–118]. The 482Ser allele was under-represented in the cohort of Polish and Russian athletes examined compared to nonathletic controls. A significantly low frequency of the 482Ser allele was observed among the endurance, strength-endurance, and sprint-strength groups of Polish athletes (which include long-, middle- and short-distance swimmers respectively), and was also less prevalent in groups of Russian endurance and strength-endurance athletes (which include long- and short-distance swimmers respectively). The authors concluded that the PPARGC1A Gly482 allele may be considered a beneficial factor for endurance performance [119]. A meta-analysis on 1,979 endurance athletes (including long-distance swimmers) and 1729 power type athletes (including short-distance swimmers) revealed that the Gly/Gly genotype and the Gly allele of the PPARGC1A Gly482Ser polymorphism may facilitate athletic performance regardless of the type of sport [120].
VEGFR2
Vascular endothelial growth factor (VEGF) is a major growth polypeptide regulating skeletal muscle angiogenesis [121]. VEGF is associated with proportion of oxidative skeletal muscle fibres (type I) and maximal rate of oxygen consumption [122, 123]. VEGF acts via two major receptors, VEGF1 and VEGFR2, localized mostly in the vascular endothelium [121].
The VEGFR2 gene is located on chromosome 4q11–q12. The rs1870377 T/A functional polymorphism in exon 11 results in the replacement of histidine (His) with glutamine (Gln) at the receptor within the extracellular region, which is important for ligand binding [124]. The frequency of the VEGFR2 472Gln allele was significantly higher in long, middle and short distance swimmers compared to controls [125]. In another study, Eider et. al [126] found that while the VEGFR2 gene Gln/Gln genotype was associated with endurance performance among Polish endurance athletes, including long-distance swimmers, when long-distance swimmers were separated from other endurance-type sports, no statistically significant difference between swimmers and controls was found.
Actinin-3 (ACTN3) gene
The human ACTN3 gene encodes α-actinin-3, an actin-binding protein with a structural role at the sarcomeric Z-line in glycolytic (type II, fast-twitch) muscle fibres, and plays an important role in muscle metabolism regulation [127]. A common genetic SNP at codon 577 of ACTN3 (rs1815739) results in the replacement of arginine (R) with a stop codon (X) [128]. The R allele is the normal functional version of the gene, whereas the X allele contains a sequence change that completely stops production of a functional protein [128]. Therefore, ACTN3 knockout (KO) mouse exhibit reduced muscle mass, mainly due to the decreased diameter of fast muscle fibres, significant decrease in grip strength, higher endurance and a shift towards increased activity of the mitochondrial oxidative metabolism compared with wild-type mice [129, 130].
A significant association between the ACTN3 genotype and athletic performance was first demonstrated by Yang et al. [131]. They showed that both male and female elite sprinters have significantly higher frequencies of the 577R allele compared to controls. Since then, ACTN3 R577X has been studied in other cohorts such as rowers [132], soccer players [133], ironman triathletes [134], basketball [135] etc. Some articles have reported a strong association between the RR genotype and elite power performance [136–139]. While ACTN3 R carriage may enhance power performance, other studies have shown that ACTN3 XX might contribute to endurance performance [129, 130], suggesting that ACTN3 is a bipotent gene (carrying the R allele enhances power performance and carrying the X allele favours endurance capacity). In contrast, reports on Asian and African runners suggested that ACTN3 deficiency was not associated with endurance performance [140, 141]. A meta-analysis of 88 articles did not confirm an association between ACTN3 RR genotype and performance (OR, 1.03; 95% CI, 0.92–1.15). However, when the analysis was restricted to power events (sprinters and jumpers in track and field and sprinters in swimming), a significant association was found (odds ratio (OR), 1.21; 95% CI, 1.03–1.42) [142]. Overall, association studies show that the ACTN3 R577X polymorphism plays a pivotal role in power athletic performance across multiple cohorts [143].
Interestingly, while the ACTN3 R577X polymorphism has been studied extensively among track and field athletes (especially runners), its contributions to performance in swimming – a sport that also relies on muscle strength [144] has received little attention. A significant association between the ACTN3 R577X polymorphism and swimming performance was not found among Caucasian, East Asians [145] or Spanish swimmers [146]. Among Taiwanese, R allele frequency was significantly higher in female sprint swimmers of international level compared to national-level swimmers or the general population [136].
In addition, it was shown previously that the ACTN3 R577X polymorphism was able to distinguish significantly between short- and long-distance runners, but was unable to differentiate between short- and long-distance swimmers [147]. Consistent with this, a strong correlation was found between 100 m and 2000 m swim times of elite short- and long-distance male swimmers [25]. Interestingly, both short- and long-distance swimmers had a higher RR genotype frequency compared to non-athletic controls. This implies that both short- and long-distance swimmers may benefit from ACTN3 R allele existence, strengthening the notion that there are no clear ACTN3 polymorphism differences between short-distance (power) and long-distance (endurance) swimmers[147]. Moreover, RR genotype and R allele frequencies (again, associated with power performance) were significantly higher among long-distance swimmers compared to long-distance runners. This may be explained by the large differences in the specific activity duration between competitive swimming and running events. The longest Olympic swimming event (1500 m ~ 15 min duration) is much shorter than the longest Olympic running race (marathon ~ 2:10 hr. duration), and therefore ACTN3 power characteristics are needed among long-distance swimmers but much less among long-distance runners. When comparing the short duration events, there is also a large difference in the specific activity duration of competitive swimming and running events. The time of the 100 m distance covered in swimming is approximately 50 s, while the same distance covered in running takes approximately 10 s. As a result, the sprint runner relies mostly on anaerobic energy sources, while the sprint swimmer also relies on aerobic energy production for his/her activity. Therefore, short- and long-distance swimmers share some common features, and the ACTN3 RR genotype may not be a suitable distinguishing genetic marker between them, as it is for runners. It is possible, however, that if open water swimmers (Olympic race – 10k) and only pure swimming sprinters (50 m) were included among the long-distance and short-distance swimmers in that study, similar R allele frequency differences would have been found between the swimmers and runners.
Lactate transport genes
Skeletal muscle is the major producer and the major user of lactate in the body. Therefore, transport of lactate across the cell membrane is of considerable importance during intense aerobic and anaerobic exercise [148, 149]. Lactate transport is mediated by a proton-linked monocarboxylate transporter (MCT1 & MCT4)[150]. Previous studies have demonstrated that MCT4 is related to lactate efflux from highly glycolytic muscle fibres, while MCT1 is associated with lactate uptake for further oxidation [150–152].
The MCT1 A1470T single nucleotide polymorphism (rs1049434) affects lactate transport across the sarcolemma [153]. The MCT1 T-allele is related to reduced lactate transport by MCT1 [154], but its implication for athletic performance is not clearly established. To the best of our knowledge, only a single study has examined the association between the MCT1 A1470T polymorphism and athletic performance. In a study of 323 Russian athletes and 467 non-athletic controls [155], the authors found that the frequencies of the A allele and AA genotype (defined in their study as the MCT1 polymorphism associated with reduced lactate transport) were significantly higher in endurance-oriented rowers compared to the control group. A previous study reported a 40% reduction of the lactate transport rate in red blood cells among T allele carriers [156]. Moreover, male carriers of the T allele had lower blood lactate levels compared to carriers of the AA genotype following different circuit exercise training protocols [154].
Participation in both sprint and endurance competitive swimming events results in lactate accumulation [157]. Repeating maximal performances (when progressing from preliminaries to the semifinal and to the final stage) is often required in local and international swimming championships. Therefore, efficient transfer of lactate from the active muscles to the blood following earlier maximal effort is necessary for effective recovery and improved repeated performance [158]. Interestingly, a significantly higher frequency of the T allele among Israeli swimmers compared to Israeli runners was reported [159]. Whether this suggests that swimmers better tolerate the decreased lactate transport associated with the presence of the T allele, or whether this polymorphism displays some advantage for swimmers, is currently not clear.
We previously demonstrated [25] that mean peak blood lactate level following a swimming repeated sprint test (RST) was lower than similar exercise and recovery time periods of running or cycling RST lactate levels [160]. Along with this, blood lactate concentration was relatively low (3 mmol/l) following forty 25 m swims at a 100 m race pace in highly-trained swimmers [161]. Moreover, previous studies have shown reduced lactate accumulation following deep-water running compared to land running [162, 163]. It was suggested that the lower lactate level following exercise in the water is related – at least partially – to smaller muscle mass recruitment during swimming compared to running, and to reduced muscle fibre contraction due to reduced weight-bearing in the water. In addition, water immersion generates hydrostatic pressure resulting in fluid shift from the muscles to the blood, facilitating effective waste clearance [164, 165]. Therefore, swimmers may benefit from enhanced blood flow caused by water immersion [166], which likely improves elimination of intramuscular metabolic elimination and enhances post-competition recovery [167]. It is possible that differences in lactate level between runners and swimmers are related to lactate transport differences due to the higher presence of the MCT1 T allele among swimmers.
The higher frequency of the MCT1 T-allele polymorphism among swimmers also raises the possibility of genetically determined unconscious sports self-selection [159]. Many talented young athletes with excellent power-sprint or endurance ability are exposed to a large variety of sports at initial phases of their career [36]. Carrying a polymorphism like the MCT1 T allele may limit their capability to excel in sports characterized by high lactate accumulation (e.g., sprint and middle-distance running) [36]. Consequently, they unconsciously choose swimming as their sport specialty, due to the unique physiological settings and relatively lower lactate level during exercise in the water [36].
The IGF axis genes
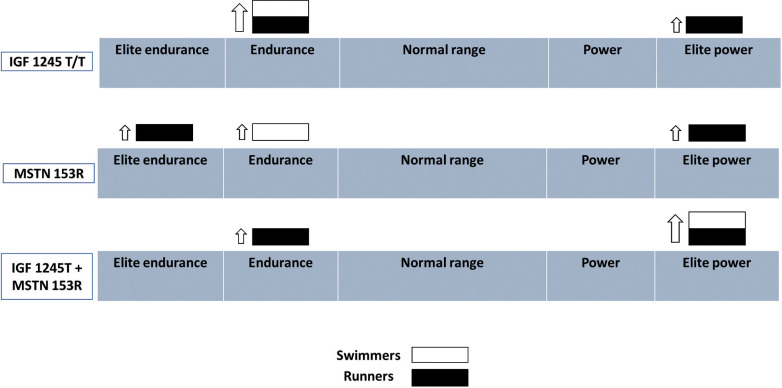
The potential use of genetic SNP’s of hormone genes as a tool to assist in predicting future athletic performance is currently an extremely challenging topic, mainly because each possible gene makes only a small contribution to the overall heritability [168]. It is now well established that an increase in insulin-like growth factor-I (IGF1) plays a pivotal role in the functional and structural muscle adaptation to exercise [169, 170]. Therefore, most previous reports related to hormonal gene polymorphism and athletic performance in professional athletes studied variations in the IGF1 polymorphism that affect its circulating levels [171, 172]. The polymorphism of IGF1 promoter frequency was significantly greater in athletes (9.2%) compared to controls (2.4%), and particularly among strength (11%) athletes compared to athletes participating in team sports (7.8%) [173]. A higher frequency of the IGF1 C1245T T/T IGF1 promoter polymorphism (associated with higher circulating IGF levels) was demonstrated among Israeli track and field athletes (4.8%), compared to controls (non-existent) [171]. Interestingly, while T/T polymorphism carriers were both endurance and power athletes, endurance athletes were of national level, but the power athletes were top international and Olympic athletes [171] (see Fig. 1). This suggests that the IGF1 T/T (rs35767) polymorphism is more beneficial for power sport performance at the elite level. Along these lines, an assessment of the frequency of another polymorphism of the IGF1 gene (i.e. IGF1 rs7136446) showed that the frequency of carrying the GG genotype was significantly greater among sprinters compared to weight lifters [174]. Altogether, this may suggest that among typical power sports, the IGF1 polymorphism is more important for speed than strength sport events. In addition, the IGF2 (rs680) GG genotype frequency was significantly greater among sprinters compared to weight lifters [175], suggesting that carrying this IGF2 polymorphism may also be beneficial mainly for speed-related and not for strength sports. Circulating IGF2 levels were lower among individuals homozygous for the G allele [176], and higher levels of circulating IGF1 levels were found in individuals carrying the IGF2 GG genotype [177]. Thus, it is possible that the beneficial effect of the IGF2 rs680 polymorphism on speed performance is not necessarily mediated through its influence on circulating IGF2, but via its effect on IGF1 levels.
FIG. 1.
IGF and myostatin gene polymorphism and competitive swimming and running.
In contrast to elite track and field athletes, SNPs of IGF1, IGF1 receptor and IGF2 were not frequent among swimmers [175, 178, 179] _ENREF_74_ENREF_74. This suggests that the insulin-like growth factor system is less significant for elite swimming than for running performance, although the mechanism for this discrepancy is currently unknown. A possible explanation for this is that swimming excellence is determined primarily by the swimming technique and the swimmers’ anthropometric characteristics (mainly limb length) [180], masking muscle mass and physiologic and metabolic differences, and enabling tall, long-armed and technically talented swimmers to excel in the majority of swimming distances. However, interestingly, and consistent with the findings of the ACTN3 polymorphism [147], IGF1 polymorphism (rs35767) frequency was also higher [171] among endurance swimmers (although not top level), emphasizing once more the importance of speed in typical long-distance swimming events.
Whether evaluation of the IGF axis gene polymorphism can be used for sports selection in young athletes, or whether a multi-potent athlete who wants to develop a competitive career and carries the beneficial IGF polymorphism should prefer track and field over swimming, is currently speculative and needs to be further studied.
Myostatin
Myostatin belongs to the transforming growth factor β super-family of proteins that control tissue growth and differentiation [181, 182]. The myostatin (MSTN) gene is expressed almost exclusively in skeletal-muscle cells, and functions as a negative regulator of muscle growth. [183–185] In contrast, gene knockout [186] and inhibition of the gene signalling [181] lead to dramatic muscle hypertrophy and/or hyperplasia. The MSTN Lys(K)-153Arg(R) polymorphism (rs1805086, 2379 A > G replacement) influences skeletal muscle phenotypes [187]. The frequency of the mutant R allele (associated with lower myostatin levels) is about 3–4% among Caucasians, with a frequency of mutant homozygote (RR) below 1% [188–190]. A previous study [191] assessed the frequency of the MSTN Lys(K)-153Arg(R) polymorphism among elite Israeli track and field athletes and swimmers, and found that the MSTN 153R allele frequency was significantly higher among top- compared to national-level runners. This suggests the possible importance of the MSTN K153R polymorphism for excellence in both long-distance and sprint running. Interestingly, while the frequency of the MSTN 153R allele was significantly greater among long-compared to short-distance swimmers, none of the long-distance swimmers who carried the MSTN 153RR genotype was top level. Only a single short-distance swimmer carried the MSTN 153R allele, indicating that this polymorphism may not be significant for success in swimming, particularly in sprints.
Since both IGF and myostatin regulate muscle hypertrophy, the combined frequency of the IGF 1245T (rs35767) and MSTN 153Arg(R) polymorphisms was also studied [172]. All carriers of both mutations among the short-distance runners and swimmers were of elite competitive calibre (Fig. 1).
IL6
The IL6 -174G/C SNP modifies the IL6 promoter activity, with carriers of the G allele demonstrating higher circulating IL6 levels [192]. The protective effect of the IL6 -174G genotype from developing exercise-induced muscle damage is related to greater muscle hypertrophy and improved glucose uptake, and to reduced exercise-associated muscle inflammation by favourably modifying the pro- and anti-inflammatory cytokine production balance [193–195]. In contrast, carrying the C allele (in particularly CC homozygotes), which is associated with reduced circulating IL6 levels, increases the probability of developing eccentric exercise-related muscle injury, possibly resulting in reduced training recovery capabilities and competitive performance [196, 197]. Interestingly, a significantly higher frequency of the CC genotype was found in long-distance swimmers compared to long-distance runners and controls [198]. It was suggested that the rarity of exercise-associated rhabdomyolysis among swimmers probably results from other sport-specific and/or water-related protective factors, and is not related to genetic features [198]. It was also proposed that swimming selection in gifted endurance athletes who are C-allele carriers possibly represents genetically dependent sports selection [198]. Both disciplines (long-distance running and long-distance swimming) require remarkable endurance skills [199, 200]. However, since carriers of the IL6 -174C allele are more susceptible to developing exercise-induced muscle damage (in particularly during land activity) [196], their recovery capabilities from intense training may be impaired. This may highlight the possibility that athletes with superior endurance abilities, but also with an unfavourable IL6 -174C polymorphism, subconsciously prefer to specialize in water-type sports that would expose their genetic disadvantages to a lesser degree. IL6 -174C represents, therefore, a genetic polymorphism that may lead to sports selection based on more effective training tolerability characteristics and not on performance qualities per se.
Interestingly, a recent study showed that carrying both the IL6C and IGFBP3C mutations was significantly more frequent among long- and short-distance swimmers compared to long-distance runners and sprinters [201]. What advantage could swimmers (both long- and short-distance) gain from carrying both mutations? Higher prevalence of the IL6 -174C polymorphism increases the potential risk of exercise-induced muscle damage and delayed onset muscle soreness, even among swimmers – mainly following ground and resistance training, both taking a much greater place in elite swimming training over the last decades [202]. As stated earlier, an attenuated genetically predisposed insulin-like growth factor system might also be disadvantageous for swimmers, mainly in the short distances [172]. Therefore, it is possible that carrying the IGFBP3C polymorphism serves as a compensation mechanism. Carriers of this polymorphism have lower circulating IGF binding protein-3 (IGFBP3) levels. Therefore, levels of free IGF1, which is the actual bioactive growth factor, increase. The hypertrophic effect of higher free IGF1 levels may assist in the repair process of exercise-associated muscle damage. Again, this possible IGF-system compensating mechanism does not operate by inducing direct changes in IGF1, but instead by indirectly modifying its binding proteins.
CONCLUSIONS
Swimming performance related genetic variants are presented in Table 1. Scientific research on the relationship between genes and competitive sports, and in particularly in swimming, is still in its infancy. Currently, the possibility of using genetic assessment to advise a young athlete what type of sports he/she should select, and to predict excellence or future success in professional sports, is only hypothetical. Whether a multi-potent athlete who wants to develop a competitive career and carries a beneficial IGF system polymorphism should prefer running, or carriers of unfavourable lactate transport and IL6 polymorphism should select swimming, is speculative at present (Table 2); any genetic counselling must be interpreted with caution. Furthermore, while a favourable genetic predisposition is important, other environmental and psychological features, such as training strategy, equipment and facilities, adequate nutrition and replacement of nutritional deficiencies, familial support, competitive characteristics and motivational issues, are also critical for top-level sports success.
TABLE 1.
Swimming performance related genetic variants
| Gene | Genetic polymorphism | Beneficial allele for swimming performance (compared to controls) |
Reference | |
|---|---|---|---|---|
| SDS | LDS | |||
| ACE | I/D rs4340 | D | I | [56] |
| ACTN3 | R/X rs1815739 | R | [147] [25] | |
| AMPD1 | c.34C > T rs17602729 | C | [79] | |
| BDKRB2 | -9/+9 rs581076 | +9 | -9 | [64] |
| IL6 | -174G/C rs1800795 | C | C | [198] |
| IGF1 | C1245T rs35767 | IGF1 1245TT+MSTN | T | [172] |
| MSTN | Lys(K)-153Arg(R) rs1805086 | 153R | R | [171] |
| MCT1 | A1470T rs104943 | T | T | [154] |
| NOS3 | 786T/C rs2070744 G894T rs1799983 |
TG894—786T | [93] | |
| PPARA | G/C rs4253778 | C | G | [104] |
| PPARGC1A | Gly482Ser rs8192678 | Gly | Gly | [119] |
| VEGFR2 | His/Gln rs1870377 | Gln | Gln | [126] |
TABLE 2.
Genotypes associated with competitive swimming.
| Endurance related alleles that may be advantageous for long distance swimming |
| –– ACE rs4340 I |
| –– BDKRB2 rs581076 -9 |
| –– NOS3 Combined rs1799983 G + rs2070744 T |
| –– PPARArs4253778 G |
| –– PPARGC1A rs8192678 Gly |
| –– VEGFR2 rs1870377 Gln |
| Muscle related alleles and genotypes that may be advantageous for long distance swimming |
| –– ACTN3 rs1815739 CC (RR) |
| –– IGF1 rs35767 TT |
| –– MSTN rs1805086 R |
| Endurance related alleles that may be advantageous for short distance swimming |
| –– ACE rs4340 D |
| –– AMPD1 rs17603739 C |
| –– BDKRB2 rs581076 +9 |
| –– PPARArs4253778 C |
| –– PPARGC1A rs8192678 Gly |
| –– VEGFR2 rs1870377 Gln |
| Muscle related Alleles and Genotypes that may be advantageous for elite short distance swimming |
| –– Combined IGF1 rs35767 TT + MSTN rs1805086 R |
| Recovery related alleles that may be advantageous for tolerating water sports |
| –– IL6 rs1800795 C (post exercise muscle damage) |
| –– MCT1 rs1049434 T (Lactate transport) |
Finally, a major problem in genetic research is the need for a large sample size, which in many studies is very difficult to achieve. To overcome this obstacle, some studies combine different types of sports. The results presented here indicate that despite seemingly similar metabolic characteristics, athletes from different sport disciplines may carry different genetic polymorphisms. Therefore, extreme caution should be taken before pooling different types of sports in genetic research.
Acknowledgments
We acknowledge Mr. Roi Mor for his technical assistance and athletes for participation.
Conflict of interest statement
No potential financial conflict or any other conflict of interest is reported by the authors.
REFERENCES
- 1.Physical Activity Council’s Overview Report on US Participation 2020. http://www.physicalactivitycouncil.com/
- 2.Scheerder J, Vandermeerschen H, Tuyckom C Van, Hoekman R, Breedveld K, Vos S. Understanding the game Sport Participation in Europe Facts, reflections and recommendationsץ Sport Policy & Management. 2011. http://hdl.handle.net/1854/LU-1932490 .
- 3.De Koning JJ, Foster C, Lucia A, Bobbert MF, Hettinga FJ, Porcari JP. Using modeling to understand how athletes in different disciplines solve the same problem: Swimming versus running versus speed skating. Int J Sports Physiol Perform. 2011;6(2):276–80. doi: 10.1123/ijspp.6.2.276. [DOI] [PubMed] [Google Scholar]
- 4.Why do swimmers break more records than runners? – BBC News. https://www.bbc.com/news/magazine-37064144 .
- 5.Holmér I. Swimming physiology. Ann Physiol Anthr. 1992;11:269–76. [PubMed] [Google Scholar]
- 6.Maron BJ. Structural features of the athlete heart as defined by echocardiography. J Am Coll Cardiol. 1986;7:190–203. doi: 10.1016/s0735-1097(86)80282-0. [DOI] [PubMed] [Google Scholar]
- 7.Barbier J, Ville N, Kervio G, Walther G, Carré F. Sports-specific features of athlete’s heart and their relation to echocardiographic parameters. Herz. 2006;31(6):531–43. doi: 10.1007/s00059-006-2862-2. [DOI] [PubMed] [Google Scholar]
- 8.Lee B-A, Oh D-J. The effects of long-term aerobic exercise on cardiac structure, stroke volume of the left ventricle, and cardiac output. J Exerc Rehabil. 2016;12:37–41. doi: 10.12965/jer.150261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatta G, Leban B, Paderi M, Padulo J, Migliaccio GM, Pau M. The development of swimming power. Muscles Ligaments Tendons J. 2014;4:438–45. [PMC free article] [PubMed] [Google Scholar]
- 10.Holmer I, Lundin A, Eriksson BO. Maximum oxygen uptake during swimming and running by elite swimmers. J Appl Physiol. 1974;36:711–4. doi: 10.1152/jappl.1974.36.6.711. [DOI] [PubMed] [Google Scholar]
- 11.Racinais S, Cocking S, Périard JD. Sports and environmental temperature: From warming-up to heating-up. Temperature. 2017;4:227–57. doi: 10.1080/23328940.2017.1356427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa MJ, Bragada JA, Mejias JE, Louro H, Marinho DA, Silva AJ, Barbosa TM. Effects of swim training on energetics and performance. Int J Sports Med. 2013 Jun;34(6):507–13. doi: 10.1055/s-0032-1327573. [DOI] [PubMed] [Google Scholar]
- 13.Barbosa TM, Bragada JA, Reis VM, Marinho DA, Carvalho C, Silva AJ. Energetics and biomechanics as determining factors of swimming performance: updating the state of the art. J Sci Med Sport. 2010 Mar;13(2):262–9. doi: 10.1016/j.jsams.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Capelli C, Pendergast DR, Termin B. Energetics of swimming at maximal speeds in humans. Eur J Appl Physiol Occup Physiol. 1998;78:385–93. doi: 10.1007/s004210050435. [DOI] [PubMed] [Google Scholar]
- 15.Barbosa T, Fernandes R, Keskinen KL, Colaço P, Cardoso C, Silva J, et al. Evaluation of the energy expenditure in competitive swimming strokes. Int J Sports Med. 2006;27:894–9. doi: 10.1055/s-2006-923776. [DOI] [PubMed] [Google Scholar]
- 16.McNarry MA, Wilson RP, Holton MD, Griffiths IW, Mackintosh KA. Investigating the relationship between energy expenditure, walking speed and angle of turning in humans. PLoS One. 2017 doi: 10.1371/journal.pone.0182333. https://pubmed.ncbi.nlm.nih.gov/28796796/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Geus B, De Smet S, Nijs J, Meeusen R. Determining the intensity and energy expenditure during commuter cycling. Br J Sports Med. 2007;41:8–12. doi: 10.1136/bjsm.2006.027615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.di Prampero PE, Osgnach C. Muscle Exerc Physiol. Elsevier; 2018. Energy Cost of Human Locomotion on Land and in Water; pp. 183–213. [Google Scholar]
- 19.Zamparo P, Cortesi M, Gatta G. The energy cost of swimming and its determinants. Eur J Appl Physiol. 2020 Jan;120(1):41–66. doi: 10.1007/s00421-019-04270-y. [DOI] [PubMed] [Google Scholar]
- 20.Barbosa T, Fernandes R, Keskinen KL, Colaço P, Cardoso C, Silva J, et al. Evaluation of the energy expenditure in competitive swimming strokes. Int J Sports Med. 2006;27:894–9. doi: 10.1055/s-2006-923776. [DOI] [PubMed] [Google Scholar]
- 21.Barbosa TM, Fernandes RJ, Keskinen KL, Vilas-Boas JP. The influence of stroke mechanics into energy cost of elite swimmers. Eur J Appl Physiol. 2008;103:139–49. doi: 10.1007/s00421-008-0676-z. [DOI] [PubMed] [Google Scholar]
- 22.Barbosa TM, Marinho DA, Costa MJ, Silva AJ. Biomechanics of Competitive Swimming Strokes. Biomech Appl. 2011. www.intechopen.com .
- 23.Pyne DB, Sharp RL. Physical and energy requirements of competitive swimming events. Int J Sport Nutr Exerc Metab. 2014;24(4):351–9. doi: 10.1123/ijsnem.2014-0047. [DOI] [PubMed] [Google Scholar]
- 24.Costa MJ, Bragada JA, Marinho DA, Silva AJ, Barbosa TM. Longitudinal interventions in elite swimming: a systematic review based on energetics, biomechanics, and performance. J Strength Cond Res. 26(7):2006–16. doi: 10.1519/JSC.0b013e318257807f. 2012l. [DOI] [PubMed] [Google Scholar]
- 25.Meckel Y, Bishop DJ, Rabinovich M, Kaufman L, Nemet D, Eliakim A. The relationship between short- and long-distance swimming performance and repeated sprint ability. J Strength Cond Res. 2012;26:3426–31. doi: 10.1519/JSC.0b013e3182473df3. [DOI] [PubMed] [Google Scholar]
- 26.Jürimäe J, Haljaste K, Cicchella A, Lätt E, Purge P, Leppik A, et al. Analysis of swimming performance from physical, physiological, and biomechanical parameters in young swimmers. Pediatr Exerc Sci. 2007;19:70–81. doi: 10.1123/pes.19.1.70. [DOI] [PubMed] [Google Scholar]
- 27.Kucia-Czyszczoń K, Dybińska E, Ambroży T, Chwała W. Factors determining swimming efficiency observed in less skilled swimmers. Acta Bioeng Biomech Orig Pap. 2013;15(4):115–124. [PubMed] [Google Scholar]
- 28.Morais JE, Garrido ND, Marques MC, Silva AJ, Marinho DA, Barbosa TM. The influence of anthropometric, kinematic and energetic variables and gender on swimming performance in youth athletes. J Hum Kinet. 2013;39:203–11. doi: 10.2478/hukin-2013-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mejias JE, Bragada J, Costa M, Reis V, Garrido N, Barbosa TM. “Young” masters vs. Elite swimmers: Comparison of performance, energetics, kinematics and efficiency. International Sportmed Journal. 2014;15(2):165–177. [Google Scholar]
- 30.McNarry G, Allen-Collinson J, Evans AB. ‘Doing’ competitive swimming: Exploring the skilled practices of the competitive swimming lifeworld. Int Rev Sociol Sport. 2020 jan 8; doi: 10.1177/1012690219894939. [DOI] [Google Scholar]
- 31.Mujika I, Chatard JC, Busso T, Geyssant A, Barale F, Lacoste L. Effects of training on performance in competitive swimming. Can J Appl Physiol. 1995;20:395–406. doi: 10.1139/h95-031. [DOI] [PubMed] [Google Scholar]
- 32.Ferrari R. Writing narrative style literature reviews. Med Writ. 2015;24:230–5. [Google Scholar]
- 33.Green BN, Johnson CD, Adams A. Writing narrative literature reviews for peer-reviewed journals: secrets of the trade. J Chiropr Med. 2006;5:101–17. doi: 10.1016/S0899-3467(07)60142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippi G, Longo UG, Maffulli N. Genetics and sports. Br Med Bull. 2010;93:27–47. doi: 10.1093/bmb/ldp007. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Zaken S, Meckel Y, Lidor R, Nemet D, Eliakim A. Genetic profiles and prediction of the success of young athletes’ transition from middle- to long-distance runs: an exploratory study. Pediatr Exerc Sci. 2013;25:435–47. doi: 10.1123/pes.25.3.435. [DOI] [PubMed] [Google Scholar]
- 36.Myer GD, Jayanthi N, DiFiori JP, Faigenbaum AD, Kiefer AW, Logerstedt D, et al. Sports Specialization, Part II: Alternative Solutions to Early Sport Specialization in Youth Athletes. Sports Health. 2016;8(1):65–73. doi: 10.1177/1941738115614811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayanthi N, Pinkham C, Dugas L, Patrick B, LaBella C. Sports Specialization in Young Athletes: Evidence-Based Recommendations. Sports Health. 2013;5:251–7. doi: 10.1177/1941738112464626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostafavifar AM, Best TM, Myer GD. Early sport specialisation, does it lead to long-term problems? Br J Sports Med. 2013;47(17):1060–1. doi: 10.1136/bjsports-2012-092005. [DOI] [PubMed] [Google Scholar]
- 39.Lätt E, Jürimäe J, Mäestu J, Purge P, Rämson R, Haljaste K, et al. Physiological, biomechanical and anthropometrical predictors of sprint swimming performance in adolescent swimmers. J Sport Sci Med. 2010;9:398–404. [PMC free article] [PubMed] [Google Scholar]
- 40.Mummery WK, Wankel LM. Training adherence in adolescent competitive swimmers: An application of the theory of planned behavior. J Sport Exerc Psychol. 1999;21:313–28. [Google Scholar]
- 41.Malina RM. Early Sport Specialization. Curr Sports Med Rep. 2010;9:364–71. doi: 10.1249/JSR.0b013e3181fe3166. [DOI] [PubMed] [Google Scholar]
- 42.Valovich McLeod TC, Decoster LC, Loud KJ, Micheli LJ, Parker JT, Sandrey MA, et al. National Athletic Trainers’ Association position statement: prevention of pediatric overuse injuries. J Athl Train. 2011;46:206–20. doi: 10.4085/1062-6050-46.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonçalves C EB, Rama L ML, Figueiredo AB. Talent identification and specialization in sport: an overview of some unanswered questions. Int J Sports Physiol Perform. 2012;7:390–3. doi: 10.1123/ijspp.7.4.390. [DOI] [PubMed] [Google Scholar]
- 44.Vaeyens R, Güllich A, Warr CR, Philippaerts R. Talent identification and promotion programmes of olympic athletes. J Sports Sci. 2009;27:1367–80. doi: 10.1080/02640410903110974. [DOI] [PubMed] [Google Scholar]
- 45.Balyi I. Long-term athlete development: trainability in childhood and adolescence Windows of Opportunity, Optimal Trainability. 2004. [Google Scholar]
- 46.Woods D, Hickman M, Jamshidi Y, Brull D, Vassiliou V, Jones A, et al. Elite swimmers and the D allele of the ACE I/D polymorphism. Hum Genet. 2001;108:230–2. doi: 10.1007/s004390100466. [DOI] [PubMed] [Google Scholar]
- 47.Williams AG, Dhamrait SS, Wootton PTE, Day SH, Hawe E, Payne JR, et al. Bradykinin receptor gene variant and human physical performance. J Appl Physiol (1985) 2004;96:938–42. doi: 10.1152/japplphysiol.00865.2003. [DOI] [PubMed] [Google Scholar]
- 48.Saunders CJ, de Milander L, Hew-Butler T, Xenophontos SL, Cariolou MA, Anastassiades LC, et al. Dipsogenic genes associated with weight changes during Ironman Triathlons. Hum Mol Genet. 2006;15:2980–7. doi: 10.1093/hmg/ddl240. [DOI] [PubMed] [Google Scholar]
- 49.Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- 50.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charbonneau DE, Hanson ED, Ludlow AT, Delmonico MJ, Hurley BF, Roth SM. ACE genotype and the muscle hypertrophic and strength responses to strength training. Med Sci Sports Exerc. 2008;40:677–83. doi: 10.1249/MSS.0b013e318161eab9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cam S, Colakoglu M, Colakoglu S, Sekuri C, Berdeli A. ACE I/D gene polymorphism and aerobic endurance development in response to training in a non-elite female cohort. J Sports Med Phys Fitness. 2007;47:234–8. [PubMed] [Google Scholar]
- 53.MacArthur DG, North KN. Genes and human elite athletic performance. Hum. Genet. 2005;116(5):331–9. doi: 10.1007/s00439-005-1261-8. [DOI] [PubMed] [Google Scholar]
- 54.Orysiak J, Zmijewski P, Klusiewicz A, Kaliszewski P, Malczewska-Lenczowska J, Gajewski J, et al. The association between ace gene variation and aerobic capacity in winter endurance disciplines. Biol Sport. 2013;30:249–53. doi: 10.5604/20831862.1077549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsianos G, Sanders J, Dhamrait S, Humphries S, Grant S, Montgomery H. The ACE gene insertion/deletion polymorphism and elite endurance swimming. Eur J Appl Physiol. 2004;92:360–2. doi: 10.1007/s00421-004-1120-7. [DOI] [PubMed] [Google Scholar]
- 56.Grenda A, Leońska-Duniec A, Kaczmarczyk M, Ficek K, Król P, Cięszczyk P, et al. Interaction between ACE I/D and ACTN3 R557X polymorphisms in polish competitive swimmers. J Hum Kinet. 2014;42:127–36. doi: 10.2478/hukin-2014-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nazarov IB, Woods DR, Montgomery HE, Shneider O V, Kazakov VI, Tomilin N V, et al. The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur J Hum Genet. 2001;9:797–801. doi: 10.1038/sj.ejhg.5200711. [DOI] [PubMed] [Google Scholar]
- 58.Wang G, Mikami E, Chiu LL, De Perini A, Deason M, Fuku N, et al. Association analysis of ACE and ACTN3 in Elite Caucasian and East Asian Swimmers. Med Sci Sports Exerc. 2013;45:892–900. doi: 10.1249/MSS.0b013e31827c501f. [DOI] [PubMed] [Google Scholar]
- 59.Prado GN, Taylor L, Zhou X, Ricupero D, Mierke DF, Polgar P. Mechanisms regulating the expression, self-maintenance, and signaling-function of the bradykinin B2 and B1 receptors. J Cell Physiol. 2002;193(3):275–86. doi: 10.1002/jcp.10175. [DOI] [PubMed] [Google Scholar]
- 60.Kammerer S, Braun A, Arnold N, Roscher AA. The human bradykinin B2 receptor gene: Full-length cDNA, genomic organization and identification of the regulatory region. Biochem Biophys Res Commun. 1995;211:226–33. doi: 10.1006/bbrc.1995.1800. [DOI] [PubMed] [Google Scholar]
- 61.Regoli D, Barabé J. Pharmacology of bradykinin and related kinins. Pharmacol Rev. 1980;32:1–46. [PubMed] [Google Scholar]
- 62.Lung CC, Chan EKL, Zuraw BL. Analysis of an exon 1 polymorphism of the B2 bradykinin receptor gene and its transcript in normal subjects and patients with C1 inhibitor deficiency. J Allergy Clin Immunol. 1997;99:134–46. doi: 10.1016/s0091-6749(97)70310-5. [DOI] [PubMed] [Google Scholar]
- 63.Braun A, Kammerer S, Maier E, Böhme E, Röscher AA. Polymorphisms in the gene for the human B2-bradykinin receptor. New tools in assessing a genetic risk for bradykinin-associated diseases. Immunopharmacology. 1996;33(1–3):32–5. doi: 10.1016/0162-3109(96)00079-3. [DOI] [PubMed] [Google Scholar]
- 64.Sawczuk M, Timshina YI, Astratenkova I V, Maciejewska-Karłowska A, Leońska-Duniec A, Ficek K, et al. The -9/+9 polymorphism of the bradykinin receptor beta 2 Gene and athlete status: A study involving two European cohorts. Hum Biol. 2013;85:741–55. doi: 10.3378/027.085.0511. [DOI] [PubMed] [Google Scholar]
- 65.Eynon N, Meckel Y, Alves AJ, Nemet D, Eliakim A. Is there an interaction between BDKRB2-9/+9 and GNB3 C825T polymorphisms and elite athletic performance? Scand J Med Sci Sport. 2011;21(6):e242–6. doi: 10.1111/j.1600-0838.2010.01261.x. [DOI] [PubMed] [Google Scholar]
- 66.Saunders CJ, Xenophontos SL, Cariolou MA, Anastassiades LC, Noakes TD, Collins M. The bradykinin β2 receptor (BDKRB2) and endothelial nitric oxide synthase 3 (NOS3) genes and endurance performance during Ironman Triathlons. Hum Mol Genet. 2006;15:979–87. doi: 10.1093/hmg/ddl014. [DOI] [PubMed] [Google Scholar]
- 67.Grenda A, Leonska-Duniec A, Cieszczyk P, Zmijewski P. Bdkrb2 gene -9/+9 polymorphism and swimming performance. Biol Sport. 2014;31:109–13. doi: 10.5604/20831862.1096047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fedotovskaya ON, Danilova AA, Akhmetov II. Effect of AMPD1 gene polymorphism on muscle activity in humans. Bull Exp Biol Med. 2013;154:489–91. doi: 10.1007/s10517-013-1984-9. [DOI] [PubMed] [Google Scholar]
- 69.Rico-Sanz J, Rankinen T, Joanisse DR, Leon AS, Skinner JS, Wilmore JH, et al. Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE Family study. Physiol Genomics. 2003;14:161–6. doi: 10.1152/physiolgenomics.00165.2002. [DOI] [PubMed] [Google Scholar]
- 70.Rubio JC, Martín MA, Rabadán M, Gómez-Gallego F, San Juan AF, Alonso JM, et al. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: Does this mutation impair performance? J Appl Physiol. 2005;98:2108–12. doi: 10.1152/japplphysiol.01371.2004. [DOI] [PubMed] [Google Scholar]
- 71.Fischer H, Esbjörnsson M, Sabina RL, Strömberg A, Peyrard-Janvid M, Norman B. AMP deaminase deficiency is associated with lower sprint cycling performance in healthy subjects. J Appl Physiol (1985) 2007;103:315–22. doi: 10.1152/japplphysiol.00185.2007. [DOI] [PubMed] [Google Scholar]
- 72.Norman B, Mahnke-Zizelman DK, Vallis A, Sabina RL. Genetic and other determinants of AMP deaminase activity in healthy adult skeletal muscle. J Appl Physiol. 1998;85:1273–8. doi: 10.1152/jappl.1998.85.4.1273. [DOI] [PubMed] [Google Scholar]
- 73.Thomaes T, Thomis M, Onkelinx S, Fagard R, Matthijs G, Buys R, et al. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: The CAREGENE study. BMC Genet. 2011;12:84. doi: 10.1186/1471-2156-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norman B, Nygren AT, Nowak J, Sabina RL. The effect of AMPD1 genotype on blood flow response to sprint exercise. Eur J Appl Physiol. 2008;103:173–80. doi: 10.1007/s00421-008-0683-0. [DOI] [PubMed] [Google Scholar]
- 75.Norman B, Sabina RL, Jansson E. Regulation of skeletal muscle ATP catabolism by AMPD1 genotype during sprint exercise in asymptomatic subjects. J Appl Physiol. 2001;91:258–64. doi: 10.1152/jappl.2001.91.1.258. [DOI] [PubMed] [Google Scholar]
- 76.Cieszczyk P, Ostanek M, Leońska-Duniec A, Sawczuk M, Maciejewska A, Eider J, et al. Distribution of the AMPD1 C34T polymorphism in Polish power-oriented athletes. J Sports Sci. 2012;30:31–5. doi: 10.1080/02640414.2011.623710. [DOI] [PubMed] [Google Scholar]
- 77.Cięszczyk P, Eider J, Ostanek M, Leoska-Duniec A, Ficek K, Kotarska K, et al. Is the C34T polymorphism of the AMPD1 gene associated with athlete performance in rowing? Int J Sports Med. 2011;32:987–91. doi: 10.1055/s-0031-1283186. [DOI] [PubMed] [Google Scholar]
- 78.Colombini A, Galeazzi IO. Athleticogenomics and elite athletes: A review of the state of the art and a possible relationship with inflammatory response. Artic Ital J Public Heal. 2011;8:275–85. [Google Scholar]
- 79.Cieszczyk P, Ostanek M, Leońska-Duniec A, Sawczuk M, Maciejewska A, Eider J, et al. Distribution of the AMPD1 C34T polymorphism in Polish power-oriented athletes. J Sports Sci. 2012;30:31–5. doi: 10.1080/02640414.2011.623710. [DOI] [PubMed] [Google Scholar]
- 80.Oliveira-Paula GH, Lacchini R, Tanus-Santos JE. Endothelial nitric oxide synthase: From biochemistry and gene structure to clinical implications of NOS3 polymorphisms. Gene. 2016;575(2 pt 3):584–99. doi: 10.1016/j.gene.2015.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao Y. The multiple actions of NO. Pflugers Arch. 2010;459:829–39. doi: 10.1007/s00424-009-0773-9. [DOI] [PubMed] [Google Scholar]
- 82.Martins KJB, St-Louis M, Murdoch GK, Maclean IM, Mcdonald P, Dixon WT, et al. Nitric oxide synthase inhibition prevents activity-induced calcineurin-NFATc1 signalling and fast-to-slow skeletal muscle fibre type conversions. J Physiol. 2012;590:1427–42. doi: 10.1113/jphysiol.2011.223370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suhr F, Gehlert S, Grau M, Bloch W. Skeletal muscle function during exercise-fine-tuning of diverse subsystems by nitric oxide. Int J Mol Sci. 2013;14(4):7109–39. doi: 10.3390/ijms14047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nadaud S, Bonnardeaux A, Lathrop M, Soubrier F. Gene structure, polymorphism and mapping of the human endothelial nitric oxide synthase gene. Biochem Biophys Res Commun. 1994;198:1027–33. doi: 10.1006/bbrc.1994.1146. [DOI] [PubMed] [Google Scholar]
- 85.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357(pt 3):593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakayama M, Yasue H, Yoshimura M, Shimasaki Y, Kugiyama K, Ogawa H, et al. T-786 → C mutation in the 5’-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation. 1999;99:2864–70. doi: 10.1161/01.cir.99.22.2864. [DOI] [PubMed] [Google Scholar]
- 87.Silva BM, Neves FJ, Negrão M V, Alves CR, Dias RG, Alves GB, et al. Endothelial nitric oxide synthase polymorphisms and adaptation of parasympathetic modulation to exercise training. Med Sci Sports Exerc. 2011;43:1611–8. doi: 10.1249/MSS.0b013e3182152197. [DOI] [PubMed] [Google Scholar]
- 88.Drozdovska S, Dosenko V, Ilyin V, Filippov M, Kuzmina L. Allelic Polymorphism of Endothelial No-Synthase (eNOS) Association with Exercise-Induced Hypoxia Adaptation. Balt J Heal Phys Act. Walter de Gruyter GmbH. 2009;1:10–14. [Google Scholar]
- 89.Gómez-Gallego F, Ruiz J, Buxens A, Artieda M, Arteta D, Santiago C, et al. The -786 T/C polymorphism of the NOS3 gene is associated with elite performance in power sports. Eur J Appl Physiol. 2009;107:565–9. doi: 10.1007/s00421-009-1166-7. [DOI] [PubMed] [Google Scholar]
- 90.Sessa F, Chetta M, Petito A, Franzetti M, Bafunno V, Pisanelli D, et al. Gene polymorphisms and sport attitude in Italian athletes. Genet Test Mol Biomarkers. 2011;15:285–90. doi: 10.1089/gtmb.2010.0179. [DOI] [PubMed] [Google Scholar]
- 91.Eynon N, Ruiz JR, Yvert T, Santiago C, Gómez-Gallego F, Lucia A, et al. The C allele in NOS3-786 T/C polymorphism is associated with elite soccer player’s status. Int J Sports Med. 2012;33:521–4. doi: 10.1055/s-0032-1306337. [DOI] [PubMed] [Google Scholar]
- 92.Gómez-Gallego F, Ruiz JR, Buxens A, Altmäe S, Artieda M, Santiago C, et al. Are elite endurance athletes genetically predisposed to lower disease risk? Physiol Genomics. 2010;41:82–90. doi: 10.1152/physiolgenomics.00183.2009. [DOI] [PubMed] [Google Scholar]
- 93.Zmijewski P, Cięszczyk P, Ahmetov II, Gronek P, Lulińska-Kuklik E, Dornowski M, et al. The NOS3 G894T (rs1799983) and -786T/C (rs2070744) polymorphisms are associated with elite swimmer status. Biol Sport. 2018;35:313–9. doi: 10.5114/biolsport.2018.76528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, et al. Regulation of Muscle Fiber Type and Running Endurance by PPARδ. In: O’Rahilly Steve., editor. PLoS Biol. Vol. 2. Public Library of Science; 2004. p. e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pilegaard H, Richter EA. PGC-1α: Important for exercise performance? J Appl Physiol. 1985;2008:1264–5. doi: 10.1152/japplphysiol.90346.2008. [DOI] [PubMed] [Google Scholar]
- 96.Levesque C, Copeland KJ, Pattie MD, Deci EL. Intrinsic and Extrinsic Motivation. Int Encycl Educ. 2010:618–23. [Google Scholar]
- 97.Schmitt B, Flück M, Décombaz J, Kreis R, Boesch C, Wittwer M, et al. Transcriptional adaptations of lipid metabolism in tibialis anterior muscle of endurance-trained athletes. Physiol Genomics. 2004;15:148–57. doi: 10.1152/physiolgenomics.00089.2003. [DOI] [PubMed] [Google Scholar]
- 98.Pacholczyk M, Ferenc T, Kowalski J. Part II: Its mechanisms of development and its complications. 2008. Zespół metaboliczny. Część II: patogeneza zespołu metabolicznego i jego powikłań The metabolic syndrome. [PubMed] [Google Scholar]
- 99.Guerre-Millo M, Gervois P, Raspé E, Madsen L, Poulain P, Derudas B, et al. Peroxisome proliferator-activated receptor α activators improve insulin sensitivity and reduce adiposity. J Biol Chem. 2000;275:16638–42. doi: 10.1074/jbc.275.22.16638. [DOI] [PubMed] [Google Scholar]
- 100.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: From genes to physiology. Recent Prog. 2001:239–63. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 101.Tural E, Kara N, Agaoglu SA, Elbistan M, Tasmektepligil MY, Imamoglu O. PPAR-α and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Mol Biol Rep. 2014;41:5799–804. doi: 10.1007/s11033-014-3453-6. [DOI] [PubMed] [Google Scholar]
- 102.Desvergne B, Wahli W. Peroxisome Proliferator-Activated Receptors: Nuclear Control of Metabolism*. Endocr Rev. 1999;20:649–88. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 103.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137:354–66. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 104.Ahmetov II, Mozhayskaya IA, Flavell DM, Astratenkova I V, Komkova AI, Lyubaeva E V, et al. PPARalpha gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97:103–8. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 105.Lopez-Leon S, Tuvblad C, Forero DA. Sports genetics: The PPARA gene and athletes’ high ability in endurance sports. A systematic review and meta-analysis. Biol Sport. 2016:3–6. doi: 10.5604/20831862.1180170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Drozdovska SB, Dosenko VE, Ahmetov II, Ilyin VN. The association of gene polymorphisms with athlete status in Ukrainians. Biol Sport. 2013;30:163–7. doi: 10.5604/20831862.1059168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheng CF, Ku HC, Lin H. Pgc-1α as a pivotal factor in lipid and metabolic regulation. Int J Mol Sci. 2018;19(11):3447. doi: 10.3390/ijms19113447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang H, Ward WF. PGC-1α: A key regulator of energy metabolism. Am J Physiol. 2006;30(4):145–51. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 109.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 110.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coaotivator PGC-1. Nature. 2001;413:131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 111.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 112.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 113.Terada S, Tabata I. Effects of acute bouts of running and swimming exercise on PGC-1α protein expression in rat epitrochlearis and soleus muscle. Am J Physiol – Endocrinol Metab. 2004;286(2):E208–16. doi: 10.1152/ajpendo.00051.2003. [DOI] [PubMed] [Google Scholar]
- 114.Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. PGC-1α mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol. 2004;96:189–94. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- 115.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, et al. Endurance Training in Humans Leads to Fiber Type-Specific Increases in Levels of Peroxisome Proliferator-Activated Receptor-γ Coactivator-1 and Peroxisome Proliferator-Activated Receptor-α in Skeletal Muscle. Diabetes. 2003;52:2874–81. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 116.Ridderstråle M, Johansson LE, Rastam L, Lindblad U. Increased risk of obesity associated with the variant allele of the PPARGC1A Gly482Ser polymorphism in physically inactive elderly men. Diabetologia. 2006;49:496–500. doi: 10.1007/s00125-005-0129-8. [DOI] [PubMed] [Google Scholar]
- 117.Barroso I, Luan J, Sandhu MS, Franks PW, Crowley V, Schafer AJ, et al. Meta-analysis of the Gly482Ser variant in PPARGC1A in type 2 diabetes and related phenotypes. Diabetologia. 2006;49:501–5. doi: 10.1007/s00125-005-0130-2. [DOI] [PubMed] [Google Scholar]
- 118.Vimaleswaran KS, Luan J, Andersen G, Muller YL, Wheeler E, Brito EC, et al. The Gly482Ser genotype at the PPARGC1A gene and elevated blood pressure: A meta-analysis involving 13,949 individuals. J Appl Physiol. 2008;105:1352–8. doi: 10.1152/japplphysiol.90423.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maciejewska A, Sawczuk M, Cieszczyk P, Mozhayskaya IA, Ahmetov II. The PPARGC1A gene Gly482Ser in Polish and Russian athletes. J Sports Sci. 2012;30:101–13. doi: 10.1080/02640414.2011.623709. [DOI] [PubMed] [Google Scholar]
- 120.Chen Y, Wang D, Yan P, Yan S, Chang Q, Cheng Z. Meta-analyses of the association between the PPARGC1A Gly482Ser polymorphism and athletic performance. Biol Sport. 2019;36:301–9. doi: 10.5114/biolsport.2019.88752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gille H, Kowalski J, Li B, LeCouter J, Moffat B, Zioncheck TF, et al. Analysis of biological effects and signaling properties of Flt-1 (VEGFR-1) and KDR (VEGFR-2): A reassessment using novel receptor-specific vascular endothelial growth factor mutants. J Biol Chem. 2001;276:3222–30. doi: 10.1074/jbc.M002016200. [DOI] [PubMed] [Google Scholar]
- 122.Richardson RS, Wagner H, Mudaliar SRD, Henry R, Noyszewski EA, Wagner PD. Human VEGF gene expression in skeletal muscle: Effect of acute normoxic and hypoxic exercise. Am J Physiol – Hear Circ Physiol. American Physiological Society. 1999;277(6):H2247–52. doi: 10.1152/ajpheart.1999.277.6.H2247. [DOI] [PubMed] [Google Scholar]
- 123.Brutsaert T, Gavin T, Fu Z, Breen E, Tang K, Mathieu-Costello O, et al. Regional differences in expression of VEGF mRNA in rat gastrocnemius following 1 hr exercise or electrical stimulation. BMC Physiol. 2002;2:1–10. doi: 10.1186/1472-6793-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim OJ, Hong SH, Oh SH, Kim TG, Min KT, Oh D, et al. Association between VEGF polymorphisms and homocysteine levels in patients with ischemic stroke and silent brain infarction. Stroke. 2011;42:2393–402. doi: 10.1161/STROKEAHA.110.607739. [DOI] [PubMed] [Google Scholar]
- 125.Ahmetov II, Popov D V, Lyubaeva E V, Missina SS, Vinogradova OL, Hakimullina AM, et al. Association of the VEGFR2 gene His472Gln polymorphism with endurance-related phenotypes. Eur J Appl Physiol. 2009;107:95–103. doi: 10.1007/s00421-009-1105-7. [DOI] [PubMed] [Google Scholar]
- 126.Eider J, Maciejewska-Karłowska A, Sawczuk M, Ficek K, Cięszczyk P, Leońska-Duniec A, et al. The VEGFR2 gene His472Gln polymorphism in Polish endurance athletes. International Sport med Journal. 2013;14(1):29–35. [Google Scholar]
- 127.Berman Y, North KN. A gene for speed: the emerging role of alpha-actinin-3 in muscle metabolism. Physiology (Bethesda) 2010;25:250–9. doi: 10.1152/physiol.00008.2010. [DOI] [PubMed] [Google Scholar]
- 128.North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat Genet. 1999;21:353–4. doi: 10.1038/7675. [DOI] [PubMed] [Google Scholar]
- 129.MacArthur DG, Seto JT, Raftery JM, Quinlan KG, Huttley GA, Hook JW, et al. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet. 2007;39:1261–5. doi: 10.1038/ng2122. [DOI] [PubMed] [Google Scholar]
- 130.MacArthur DG, Seto JT, Chan S, Quinlan KGR, Raftery JM, Turner N, et al. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum Mol Genet. 2008;17:1076–86. doi: 10.1093/hmg/ddm380. [DOI] [PubMed] [Google Scholar]
- 131.Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73:627–31. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tharabenjasin P, Pabalan N, Jarjanazi H. Association of the ACTN3 R577X (rs1815739) polymorphism with elite power sports: A meta-analysis. PLoS One. 2019:14. doi: 10.1371/journal.pone.0217390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Domańska-Senderowska D, Szmigielska P, Snochowska A, Jastrzębski Z, Jegier A, Kiszałkiewicz J, et al. Relationships between the expression of the ACTN3 gene and explosive power of soccer players. J Hum Kinet. 2019;69:79–87. doi: 10.2478/hukin-2019-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saunders CJ, September AV, Xenophontos SL, Cariolou MA, Anastassiades LC, Noakes TD, et al. No association of the ACTN3 gene R577X polymorphism with endurance performance in Ironman Triathlons. Ann Hum Genet. 2007;71:777–81. doi: 10.1111/j.1469-1809.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 135.Lima GHO, Silva ED, Rosa JPP, Almeida SS, Correia PR, Costa C, et al. Association Between Gene ACTN3 and Basketball Position in Elite Athletes of Brazilian League. Med Sci Sport Exerc. 2015;47:424. [Google Scholar]
- 136.Chiu LL, Wu YF, Tang MT, Yu HC, Hsieh LL, Hsieh SSY. ACTN3 genotype and swimming performance in Taiwan. Int J Sports Med. 2011;32:476–80. doi: 10.1055/s-0030-1263115. [DOI] [PubMed] [Google Scholar]
- 137.Eynon N, Duarte JA, Oliveira J, Sagiv M, Yamin C, Meckel Y, et al. ACTN3 R577X polymorphism and Israeli top-level athletes. Int J Sports Med. 2009;30:695–8. doi: 10.1055/s-0029-1220731. [DOI] [PubMed] [Google Scholar]
- 138.Papadimitriou ID, Papadopoulos C, Kouvatsi A, Triantaphyllidis C. The ACTN3 gene in elite Greek track and field athletes. Int J Sports Med. 2008;29:352–5. doi: 10.1055/s-2007-965339. [DOI] [PubMed] [Google Scholar]
- 139.Paparini A, Ripani M, Giordano GD, Santoni D, Pigozzi F, Romano-Spica V. ACTN3 genotyping by real-time PCR in the Italian population and athletes. Med Sci Sports Exerc. 2007;39:810–5. doi: 10.1097/mss.0b013e3180317491. [DOI] [PubMed] [Google Scholar]
- 140.Shang X, Huang C, Chang Q, Zhang L, Huang T. Association between the ACTN3 R577X polymorphism and female endurance athletes in China. Int J Sports Med. 2010;31:913–6. doi: 10.1055/s-0030-1265176. [DOI] [PubMed] [Google Scholar]
- 141.Yang N, MacArthur DG, Wolde B, Onywera VO, Boit MK, Lau SYM-A, et al. The ACTN3 R577X polymorphism in East and West African athletes. Med Sci Sports Exerc. 2007;39:1985–8. doi: 10.1249/mss.0b013e31814844c9. [DOI] [PubMed] [Google Scholar]
- 142.Ma F, Yang Y, Li X, Zhou F, Gao C, Li M, et al. The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS One. 2013;8:e54685. doi: 10.1371/journal.pone.0054685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, et al. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013;43:803–17. doi: 10.1007/s40279-013-0059-4. [DOI] [PubMed] [Google Scholar]
- 144.Bloomfield J, Blanksby BA, Ackland TR, Elliott BC. The anatomical and physiological characteristics of pre-adolescent swimmers, tennis players and non-competitors. Aust J Sci Med Sport. 1985;17:19–23. [Google Scholar]
- 145.Wang G, Mikami E, Chiu L-L, DE Perini A, Deason M, Fuku N, et al. Association analysis of ACE and ACTN3 in elite Caucasian and East Asian swimmers. Med Sci Sports Exerc. 2013;45:892–900. doi: 10.1249/MSS.0b013e31827c501f. [DOI] [PubMed] [Google Scholar]
- 146.Ruiz JR, Santiago C, Yvert T, Muniesa C, Díaz-Ureña G, Bekendam N, et al. ACTN3 genotype in Spanish elite swimmers: no “heterozygous advantage”. Scand J Med Sci Sports. 2013;23:e162–7. doi: 10.1111/sms.12045. [DOI] [PubMed] [Google Scholar]
- 147.Ben-Zaken S, Eliakim A, Nemet D, Rabinovich M, Kassem E, Meckel Y. ACTN3 Polymorphism: Comparison Between Elite Swimmers and Runners. Sports Med Open. 2015;1(1):13. doi: 10.1186/s40798-015-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brooks GA. Intra- and extra-cellular lactate shuttles. Med Sci Sports Exerc. 2000;32:790–9. doi: 10.1097/00005768-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 149.Juel C. Symmetry and pH dependency of the lactate/proton carrier in skeletal muscle studied with rat sarcolemmal giant vesicles. Biochim Biophys Acta. 1996;1283:106–10. doi: 10.1016/0005-2736(96)00084-3. [DOI] [PubMed] [Google Scholar]
- 150.Workman P. Inhibiting the phosphoinositide 3-kinase pathway for cancer treatment. Biochem Soc Trans. 2004;32:393–6. doi: 10.1042/bst0320393. [DOI] [PubMed] [Google Scholar]
- 151.Halestrap AP, Price NT. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J. 1999;343(Pt 2):281–99. [PMC free article] [PubMed] [Google Scholar]
- 152.Bonen A. Lactate transporters (MCT proteins) in heart and skeletal muscles. Med Sci Sports Exerc. 2000;32:778–89. doi: 10.1097/00005768-200004000-00010. [DOI] [PubMed] [Google Scholar]
- 153.Juel C, Halestrap AP. Lactate transport in skeletal muscle – role and regulation of the monocarboxylate transporter. J Physiol. 1999;517(Pt 3):633–42. doi: 10.1111/j.1469-7793.1999.0633s.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cupeiro R, González-Lamuño D, Amigo T, Peinado AB, Ruiz JR, Ortega FB, et al. Influence of the MCT1-T1470A polymorphism (rs1049434) on blood lactate accumulation during different circuit weight trainings in men and women. J Sci Med Sport. 2012;15:541–7. doi: 10.1016/j.jsams.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 155.Fedotovskaya ON, Mustafina LJ, Popov DV, Vinogradova OL, Ahmetov II. A Common Polymorphism of the MCT1 Gene and Athletic Performance. Int J Sports Physiol Perform. 2014;9(1):173–80. doi: 10.1123/ijspp.2013-0026. [DOI] [PubMed] [Google Scholar]
- 156.Merezhinskaya N, Fishbein WN, Davis JI, Foellmer JW. Mutations in MCT1 cDNA in patients with symptomatic deficiency in lactate transport. Muscle Nerve. 2000;23:90–7. doi: 10.1002/(sici)1097-4598(200001)23:1<90::aid-mus12>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 157.Bonifazi M, Martelli G, Marugo L, Sardella F, Carli G. Blood lactate accumulation in top level swimmers following competition. J Sports Med Phys Fitness. J Sports Med Phys Fitness. 1993;33:13–8. [PubMed] [Google Scholar]
- 158.Kenney WL, Wilmore JH, Costill DL. Physiology of Sport and Exercise. 6th ed. Champaign, IL: Human Kinetics; 2015. [Google Scholar]
- 159.Ben-Zaken S, Eliakim A, Nemet D, Rabinovich M, Kassem E, Meckel Y. Differences in MCT1 A1470T polymorphism prevalence between runners and swimmers. Scand J Med Sci Sports. 2015;25(3):365–71. doi: 10.1111/sms.12226. [DOI] [PubMed] [Google Scholar]
- 160.Meckel Y, Machnai O, Eliakim A. Relationship among repeated sprint tests, aerobic fitness, and anaerobic fitness in elite adolescent soccer players. J Strength Cond Res. 2009;23:163–9. doi: 10.1519/JSC.0b013e31818b9651. [DOI] [PubMed] [Google Scholar]
- 161.Gullstrand L, Lawrence S. Heart rate and blood lactate response to short intermittent work at race pace in highly trained swimmers. Aust J Sci Med Sport. 1987;19:10–4. [Google Scholar]
- 162.Nakanishi Y, Kimura T, Yokoo Y. Maximal physiological responses to deep water running at thermoneutral temperature. Appl Human Sci. 1999;18:31–5. doi: 10.2114/jpa.18.31. [DOI] [PubMed] [Google Scholar]
- 163.Town GP, Bradley SS. Maximal metabolic responses of deep and shallow water running in trained runners. Med Sci Sports Exerc. 1991;23:238–41. [PubMed] [Google Scholar]
- 164.Tomasik M. Effect of hydromassage on changes in blood electrolyte and lactic acid levels and haematocrit value after maximal effort. Acta Physiol Pol. 1983;34:257–61. [PubMed] [Google Scholar]
- 165.Nakamura K, Takahashi H, Shimai S, Tanaka M. Effects of immersion in tepid bath water on recovery from fatigue after submaximal exercise in man. Ergonomics. 1996;39:257–66. doi: 10.1080/00140139608964456. [DOI] [PubMed] [Google Scholar]
- 166.Wilcock IM, Cronin JB, Hing WA. Water immersion: does it enhance recovery from exercise? Int J Sports Physiol Perform. 2006;1:195–206. doi: 10.1123/ijspp.1.3.195. [DOI] [PubMed] [Google Scholar]
- 167.Park KS, Choi JK, Park YS. Cardiovascular regulation during water immersion. Appl Human Sci. 1999;18:233–41. doi: 10.2114/jpa.18.233. [DOI] [PubMed] [Google Scholar]
- 168.Lucía A, Morán M, Zihong H, Ruiz JR. Elite athletes: are the genes the champions? Int J Sports Physiol Perform. 2010;5:98–102. doi: 10.1123/ijspp.5.1.93. [DOI] [PubMed] [Google Scholar]
- 169.Eliakim A, Brasel JA, Mohan S, Barstow TJ, Berman N, Cooper DM. Physical fitness, endurance training, and the growth hormone-insulin-like growth factor I system in adolescent females. J Clin Endocrinol Metab. 1996;81:3986–92. doi: 10.1210/jcem.81.11.8923848. [DOI] [PubMed] [Google Scholar]
- 170.Eliakim A, Scheett TP, Newcomb R, Mohan S, Cooper DM. Fitness, training, and the growth hormone-- > insulin-like growth factor I axis in prepubertal girls. J Clin Endocrinol Metab. 2001;86:2797–802. doi: 10.1210/jcem.86.6.7560. [DOI] [PubMed] [Google Scholar]
- 171.Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. Can IGF-I polymorphism affect power and endurance athletic performance? Growth Horm IGF Res. 2013;23(5):175–8. doi: 10.1016/j.ghir.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 172.Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. The combined frequency of IGF and myostatin polymorphism among track & field athletes and swimmers. Growth Horm IGF Res. 2017;32:29–32. doi: 10.1016/j.ghir.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 173.Krych-Garsztka K, Mizgajska-Wiktor H, Goździcka-Józefiak A. An Analysis of the Regulatory Region of the IGF1 Gene in Professional Athletes in Youth Sports Teams. Hum Mov. 2011;12(3) [Google Scholar]
- 174.Ben-Zaken S, Malach S, Meckel Y, Nemet D, Eliakim A. Frequency of the IGF A/G rs7136446 polymorphism and athletic performance. Acta Kinesiol Univ Tartu. 2016;22:36–49. [Google Scholar]
- 175.Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. High prevalence of the IGF2 rs680 GG polymorphism among top-level sprinters and jumpers. Growth Horm IGF Res. 2017;37:26–30. doi: 10.1016/j.ghir.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 176.O’Dell SD, Miller GJ, Cooper JA, Hindmarsh PC, Pringle PJ, Ford H, Humphries SE, Day IN. Apal polymorphism in insulin-like growth factor II (IGF2) gene and weight in middle-aged males. Int J Obes Relat Metab Disord. 1997 Sep;21(9):822–5. doi: 10.1038/sj.ijo.0800483. [DOI] [PubMed] [Google Scholar]
- 177.Gatford KL, Heinemann GK, Thompson SD, Zhang J V, Buckberry S, Owens JA, et al. Circulating IGF1 and IGF2 and SNP genotypes in men and pregnant and non-pregnant women. Endocr Connect. 2014;3:138. doi: 10.1530/EC-14-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ben Zaken S, Meckel Y, Dror N, Nemet D, Eliakim A. IGF-I and IGF-I receptor polymorphisms among elite swimmers. Pediatr Exerc Sci. 2014;26:470–6. doi: 10.1123/pes.2014-0158. [DOI] [PubMed] [Google Scholar]
- 179.Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. IGF-I receptor 275124A > C (rs1464430) polymorphism and athletic performance. J Sci Med Sport. 2015 May;18(3):323–7. doi: 10.1016/j.jsams.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 180.Maglischo E. Swimming Fastest. Human Kinetics; 2003. [Google Scholar]
- 181.Li ZB, Kollias HD, Wagner KR. Myostatin directly regulates skeletal muscle fibrosis. J Biol Chem. 2008;283:19371–8. doi: 10.1074/jbc.M802585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Morissette MR, Cook SA, Buranasombati C, Rosenberg MA, Rosenzweig A. Myostatin inhibits IGF-I-induced myotube hypertrophy through Akt. Am J Physiol Cell Physiol. 2009;297:C1124–32. doi: 10.1152/ajpcell.00043.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A. 1997;94:12457–61. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–4. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 185.Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–8. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 187.Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, Hurley BF. Frequent sequence variation in the human myostatin (GDF8) gene as a marker for analysis of muscle-related phenotypes. Genomics. 1999;62:203–7. doi: 10.1006/geno.1999.5984. [DOI] [PubMed] [Google Scholar]
- 188.Fedoruk MN, Rupert JL. Myostatin inhibition: a potential performance enhancement strategy? Scand J Med Sci Sports. 2008;18:123–31. doi: 10.1111/j.1600-0838.2007.00759.x. [DOI] [PubMed] [Google Scholar]
- 189.Kostek MA, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, et al. Myostatin and follistatin polymorphisms interact with muscle phenotypes and ethnicity. Med Sci Sports Exerc. 2009;41:1063–71. doi: 10.1249/MSS.0b013e3181930337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Corsi AM, Ferrucci L, Gozzini A, Tanini A, Brandi ML. Myostatin polymorphisms and age-related sarcopenia in the Italian population. J Am Geriatr Soc. 2002;50:1463. doi: 10.1046/j.1532-5415.2002.50376.x. [DOI] [PubMed] [Google Scholar]
- 191.Ben-Zaken S, Meckel Y, Nemet D, Rabinovich M, Kassem E, Eliakim A. Frequency of the MSTN Lys(K)-153Arg(R) polymorphism among track & field athletes and swimmers. Growth Horm IGF Res. 2015;25:196–200. doi: 10.1016/j.ghir.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 192.Galimudi RK, Spurthi MK, Padala C, Kumar KG, Mudigonda S, Reddy SG, et al. Interleukin 6(-174G/C) variant and its circulating levels in coronary artery disease patients and their first degree relatives. Inflammation. 2014;37:314–21. doi: 10.1007/s10753-013-9742-8. [DOI] [PubMed] [Google Scholar]
- 193.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–44. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 195.Bennermo M, Held C, Stemme S, Ericsson C-G, Silveira A, Green F, et al. Genetic predisposition of the interleukin-6 response to inflammation: implications for a variety of major diseases? Clin Chem. 2004;50:2136–40. doi: 10.1373/clinchem.2004.037531. [DOI] [PubMed] [Google Scholar]
- 196.Yamin C, Duarte JAR, Oliveira JMF, Amir O, Sagiv M, Eynon N, et al. IL6 (-174) and TNFA (-308) promoter polymorphisms are associated with systemic creatine kinase response to eccentric exercise. Eur J Appl Physiol. 2008;104:579–86. doi: 10.1007/s00421-008-0728-4. [DOI] [PubMed] [Google Scholar]
- 197.Funghetto SS, Prestes J, Silva A de O, Farias DL, Teixeira TG, Vieira DCL, et al. Interleukin-6 -174G/C gene polymorphism affects muscle damage response to acute eccentric resistance exercise in elderly obese women. Exp Gerontol. 2013;48:1255–9. doi: 10.1016/j.exger.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 198.Ben-Zaken S, Meckel Y, Nemet D, Kassem E, Eliakim A. Increased Prevalence of the IL-6 -174C Genetic Polymorphism in Long Distance Swimmers. J Hum Kinet. 2017;58:121–30. doi: 10.1515/hukin-2017-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Vasile L. Endurance Training in Performance Swimming. Procedia – Soc Behav Sci. Elsevier BV. 2014;117:232–7. [Google Scholar]
- 200.Hausswirth C, Lehénaff D. Growth Horm IGF Res. Springer; 2001. Physiological demands of running during long distance runs and triathlons; pp. 679–89. [DOI] [PubMed] [Google Scholar]
- 201.Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. The combined frequency of IGF and myostatin polymorphism among track & field athletes and swimmers. Growth Horm IGF Res. 2017;32:29–32. doi: 10.1016/j.ghir.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 202.Crowley E, Harrison AJ, Lyons M. The Impact of Resistance Training on Swimming Performance: A Systematic Review. Springer International Publishing; 2017. pp. 2285–307. [DOI] [PubMed] [Google Scholar]