Abstract
MicroRNAs are important coordinators of circadian regulation that mediate the fine-tuning of gene expression. Although many studies have shown the effects of individual miRNAs on the circadian clock, the global functional miRNA-mRNA interaction network involved in the circadian system remains poorly understood. Here, we used CLEAR (Covalent Ligation of Endogenous Argonaute-bound RNAs)-CLIP (Cross-Linking and Immuno-Precipitation) to explore the regulatory functions of miRNAs in the circadian system by comparing the miRNA-mRNA interactions between Drosophila wild-type strain W1118 and a mutant of the key circadian transcriptional regulator Clock (Clkjrk). This experimental approach unambiguously identified tens of thousands of miRNA-mRNA interactions in both the head and body. The miRNA-mRNA interactome showed dramatic changes in the Clkjrk flies. Particularly, among ~300 miRNA-mRNA circadian relevant interactions, multiple interactions involving core clock genes pdp1, tim and vri displayed distinct changes as a result of the Clk mutation. Based on the CLEAR-CLIP analysis, we found a novel regulation of the circadian rhythm and sleep by the miR-375-timeless interaction. The results indicated that Clk disruption abolished normal rhythmic expression of miR-375 and the functional regulation occurred in the l-LNv neurons, where miR-375 modulated circadian rhythm and sleep via targeting timeless. This work provides the first global view of miRNA regulation in the circadian rhythm.
Keywords: CLEAR-CLIP, miRNAs, circadian rhythm, sleep, miR-375
Introduction
In animals, the intrinsic circadian clock regulates daily rhythms in physiology and behavior, which are entrained by environmental stimuli such as light and temperature (1, 2). This robust timing system maintains rhythmic oscillation even under constant darkness conditions in the fruit fly Drosophila melanogaster.
Sleep is one of the established circadian behavior, which is highly associated with health status (3). Sleep in Drosophila has been well characterized by certain parameters such as total sleep, sleep bout duration, and sleep bout number (1, 4, 5). Throughout the daily sleep-wake cycle, the flies exhibit two peaks of activity - one before lights on and one before lights off. Genetic dissection in Drosophila indicates that sleep is regulated by the circadian clock. The mutations of the core clock genes result in abnormal sleep (6, 7), while the peptidergic clock neurons that produce the neuropeptide pigment dispersing factor (PDF) regulate arousal as well as sleep stability (8).
The Drosophila rhythmic behavior is maintained by a central clock network in the brain, which is consisted of ~150 circadian neurons (9). These clock neurons include ventral lateral neurons (LNvs) (I-LNvs, s-LNvs, and 5th s-LNvs), dorsal lateral neurons (LNds), lateral posterior neurons (LPN) and dorsal neurons (DN1, DN2, and DN3), classified based on anatomical locations and varied expression of core clock genes (9–12). Among them, the PDF-positive neurons (I-LNvs and s-LNvs) are essential for normal circadian activity and daily sleep. Their ablation results in arrhythmic flies (4).
The circadian clocks are modulated by a complex transcriptional and translational feedback loop (TTFL) in both mammals and flies (1, 5, 6). In D. melanogaster, two interlocked clock feedback loops are known to regulate the circadian rhythm. They consist of period (per), timeless (tim), clock (clk), cycle (cyc), vrille (vri) and par domain protein 1e (PDP1e). The transcription factors CLK and CYC form a heterodimer that binds to the E-box of per and tim promoters, thereby promoting their transcription (7, 8). The cycling expression of tim mRNA with a peak at Zeitgeber time (ZT) 14 and a trough at ZT 2 is vital for circadian output (9, 10). Another feedback loop consists of the clk, cyc, vri, and PDP1e (7, 8, 11–13). However, the mechanism of how these feedback loops sustain ~24 h rhythm is still unclear, since multi-layered regulation involving post-transcriptional and post-translational regulation is imposed to produce the daily rhythm (14–16).
The highly conserved and widespread microRNAs (miRNAs) play a critical role in post-transcriptional gene regulation (17–21). They are loaded into Argonaute (AGO) proteins to mediate mRNA cleavage or translational inhibition via base-pairing with target mRNAs (19, 22–24). Disruption of the miRNA biogenesis pathway considerably weakens the rhythm in locomotor activity (25). Although the rhythmically expressed miRNAs have been identified (26–28), only a limited set of miRNA-target pairs have been verified to affect circadian rhythms (25, 29–32). The decoding of the miRNA-mRNA interaction network is difficult because of incomplete complementary pairing between miRNAs and targets as well as context-dependent dynamics of the miRNA-target interactions. As a result, some miRNAs such as miR-124 and miR-959-964 have been shown to be involved in circadian rhythms but without known targets (28, 33, 34). Thus, global identification of circadian-related miRNA-target interactions will greatly improve our understanding of the role of miRNAs in circadian regulation.
The effort to accurately predict miRNA targets is compromised by the complex and dynamic cellular interaction networks (35, 36). Immunoprecipitation of the miRNA effector protein Argonaute followed by microarray (25) or RNA-seq analysis (37) narrows down the search scope of potential targets but still relies on bioinformatics analysis to predict miRNA-mRNA pairing. The recently improved experimental method of the CLEAR (Covalent Ligation of Endogenous Argonaute-bound RNAs)–CLIP (Cross-Linking and Immuno-Precipitation), which isolates miRNA-mRNA chimeras from endogenous AGO-miRNA-mRNA complexes, permits unambiguous identification of miRNAs’ targets and provides a snapshot of true, physiological miRNA-mRNA interactions in vivo (23, 38–40).
To systematically investigate the role of miRNAs in circadian rhythms, we used the CLEAR-CLIP assay in Drosophila. Thousands of miRNA-mRNA interactions were detected highlighting the well-established miRNAs targeting features. Our data indicate that miRNAs extensively fine-tune the expression of circadian-relevant genes. In addition, miRNA-target chimeras revealed Clk-dependent miRNA-mRNA interaction patterns and the miRNA changes induced by Clk disruption may lead to the alteration of miRNA-mRNA interactome. Importantly, we show here that the regulation of tim by miR-375 plays an important role in locomotor activity and sleep. This work provides a global view of the miRNA-mRNA interactions in circadian rhythms and a solid foundation for future functional studies.
Materials and Methods
Fly strains
Fly strains were maintained on standard molasses-cornmeal-yeast food in a 12L:12D cycle at 25°C and 60% humidity. The fly lines were used in this study as follows: w1118, ClkJrk, USA-LUC-miR-305/TM3, UAS-LUC-miR-275.mir-305, UAS-LUC-miR-375, UAS-LUC-miR-9c, UAS-Mcd8::GFP, TI{GAL4}miR-375[KO], C929-gal4,w1118; pdf-gal4 which were purchased from the Bloomington Drosophila Stock Center. The iso:tim-gal4 was obtained from Yirao’s lab, UAS-tim2-5 from Jeffrey L. Price’s lab and was originally generated by Amita Sehgal’s lab.
Ago1 CLEAR-CLIP library construction
The protocol was adapted from previous reports (23, 38). The flies (3–5d) were collected at ZT3 (Zeitgeber time) and frozen in liquid nitrogen. The heads were separated from the rest of body by rigorous vibrating and collected using a colander. The collected tissue samples were ground with a mortar and pestle in liquid nitrogen before ultraviolet irradiation (Fig. 1). The purified Ago1-RNA complexes from Drosophila lysates were subjected to stringent wash to reduce nonspecific binding. T4 RNA ligase 1 was added into the complex to promote the formation of miRNA-target chimeras. To track the size of the Ago1-RNA complex, RNA was labeled through ligation with a 32P-labeled 3’ DNA linker. A subsequent size selection on an SDS polyacrylamide gel was used to isolate the Ago1-miRNA-mRNA complexes (>130 kDa) and recover authentic target RNAs for library construction and sequencing.
Fig. 1. Transcriptome-wide miRNA-mRNA interactome in Drosophila.
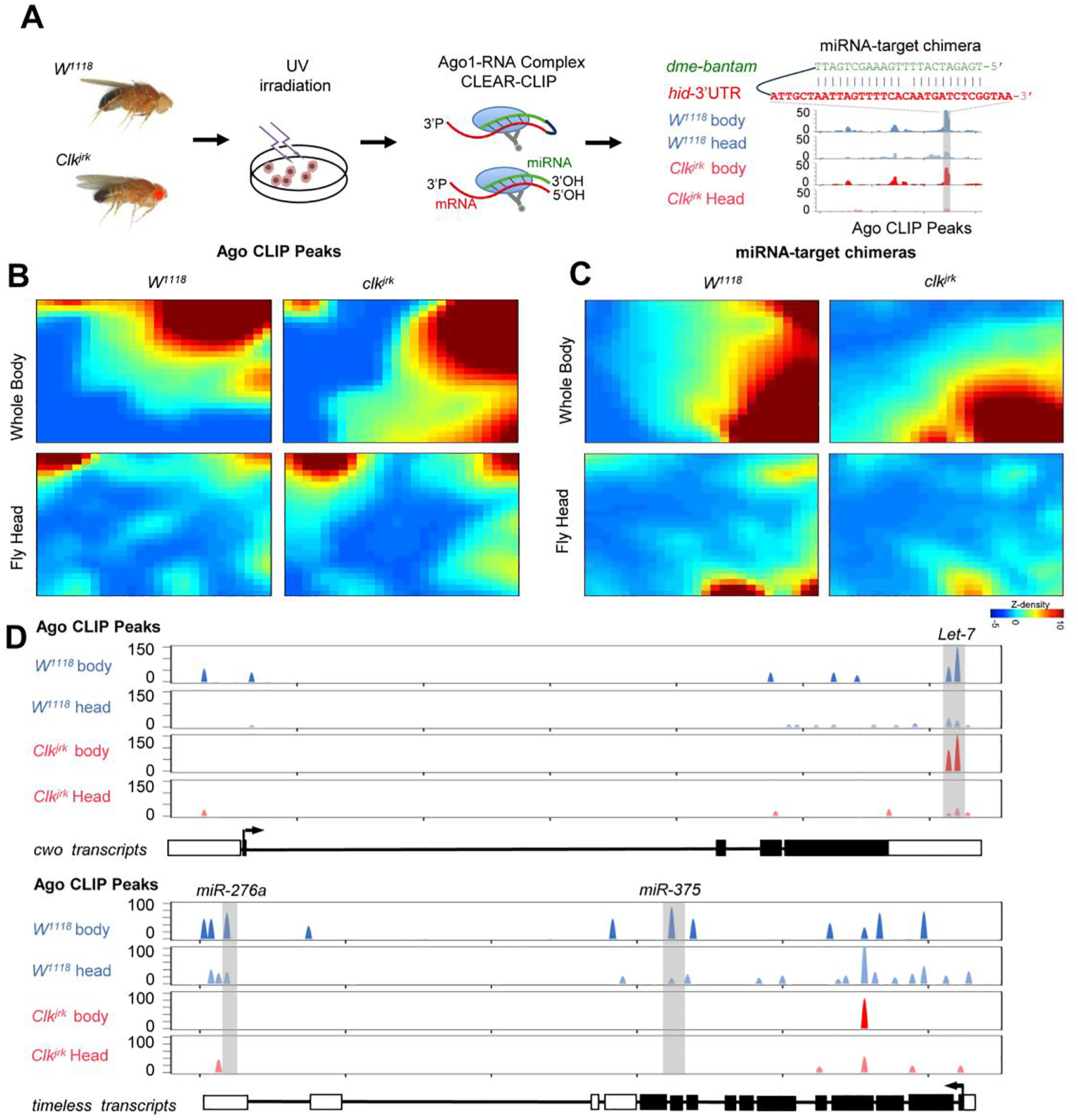
(A) Schematics of CLEAR-CLIP to detect miRNA-mRNA interactions in Drosophila. Heads or whole body flies were collected from wild-type W1118 and Clk mutant clkjrk flies with arrhythmic phenotype. The UV-crosslinked sample was homogenized in lysis buffer. Ago1-RNA complex was immunoprecipitated and incubated with T4 RNA ligase to promote the direct ligation of miRNA with its target mRNA fragments. The recovered RNAs were subjected to library construction and sequencing. After bioinformatics analysis, chimeric reads could be identified along with the Ago1 binding sites (CLIP peaks), exemplified by the known regulation of hid by bantam. (B) Globally distinct Ago1 CLIP peaks in the wild type W1118 and Clkjrk mutant flies. The interaction data were clustered and transformed into a two-dimensional self-organizing map for individual conditions. Each tile at the same position in all the maps corresponds to a group of miRNA-target interactions that share similar patterns. The color scale denotes the heights of Ago1 CLIP peaks. The four panels were generated together as described by Eichler et al.(2003). The color scales have already been normalized. (C) Globally distinct miRNA-target chimeras that were supported by Ago1 CLIP peaks. (D) Distribution of Ago1 CLIP peaks along the timeless transcripts. The regulation of timeless by miR-276a has been reported by Chen et al. (2016).
Behavior analysis
Adult male flies (2–5 d old) were used to test locomotor activity rhythms. Flies were entrained under LD for 3 d and released into constant darkness (DD) for at least 6 d at 25°C. Locomotor activity was recorded with Drosophila activity monitors (Trikinetics). Sleep was analyzed by using pysolo and GraphPad software. FaasX software was used to analyze behavioral data. Locomotor behavior was analyzed in MATLAB. The details for the experimental protocol and data analysis were described by Chen et al. (29).
Confocal microscopy
Adult male flies of 5–10 days were collected and their brains were dissected in phosphate-buffered saline (PBS). The brains were subjected to 4% paraformaldehyde in PBS for 1 h and then washed three times with the wash buffer (0.05% Triton X-100 in PBS) for 10 min at room temperature. Samples were transferred to Blocking buffer (2% Triton-100, 10% normal goat serum in PBS) for overnight incubation at 4 °C and incubated with primary antibodies (diluted in blocking buffer) overnight at 4°C. The following primary antibodies were diluted in the blocking buffer: rat-anti-TIM (1:1000, from Jeffrey L. Price), mouse-anti-PDF (1:400, from DSHB). After washing samples three times for 15 min at room temperature, the samples were incubated with secondary antibody (1:200) at 4°C overnight. The samples were imaged on a Leica SP8 confocal microscope. ImageJ software was used for TIM and PDF quantification.
RNA extraction and qRT-PCR analysis
Fly heads were collected at the indicated time points and stored at −80°C before RNA extraction. Total RNA was extracted from the heads by using Trizol reagent (TIANGEN) following the supplier’s instruction. For reverse transcription and real-time PCR of timeless, we used the PrimeScript RT reagent kit with gDNA Eraser (TAKARA) and superreal premix plus (SYBR Green) (TIANGEN). The sequences of primers are shown in Table S2. For quantitative analysis of timeless, miR-375, and 2s rRNA, we used a miRcute miRNA first-strand cDNA synthesis kit and a miRcute miRNA qRT-PCR detection kit (SYBR Green) (TIANGEN). The miRNA-specific forward primers used for qRT-PCR are also shown in Table S2.
Western blot
Fly heads were collected at the indicated time points and homogenized in a 1.5 ml microtube with RIPA lysis buffer (strong) supplemented with proteinase inhibitors. For immunoblot analysis, proteins were transferred to PVDF membrane. After blocking, the membrane was incubated with actin antibody (1:10000) and TIM antibody (1:4000) for 2 h at room temperature. Image J was used to calculate band intensity.
Ago1 immunoprecipitation
Fly heads were homogenized in 500 μl lysis buffer. The lysates were incubated on ice for 10 min and then centrifuged at 13000 rpm for 30 min, the supernatant was collected. Protein A Dynabeads (Invitrogen) were washed twice and resuspended in Tris-PNK buffer (20 mM tris PH 7.5, 10 mM MgCl2, 50 mM NaCl, 0.5% NP-40). For each sample, 20 μL of beads were used. Beads were rotated with 4 μg of Ago1 antibody (ab5070, abcam) in Tris-PNK buffer for 1 h at 4 °C. Beads were washed twice in Tris-PNK buffer. The input and antibody-bound beads were incubated for 2 h at 4°C, then washed twice in Tris-PNK buffer and followed by RNA extraction.
Statistics analysis
Statistics analysis for all indicated data in this study were performed with t-test and p values were considered significant at *p < 0.05, **p < 0.01 and ***p < 0.001.
Bioinformatic analysis of miRNA-mRNA chimeras
Ago1 CLEAR-CLIP sequencing reads were preprocessed to remove low-quality reads and 3’ adapter sequences using Flexbar with the setting -ao 4-u 3-m 16-n6 (41). Shorter reads (less than 16 nt) were discarded. Reads with identical sequences were collapsed. The unique molecular identifiers (UMIs) bearing in the 5’end were also trimmed before mapping using Flexbar with the setting -umi-tags.
To identify the chimeric reads, we first defined the miRNA part through reverse-mapping mature Drosophila miRNA sequences (miRbase) against sample libraries using BLAST (v2.3.0) with an e-value cutoff at 0.4 (42). The non-miRNA sequences in the chimeric reads were extracted for mapping to Drosophila transcripts using BLAST with e-value cutoff equal to 0.2. The unique aligned reads were retained. For reads that were mapped to more than one transcript with the same e-value, we assigned the read to the transcript with most BLAST hits as described by Helwak et al. (43). Those reads mapped to rRNA, tRNA, and miRNA genes were removed. The reads containing both mature miRNA and mRNA with less than 4-nt gap were retained as chimeras for potential interactions (38).
Peak calling of CLIP cluster
We followed the analysis pipeline as previously reported (37, 44). The background was estimated using Poisson distribution with lambda equal to 1+z/(45*L). Z was the total reads mapped to the Drosophila transcript; L was the total length of the transcript; 45 was the expected Ago1 footprint length. We then compared the mapped cluster to the background and used p-values to determine the existence of Ago binding peaks. At the same time, we used the Cubic spline interpolation to calculate the peak height and the corresponding locus. When Ago1 CLIP peaks were compared, the p-value was the first filter and we only kept peaks with p <0.01.
Dynamic analysis of miRNA-mRNA interactions
To quantitatively analyze the profile of miRNA-mRNA interactions, the chimera-supported Ago1 binding peaks were normalized to the total unique reads (Tags per million) in each condition (45). In order to visualize the pattern of difference between each time point, the data were loaded into Gene Expression Dynamics Inspector (GEDIv2.1) with the default parameter (26×25 grids) to perform the dynamic analysis (46). GEDI employs the SOM algorithm to assign the interactions with similar trends into close tiles.
Motif analysis
The analysis of overrepresented motifs of Ago1 CLEAR-CLIP (Fig. S2A, Fig. S3A) was performed using MEME on the whole chimeras data from head and body as previously described (23) with the settings: -dna -mod zoops -max w7 -n motifs 1. FIMO was used to position the enriched motifs against miRNAs’ sequence (47), with the setting-output-p thresh 0.01.
RNA duplex structure prediction and binding free energy
The predictions of duplex structure for miRNAs and their target sequences were performed on the whole chimeras data from heads and bodies by RNAhybrid (48). Mutations in chimeras sequences for both miRNA and mRNA were omitted by comparing them with the original annotation. The target sequences were extended to 100 nt according to the miRNA seed site distribution in D. melanogaster. The setting of RNAhybrid was modified as previously described to favor the seed pairing (23). The minimum free energy of each interaction was calculated by RNAhybrid with default settings (48). The shuffled interactions served as controls for the comparison of free energy distribution (Fig. 2B). The median free energy for each profile was calculated in R for comparison.
Fig. 2. Validation of the miRNA-target interactions identified by CLEAR-CLIP.
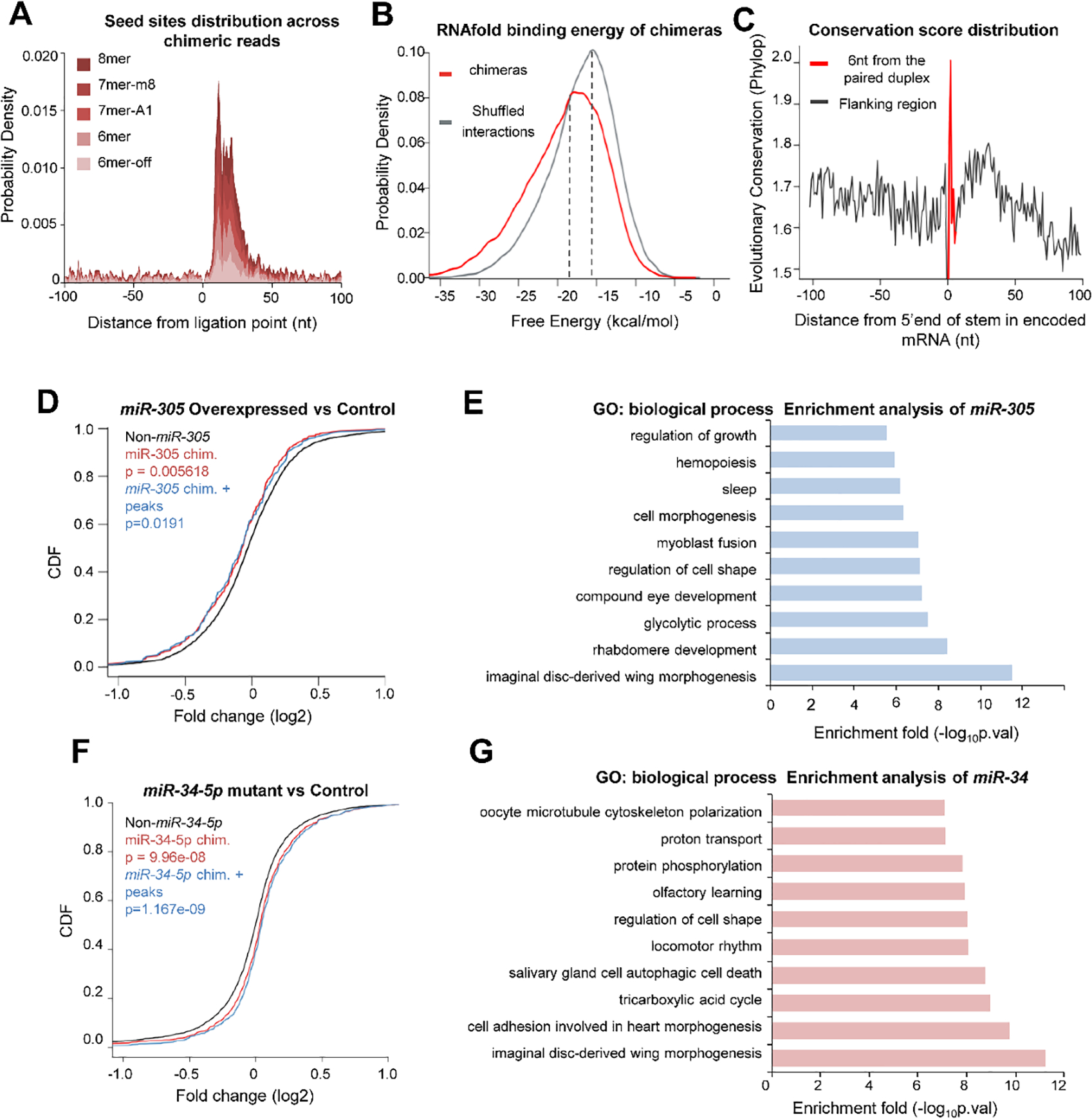
(A) Density plot of canonical miRNA seed matches in target regions relative to the ligation site for all the chimeras. (B) The median predicted binding energy between miRNAs and matching target mRNAs found in chimeras was 2.8 kcal mol−1 lower than in randomly matched pairs. (C) Average conservation score of seed matches in the targets identified from the chimeras. The Phylop conservation score for 27 insect species was downloaded from UCSC. (D) The effect of miR-305 overexpression on the expression of its target genes. The analysis was performed using the transcriptome measurement in the miR-305-overexpressed flies versus the wild type control. The cumulative distribution function (CDF) plot shows log2-transformed expression differences for mRNAs that were identified in the miR-305 chimeras and supported by Ago1 CLIP peaks, compared with other mRNAs that were not defined as miR-305 targets. P-values were calculated from Kolmogorov–Smirnov testing. (E) GO enrichment analysis of the chimeras-defined targets of miR-305. The top ten biological process terms were plotted. (F) CDF plot shows the change in transcript levels of the target genes after knocking out miR-34-5p. (G) GO enrichment analysis of the chimeras-defined targets of miR-34.
Analysis of chimera targets in miRNA perturbation experiments
The gene expression data after miRNA perturbation was downloaded (49, 50). The log2 fold-change was used to perform cumulative distribution function (CDF) analysis comparing transcripts bearing the whole chimeras-derived miRNA target sites (head and bodies) versus those without target sites. The CDFs were also plotted for transcripts in which the chimera-derived miRNA target sites overlap with Ago1 CLIP peaks (Fig. 2D, F). We used the Kolmogorov–Smirnov test in R to evaluate the statistical significance between groups.
GO and KEGG enrichment analysis
The selected gene list was taken as an input for the functional enrichment analysis with DAVID (51). The top-ranked GO terms of biological functions were chosen. Heatmaps with the enriched p-value of the GO terms were created using R heatmap package. The circadian relevant genes were defined by the described circadian rhythm defective phenotypes in the FlyBase.
Data availability
CLEAR-CLIP sequence data are available at the NCBI SRA database (Accession Number: SRP148458). The scripts for the bioinformatic analysis were uploaded to Github (https://github.com/XiaonanFu/zhulab).
Results
Transcriptome-wide miRNA-mRNA interactome in Drosophila
The brain clock neurons are the regulatory center of circadian rhythm (4), in which the transcription factor Clk is essentially required for rhythmic behavior. The ablation of Clk expression in the mutant Clkjrk makes the oscillation of miRNAs disappear in post-transcriptional regulation (28). To uncover the physiological miRNA-mRNA interactions in response to the circadian changes, the head from both Clkjrk and w1118 flies were separately analyzed by a modified Ago1 CLEAR-CLIP (Fig. 1A, Fig. S1). Although CLK is also known to regulate many non-cycling genes, a large portion of the miRNA-mRNA interactions that are affected in the head by the Clk mutation is expected to be relevant to circadian rhythms. The identified miRNA-mediated gene regulations will await further functional studies to assess their actual contributions to circadian control. The inclusion of the whole body in this study is to validate the interactions discovered in the heads and also capture additional interactions that may take place only in the thorax and abdomen (Fig. 1A, Fig. S1). RNAs in the purified Ago1-miRNA-mRNA complexes were either self-ligated as chimeric reads or remain separated mapping to a single locus (CLIP peaks). In total, we obtained 213,404,300 reads from 8 libraries (Table S1A), of which 88.32% could be mapped to mRNAs and 1.92% were miRNA-target chimeras. The recovery of well-established miRNA-mRNA interactions in Drosophila, exemplified by the verified regulation of hid by bantam (52) (Fig. 1A), lent support to the reliability of the obtained dataset (Table S3B).
In this study, we identified 45,597 unique Ago1 CLIP peaks in total and recovered 15,825 miRNA-target interactions involving 239 mature miRNAs and 6,540 mRNA transcripts in Drosophila (Table S3). To explore the whole miRNA-mRNA interaction networks in the circadian system, we first visualized the Ago1 CLIP peaks with SOM (Self-Organized Map) clustering (Fig. 1B). The SOM algorithm groups together interactions with similar patterns. Each tile at the same position in all the maps corresponds to a group of miRNA-target interactions that share similar patterns. The Ago1 binding profiles presented considerable pattern changes in Clkjrk and w1118 flies, with around 80% of them displayed more than 2-fold changes in the head or whole body (Fig.1B). Moreover, 9,325 Ago1 CLIP peaks supported by miRNA-target chimeras also showed shifted interaction patterns in Clkjrk and w1118 flies, implying widespread circadian impact by CLK (Fig. 1C).
Among the Ago1 CLIP peaks, we found multiple peaks in the core clock components tim and cwo, where they displayed different context-dependent patterns. The Ago1 CLIP peaks in the 3’UTR of cwo coincided with the previously verified let-7 binding sites (29), appearing in all the conditions (Fig. 1D). In contrast, multiple miRNA-timeless interactions disappeared in Clkjrk flies, including the well-established miR-276: timeless (30) (Fig. 1D). These results showed distinct miRNA regulatory patterns with a high-resolution transcriptome-wide miRNA-target interaction map.
Context-specific Ago1 CLIP peaks under Clk disruption
To further evaluate the impact of Clk disruption on miRNA-mRNA interactome, we analyzed the Ago1 binding peaks in the head and whole body. We defined Clk-dependent and independent peaks at different conditions (Fig. S2A). Interestingly, the scatter plot of Ago1 binding peaks (Clkjrk vs W1118) yielded 9 clearly separated groups with a 2-fold change as the cutoff. Around 80% of the Ago1 binding peaks were classified in the differential groups, in which 12% and 17% of the Ago1 binding peaks were decreased, while 20% and 31% of those were increased in the fly body and the head, respectively. It is conceivable that not all the alterations were related to the circadian rhythm. Nevertheless, this observation implied that considerable miRNA-mRNA interactions in the head are under the control of CLK.
To assess the functional impact of Clk changes on the miRNAs-regulated genes, we calculated the enrichment p-values according to gene ontology (GO) (Fig. S2B). The top enriched GO terms from all groups were related to circadian rhythms, such as “locomotor rhythm”, “phototransduction”, and “sleep”, suggesting that Clk governs the miRNA regulation of the circadian system. Body-specific enriched terms were more likely to be “cell cycle” and “germ cell development”. In contrast, we observed striking head-specific enriched terms associated with circadian regulation (“adult locomotory behavior” and “response to light stimulus”). Collectively, these results showed that the miRNA-mRNA interaction network was altered as a result of CLK malfunction.
miRNA-target chimeras preserve physiological relevant interactions
The Ago1 binding peaks were just one part of the miRNA-mRNA interaction network. We then turned to the recovered miRNA-mRNA chimeric reads and conducted bioinformatics analyses of the chimeras from heads and whole bodies. The target sequences in the chimeras were enriched with canonical seeds which matched within 50 nt of the ligation sites (Fig. 2A). The miRNAs and targets in the chimeras normally formed stable RNA duplexes. The mean of predicted free energy between miRNAs and the matched target mRNAs found in chimeras was lower by 2.8 kcal mol−1 than that in randomly matched pairs (p<0.001) (Fig. 2B). In addition, we found that the chimera-defined miRNA target sites were generally more evolutionarily conserved in different insect species than their flanking sequences in 3’ UTR (Fig. 2C), implying the functional importance of those sites. These data collectively indicated that the CLEAR-CLIP chimeras define a reliable and high-resolution miRNA-target interaction map.
To evaluate the physiological functions of miRNA-target interactions, we analyzed the identified targets of let-7-5p (29), miR-34-5p (49), miR-305-5p (50), and miR-276a (30), which have been previously investigated in circadian regulation. The enriched 7-mer motifs in the targets of these miRNA were reverse complementary to the miRNA seed regions (Fig. S3). In the CLEAR-CLIP dataset, miR-305 bound to 598 target sites within 555 mRNA transcripts. Overexpression of miR-305 in Drosophila considerably lowered the abundance of mRNAs bearing the chimeras-defined miR-305 target sites, compared with the mRNAs that had no miR-305 target site (p=0.005618). The chimeras-defined miR-305 targets that were supported by Ago1 CLIP peaks were also significantly repressed (p=0.0191) by the miR-305 overexpression (Fig. 2D). Functional annotation of the miR-305 targets implicated miR-305 in the control of sleep (Fig. 2E), which has been observed in the miR-305 mutant strain (50). Similarly, the mRNA levels of the miR-34 targets were significantly elevated in the miR-34 (miR-34-5p) null mutant, compared with w1118 (p<0.001) (Fig. 2F). In this study, GO enrichment analysis of the chimera-defined miR-34 targets linked miR-34 to aging and brain disease (Fig. 2G), consistent with the previous report that miR-34 is associated with aging and neurodegeneration (49). These results thus validated the physiological significance of the miRNA-mRNA interactions identified by CLEAR-CLIP.
miRNA-target chimeras reveal Clk-dependent miRNA-mRNA interaction patterns
With the guide of miRNA-mRNA chimeras, we constructed the miRNA-mRNA interaction network implicated in the circadian phenotype. The 299 miRNA-mRNA interactions involving 9 core circadian genes were plotted as a miRNA-regulated network (Fig. S4A), in which 35 interactions appeared to target the three Clk-downstream genes pdp1, vri, and tim (Fig. S4C).
To compare the miRNA regulations of these three genes, we retrieved the interaction patterns under different conditions. Results displayed distinct regulatory features in response to Clk disruption (Fig. 3A). The miRNAs targeting pdp1 acted in a Clk-independent manner. Conversely, the miRNA-tim interactions that occurred in w1118 were mostly not detectable in Clkjrk, with exceptions of miR-193-3p and miR-2c-3p. Our data indicated that miR-375-3p and miR-305-5p only targeted tim in wild type flies (Fig. 3B), and miR-305-5p had been reported to modulate the expression of tim in several circadian screens (25, 54, 55). Binding of miRNAs to the vri transcript exhibited a mixed pattern of both Clk-dependence and Clk-independence. These findings suggested that gene regulation by miRNAs has a profound impact on circadian system stability.
Fig. 3. Clk disruption is associated with the changes of miRNA-mRNA interactome.
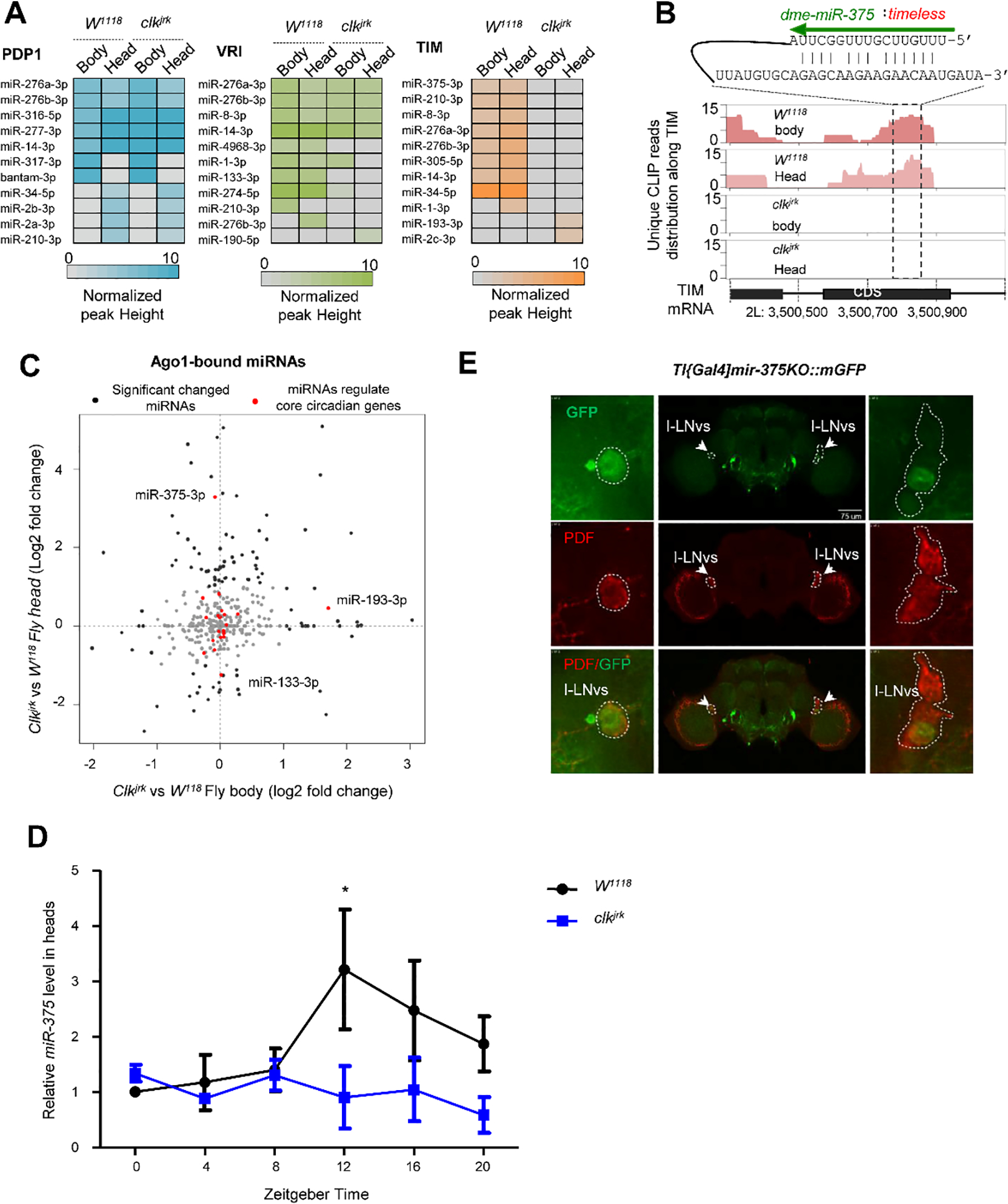
(A) The heatmap shows the effect of Clk disruption on the miRNA interactions with the transcripts of pdp1, tim, and vir. (B) The Clk-dependent interaction of miR-375 and timeless. The duplex defined in the chimeras was predicted by RNAhybrid. (C) Comparison of the Ago1-bound miRNA abundance in the head and whole body between the Clkjrk and W1118 flies. (D) The RNA levels of miR-375, measured by qRT-PCR, were reduced in the head of Clkjrk flies, compared with W1118 flies. (E) miR-375 was expressed in PDF neurons, which were detected by the GFP reporter driven by tim-gal4 (w*; TI{GAL4}miR-375KO×UAS-mGFP). The images in the insets were from the brain shown in the middle. The left panel reflects the LNv neurons in the left hemisphere, and the right panel reflects the LNv neurons in the right hemisphere. These images were obtained from the z-stack of confocal microscopy images.
MiR-375 expression oscillations and location
The Ago1 binding peaks presented dramatic shifts in Clkjrk strain. To test whether miRNA expression was affected by the Clk mutation, we analyzed the sequence reads of mature miRNAs from the CLEAR-CLIP dataset, which have been suggested as a reliable abundance measurement of functional miRNAs (56). Our results showed that miR-375 substantially increased with a 3.28 log fold in the head of ClkJrk compare to W1118. (Fig. 3C). We further quantified the daily expression of miR-375-3p in the fly head by qRT-PCR at six time points (ZT0, 4, 8, 12, 16, 20) under LD condition. miR-375-3p was rhythmically expressed with its peak at ZT12 (3-fold higher than ZT0, p<0.05) in w1118, but it disappeared in Clkjrk (Fig. 3D), implying that its expression is under the control of Clk. To investigate the spatial expression of miR-375-3p, we utilized a reporter strain (TI{GAL4}miR-375[KO] ×UAS-Mcd8::GFP) to monitor the expression of miR-375. The results showed that it was expressed in the PDF-expressing I-LNv neurons and co-localized with TIM (Fig. 3E).
Overexpression of miR-375 affects normal circadian rhythm and sleep
To investigate the role of miR-375 in circadian behavior, we overexpressed miR-375 in tim neurons by using iso:tim-gal4 driver. The amount of miR-375 in the Ago1-RNA complex was significantly higher (p<0.001) in the iso:tim-gal4/UAS-LUC-miR-375 flies than the control (Fig. S5). The flies with overexpression of miR-375 in tim neurons lost daily locomotor rhythm (100% arrhythmic) under both LD and DD conditions (iso:tim-gal4/UAS-LUC-miR-375: 0% rhythmic, n=65). Both morning and evening anticipations disappeared in the overexpression flies (Table 1, Fig. 4A–C), while control flies exhibited normal behavioral rhythms (UAS-LUC-miR-375/+: 95.74% rhythmic, tau=24.5 h, n=47; iso:tim-gal4/+: 100% rhythmic, tau=24.6 h, n=75). In addition, flies with overexpressed miR-375 showed significant increases in total sleep at daytime caused by sleep bout number (p<0.001) and decreases at nighttime caused by sleep bout duration (p<0.001), compared with the control (Fig. 4D & E). Overexpression of miR-375 in pdf neurons (pdf-gal4 × uas-miR-375) caused 43.3% of the flies to become arrhythmic (p<0.001) (Fig. 4A). Since miR-375 and timeless co-expressed in I-LNVs neurons, we also overexpressed miR-375 in I-LNVs neurons by using the C929-gal4 driver, and found that 28.15% of the flies were arrhythmic (Table 1, Fig. 4A, p<0.001). These results suggest that in addition to targeting tim, miR-375 might also regulate other genes to modulate circadian locomotor rhythm.
Table 1.
Locomotor activities of flies in DD
| Genotype | Total flies | Rhythmic flies (%) | Period (h) | Power |
|---|---|---|---|---|
|
| ||||
| Iso:tim-gal4/+ | 75 | 100 | 24.6±0.09 | 56.2±6.2 |
| UAS-LUC-mir-375/+ | 47 | 95.74 | 24.5±0.08 | 65.2±7.22 |
| Iso:tim-gal4/UAS-LUC-mir-375 | 62 | 0 | * | * |
| UAS-mir-305/+ | 45 | 93.3 | 24.3±0.09 | 97.5±9.62 |
| Iso:tim-gal4/uas-luc-mir-305 | 47 | 91.5 | 23.7±0.13 | 82.8±9.33 |
| uas-luc-mir-9c/+ | 47 | 100 | 23.4±0.07 | 97.9±6.76 |
| Iso:tim-gal4/uas-luc-mir-9c | 29 | 0 | * | * |
| w 1118 | 56 | 98.83 | 24.03±0.06 | 97.1±12.1 |
| w 1118 ; TI{GAL4}mir-375KO | 70 | 67 | 21.8±0.3 | 58.27±2.37 |
| pdf-gal4/+ | 32 | 100 | 24.03±0.05 | 127.1±39.85 |
| pdf-gal4/UAS-LUC-mir-375 | 32 | 56.65 | 24±0.03 | 77.9±5.85 |
| C929-gal4/+ | 54 | 100 | 24±0.06 | 130.33±46.38 |
| C929-gal4/UAS-LUC-mir-375 | 46 | 70.93 | 24.03±0.06 | 75.37±15.85 |
Fig. 4. Overexpression of miR-375 regulates circadian rhythm and sleep.
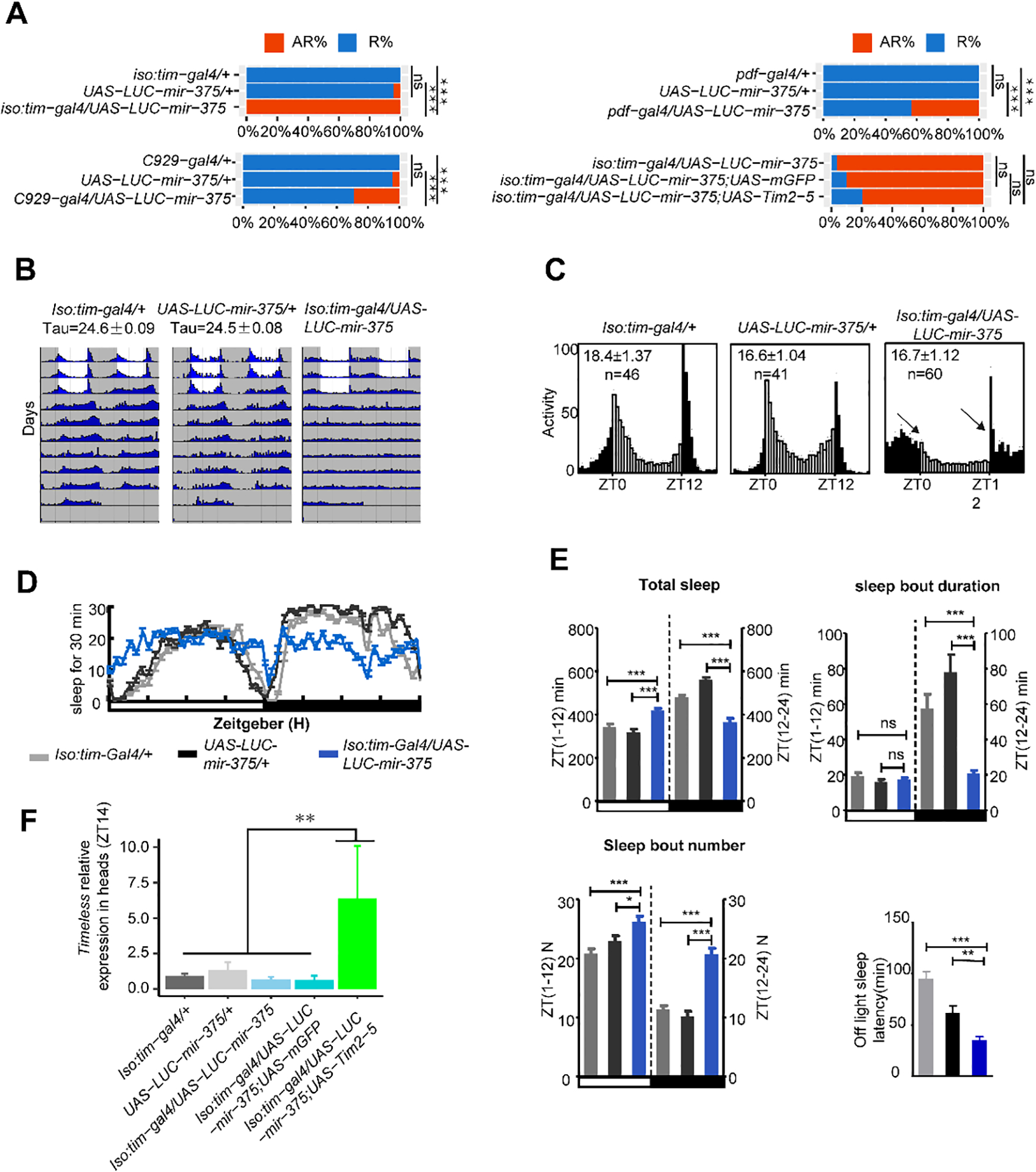
(A) miR-375 levels affect arrhythmicity in flies. AR% represents the percentage of arrhythmic flies, R% represents the percentage of rhythmic flies. *, p< 0.05; **, p < 0.01; ****, p< 0.0001 (two-tailed t-test).
(B) Locomotor behavior under LD and DD in the flies with miR-375 OE in time neurons. White indicates the light phase; gray indicates the dark phase.
(C) Comparison of the circadian locomotor activity in light/dark cycles of controls and miR-375 OE flies. Histograms represent the distribution of activity through 24 h, averaged over three LD days. Morning and evening anticipation was lost in the flies with miR-375 OE in time neurons.
(D) Sleep traces for male flies with overexpression of miR-375 driven by iso-tim-gal4. Gray dark represents tim-gal4, light dark represents UAS-miR-375, and blue represents tim-gal4/UAS-miR-375.
(E) Sleep profiles of male flies entrained under three LD cycles. Data represent mean ± SEM. **, p<0.01; ***, p< 0.001 (student’s t-test).
(F) Comparison of timeless mRNA levels in the head at ZT14 in flies in the rescue assay. Data represent mean ± SD from three independent experiments. ns, nonsignificant; *, p<0.05; **, p<0.01; ***, p< 0.001 (one way ANOVA test).
The flies with up-regulation of miR-375 had a similar phenotype of circadian locomotor rhythm as the tim null mutants (Fig. 4C and Fig. S4D), and they both expressed miR-375 in l-LNv neurons. In this study, 11 chimera-defined miRNA target sites were founded across the tim transcript, with the miR-375 targeting site located at the CDS region (Fig. S4C). To determine whether miR-375 modulates the expression of tim, the mRNA levels of tim were measured at daily six time points (ZT2, 6, 10, 14, 18, 22) in tim neurons of both miR-375 OE and w1118 flies. The amounts of tim mRNA were reduced by 1.78-fold (p<0.05), 1.77-fold (p<0.05), and 1.59-fold (p<0.05) at ZT 10, 14, and 18 in miR-375 OE flies, respectively, compared with the control flies (Fig. 5A). Similarly, TIM protein levels in I-LNvs neurons, detected at ZT18 and ZT24 by confocal microscopy, also showed significant decreases [31.1% at ZT 18 (p<0.01) and 54.37% at ZT24 (p<0.001)] in miR-375 OE flies compared with control flies (Fig. 5B). The decrease in TIM was further verified by Western blot analysis at ZT18 (Fig. 5C). Furthermore, we also detected the TIM levels in pdf neurons in the flies of miR-375 OE and control flies of the same background. Results showed that the miR-375 level at ZT3 was increased by 5.1 folds (Fig.5D) and the TIM level at ZT18 was decreased by 26.58% (p<0.05) in pdf neurons in the flies of overexpressed miR-375, compared to those of control flies (Fig.5E). These results indicate that TIM in the brain was significantly lower in the miR-375 OE flies than in the control flies, confirming that timeless is a target of miR-375.
Fig. 5. Interactions between miR-375 and tim/TIM.
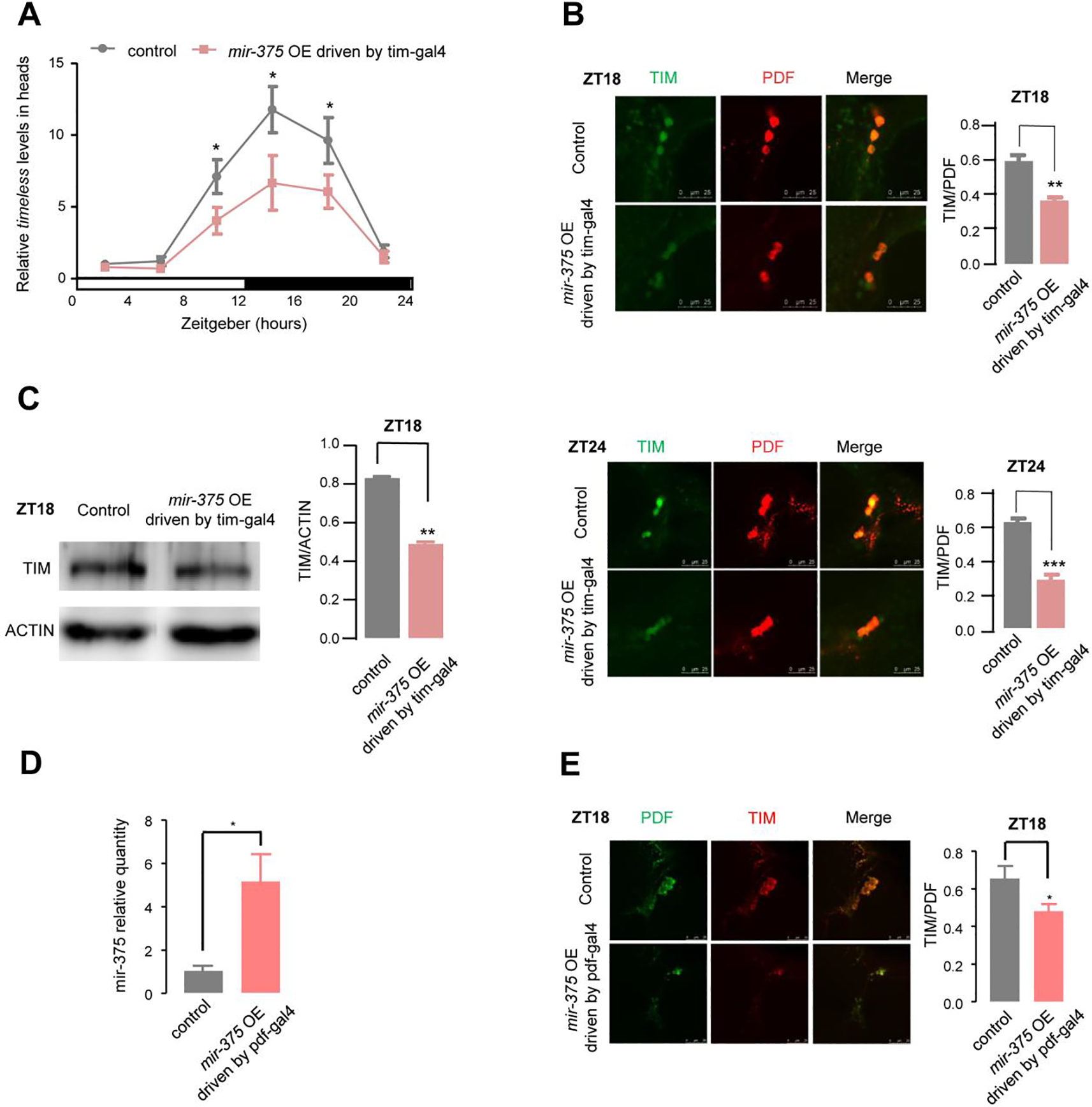
(A) The levels of timeless mRNA and miR-375 were altered in the flies of the miR-375 OE in the tim neurons. The relative expression levels were normalized to actin levels and were further normalized to control at ZT2. The error bar indicates SEM. *p < 0.05 (two-tailed t-test).
(B) Immunostaining analysis of TIM levels at ZT18 (upper) and ZT24 (lower) in the flies of control (iso:tim-gal4/+) and miR-375 OE in the tim neurons (iso:tim-gal4/UAS-LUC-mir-375), with detected TIM in green and PDF in red. TIM signal was normalized to the PDF signal (n=10–17 hemisphere). Error bar indicates SEM. **, p < 0.01; ***, p < 0.001 (two-tailed t-test). (C) Western blot analysis of TIM at ZT18 in the flies of control (iso:tim-gal4/+) and miR-375 OE in the tim neurons (iso:tim-gal4/UAS-LUC-mir-375). ACTIN was used as a loading control, TIM levels were normalized to ACTIN levels. Protein signals were quantitated by using Image J. Error bar indicates SEM (n=4). **, p < 0.01 (two-tailed t-test).
(D) Relative quantity analysis of the miR-375 at ZT3 in the pdf neurons in the flies of miR-375 OE (pdf-gal4/UAS-LUC-mir-375) and control flies (pdf-gal4/+). Error bar indicates SEM, *, p< 0.05; (two-tailed t test).
(E) Immunostaining analysis of TIM levels at ZT18 in the flies of control (pdf-gal4/+) and miR-375 OE in the pdf neurons (pdf-gal4/UAS-LUC-mir-375), with detected TIM in green and PDF in red. TIM signal was normalized to the PDF signal. Error bar indicates SEM, *p < 0.05 (two-tailed t test).
To explore the role of TIM in the arrhythmic phenotypes caused by overexpressing miR-375 in tim-staining neurons, we simultaneously used the UAS-tim2-5 to rescue this phenotype while the UAS-mCD8-GFP served as a control. Results showed that the UAS-tim2-5 could reduce arrhythmicity only to 80% compared with the UAS-mCD8-GFP control (Fig. 4A). Thus, we further examined tim expression in these flies. Surprisingly, the tim RNA levels in the flies (iso:tim-gal4/UAS-LUC-miR-375/UAS-tim2-5) were over-rescued with much greater increases of tim compared with those in controls (p<0.01) (Fig. 4F), suggesting that tim overexpression was much stronger than miR-375 overexpression in the iso:tim-gal4/UAS-LUC-miR-375/UAS-tim2-5 flies. Further, we also rescued the heterozygous tim01 mutant in TIM neurons by down-regulating miR-375 (UAS-mir375sp). The results showed that knockdown of miR-375 in tim01 (tim-gal4/tim01;UAS-mir375sp) could effectively rescue tim expression and rhythmic behavior, compared with their controls (Fig. S6).
miR-375 KO mutant influences circadian rhythm and sleep
In order to elucidate the circadian role of miR-375, we further analyzed the circadian behavior of miR-375 KO mutant (w1118; TI{GAL4}miR-375KO). Results showed that 33% of the miR-375 KO mutant were arrhythmic compared with 1.17% of w1118 control (p<0.05) (Fig. 6A), and the activities significantly reduced to 58.27 compared with 97.1 in w1118 control (Table 1, Fig. 6B & D). Moreover, the total sleep at daytime (p<0.001) and nighttime (p<0.01) both decreased significantly compared to those in control, caused by sleep bout duration (Fig. 6E & F). In addition, the mRNA levels of tim were also detected at daily six time points (ZT2, 6, 10, 14, 18, 22). The amounts of tim mRNA in the miR-375 KO mutant flies were significantly higher at ZT10 (p<0.01) and ZT14 (p<0.001) than in w1118 flies (Fig. 6G). Furthermore, we purified the Ago1-RNA complexes from flies and measured the amounts of miR-375 and tim mRNA in the complexes. Compared with w1118 flies, the RNA levels of tim decreased significantly in the Ago1-RNA complex of miR-375 KO mutant (w1118; TI{GAL4}miR-375KO) (p<0.05) (Fig. 6C). These results further demonstrated that miR-375 is a critical regulator for circadian rhythm and sleep and it exerts this function, at least in part, by targeting timeless.
Fig. 6. MiR-375 KO mutation and behavioral phenotypes.
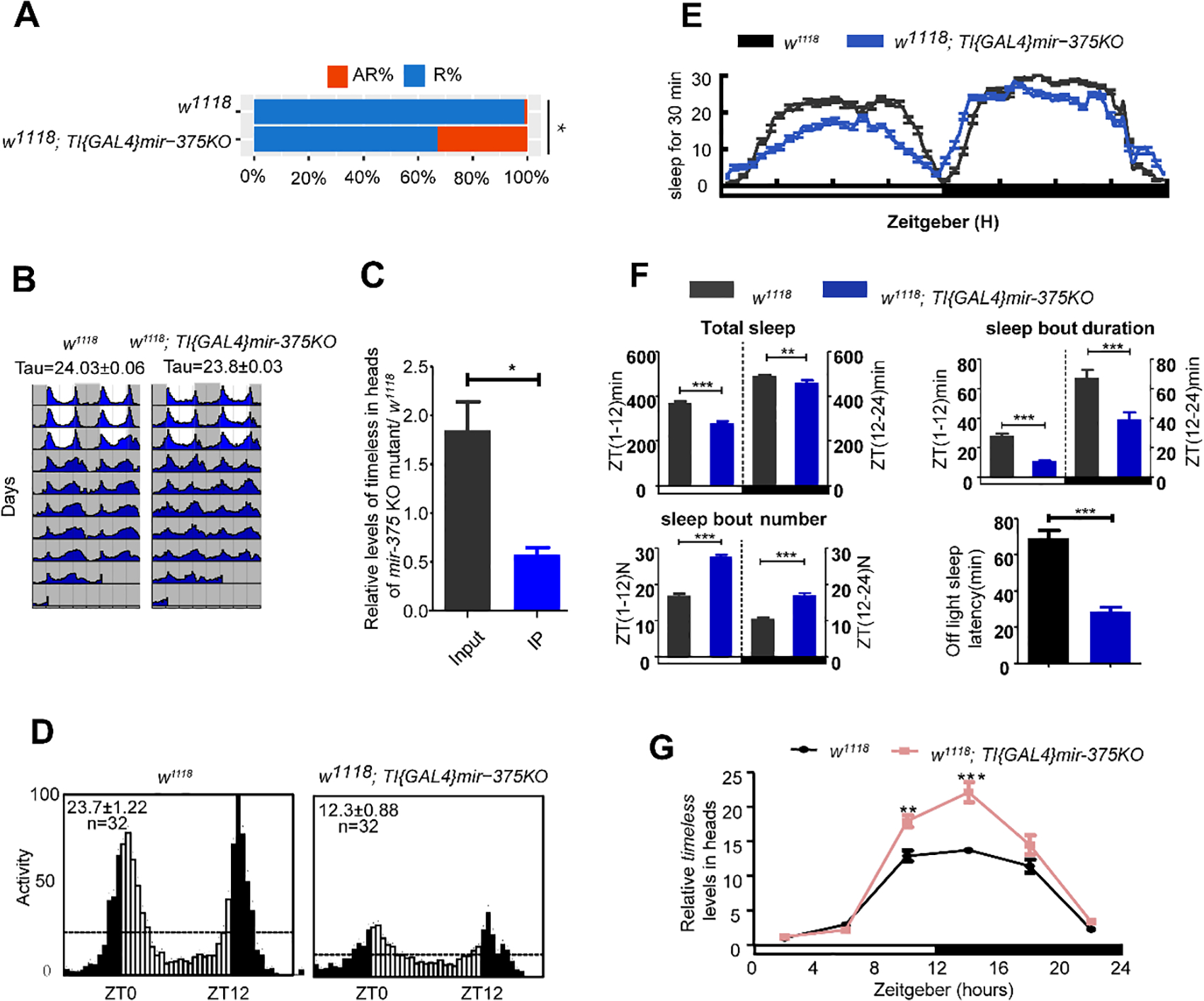
(A) Comparison of circadian rhythm in miR-375 KO mutant (w1118; TI{GAL4}mir-375KO) and control (w1118). Data represent mean±SEM (n = 50–70). ns, nonsignificant; *, p<0.05; **, p<0.01; ***, p< 0.001 (one-way ANOVA test).
(B) Locomotor behavior under LD cycle and constant darkness. Representative double plotted actograms of w1118 and miR-375 KO mutant (w1118; TI{GAL4}mir-375KO) flies. White indicates the light phase; gray indicates the dark phase.
(C) Analysis of timeless levels in the Ago1-RNA complex in heads of miR-375 KO mutant and w1118. Error bar indicates SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed t-test).
(D) Locomotor behavior under LD cycle and constant darkness. Representative double plotted actograms of w1118 and miR-375 KO mutant (w1118; TI{GAL4}mir-375KO) flies. White indicates the light phase; gray indicates the dark phase.
(E) Sleep analysis of male flies with overexpression of miR-375 driven by iso-tim-gal4. The colors of gray dark, light dark, and blue represent w1118 and miR-375 KO mutant (w1118; TI{GAL4}mir-375KO) flies sleep traces.
(F) Sleep profiles of male flies entrained under three LD cycles in miR-375 KO mutant (w1118; TI{GAL4}mir-375KO) and control (w1118). Data represent mean ± SEM. **, p<0.01; ***, p< 0.001 (Student’s t-test).
(G) Levels of timeless mRNA were altered in miR-375 KO (w1118; TI{GAL4}mir-375KO) flies. The relative expression levels were normalized to actin levels and were further normalized to control at ZT2. Error bar indicates SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.01 (two-tailed t-test).
Discussion
The significance of post-transcriptional regulation in circadian rhythms has been demonstrated by monitoring the core clock gene expression (14, 57) and screening miRNAs involved in rhythmic behavior (27–30, 32–34, 53, 54, 55). However, elucidation of the underlying molecular mechanism is impeded because only a few of the predicted miRNA-mRNA interactions have been verified. In this study, we captured miRNA-mRNA interaction pairs directly from fly tissue using the recently developed Ago1 CLEAR-CLIP (23, 38), leading to the detection of thousands of miRNA-target interactions. Of these interactions, 75 circadian genes are regulated by 61 miRNAs in the network of 299 interactions. Because of the central roles of the head in the circadian system, we did the CLEAR-CLIP using the heads firstly. To validate the head-enriched miRNA-target interactions, we also carried out the CLEAR-CLIP experiments with the whole body for comparison (Fig. S7). We expected that strong miRNA-mRNA interactions in the head might also be detectable in the whole-body samples while less abundant interactions in the head were less likely to be detected in the whole body samples. The whole-body data indeed retrieved some miRNA-target interactions detected in the head, such as the miR-375-timeless interaction (Figure 3A). Of course, many miRNA-target interactions are different.
We performed multiple lines of evaluation to assess the quality of the first global miRNA-mRNA interaction profiles in the head of Drosophila. First, CLEAR-CLIP recovered many known miRNA-target interactions such as bantam:hid (52) and miR-276a:tim (30). Second, the analysis of chimeras-defined interactions revealed general bioinformatics features for miRNA targeting, including seed enrichment, stable binding energy, and evolutionary conservation of the target sites. Third, the chimeras-defined mRNA targets exhibited significant changes in abundance after the manipulation of the corresponding miRNA. Fourth, functional enrichment analysis of the targets of miR-305 and miR-34 are consistent with the previous phenotypic study of those two miRNAs (49, 50). These together substantiate the reliability of the datasets for miRNA-mRNA interactions, which lay the groundwork for further functional studies of miRNAs.
This study reports for the first time that miR-375 is involved in circadian rhythm and sleep. Clk disruption abolished normal rhythmic expression of miR-375 and the functional regulation occurred in the l-LNv neurons, where miR-375 modulated circadian rhythm and sleep via targeting timeless. The disruption of Clk has a broad impact on the miRNA-mRNA interactome. Clkjrk is a classic arrhythmic fly line that is commonly used in circadian rhythm assay. Many studies have used clkjrk as a model to find genes involved in circadian-regulated behaviors (29, 31, 49), although this mutant also causes changes in other types of genes. Luo et.al (31) reported that wake mediates the circadian timing of sleep onset by comparing the expression of wake between clkjrk and w1118 flies. Although some CLK-dependent miRNA-mRNA interactions may not be relevant to circadian rhythms, we focused on the interactions between miRNAs and other core clock genes, like tim, pdp1, and vri. In this study, the CLEAR-CLIP assay was selected at ZT3 based on a previous report, at which ZT3 is the most important time point for miRNA-participated regulation (25). The miRNA-mRNA interactions throughout the whole day will be investigated in subsequent studies.
To rescue the arrhythmicity caused by miR-375 overexpression, we used the tim-gal4 driver to overexpress tim in all clock neurons. The expression of tim was much stronger than that in w1118 flies but the arrhythmic phenotype was only weakly reversed. A similar partial rescue was reported in the study of the regulation of tim by miR-276a, in which overexpression of miR-276a causes a 50% decrease in TIM and leads to 81% arrhythmicity (30). This means that a steady TIM level is important in maintaining circadian rhythm. The partial rescue in our study might result from several reasons. First, tim overexpression was much stronger than the suppression effect of miR-375 overexpression in the iso:tim-gal4/UAS-LUC-miR-375/UAS-tim2-5 flies. Second, abnormally high levels of TIM have been shown to lead to arrhythmic phenotype (58). Third, miR-375 may have other targets, which also contribute to circadian regulation.
In addition to miR-375, we also identified some other miRNAs and related targets, such as miR-4968-cwo, miR-1-timeless, miR-1-vri, miR-9c-clock, and so on. Our identification of the global function of the miRNA-mRNA interaction network involved in circadian systems will supply a crucial basis for future functional studies.
Supplementary Material
Supplemental_Table_S1A. Read statistics for 8 drosophila CLEAR-CLIP samples.
Supplemental Table_S1B. Experimentally Validated miRNA Targets Found in chimeras.
Supplemental_Table_S1C. The miRNA-first and miRNA-last distribution found in CLEAR-CLIP.
Supplemental_Table_S2A Ago1 CLIP peaks.
Supplemental_Table_S2B Ago1 CLASH chimeras.
Supplemental_Table_S3. The primers are used in this study.
Acknowledgments
We thank Jeffery Price (University of Missouri at Kansas City) for the revision of this manuscript. This work is supported by grants from the National Nature Science Foundation of China (Grant numbers 31730076 and 31970458) to Z. Z., and National Institutes of Health (Grant number R01 AI122743) to J.Z.
Nonstandard abbreviations
- miRNA
microRNA
- CLEAR-CLIP
Covalent Ligation of Endogenous Argonaute-bound RNAs-Cross-Linking and Immuno-Precipitation
pigment dispersing factor
- LNvs
ventral lateral neurons
- LNds
dorsal lateral neurons
- LPN
lateral posterior neurons
- DNs
dorsal neurons
- TTFL
transcriptional and translational feedback loop
- AGO
Argonaute
- clk
clock
- tim
timeless
- per
period
- cyc
cycle
- vri
vrille
- PDP1e
par domain protein 1e
- KO
knock out
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Allada R, Chung BY, Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72:605–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CH, Elliott JA, Foster R, Entrainment of circadian programs. Chronobiol Int 20(5):741–74 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Buysse DJ, Sleep health: can we define it? Does it matter? Sleep 37(1):9–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH, A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila 99(7):791–802 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Zheng X, Sehgal A, Probing the relative importance of molecular oscillations in the circadian clock. Genetics 178(3):1147–55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosbash M, Bradley S, Kadener S, Li Y, Luo W, Menet JS, Nagoshi E, Palm K, Schoer R, Shang Y, Tang CH, Transcriptional feedback and definition of the circadian pacemaker in Drosophila and animals. Cold Spring Harb Symp Quant Biol 72:75–83 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Allada R, White NE, So WV, Hall JC, Rosbash M, A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93(5):791–804 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC, CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93(5):805–14 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M, Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A 91(6):2260–4 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter-Ensor M, Ousley A, Sehgal A, Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84(5):677–85 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M, Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21(13):1675–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J, vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112(3):329–41 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE, VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37(2):249–61 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Lim C, Allada R, Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci 16(11):1544–50 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Reischl S, Kramer A, Kinases and phosphatases in the mammalian circadian clock. FEBS Lett 585(10):1393–9 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Bae K, Edery I, Regulating a circadian clock’s period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem 140(5):609–17 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Bushati N, Cohen SM, microRNA functions. Annu Rev Cell Dev Biol 23:175–205 (2007). [DOI] [PubMed] [Google Scholar]
- 18.He L, Hannon GJ, MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5(7):522–31 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Fabian MR, Sonenberg N, Filipowicz W, Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351–79 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Hausser J, Zavolan M, Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat Rev Genet 15(9):599–612 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, Tsai TR, Ho SY, Jian TY, Wu HY, Chen PR, Lin NC, Huang HT, Yang TL, Pai CY, Tai CS, Chen WL, Huang CY, Liu CC, Weng SL, Liao KW, Hsu WL, Huang HD, miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 44(D1):D239–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP, MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB, miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6:8864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorski SA, Vogel J, Doudna JA, RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol 18(4):215–28 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M, A role for microRNAs in the Drosophila circadian clock. Genes Dev 23(18):2179–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M, Lee JE, Padgett RW, Edery I, Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics 9:83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Rosbash M, MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat Commun 8:14707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vodala S, Pescatore S, Rodriguez J, Buescher M, Chen YW, Weng R, Cohen SM, Rosbash M, The oscillating miRNA 959–964 cluster impacts Drosophila feeding time and other circadian outputs. Cell Metab 16(5):601–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, Bai W, Zhou D, Zhao Z, Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat Commun 5:5549 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Rosbash M, mir-276a strengthens Drosophila circadian rhythms by regulating timeless expression. Proc Natl Acad Sci U S A 113(21):E2965–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W, Sehgal A, Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148(4):765–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y, Zhang Y, Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci 19(1):1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garaulet DL, Sun K, Li W, Wen J, Panzarino AM, O’Neil JL, Hiesinger PR, Young MW, Lai EC, miR-124 Regulates Diverse Aspects of Rhythmic Behavior in Drosophila. J Neurosci 36(12):3414–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Lamba P, Guo P, Emery P, miR-124 Regulates the Phase of Drosophila Circadian Locomotor Behavior. J Neurosci 36(6):2007–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnall-Levin M, Zhao Y, Perrimon N, Berger B, Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3’UTRs. Proc Natl Acad Sci U S A 107(36):15751–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan X, Kurgan L, Comprehensive overview and assessment of computational prediction of microRNA targets in animals. Brief Bioinform 16(5):780–94 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Chi SW, Zang JB, Mele A, Darnell RB, Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460(7254):479–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helwak A, Tollervey D, Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH). Nat Protoc 9(3):711–28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheel TKH, Moore MJ, Luna JM, Nishiuchi E, Fak J, Darnell RB, Rice CM, Global mapping of miRNA-target interactions in cattle (Bos taurus). Sci Rep 7(1):8190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N, Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54(6):1042–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodt M, Roehr JT, Ahmed R, Dieterich C, FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology (Basel) 1(3):895–905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J Mol Biol 215(3):403–10 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 153(3):654–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17(2):173–U6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hafner A, Stewart-Ornstein J, Purvis JE, Forrester WC, Bulyk ML, Lahav G, p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat Struct Mol Biol 24(10):840–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichler GS, Huang S, Ingber DE, Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics 19(17):2321–2 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Grant CE, Bailey TL, Noble WS, FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7):1017–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruger J, Rehmsmeier M, RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34(Web Server issue):W451–4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM, The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482(7386):519–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueda M, Sato T, Ohkawa Y, Inoue YH, Identification of miR-305, a microRNA that promotes aging, and its target mRNAs in Drosophila. Genes Cells 23(2):80–93 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM, Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Cusumano P, Biscontin A, Sandrelli F, Mazzotta GM, Tregnago C, De Pitta C, Costa R, Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster. PLoS Genet 14(7):e1007500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You S, Fulga TA, Van Vactor D, Jackson FR, Regulation of Circadian Behavior by Astroglial MicroRNAs in Drosophila. Genetics 208(3):1195–207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodwin PR, Meng A, Moore J, Hobin M, Fulga TA, Van Vactor D, Griffith LC, MicroRNAs Regulate Sleep and Sleep Homeostasis in Drosophila. Cell Rep 23(13):3776–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flores O, Kennedy EM, Skalsky RL, Cullen BR, Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic acids research 42(7):4629–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lerner I, Bartok O, Wolfson V, Menet JS, Weissbein U, Afik S, Haimovich D, Gafni C, Friedman N, Rosbash M, Kadener S, Clk post-transcriptional control denoises circadian transcription both temporally and spatially. Nat Commun 6:7056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang ZH, Sehgal A, Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29(2):453–67 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental_Table_S1A. Read statistics for 8 drosophila CLEAR-CLIP samples.
Supplemental Table_S1B. Experimentally Validated miRNA Targets Found in chimeras.
Supplemental_Table_S1C. The miRNA-first and miRNA-last distribution found in CLEAR-CLIP.
Supplemental_Table_S2A Ago1 CLIP peaks.
Supplemental_Table_S2B Ago1 CLASH chimeras.
Supplemental_Table_S3. The primers are used in this study.
Data Availability Statement
CLEAR-CLIP sequence data are available at the NCBI SRA database (Accession Number: SRP148458). The scripts for the bioinformatic analysis were uploaded to Github (https://github.com/XiaonanFu/zhulab).


