Abstract
Real-time PCR methods with primers and a probe targeting conserved regions of the bacterial 16S ribosomal DNA (rDNA) revealed a larger amount of rDNA in blood specimens from healthy individuals than in matched reagent controls. However, the origins and identities of these blood-associated bacterial rDNA sequences remain obscure.
Cultivation-independent laboratory approaches have revealed previously unsuspected degrees of diversity in the environment. These approaches are now being directed toward the human host (12). As a result, the complexity and distribution of the bacterial flora found in the human body have received increasing attention. The ability to detect and identify microorganisms at sites of disease and on skin and mucosal surfaces is an obvious but critical requirement for understanding the etiology of disease and the maintenance of health. Cultivation-independent approaches provide a more sensitive means for detection (7) and have provided evidence of microbial “contamination” within anatomically privileged compartments of the human body. For example, there is growing evidence that bacteria or parts thereof may circulate in the blood of healthy individuals (9). In fact, culture-positive bacteremia is known to occur after toothbrushing in the context of periodontal disease (2). The presence of bacterial DNA in circulating blood has important implications for immune system surveillance and development and for a possible, previously uncharacterized role of bacteria in idiopathic systemic disease.
Primers that target conserved regions of the genes encoding the small- and large-subunit rRNAs (rDNAs) can be used to detect and characterize known and previously unrecognized microbial pathogens and commensals (12, 20, 21). The approach has been shown to facilitate the routine diagnosis of bacterial diseases (1, 19, 25) and may allow direct identification of infectious agents in blood (6, 13). However, a number of factors limit the direct application of broad-range rDNA PCR to clinical specimens. Among the most important of these is bacterial DNA contamination of PCR reagents and other laboratory materials (8). This poses a challenge especially when trying to distinguish between low copy numbers of bacterial rDNA in clinical specimens and the bacterial rDNA that contaminates the specimen processing and PCR procedures. We approached this problem by developing a bacterial broad-range semiquantitative PCR method based on the use of a fluorescent rDNA reporter probe and real-time sequence detection. We then used this method to analyze peripheral blood specimens from persons without clinical signs of disease.
Venous blood from four healthy individuals (subjects 1 to 4) was drawn into Vacutainer tubes containing either EDTA, citrate, or heparin (Becton Dickinson, Franklin Lakes, N.J.) after rubbing of the skin with a sterile pad containing 70% isopropyl alcohol. As controls, sterile water (Abbott Laboratories, North Chicago, Ill.) was drawn into Vacutainer tubes containing one of each of the anticoagulants. DNA from the samples described above and from a B-cell-lymphoma cell line (DHL-4) was isolated with the IsoQuick kit (ORCA Research, Bothell, Wash.).
The broad-range bacterial rDNA primers fD1mod and 16S1RR-B and a 6-carboxy-fluorescein labeled probe, probe 516F, were used for real-time PCR (3, 11, 21). Amplification profiles were measured as the fluorescence emitted by the probe in an ABI PRISM 7700 sequence detector (Applied Biosystems, Foster City, Calif.). The final incubation volume was 25 or 50 μl and contained 50 mM Tris-HCl (pH 8.0), 4.0 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 60 nM Rox (Applied Biosystems), 0.01% Tween 20, 0.05 U of AmpliTaq Gold polymerase (Applied Biosystems) per μl, 0.9 pmol of each primer and 0.2 pmol of probe per μl, and 2.5 or 5.0 μl of experimental sample. After an initial step of 5 min at 95°C, 40 cycles were performed, each with steps at 95°C for 15 s, 54 or 56°C for 10 s, and 60°C for 1 min. Amplification of human β-actin sequences was performed with the TaqMan PCR reagent kit (Applied Biosystems). The sensitivity of the rDNA assay with purified DNA was 15 fg of Escherichia coli DNA (corresponding to approximately 25 E. coli rDNA copies) and 4 fg of Micrococcus luteus DNA.
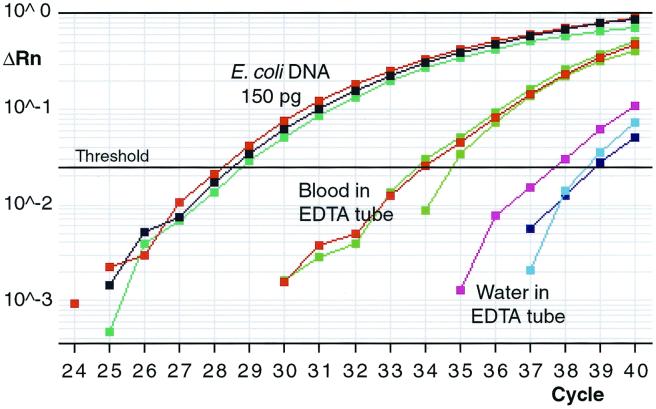
When DNA was isolated from EDTA-anticoagulated blood of four healthy individuals and used as the experimental sample, the threshhold cycle (Ct) was 4.2 to 5.9 cycles less (average, 4.9 cycles; standard deviation, 0.68 cycles between specimens P = 0.0017 by the paired t -test) than the Ct observed with control samples (sterile water drawn into an identical EDTA-containing tube and processed by the same procedure and with the same reagents used for the human blood samples) (Fig. 1). These four blood specimens were collected from persons without clinical signs or symptoms suggesting infection or other disease. When DNA from DHL-4 cells grown in RPMI (Irvine Scientific, Santa Ana, Calif.) culture medium and 10% fetal bovine serum (Sigma, St. Louis, Mo.) was used as the template, there were no differences in the Ct values compared with those for the water-EDTA controls, while abundant levels of β-actin gene DNA were detected. The experiments with purified human genomic DNA were performed in order to determine whether the differences in the Ct values between the blood-derived and corresponding control specimens might be caused by nonspecific probe hybridization to human DNA sequences or by the carrier effect of excess DNA in the DNA isolation process.
FIG. 1.
A representative logarithmic broad-range bacterial 16S rDNA amplification plot obtained with 150 pg of E. coli DNA, as well as DNA isolated from the blood of a healthy individual (subject 1) and similarly processed sterile water, both of which were drawn into EDTA-containing tubes, as templates. All samples were analyzed in triplicate and for 40 cycles. Each plot line represents independent measurements for each of the triplicates. The average Ct values in this experiment were 34.1 (range, 33.7 to 34.7) for the blood sample and 38.3 (range, 37.6 to 38.8) for the reagent control. PCR products from reactions with EDTA-anticoagulated blood and reagent control (water in an EDTA-containing tube) as templates were further characterized by cloning and sequencing. ΔRn, relative fluorescence.
Amplified PCR product was ligated into the pCR2.1 vector and transformed into E. coli cells (Invitrogen, Carlsbad, Calif.). Three clone libraries were created: two from the PCR product amplified from EDTA-anticoagulated blood from subject 1 (131 clones) and one from the PCR product obtained from a matched control reaction with sterile water drawn into an identical EDTA-containing tube and processed in an identical manner (77 clones). Sequencing of these 208 clone inserts was performed with ABI PRISM 377 or 373 DNA sequencers and by BigDye chemistry (Applied Biosystems). The DNA sequences were processed, aligned, and manually reviewed with the Factura and AutoAssembler programs (Applied Biosystems) and divided into phylotype groups with ≤1% dissimilarity (12). By this process, 192 rDNA sequences were assigned to eight bacterial phylogenetic groups. Each group was named after the most closely related well-characterized bacterium in the GenBank and MicroSeq (Applied Biosystems) (22) databases identified with the BLAST search tool. The automated 16S rRNA sequence alignment tool and phylogenetic analysis programs from the ARB software package (Technical University of Munich, Munich, Germany) were used to infer the phylogenetic relationships among these sequences (Fig. 2). Five blood-associated clone sequences were similar (>98%) to published sequences of human origin, and 11 were of unknown origin but lacked similarity with published bacterial 16S rDNA sequences. None of the human or unknown sequences contained the sequence of the probe (probe 516F) used in the real-time assay.
FIG. 2.
Phylogenetic relationships inferred from bacterial 16S rDNA sequences detected in a blood specimen of a healthy individual and from control clone library sequences. This unrooted tree was constructed by using 425 homologous sequence positions and a maximum-likelihood algorithm (17). The sequences associated with the blood specimen (study groups) are shown in red, and those generated in a control PCR with water as template are shown in green. Representative percentages for each group from the rDNA sequences from the total clone library are shown in parentheses. The blood-associated clone libraries and the control clone library included 115 and 77 rDNA sequences, respectively. Well-characterized reference sequences most similar to the study sequences are given in blue; and those of Staphylococcus epidermidis, Pseudomonas aeruginosa, and E. coli are shown as additional references. BCF, Bacteroides-Cytophaga-Flexibacter group; Low GC, low G+C content.
Members of seven phylogenetic groups and five bacterial divisions or subdivisions were detected in the blood specimen-associated clone libraries (Fig. 2, study groups). In our limited survey of control reagent-associated sequences, no members were identified for four of these groups. Of interest was a sequence that was detected in a blood-associated library closely corresponding to the Riemerella anatipestifer 16S rDNA sequence. However, the majority of the 16S rDNA sequences in the libraries from both blood- and reagent-associated PCR products were highly similar to that of Pseudomonas fluorescens (158 of the 192 rDNA clones); pseudomonads are common water-associated organisms. Furthermore, when we used primers specific for the P. fluorescens 16S rDNA, this sequence was detected in PCRs with AmpliTaq LD and no added template (data not shown). Previous reports have revealed sequences from Pseudomonas-like or other organisms as possible contaminants of Taq (and AmpliTaq) polymerases (10, 16, 18).
Environmental contaminants become disproportionately well represented in recombinant bacterial 16S rDNA clone libraries when specimens containing low concentrations of bacterial DNA are studied (24). Some sequences amplified under these conditions with broad-range bacterial 16S rDNA primers and reported in the literature as being specimen associated may in fact originate in the experimental reagents. Our most abundant reagent-associated sequence from P. fluorescens shares 99 to 100% similarity with 12 GenBank entries from amplicon clone libraries published by six different groups. The origins of these sequences are presumed to have been from the experimental specimens (4, 14, 23, 26, 27; R. M. Goodman et al., unpublished data [GenBank accession no. AF010025]). Similar GenBank findings were found for our Propionibacterium acnes-like and Microbacterium schleiferi-like reagent-associated sequences. In contrast, no such sequences that are highly similar (>99%) to our blood specimen-associated sequences from the Riemerella, Stenotrophomonas, and Pseudomonas putida groups are available in GenBank. Further details of the comparisons of the study and control clone library sequences with database entries, as well as primary data for our contaminant sequences, are available at http://relman.stanford.edu. While one might be tempted to consider the origin of these sequences to be the blood itself, we cannot rule out the possibility that they were introduced into the specimen from the skin during phlebotomy. Furthermore, one would need a larger number of clone libraries and clone sequences in order to establish a statistically meaningful association of sequences with blood specimens.
Our results suggest that blood specimens contain bacterial DNA. We cannot conclude whether the origins of this DNA are the skin or blood, or both. Our identification of a small number of human-derived sequences using the real-time broad-range bacterial 16S rDNA assay could explain some portion of this apparent bacterial DNA burden, despite our negative results with purified human genomic DNA from cell culture. Nevertheless, these findings have important implications for studies of blood-associated bacterial pathogens. They raise the possibility that there is a “normal” population of bacterial DNA sequences in this anatomic compartment that has previously been considered sterile most of the time. As a practical suggestion, investigators who perform broad-range bacterial PCR, and particularly those who screen bacterial rDNA clone libraries for diagnostic purposes, should first identify the most abundant sequences present in their PCR reagents. UV irradiation and DNase treatment have been proposed as methods that can be used to decrease the level of background bacterial DNA contamination of PCR reagents (15); however, these attempts have also led to a decrease in the detection sensitivity of the assay (5).
The broad-range rDNA PCR and real-time product detection approach may prove to be a useful tool in the analysis of complex specimens in which the presence of “background” sequences is expected. Automation of specimen preparation, PCR, and sequence detection may improve the value of this approach for diagnostic applications. The possibility that multiple clinically relevant bacterial sequence types are present in a clinical specimen will require that PCR products be analyzed either as separate cloned molecules in a high-throughput manner or with a complex (presumably, high-density) array of DNA probes. Our analysis is an early step toward a more comprehensive assessment of the “normal” DNA composition of human blood. Above all, our findings highlight the need for caution in interpreting the results obtained by cloning PCR products from clinical specimens with small amounts of microbial DNA.
Acknowledgments
We thank Paul Lepp for invaluable help with the phylogenetic analyses and for useful insights. Caroline Heckman and Linda Boxer kindly provided DHL-4 cells. Ken Livak provided invaluable help in the development of the real-time PCR assay. Shirley Kwok is acknowledged for critical reading of the manuscript.
We also acknowledge the generous support of the Emerging Infections Program of the Centers for Disease Control and Prevention and the Unexplained Deaths and Critical Illnesses Project (Brad Perkins, Centers for Disease Control and Prevention, Atlanta, Ga.).
REFERENCES
- 1.Anthony R M, Brown T J, French G L. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol. 2000;38:781–788. doi: 10.1128/jcm.38.2.781-788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger S A, Weitzman S, Edberg S C, Casey J I. Bacteremia after the use of an oral irrigation device. A controlled study in subjects with normal-appearing gingiva: comparison with use of toothbrush. Ann Intern Med. 1974;80:510–511. doi: 10.7326/0003-4819-80-4-510. [DOI] [PubMed] [Google Scholar]
- 3.Bergmans A M, Groothedde J W, Schellekens J F, van Embden J D, Ossewaarde J M, Schouls L M. Etiology of cat scratch disease: comparison of polymerase chain reaction detection of Bartonella (formerly Rochalimaea) and Afipia felis DNA with serology and skin tests. J Infect Dis. 1995;171:916–923. doi: 10.1093/infdis/171.4.916. [DOI] [PubMed] [Google Scholar]
- 4.Cho J C, Kim S J. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl Environ Microbiol. 2000;66:956–965. doi: 10.1128/aem.66.3.956-965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corless C E, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski E B, Fox A J. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cursons R T, Jeyerajah E, Sleigh J W. The use of polymerase chain reaction to detect septicemia in critically ill patients. Crit Care Med. 1999;27:937–940. doi: 10.1097/00003246-199905000-00029. [DOI] [PubMed] [Google Scholar]
- 7.Fredricks D N, Relman D A. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin Infect Dis. 2000;29:475–486. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 8.Fredricks D N, Relman D A. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granfors K, Merilahti P R, Luukkainen R, Möttönen T, Lahesmaa R, Probst P, Märker-Hermann E, Toivanen P. Persistence of Yersinia antigens in peripheral blood cells from patients with Yersinia enterocolitica O:3 infection with or without reactive arthritis. Arthritis Rheum. 1998;41:855–862. doi: 10.1002/1529-0131(199805)41:5<855::AID-ART12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 10.Hughes M S, Beck L A, Skuce R A. Identification and elimination of DNA sequences in Taq DNA polymerase. J Clin Microbiol. 1994;32:2007–2008. doi: 10.1128/jcm.32.8.2007-2008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotilainen P, Jalava J, Meurman O, Lehtonen O P, Rintala E, Seppälä O P, Eerola E, Nikkari S. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998;36:2205–2209. doi: 10.1128/jcm.36.8.2205-2209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroes I, Lepp P W, Relman D A. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley B E, Linton C J, Bennett D M, Jalal H, Foot A B, Millar M R. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 15.Lyons S R, Griffen A L, Leys E J. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–2365. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiwald M, Ditton H J, Sonntag H G, von Knebel Doeberitz M. Characterization of contaminating DNA in Taq polymerase which occurs during amplification with a primer set for Legionella 5S ribosomal RNA. Mol Cell Probes. 1994;8:11–14. doi: 10.1006/mcpr.1994.1002. [DOI] [PubMed] [Google Scholar]
- 17.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 18.Rand K H, Houck H. Taq polymerase contains bacterial DNA of unknown origin. Mol Cell Probes. 1990;4:445–450. doi: 10.1016/0890-8508(90)90003-i. [DOI] [PubMed] [Google Scholar]
- 19.Rantakokko-Jalava K, Nikkari S, Jalava J, Eerola E, Skurnik M, Meurman O, Ruuskanen O, Alanen A, Kotilainen E, Toivanen P, Kotilainen P. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000;38:32–39. doi: 10.1128/jcm.38.1.32-39.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 21.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y W, Ellis N M, Hopkins M K, Smith D H, Dodge D E, Persing D H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol. 1998;36:3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner M A, Everett C L, Youvan D C. Molecular phylogenetic evidence for noninvasive zoonotic transmission of Staphylococcus intermedius from a canine pet to a human. J Clin Microbiol. 2000;38:1628–1631. doi: 10.1128/jcm.38.4.1628-1631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turenne C Y, Witwicki E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of bacteria from positive blood cultures by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 2000;38:513–520. doi: 10.1128/jcm.38.2.513-520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weidner S, Arnold W, Puhler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagibayashi M, Nogi Y, Li L, Kato C. Changes in the microbial community in Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiol Lett. 1999;170:271–279. doi: 10.1111/j.1574-6968.1999.tb13384.x. [DOI] [PubMed] [Google Scholar]