Abstract
The continuous emergence and reemergence of diverse subtypes of influenza A viruses, which are known as “HxNy” and are mediated through the reassortment of viral genomes, account for seasonal epidemics, occasional pandemics, and zoonotic outbreaks. We summarize and discuss the characteristics of historic human pandemic HxNy viruses and diverse subtypes of HxNy among wild birds, mammals, and live poultry markets. In addition, we summarize the key molecular features of emerging infectious HxNy influenza viruses from the perspectives of the receptor binding of Hx, the inhibitor-binding specificities and drug-resistance features of Ny, and the matching of the gene segments. Our work enhances our understanding of the potential threats of novel reassortant influenza viruses to public health and provides recommendations for effective prevention, control, and research of this pathogen.
The Orthomyxoviridae family includes seven genera, which are all segmented negative-strand RNA viruses (Perez 2011): Four of these are different types of influenza viruses, Influenza A, B, C, and D viruses, and the other three are Thogotovirus, Quaranjavirus, and Isavirus. As a highly infectious pathogen in this family, influenza A viruses (IAVs) are a significant threat to global public health. The genomic evolution has enabled IAVs to reinvent themselves continuously, and it accounts for seasonal epidemics/endemics, occasional pandemics, and zoonotic outbreaks. The evolution of IAVs is primarily mediated through the genome mutation and/or the reassortment of viral genome segments derived from various strains when they coinfect the same host. Together with the bat-derived influenza-like viruses, IAVs of subtype HxNy are classified depending on the 18 hemaglutinin (HA) serotypes or subtypes on the viral surface, which are input as variable x, and the 11 subtypes of neuraminidase (NA) on the viral surface, as variable y (Wu et al. 2014).
Until now, influenza viruses that caused human pandemics have been the result of genetic reassortment between avian, human, and swine influenza viruses (Guan et al. 2010; Liu et al. 2018). Although IAVs have a wide range of host species, wild aquatic bird populations have long been considered the natural reservoir for all the identified HxNy viruses (except bat-derived H17N10 and H18N11), and they have been known to transfer viruses to other avian and mammalian hosts, including humans (Webster et al. 1992). In addition, considering the recently identified bat influenza-like viruses, other reservoir species may also exist (Wu et al. 2014). Furthermore, for epidemic/pandemic human influenza strains, swines have been postulated to be the “mixing vessels” for new HxNy viruses because of the presence of both α-2,3 and α-2,6 sialic acids (SAs) in their respiratory tracts (Ma et al. 2008). Recently, with the emergence of diverse avian influenza viruses (AIVs) in poultry, live poultry markets (LPMs) have also been considered “mixing vessels” for new HxNy viruses (Gao 2014).
For a specific HxNy subtype influenza virus, the characteristics of receptor binding of Hx and the enzymatic activity of Ny, including its inhibitor binding specificity or drug resistance feature, could be two of the key determinants for the transmission of a new virus in the target host. In addition, the mechanism for matching the HA segment, NA segment, and internal gene segments is crucial for determining whether an HxNy virus will emerge and successfully spread among humans.
We summarize the known emerging HxNy viruses in humans during influenza pandemics and diverse subtypes of HxNy among wild birds, mammals, and LPMs. Furthermore, we summarize the molecular basis for the characteristics of the receptor binding of Hx and the inhibitor binding specificity and drug resistance features of Ny. Moreover, we also review knowledge on the mechanism for the matching of the HA segment, NA segment, and internal gene segments. The summarization increases our understanding of the potential threats of novel reassortant influenza viruses to public health and provides recommendations for effective prevention and control strategies of this pathogen.
THE EMERGING PANDEMIC INFLUENZA VIRUSES
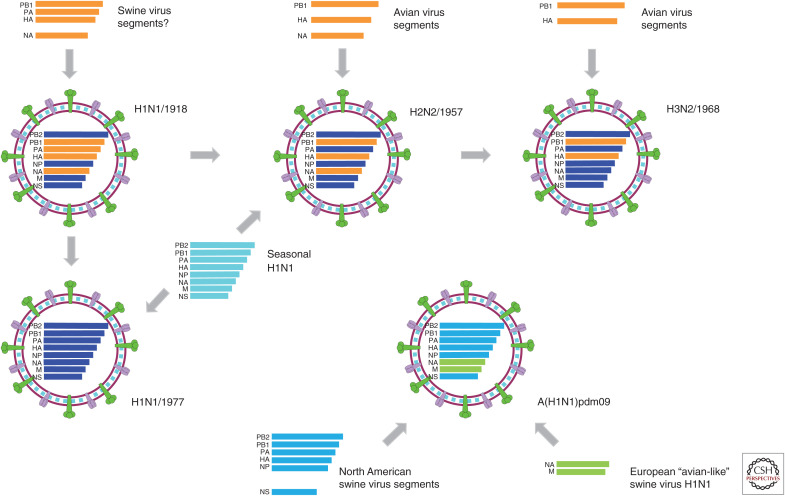
Since the beginning of the last century, the 1918 H1N1, 1957 H2N2, 1968 H3N2, and 2009 H1N1 influenza viruses have caused influenza pandemics, with the reemergence of H1N1 in 1977 considered as a “pseudo epidemic” (Liu et al. 2018). Generally, the pandemics were initiated by a successful reassortant virus bearing genes of human and avian and/or swine influenza viruses through the introduction and adaptation of a novel HA segment, which is known as the antigenic shift (Fig. 1). Afterward, these pandemic viruses become the cause of seasonal influenza viruses that are disseminated among humans for years until being replaced by or cocirculating with the next pandemic virus.
Figure 1.
Graphical representation of emerging pandemic influenza viruses. The possible reassortant pathway of pandemic influenza viruses. Gene segments, such as PB2, PB1, PA, HA, NP, NA, M, and NS, are ordered according to the gene lengths from top to bottom within the virus particle diagrams. For the gene segments of H1N1/1918, H2N2/1957, H3N2/1968, and H1N1/1977, the human influenza–derived segments are shown in blue, whereas the animal-, including swine- and avian-, derived segments are shown in orange. For H2N2/1957 and H1N1/1977, distinct seasonal H1N1 viruses (represented in cyan) at that time were also involved for the possible reassortment.
It has been more than a century since the Spanish flu (H1N1) outbreak of 1918, which is recognized as the most lethal natural event in recent history (Guan et al. 2010; Liu et al. 2018). The origin of the 1918 H1N1 genes remains ambiguous. Before the decoding of the 1918 virus genome, the virus was widely considered to be derived directly from an avian origin (Webster et al. 1992). This theory was later supported by the similarity between the consensus amino acid sequences (Tumpey et al. 2005) of AIVs and the available genetic information on the 1918 pandemic strain, A/Brevig Mission/1/1918 (Reid et al. 1999). Smith et al. (2009a) dated the pandemic influenza viruses using Bayesian relaxed clock methods and showed that the 1918 virus was generated over a period of years by reassortment between introduced avian viruses and previously circulating swine and human strains. Each gene segment of the 1918 H1N1 virus was identified to have been circulating in swine and humans at least 2–15 yr before its occurrence in 1918 (Fig. 1).
In 1957, a pandemic known as the “Asian flu” resulted in millions of deaths caused by a new reassortant influenza A virus H2N2 that arose from the circulating human H1N1 virus with the novel H2, N2, and PB1 genes derived from Eurasian avian virus sources (Fig. 1). Dating of pandemic influenza viruses using Bayesian relaxed clock methods suggests that the introduction of these three genes into the pandemic H2N2 occurred 2–6 yr before the pandemic (Smith et al. 2009a).
In 1968, another reassortant influenza virus, H3N2, emerged, causing the “Hong Kong flu.” The virus was the result of the H3 and PB1 gene segments (Fig. 1) of an avian flu virus mixing with the H2N2 virus between 1966 and 1968 (Smith et al. 2009a; Guan et al. 2010).
“Russian flu” was a small but notorious pandemic with H1N1 reemergence in Russia reported in 1977. Phylogenetic analyses of the reemergent H1N1/1977 virus confirmed that all of the eight genes were directly derived from the H1N1 viruses circulating in the 1950s (Fig. 1; Nakajima et al. 1978), and gene segment dating analysis showed a prior circulation of 2–3 yr before its detection. These results may support the hypothesis that the reemergence of H1N1/1977 could have resulted from accidental laboratory reintroduction. However, the possibility also exists that the virus was reintroduced as a result of unintentional contact with frozen corpses of patients or infected animals in the permafrost.
In 2009, a new strain of H1N1 emerged, which carried gene segments derived from swine influenza viruses, causing what was initially called “Swine flu” (Liu et al. 2018). This 2009 pandemic virus (A(H1N1)pdm09) replaced the previous seasonal H1N1 viruses that circulated for three decades until 2009; since 2009, the A(H1N1)pdm09 virus has cocirculated in humans with H3N2 viruses. Gene segment sequencing data show that all the genetic components of this new virus have descended from influenza viruses known to have naturally occurred in swine populations, which were circulating for more than 10 years before the outbreak in humans (Neumann et al. 2009; Smith et al. 2009b). The North American pig-derived H3N2 and/or H1N2 triple reassortant viruses, which were first reported in 1998, contributed the majority of the genes of the 2009 A/H1N1 viruses, and the NA and M gene segments potentially originated from Eurasian avian-like swine H1N1 viruses, which were initially identified in European pigs in 1979 (Fig. 1).
Existing evidence points to pandemic influenza viruses having diverse reassortment origins. However, at least one virus of animal origin was involved in each of the reassortments before their circulation in humans. The novel gene segments for the H2N2/1957 and H3N2/1968 pandemics seem to have originated from avian hosts, and A(H1N1)pdm09 originated from swine hosts, whereas the zoonotic sources of the introduced viral gene segments for the 1918 pandemic are uncertain. Considering the recent H7N9 and other human-infecting AIVs (Huo et al. 2018a), poultry also plays a key role in facilitating reassortment events in addition to swine and wild birds and may provide a new source for the development of the next influenza pandemic. Thus, there is a need for systematic surveillance of influenza in wild and domestic animals that will provide early evidence for the emergence of viruses with pandemic potential in humans (Quan et al. 2018a).
EMERGING HxNy IN BIRDS, WILD MAMMALS, AND LPMs
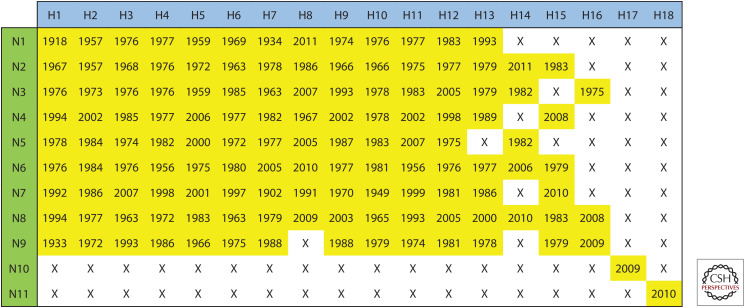
It is well-established that wild aquatic birds are the natural hosts for most subtypes of IAVs, which are known as AIVs. Until now, IAVs have been detected in more than 100 wild bird species. Of 131 known subtypes of IAV (Fig. 2), most were circulating in wild birds, except for H17N10 and H18N11, which were influenza-like viruses for which genetic material has only been detected in bats thus far (Tong et al. 2013). Among all the AIVs, only 11 subtypes, including H5N1 (deJong et al. 1997), H5N6 (Bi et al. 2019), H6N1 (Wei et al. 2013), H7N2 (Marinova-Petkova et al. 2017), H7N3 (Tweed et al. 2004), H7N4 (Tong et al. 2018), H7N7 (Puzelli et al. 2014), H7N9 (Liu et al. 2013), H9N2 (Butt et al. 2005), H10N7 (Organization 2004), and H10N8 (Chen et al. 2014) are known to infect humans (Fig. 3). The H5N1 subtype was first confirmed to infect humans in Hong Kong, China in 1997 (Claas et al. 1998; Subbarao et al. 1998), and since then it has frequently been reported in other countries of Asia, Europe, and Africa. To date, H5N1 AIV has infected 861 people worldwide, of which 455 have died (WHO 2019a), and it can be isolated from humans, pigs, tigers, cats, domestic poultry, and wild birds (Yee et al. 2009). Following the identification of H5N1, several novel AIV subtypes, such as H9N2, H5N6, H6N1, H10N8, H7N9, and H7N4, have been reported to infect humans (Quan et al. 2018a). Unlike other subtypes of AIVs, which only led to sporadic cases of human infections, since 2013, there have been five waves of H7N9 AIV, causing a total of 1568 laboratory-confirmed human cases and 616 deaths (WHO 2019b) as well as sporadic cases later. Notably, H7N9 was identified as a low pathogenic AIV (LPAIV) (Gao et al. 2013; Liu et al. 2013) initially in 2013, but it caused severe disease in humans (Yang et al. 2019). Unfortunately, the H7N9 LPAIV later mutated into highly pathogenic AIV (HPAIV) by insertion of four amino acid residues (Zhang et al. 2017; Quan et al. 2018b), generating multiple basic amino acids at the cleavage site of the HA protein. Importantly, H7N9 HPAIV infection caused a higher case fatality rate of 50% than that caused by H7N9 LPAIV (Yang et al. 2019). Because of the high human mortality rates of some AIVs, the highly pathogenic H5N1 and H7N9 AIVs pose a huge threat to public health and cause huge economic losses worldwide (Liu et al. 2014).
Figure 2.
Currently known subtypes of influenza A viruses and their earliest year of identification. To show the influenza A viruses that have naturally occurred with different pairing of HA and NA, H1-H18 and N1-N11 were paired in a matrix. The earliest year of identification for each natural influenza A virus is shown in yellow cells. The influenza A viruses that have not been identified are indicated by an “X.”
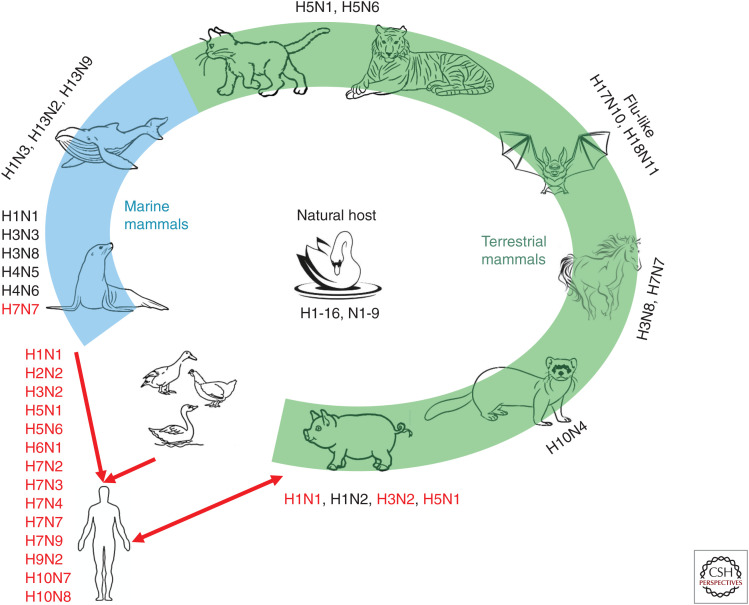
Figure 3.
Natural reservoir and mammalian hosts of influenza A viruses. Animals with green and blue backgrounds are terrestrial and marine mammals, respectively. Subtypes of influenza marked next to the animal are currently known to infect that animal. The red font indicates that the virus can infect humans.
In addition to wild aquatic birds, only a limited number of IAV subtypes circulate in some mammals other than humans, such as pigs, cats, dogs, horses, cows, ferrets, bats, whales, and seals. The most common subtypes of IAVs are H1N1, H1N2, and H3N2, which continuously circulate in swine. As swine are susceptible to influenza viruses from both humans and avians, they have been considered as a “mixing vessel,” in which novel subtypes of IAVs can be generated by massive reassortments and mutations (Ma et al. 2008).
Other mammals, such as horses, can be infected by H3N8 and H7N7, and H5N1 and H5N6 have also been isolated from cats and tigers. In addition to terrestrial mammals, IAVs have also been detected in marine mammals (Fig. 3). The first reported case of marine mammals infected by an IAV was in seals, which were infected by H7N7 IAV in 1979 (Webster et al. 1981b). The humans who handled these dead seals subsequently developed purulent conjunctivitis with a high titer of H7N7 AIV in their conjunctival membranes (Webster et al. 1981a), which raised concern regarding AIV transmission from marine mammals to humans. Since then, H4N5 (Hinshaw et al. 1984), H4N6 (Callan et al. 1995), H3N3 (Callan et al. 1995), H1N1 (Goldstein et al. 2013), H3N8 (Anthony et al. 2012), H10N7 (Zohari et al. 2014), and influenza B virus (Osterhaus et al. 2000) were isolated from seals in 1982, 1991, 1992, 2010, 2011, 2014, and 1999, respectively. Whales are another marine mammal from which H13N2 (Hinshaw et al. 1986), H13N9 (Hinshaw et al. 1986), and H1N3 (Lvov et al. 1978) viruses have been detected.
The exposure of poultry to wild birds increases the risk of novel subtypes of influenza viruses being transmitted from wild birds to poultry. Recently, LPMs have displayed an important role for the emergence and evolution of novel AIVs, such as H7N9, H10N8, and H5N6 (Wei et al. 2013; Chen et al. 2014; Gao 2014; Ma et al. 2015; Bi et al. 2016; Huo et al. 2018b). Therefore, LPMs, with chickens sold there, also play a role as a novel “mixing vessel” or “incubator” for novel influenza viruses (Su et al. 2015). Furthermore, the extensive reassortment of AIVs in live poultry increases the likelihood of the generation of novel types of IAVs and the risk of being adapted to humans. Several studies have revealed that a higher percentage of people infected by AIVs had a history of exposure to live poultry or LPMs (Cowling et al. 2013; Li et al. 2014). In China, H5N1 and H9N2 AIVs emerged during the 1990s, and since then they have been cocirculating in LPMs. The H7N9 subtype has appeared in LPMs in China since 2013 (Liao et al. 2016). After that, H7N9 AIV was documented to continuously undergo dynamic reassortments (Cui et al. 2014; Lam et al. 2015). Surveillance of LPMs in China from 2014 to 2016 revealed that H5N6, which arose from reassortments of H5 and H6N6 viruses, replaced H5N1 as one of the dominant AIV subtypes in southern China, especially in ducks (Bi et al. 2016). Until now, at least 24 human H5N6 virus infections have been reported with a case fatality rate of ∼67%, which seems higher than that of H5N1-infected cases (Bi et al. 2019; WHO 2019c).
The mounting evidence suggests that animals, including wild birds, poultry in LPMs, and pigs, play an important role in the emergence, evolution, and transmission of novel IAVs, and close contact among birds, swine, humans, and other mammals poses a potential threat to both agriculture and public health.
RECEPTOR BINDING SPECIFICITY OF Hx
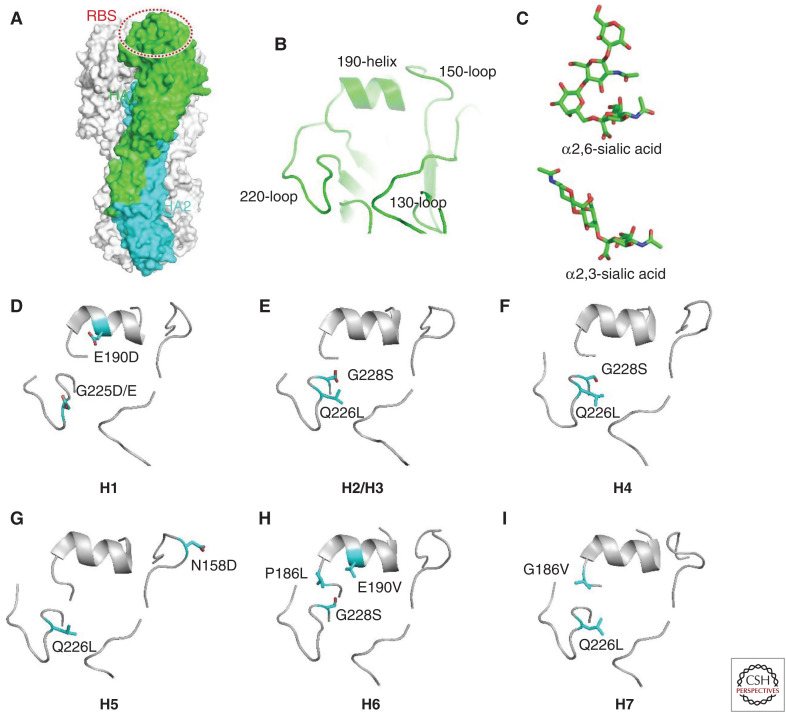
The receptor binding property of HA protein is a crucial determinant of interspecies transmission of influenza viruses. To enable “host jumping,” viruses need to be able to bind to the receptors of host cells, which is the first step of virus entry (Wiley and Skehel 1987; Sun et al. 2013; de Graaf and Fouchier 2014; Shi et al. 2014; Wu et al. 2014). Typically, avian-specific influenza viruses bind only α-2,3-linked SA glycans (avian receptor), whereas the human influenza viruses preferentially bind the α-2,6-linked SA receptors (human receptor) (Fig. 4). It is thought that human influenza viruses originated from avian-specific viral strains, which gradually evolved to obtain human-receptor binding capacity (Taubenberger and Kash 2010).
Figure 4.
Molecular determinants for receptor binding properties of different hemagglutinin (HA) subtypes. (A) Structure of HA trimer. One of the protomers is colored by subunits with HA1 in green and HA2 in cyan. The receptor binding site (RBS) is indicated by a dashed oval. (B) Close-up view of the structural motifs within RBS. (C) Structures of different sialic acid receptors. (D–I) The key determinant residues (sticks) for the receptor binding properties of different HA subtypes.
The receptor-binding site (RBS) of HA comprises three critical secondary elements, 130-loop, 190-helix, and 220-loop (Fig. 4). The key interacting residues within each element display quite remarkable variations among different subtypes of influenza viruses, resulting in distinct receptor binding properties (Shi et al. 2014). For different HA subtypes of influenza viruses, the key determinant residues for human-receptor binding capacity or human/ avian-receptor preference could also be significantly different. There could be more than one path for a specific HA subtype to evolve from avian-receptor specificity to dual-receptor tropism and even subverting to human-receptor specificity (Xiong et al. 2014). Based on biochemical and structural studies, we have gained substantial understanding of the determinants of receptor binding properties for a few HA subtype influenza viruses, including the H1, H2, H3, H4, H5, H6, and H7 subtypes. However, for the H10 subtype, occasional human infection cases have been reported, but the virus has typical avian-receptor binding preference (Wang et al. 2015b), which could be explained by the presence of α-2,3-linked SA receptors in the lower respiratory tract of humans.
Receptor Binding Determinants of H1
For the H1 subtype, two sites, 190 and 225 (Fig. 4), play critical roles in determining receptor binding specificity (Matrosovich et al. 2000; Gamblin et al. 2004; Xu et al. 2012a; Zhang et al. 2013b). Most of the H1 subtype influenza viruses can bind to both human and avian receptors but with different preference for each receptor. The viral isolates from avian hosts normally possess E190 and G225 feature residues and preferentially bind to avian receptors. Substitutions with D190 and G225 switch the binding property from avian- to human-receptor preference, but they retain avian-receptor binding capacity. In addition, the combination of D190 with 225D/E could enable H1 subtype HA to specifically bind to human receptors (Matrosovich et al. 2000; Gamblin et al. 2004; Xu et al. 2012a; Zhang et al. 2013b). The two residues D190 and 225D/E directly interact with the second (Gal-2) and third (GlcNAc3) glycan rings in α-2,6-linked SA receptors, which favorably bind to human receptors. As the glycan residues in α-2,3-linked SA receptors adopt a different configuration, similar interactions do not occur for binding avian receptors, resulting in a specificity for human receptors in this context (Zhang et al. 2013b). In addition, the residue at position 138 also affects the receptor binding property of H1 subtype HA (Rogers and D'Souza 1989), for which the structural basis has not been elucidated.
Receptor Binding Determinants of H2 and H3
In the adaptation process of H2 and H3 subtype influenza viruses, Q226L and G228S substitutions are key determinants (Fig. 4), and the Q226L substitution appears to be more critical for human-receptor recognition (Rogers et al. 1983; Eisen et al. 1997; Matrosovich et al. 2000; Ha et al. 2003; Liu et al. 2009; Pappas et al. 2010; Xu et al. 2010). The H2/H3 subtype HA proteins of avian origin can bind to both human and avian receptors, whereas human-adapted viral strains preferentially recognize human receptors. Structures of HA bound to receptor analogs have revealed that residue L226 provides a hydrophobic platform to accommodate the nonpolar C6 atom of Gal-2, which favors human-receptor binding, but is incompatible with the hydrophilic oxygen atom of the glycan residues in α-2,3-linked avian receptors (Eisen et al. 1997; Liu et al. 2009). However, cooperative residue substitution at positions 222 and 225 and similar hydrophobic substitutions for L226 by Val/Ile have also been observed. These substitutions alter the flexibility of the 220-loop, and therefore, they affect the binding affinity to receptors (Lin et al. 2012).
Receptor Binding Determinants of H4
Recent research has shown that the Q226L substitution plays a pivotal role in altering receptor binding specificity for H4 HAs, and the G228S substitution decreases the affinity for avian receptors, whereas it enhances binding to human receptors (Fig. 4; Song et al. 2017). The H4 subtype HA with Q226/G228 combination only binds to avian receptors. A single substitution of G228S could enable human-receptor binding but does not influence the affinity to bind to avian receptors. Moreover, further replacement of Q226L renders the H4 subtype HA to specifically recognize human receptors (Song et al. 2017). According to the structures of H4 in a complex with avian/human-receptor analogs, the G228S mutation contributes two additional hydrogen bonds to the SA residue, which allows binding to both human and avian receptors. The substitution of Q226L changes the microenvironment of the 220-loop, which alters the conformation of Gal-2 and favors the binding of human receptors but does not successfully accommodate avian receptors (Song et al. 2017). Although there is no direct evidence for human infection yet, the adaptation for human receptor preference in swine H4 isolates implicates the potential threat to humans.
Receptor Binding Determinants of H5
The H5 subtype influenza viruses are typical avian-specific viruses, despite the highly pathogenic H5N1 virus having caused hundreds of cases of human infection since the outbreak in 1997. No evidence has revealed that H5 subtype influenza viruses have evolved to human-receptor-binding preferences in nature, but several artificial tropism-switching substitutions have been identified in laboratories (Herfst et al. 2012; Imai et al. 2012; Russell et al. 2012). The key determinants for the receptor tropism of H5 subtype HA primarily include the Q226L substitution and the loss of glycosylation at position 158 (Fig. 4). The Q226L substitution plays a similar role to that of the H2/H3 subtype HAs, which alter the hydrophobicity of the 220-loop region to favor human-receptor binding. The substitution at residue 158 or 160 leads to a loss of glycan chain modification in the 150-loop, which avoids the potential steric hindrance for binding human receptors and further favors its preference (Lu et al. 2013; Xiong et al. 2013a; Zhang et al. 2013a). In addition, some other residue substitutions outside the RBS have also been shown to affect the thermostability of HA molecules, which may facilitate airborne transmission between mammalian hosts (Imai et al. 2012; Xiong et al. 2013a; Zhang et al. 2013a; Linster et al. 2014).
Receptor Binding Determinants of H6
In 2013, the first human case of infection with AIV H6N1 was reported in Taiwan (Wei et al. 2013). Biochemical analysis revealed that the HA of this viral isolate was dual-receptor tropic with a remarkable preference to human receptors (Wang et al. 2015a). Sequence analyses for H6 subtype HAs over the past decades have indicated the adaptation process from avian receptor specificity to dominant human-receptor preference, suggesting highly evolved human adaptation features (Shi et al. 2013a; Wang et al. 2015a). The receptor binding property of H6 subtype HA is primarily determined by two residues at 190 and 228 (Fig. 4). The combination of E190 and G228 binds to the avian receptor only in the context of a highly conserved Q226. Substitutions of E190V and G228S enable human-receptor binding but retain good binding to avian receptors (Wang et al. 2015a). Further substitution of P186L severely reduces the binding affinity to avian receptors, leading to the adaptation of human-specific viruses (Wang et al. 2015a). Structures of human/avian H6 bound to different receptor analogs showed that the G228S mutation further strengthens binding to both avian and human receptors by introducing two additional hydrogen bonds to the SA residue. The E190V substitution cooperates with P186 to create a hydrophobic environment for contacting the Gal-2 residue, which enables human-receptor binding. This context may change the conformation of the Gal-2 of the avian receptor, but it does not significantly influence its binding affinity (Wang et al. 2015a). The replacement of P186L further enhances the hydrophobicity in this region and repulses the Gal-2 of the avian receptor with cis conformation because of the longer side chain of leucine, which destabilizes the binding of avian receptors (Wang et al. 2015a).
Receptor Binding Determinants of H7
The H7N9 epidemic since 2013 and the more recent H7N4 human infection cases have highlighted the potential of H7 subtype influenza viruses to adapt toward human hosts (Bao et al. 2013; Liu et al. 2014). Although some studies have revealed that H7N9 subtype influenza viruses have not obtained efficient human-to-human transmission capacity, evident human-receptor adaptation of H7 subtype HA was observed before and after the outbreak in 2013 (Xu et al. 2019). Based on current knowledge, the receptor binding property of H7 subtype HA is primarily determined by residues at two positions, 186 and 226 (Fig. 4; Kageyama et al. 2013; Shi et al. 2013b; Xiong et al. 2013b). Two other sites, 138 and 221, play minor roles in cooperatively affecting binding affinities (Xu et al. 2019). Different from other HA subtypes, the canonical signature substitution of Q226L is not the key to enabling human-receptor binding of H7 subtype HA. Instead, a single G186V mutation is the determinant of avian-specific H7 subtype HA gaining human-receptor binding capacity, which might emerge earlier in its evolution (Xu et al. 2019). In contrast, the Q226L replacement might be selected later to further regulate its affinity for different receptors. More importantly, L226 is only favorable for both human and avian receptors when paired with all the hydrophobic residues at the 138, 186, and 226 positions. Structural evidence has revealed that the G186V mutation subverts the local hydrophobicity to affect the side-chain conformation of E190, which directly interacts with SA receptors. However, the Q226L substitution alters the stability of the 220-loop, affecting binding to both human and avian receptors (Xu et al. 2019). Bioinformatics analysis have shown that the current circulating H7 subtype influenza viruses are dual-receptor tropic with a preference for human receptors (Quan et al. 2018b; Xu et al. 2019), but engineered mutants with human-receptor specificity have also been developed in laboratories (de Vries et al. 2017), which might be indicative of the next stage of viral adaptation.
MOLECULAR FEATURES OF Ny AND ITS INHIBITORS
Enzymatic Activity and Inhibitor Binding Specificity of Ny
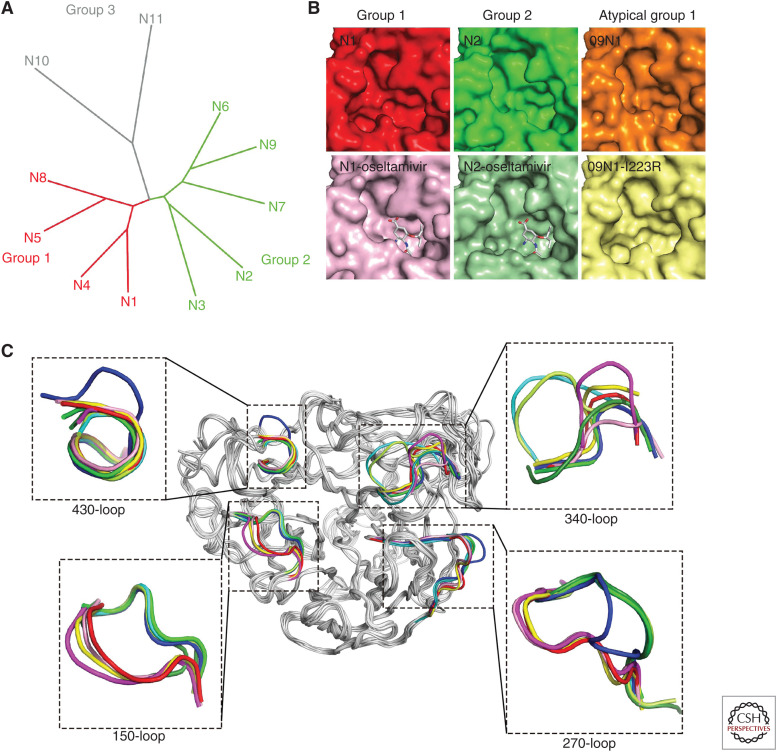
Neuraminidase is one of the major envelope proteins on the surface of influenza viruses, and it is responsible for the cleavage of SAs from glycans on the host cell surface to release progeny viruses (Palese et al. 1974). So far, 11 subtypes of NAs of IAVs have been discovered (Fig. 5). Apart from two NA-like (N10 and N11) genes, which were derived from bats, nine subtypes (N1-N9) of canonical NAs are classified into two groups based on their primary sequences: Group 1 consists of N1, N4, N5, and N8, and group 2 consists of N2, N3, N6, N7, and N9 (Air and Laver 1989; Tong et al. 2012, 2013). The head domain structures of these eleven subtypes of NAs have been determined (Varghese et al. 1992; Russell et al. 2006; Wang et al. 2011; Imai et al. 2012; Zhu et al. 2012; Tong et al. 2013; Wu et al. 2013b; Sun et al. 2014). The eight functional residues, R118, D151, R152, R224, E276, R292, R371, and Y406 in the N2 numbering, directly interacting with the substrate and the 11 framework residues, E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425, supporting the catalytic sites are conserved among N1–N9, whereas, for N10 and N11, only three functional residues and three (N10) or four (N11) framework residues are conserved. In addition, the N10 and N11 proteins exhibit no sialidase enzymatic activities in vitro, which can be explained by the structural analysis.
Figure 5.
Comprehensive analysis of 11 known neuraminidases of influenza A virus. (A) The phylogenetic tree of neuraminidases (NAs) from 11 known influenza A virus subtypes. (B) Comparison of the active sites of typical group 1, group 2, and atypical group 1 NAs. All the NA molecules are presented in surface representation. Free VN04N1 (PDB code 2HTY, red), free N2 (PDB code 4K1H, green), 09N1 (PDB code 3NSS, orange), oseltamivir carboxylate-bound VN04N1 (PDB code 2HU4, pink), oseltamivir carboxylate-bound N2 with half-open form of 150-loop (PDB code 4K1K, light green), and 09N1-I223R (PDB code 4B7M, light yellow). (C) Superimposition of NA monomers with emphasis on the 150-loop, 270-loop, 340-loop, and 430-loop. The colors of different NAs are VN04N1 (PDB code 2HTY, red), N2 (PDB code 4K1H, green), N3 (PDB code 4HZV, blue), N4 (PDB code 2HTV, yellow), N5 (PDB code 3SAL, magenta), N6 (PDB code 4QN4, cyan), N7 (PDB code 4QN3, lemon), N8 (PDB code 2HT5, pink), and N9 (PDB code 4MWJ, deep green).
It is clear to observe the additional cavity (150-cavity) adjacent to the active site in group 1 NAs (Vavricka et al. 2011). The 150-cavity stems from the open form of the 150-loop (residues 147–152), which plays an essential role in substrate/inhibitor binding (Fig. 5). Compared with other group 1 members, N5 has an extended 150-cavity due to the residue N147 (Wang et al. 2011). All the group 2 NA members display a closed form of the 150-loop, which results in the deficiency of the 150-cavity. Interestingly, the pandemic 2009 H1N1 neuraminidase (p09N1) exhibits a closed 150-loop, which is the same as group 2 NA members (Li et al. 2010). Therefore, p09N1 was defined as an atypical group 1 NA.
Oseltamivir carboxylate can induce the rigid N2 150-loop to open, and this half-open form of the 150-loop can be stabled by the neighboring NA molecule (Wu et al. 2013c). In contrast, zanamivir guanidino has stronger interactions with the 150-loop for maintaining its closed conformation. Moreover, drug-resistant mutation also has an effect on the conformation of the 150-loop. p09N1-I223R adopts an open conformation compared with the closed form in wild-type p09N1 (van der Vries et al. 2012). The I223R mutation has a markedly more substantial effect on NA inhibition by oseltamivir than by zanamivir, which indicates that the interactions provided by the 4-guanidino group of zanamivir may compensate for lost interactions resulting from drug-resistant mutations. This hypothesis was proved by a hybrid inhibitor, MS-257, which combines the advantageous groups from both oseltamivir and zanamivir. This small molecule exhibits potent inhibition to N2 with drug-resistant mutations (E119V and I222L) compared with oseltamivir. The N2-inhibitor complex structure indicates that the 4-guanidino group interacts with the residues on the 150-loop (Liu et al. 2016). Group-specific inhibitor is one of the options of drug development. Inhibitors targeting the 150-cavity have been reported, which display the inhibition ability to group 1 NAs (Rudrawar et al. 2010; Wu et al. 2015).
Apart from the 150-loop, the 270-loop (resides 267–276), 430-loop (429–433), and 340-loop (342–347) surrounding the active site are diversified in different subtypes of NAs (Fig. 5), and the 430-loop coordinates the conformation of the 150-loop. A series of explorations targeting these two sites has been reported (Feng et al. 2013; Xie et al. 2014). Structural analysis indicates that the 270-loop of group 1 NAs and group 2 NAs (without N3) are located on opposite sides, whereas the 270-loop of N3 is in the middle. This unique 270-loop results from the specific interactions between Q273, Y252, K270, and Y316. An additional residue, K271, forms hydrogen bonds with the carboxyl oxygen of T314, which also contributes to stabilize the 270-loop. The 340-loop plays an essential role in the conserved NA calcium-binding. Compared with the NA structures of other known influenza virus, the 340-loop of N6 and N7 are oriented further away from the conserved calcium ion because of the hydrogen bond with specific residue S326. Residue P347 and the additional proline, P345 in N6 and P344 in N7, may be another essential factor in determining the unique conformation of the 340-loop. Importantly, these unique 340-loops result in a distinct 340-cavity adjacent to the active site in both N6 and N7, which may be a potential inhibitor target.
The Drug Resistance Profile of Ny
Currently, NA inhibitors (NAIs) are widely used as antiviral drugs for the treatment and chemoprophylaxis of influenza infections. Four NAIs, oseltamivir, zanamivir, peramivir, and laninamivir, are commercially available (Webster and Govorkova 2014). These NAIs have been developed to mimic the natural substrate of NA, SA, thereby preventing the cleavage function of NA through competition (Kim et al. 1998). The emergence of NAI-resistant influenza viruses is mainly related to the use of NAIs, although in some cases it is difficult to find direct evidence. Through sequence comparison, NAI resistance-associated amino acid substitutions were determined, which affected the binding affinity between the NA enzyme and the inhibitor and allowed influenza viruses to evade the action of NAIs (Gubareva 2004). Therefore, it is important to classify and monitor NAI resistance-associated substitutions in circulating human seasonal influenza viruses and pandemic potential zoonotic influenza viruses.
Because of the highly conserved active sites in IAV subtypes (Colman 1994; Gamblin and Skehel 2010), viruses carrying substitutions at or nearby the eight catalytic residues and 11 framework residues show a various susceptibility to different NAIs, from susceptibility, normal inhibition, and reduced inhibition to highly reduced inhibition (HRI) (WHO 2012). In 2013–2014, global surveillance of the susceptibility of human influenza viruses performed by the World Health Organization (WHO) Collaborating Centres reported that A(H1N1)pdm09 with NA H274Y, A(H3N2) with NA E119V, B/Victoria-lineage with NA E119G, and B/Yamagata-lineage with NA H274Y, showed HRI against at least one of the four NAIs (Takashita et al. 2015). In a recent WHO report in September 2019, 33 of 4626 tested influenza A(H1N1)pdm09 viruses, 1 of 3424 H3N2 viruses, and 18 of the 1727 influenza B viruses showed reductions in susceptibility to one or more of the inhibitors (https://www.who.int/influenza/vaccines/en/). NA inhibitor resistance-associated amino acid substitutions have also been detected in zoonotic influenza viruses isolated from humans, such as A(H5N1) with NA H274Y (Le et al. 2005) and A(H7N9) with NA R292K (Zhu et al. 2018).
Evidently, each NA type/subtype may prefer different NAI resistance markers. This reflects differences in the intrinsic chemical properties of drugs and differences in the structures of viral enzymes (Reece 2007). Because of widespread use in clinics against seasonal influenza, NAI resistance markers of IAVs of N1 and N2 subtypes and influenza B viruses have been well studied and summarized (Nguyen et al. 2012). Some of the other subtypes, such as N3-N9, sporadically spill over from birds and infect humans, indicating their pandemic potential (Table 1). Song et al. developed an NA gene-fragmented random mutagenesis method for screening mutations and conferring reduced susceptibility to NAIs among avian-origin N3-N9 (Song et al. 2015; Choi et al. 2018). Some NAI markers are only detected in one type/subtype, such as 117V, 149A and 155H in A(N1), 151E/V/D in A(N2), 293N in A(N7), and 402S in influenza B. The NAI markers at 119, 274, and 292 could be functional among type/subtypes.
Table 1.
Reported molecular markers that reduce the susceptibility of neuraminidase inhibitors for each influenza type/subtype
| Site in N2 numbering | Influenza A subtypes | Influenza B | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N3 | N4 | N5 | N6 | N7 | N8 | N9 | ||
| 116 | Aa | D | ||||||||
| 117 | V | |||||||||
| 119 | V/A/G | D/I/V | G | V | D | V/G/D | V/A/G/D | V/A/G/D | ||
| 136 | K/L/R | K/L | K | K | ||||||
| 147 | I | V | ||||||||
| 149 | A | |||||||||
| 151 | E/V/D | |||||||||
| 152 | W | K | K | |||||||
| 155 | H | |||||||||
| 198 | G | E/N/Y | ||||||||
| 222 | K/R/V/M | M/T/R/K | L/T/V | |||||||
| 224 | K | |||||||||
| 246 | N/G | V | T | |||||||
| 247 | P | |||||||||
| 274 | Y | Y | Y | Y | Y | Y | Y | Y | ||
| 276 | D | D | D | |||||||
| 292 | K | K | K | K | K | K | K | K | ||
| 293 | N | |||||||||
| 294 | S | S | S | |||||||
| 371 | K | K | K | |||||||
| 402 | S | |||||||||
| 427 | T | L | ||||||||
aCapital letters are amino acid abbreviations.
The NAI resistance mutation, such as H274Y on NA of A(H1N1)pdm09, could impair enzyme activity and the balance between HA and NA proteins, resulting in lower viral fitness (Brookes et al. 2011). However, substitutions in HA elicited simultaneously to changes in the NA protein may function as permissive substitutions to maintain the HA/NA balance. Thus, different combinations of HA and NA subtypes or their continuing evolution may affect the resistance of NAI markers and permissive mutations. Therefore, the assessment of new emerging influenza viruses needs to be sustained.
THE MATCHING PATTERN OF THE GENE SEGMENTS IN HxNy
Reassortment is the process in which viruses possessing segmented genomes, such as IAVs, exchange gene segments, leading to the emergence of a new subtype/genotype of viruses. The reshuffling of genetic materials provides the possibility of rapidly forming variable progeny viruses, which is essential to increase genetic diversity and promotes the evolution of IAVs, especially when it occurs between avian- and mammalian-origin IAVs (Marshall et al. 2013). Some of these descendants may acquire both antigenic novelty, likely leading to antigenic shift, and the ability to spread efficiently through a human population, which could become epidemiologically privileged strains and have the potential to cause influenza pandemics (White and Lowen 2018).
The genome of AIVs shows high heterogenicity and flexibility, and accordingly, frequent genetic reassortment has been observed among the AIVs in wild birds and LPMs. In tracing the transmission and evolution of the novel human-infecting H7N9 influenza virus, a dynamic reassortment model was proposed to illustrate the evolutionary pattern of H7N9 AIVs (Liu et al. 2013). In this model, the two surface glycoprotein genes, HA and NA, were relatively stable. However, after an H7N9 virus was transmitted to a new region, it would reassort with H9N2 AIVs circulating locally and obtain one to six internal gene segments from H9N2, generating a series of novel H7N9 genotypes (Fig. 6). Meanwhile, dynamic reassortment was also found in H4N2, H5N6, H10N8, and HPAIV H7N9 influenza viruses (Wu et al. 2013a; Zhang et al. 2014; Bi et al. 2016; Gao et al. 2016; Quan et al. 2018a,b). Apart from the internal genes, interlineage reassortment of the NA gene segment was also found in the HPAIV H7N9 variants. Therefore, the dynamic reassortment process, especially among the internal genes, is quite flexible and accounts for the major evolutionarily driving force of AIVs.
Figure 6.
The matching of internal gene segments within H7N9 and H9N2. Poultry act as a “mixing vessel” (in the blue box) for the emergence of different strains of H7N9. When H7N9 and H9N2 avian influenza viruses coinfect the same cell via HA glycoproteins their genomes replicate synchronously. If the packaging signals from viral ribonucleoprotein complexes (vRNPs) of the same segments are the same, the genomic segments from different viruses may assemble together. The progeny viruses with appropriate adaptability, transmissibility, and compatibilities will successfully survive and jump to the new host. Different colors represent different gene segments of H7N9 and H9N2 avian influenza viruses. (Magenta) Internal genes of H7N9, (green) HA and NA of H7N9, (blue) internal genes of H9N2, (gray) HA and NA of H9N2.
However, being a virus with eight separate gene segments, an infectious virion requires a constellation of all the segments to constitute the virus genome. Many studies show that this constellation is specific because of the potential segment mismatch at the RNA and protein levels, although some researchers believe that genetic reassortment may be a random process (Lowen 2017; White and Lowen 2018). As most mutations are deleterious, most reassortment events result in genetic incompatibilities and fitness defects of the progeny viruses. Therefore, most of the reassorted influenza viruses may be negatively selected by the incompatibility among viral genes/proteins.
First of all, the interaction and functional balance between the two surface glycoprotein genes, HA and NA, in AIVs, have been found to be closely related to viral replication efficiency and fitness (Mitnaul et al. 2000). Lineage-specific HA-NA matching patterns have also been reported in H5N6 AIVs (Bi et al. 2016). In terms of human influenza, Xu and colleagues (Xu et al. 2012b) found that both A/Japan/305+/1957 (H2N2) and A/Hong Kong/68 (H3N2) had strong HA binding and high NA activity, whereas A/California/04/2009 (H1N1) displayed weak HA receptor binding with weak NA activity, which demonstrated a remarkable correlation between HA and NA in these pandemic viruses. In addition, despite apparently ample opportunity for intersubtype reassortment among seasonal IAVs, epidemiological evidence shows that only a few reassortant viruses achieve sustained circulation. All of this evidence suggests that evolutionary constraints lead to negative selection of the reassortants. However, the disrupted balance and fitness between HA and NA functions can be recovered through mutations in HA, NA, or both (Mitnaul et al. 2000).
Reassortment between influenza viruses is also restricted by limited compatibility among the viral ribonucleoprotein (vRNP) complex subunits, including PB2, PB1, PA, and NP. The amino terminus of PB1 (amino acid positions 1–15) interacts with PA, and its carboxyl terminus (amino acid positions 685–756) has PB2-binding domains, which interact with polymerase and oligomeric NP to form vRNP complexes (Oldstone and Compan 2014). To exert biological function, PB2 accurately captures the cellular pre-mRNAs to generate 5' capped RNA fragments, which are cleaved by the amino-terminal endonuclease domain of PA, and then PB1 catalyzes RNA synthesis. Therefore, the interaction among the three polymerase subunits is intimately linked, which is critical to virus replication efficiency and pathogenicity.
Octaviani and colleagues designed an experiment in which every segment of a seasonal H1N1 virus, A/Kawasaki/UT-SAI-23/2008(H1N1), was substituted individually by a pandemic A(H1N1)pdm09 virus using reverse genetics. They found that reassortants containing PB2, PA, or NP genes derived from the pandemic virus could not be rescued. In particular, reassortants with the A(H1N1)pdm09 PB2 had the lowest polymerase activity (Octaviani et al. 2011). Although more than half (58/95) of the A/California/4/2009 (A(H1N1)pdm09) variants that contained the polymerase genes PB2, PB1, and PA from seasonal H3N2 and avian H5N1 could be rescued, 23 of the 50 available reassortants exhibited low or impaired replication and transcription activities. Moreover, in most of the reassortants, PB2 and PB1 were reassorted together (Song et al. 2011). However, when the avian H5N1 virus genome was replaced by at least one internal gene from human H3N2 influenza, most chimeric constellations were deleterious for viral replication in cell culture. Yet, in vitro, the remaining reassortants, most of which contained original avian-like PB1, showed high replication efficiency (Chen et al. 2008).
CONCLUDING REMARKS
The unpredictable occurrence of new subtype HxNy influenza may lead to the next pandemic, posing a continuous threat to global public health. Based on the main points raised here, we suggest two primary strategies: global cooperation-based surveillance and applied basic research to enhance the existing cross-department and multidisciplinary measures to prevent and control influenza epidemics and pandemics. Continuous prevalence of diverse subtypes of IAVs among wild birds, mammals, and especially LPMs provides a range of possibilities for the emerging of human-infecting viruses with the potential for a pandemic. Thus, one-health-based surveillance, which concerns both human and animal hosts, is a basic strategy and prerequisite for influenza control and prevention. Meanwhile, applied basic research would enable us to understand the mechanisms and patterns of the emergence of new HxNy and the development and utilization of corresponding vaccines and drugs. HA is one of the primary determinants of whether the influenza virus binds to different hosts and is also the target of antibodies. Thus, the investigation of the molecular features of Hx would benefit the development of vaccines and monoclonal antibody-based drugs. Considering the suboptimal performance of the current HA-oriented influenza vaccine, the understanding of the reassortment mechanism for the gene segments, especially the six internal segments, may shed light on the development of a universal vaccine. Meanwhile, as the target of globally used anti-flu drugs, NA-oriented drug resistance surveillance and investigation of the underpinned molecular mechanism would benefit next-generation drug development for preparedness of pandemics. The combined evidence suggests that integrated anti-influenza activity provides a preeminent model for the surveillance network and basic research against other infectious diseases.
ACKNOWLEDGMENTS
We thank Drs. Chuansong Quan, Hao Song, and Ying Xu for their contributions during the preparation of this manuscript.
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Air GM, Laver WG. 1989. The neuraminidase of influenza virus. Proteins 6: 341–356. 10.1002/prot.340060402 [DOI] [PubMed] [Google Scholar]
- Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, et al. 2012. Emergence of fatal avian influenza in New England harbor seals. MBio 3: e00166-12. 10.1128/mBio.00166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao CJ, Cui LB, Zhou MH, Hong L, Gao GF, Wang H. 2013. Live-animal markets and influenza A (H7N9) virus infection. N Engl J Med 368: 2337–2339. 10.1056/NEJMc1306100 [DOI] [PubMed] [Google Scholar]
- Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, et al. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20: 810–821. 10.1016/j.chom.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Bi YH, Tan SG, Yang Y, Wong G, Zhao M, Zhang QC, Wang Q, Zhao XN, Li LQ, Yuan J, et al. 2019. Clinical and immunological characteristics of human infections with H5N6 avian influenza virus. Clin Infect Dis 68: 1100–1109. 10.1093/cid/ciy681 [DOI] [PubMed] [Google Scholar]
- Butt KM, Smith GJD, Chen HL, Zhang LJ, Leung YHC, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JSM, et al. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43: 5760–5767. 10.1128/JCM.43.11.5760-5767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan RJ, Early G, Kida H, Hinshaw VS. 1995. The appearance of H3 influenza viruses in seals. J Gen Virol 76: 199–203. 10.1099/0022-1317-76-1-199 [DOI] [PubMed] [Google Scholar]
- Chen LM, Davis CT, Zhou H, Cox NJ, Donis RO. 2008. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog 4: e1000072. 10.1371/journal.ppat.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Yuan H, Gao RB, Zhang JX, Wang DY, Xiong Y, Fan GY, Yang F, Li XD, Zhou JF, et al. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383: 714–721. 10.1016/S0140-6736(14)60111-2 [DOI] [PubMed] [Google Scholar]
- Choi WS, Jeong JH, Kwon JJ, Ahn SJ, Lloren KKS, Kwon HI, Chae HB, Hwang J, Kim MH, Kim CJ, et al. 2018. Screening for neuraminidase inhibitor resistance markers among avian influenza viruses of the N4, N5, N6, and N8 neuraminidase subtypes. J Virol 92: e01580-17. 10.1128/JVI.01580-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351: 472–477. 10.1016/S0140-6736(97)11212-0 [DOI] [PubMed] [Google Scholar]
- Colman PM. 1994. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci 3: 1687–1696. 10.1002/pro.5560031007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BJ, Jin LM, Lau EHY, Liao QH, Wu P, Jiang H, Tsang TK, Zheng JD, Fang VJ, Chang ZR, et al. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382: 129–137. 10.1016/S0140-6736(13)61171-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, et al. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5: 3142. 10.1038/ncomms4142 [DOI] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RA. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33: 823–841. 10.1002/embj.201387442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deJong JC, Claas ECJ, Osterhaus ADME, Webster RG, Lim WL. 1997. A pandemic warning? Nature 389: 554. 10.1038/39218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Peng W, Grant OC, Thompson AJ, Zhu X, Bouwman KM, de la Pena ATT, van Breemen MJ, Ambepitiya Wickramasinghe IN, de Haan CAM, et al. 2017. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog 13: e1006390. 10.1371/journal.ppat.1006390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Sabesan S, Skehel JJ, Wiley DC. 1997. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 232: 19–31. 10.1006/viro.1997.8526 [DOI] [PubMed] [Google Scholar]
- Feng E, Shin WJ, Zhu X, Li J, Ye D, Wang J, Zheng M, Zuo JP, No KT, Liu X, et al. 2013. Structure-based design and synthesis of C-1- and C-4-modified analogs of zanamivir as neuraminidase inhibitors. J Med Chem 56: 671–684. 10.1021/jm3009713 [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Skehel JJ. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285: 28403–28409. 10.1074/jbc.R110.129809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, et al. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303: 1838–1842. 10.1126/science.1093155 [DOI] [PubMed] [Google Scholar]
- Gao GF. 2014. Influenza and the live poultry trade. Science 344: 235. 10.1126/science.1254664 [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368: 1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- Gao R, Pan M, Li X, Zou X, Zhao X, Li T, Yang H, Zou S, Bo H, Xu J, et al. 2016. Post-mortem findings in a patient with avian influenza A (H5N6) virus infection. Clin Microbiol Infect 22: 574.e1–574.e5. 10.1016/j.cmi.2016.03.017 [DOI] [PubMed] [Google Scholar]
- Goldstein T, Mena I, Anthony SJ, Medina R, Robinson PW, Greig DJ, Costa DP, Lipkin WI, García-Sastre A, Boyce WM. 2013. Pandemic H1N1 influenza isolated from free-ranging Northern Elephant Seals in 2010 off the central California coast. PLoS One 8: e62259. 10.1371/journal.pone.0062259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Vijaykrishna D, Bahl J, Zhu H, Wang J, Smith GJ. 2010. The emergence of pandemic influenza viruses. Protein Cell 1: 9–13. 10.1007/s13238-010-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV. 2004. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res 103: 199–203. 10.1016/j.virusres.2004.02.034 [DOI] [PubMed] [Google Scholar]
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2003. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309: 209–218. 10.1016/S0042-6822(03)00068-0 [DOI] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336: 1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw VS, Bean WJ, Webster RG, Rehg JE, Fiorelli P, Early G, Geraci JR, Staubin DJ. 1984. Are seals frequently infected with avian influenza viruses. J Virol 51: 863–865. 10.1128/JVI.51.3.863-865.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw VS, Bean WJ, Geraci J, Fiorelli P, Early G, Webster RG. 1986. Characterization of two influenza A viruses from a pilot whale. J Virol 58: 655–656. 10.1128/JVI.58.2.655-656.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X, Cui LB, Chen C, Wang D, Qi X, Zhou MH, Guo X, Wang F, Liu WJ, Kong W, et al. 2018a. Severe human infection with a novel avian-origin influenza A(H7N4) virus. Science Bulletin 63: 1043–1050. 10.1016/j.scib.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486: 420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveil 18: 20453. [PMC free article] [PubMed] [Google Scholar]
- Kim CU, Lew W, Williams MA, Wu H, Zhang L, Chen X, Escarpe PA, Mendel DB, Laver WG, Stevens RC. 1998. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J Med Chem 41: 2451–2460. 10.1021/jm980162u [DOI] [PubMed] [Google Scholar]
- Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, et al. 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522: 102–105. 10.1038/nature14348 [DOI] [PubMed] [Google Scholar]
- Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, Nguyen KH, Pham ND, Ngyen HH, Yamada S, Muramoto Y, et al. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437: 1108. 10.1038/4371108a [DOI] [PubMed] [Google Scholar]
- Li Q, Qi J, Zhang W, Vavricka CJ, Shi Y, Wei J, Feng E, Shen J, Chen J, Liu D, et al. 2010. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol 17: 1266–1268. 10.1038/nsmb.1909 [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou L, Zhou MH, Chen ZP, Li FR, Wu HY, Xiang NJ, Chen EF, Tang FY, Wang DY, et al. 2014. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med 370: 520–532. 10.1056/NEJMoa1304617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao QY, Wu P, Lam WWT, Fang VJ, Wu JT, Leung GM, Fielding R, Cowling BJ. 2016. Public risk perception and attitudes towards live poultry markets before and after their closure due to influenza A(H7N9), Hong Kong, January–February 2014. J Public Health 38: 34–43. 10.1093/pubmed/fdv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Xiong XL, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu JF, Skehel JJ, et al. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci 109: 21474–21479. 10.1073/pnas.1218841110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster M, van Boheemen S, de Graaf M, Schrauwen EJA, Lexmond P, Manz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, et al. 2014. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157: 329–339. 10.1016/j.cell.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci 106: 17175–17180. 10.1073/pnas.0906849106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi WF, Shi Y, Wang DY, Xiao HX, Li W, Bi YH, Wu Y, Li XB, Yan JH, et al. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381: 1926–1932. 10.1016/S0140-6736(13)60938-1 [DOI] [PubMed] [Google Scholar]
- Liu J, Xiao H, Wu Y, Liu D, Qi X, Shi Y, Gao GF. 2014. H7N9: a low pathogenic avian influenza A virus infecting humans. Curr Opin Virol 5: 91–97. 10.1016/j.coviro.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Tan S, Zhao M, Quan C, Bi Y, Wu Y, Zhang S, Zhang H, Xiao H, Qi J, et al. 2016. Cross-immunity against avian influenza A(H7N9) virus in the healthy population is affected by antigenicity-dependent substitutions. J Infect Dis 214: 1937–1946. 10.1093/infdis/jiw471 [DOI] [PubMed] [Google Scholar]
- Liu WJ, Bi Y, Wang D, Gao GF. 2018. On the centenary of the Spanish flu: being prepared for the next pandemic. Virol Sin 33: 463–466. 10.1007/s12250-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC. 2017. Constraints, drivers, and implications of influenza A virus reassortment. Annu Rev Virol 4: 105–121. 10.1146/annurev-virology-101416-041726 [DOI] [PubMed] [Google Scholar]
- Lu X, Shi Y, Zhang W, Zhang Y, Qi J, Gao GF. 2013. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4: 502–511. 10.1007/s13238-013-3906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lvov DK, Zdanov VM, Sazonov AA, Braude NA, Vladimirtceva EA, Agafonova LV, Skljanskaja EI, Kaverin NV, Reznik VI, Pysina TV, et al. 1978. Comparison of influenza viruses isolated from man and from whales. Bull World Health Organ 56: 923–930. [PMC free article] [PubMed] [Google Scholar]
- Ma W, Kahn RE, Richt JA. 2008. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med 3: 158–166. [PMC free article] [PubMed] [Google Scholar]
- Ma C, Lam TTY, Chai YJ, Wang J, Fan XH, Hong WS, Zhang Y, Li LF, Liu YM, Smith DK, et al. 2015. Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol 89: 3534–3541. 10.1128/JVI.03167-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova-Petkova A, Laplante J, Jang YH, Lynch B, Zanders N, Rodriguez M, Jones J, Thor S, Hodges E, De la Cruz JA, et al. 2017. Avian influenza A(H7N2) virus in human exposed to sick cats, New York, USA, 2016. Emerg Infect Dis 23: 2046–2049. 10.3201/eid2312.170798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. 2013. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog 9: e1003421. 10.1371/journal.ppat.1003421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74: 8502–8512. 10.1128/JVI.74.18.8502-8512.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74: 6015–6020. 10.1128/JVI.74.13.6015-6020.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Desselberger U, Palese P. 1978. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature 274: 334–339. 10.1038/274334a0 [DOI] [PubMed] [Google Scholar]
- Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459: 931–939. 10.1038/nature08157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Fry AM, Gubareva LV. 2012. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 17: 159–173. 10.3851/IMP2067 [DOI] [PubMed] [Google Scholar]
- Octaviani CP, Goto H, Kawaoka Y. 2011. Reassortment between seasonal H1N1 and pandemic (H1N1) 2009 influenza viruses is restricted by limited compatibility among polymerase subunits. J Virol 85: 8449–8452. 10.1128/JVI.05054-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MBA, Compan RW. 2014. Influenza pathogenesis and control—Volume I. Springer, New York. [Google Scholar]
- Organization Pan American Health. 2004. Avian influenza virus A (H10N7) circulating among humans in Egypt. Emerging Infectious Diseases weekly updates. [EB/OL]. http://new.paho.org/hq/dmdocuments/2010/Avian_Influenza_Egypt_070503.pdf.
- Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. 2000. Influenza B virus in seals. Science 288: 1051–1053. 10.1126/science.288.5468.1051 [DOI] [PubMed] [Google Scholar]
- Palese P, Tobita K, Ueda M, Compans RW. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61: 397–410. 10.1016/0042-6822(74)90276-1 [DOI] [PubMed] [Google Scholar]
- Pappas C, Viswanathan K, Chandrasekaran A, Raman R, Katz JM, Sasisekharan R, Tumpey TM. 2010. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS One 5: e11158. 10.1371/journal.pone.0011158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez D, Rimstad E, Smith G, Kochs G, McCauley JW, et al. 2011. Orthomyxoviridae. In Virus taxonomy (eds. King AMQ, Adams MJ, Carstens EB, et al. ), pp. 749–762. Elsevier, Oxford. [Google Scholar]
- Puzelli S, Rossini G, Facchini M, Vaccari G, Di Trani L, Di Martino A, Gaibani P, Vocale C, Cattoli G, Bennett M, et al. 2014. Human infection with highly pathogenic A(H7N7) avian influenza virus, Italy, 2013. Emerg Infect Dis 20: 1745–1749. 10.3201/eid2010.140512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan C, Huang T, Chen X, Zhang J, Wang Q, Zhang C, Zhang T, Zhou L, Shu L, Long C, et al. 2018a. Genomic characterizations of H4 subtype avian influenza viruses from live poultry markets in Sichuan province of China, 2014-2015. Sci China Life Sci 61: 1123–1126. 10.1007/s11427-018-9327-4 [DOI] [PubMed] [Google Scholar]
- Quan C, Shi W, Yang Y, Yang Y, Liu X, Xu W, Li H, Li J, Wang Q, Tong Z, et al. 2018b. New threats from H7N9 influenza virus: spread and evolution of high- and low-pathogenicity variants with high genomic diversity in wave five. J Virol 92: e00301-18. 10.1128/JVI.00301-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece PA. 2007. Neuraminidase inhibitor resistance in influenza viruses. J Med Virol 79: 1577–1586. 10.1002/jmv.20951 [DOI] [PubMed] [Google Scholar]
- Reid AH, Fanning TG, Hultin JV, Taubenberger JK. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci 96: 1651–1656. 10.1073/pnas.96.4.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173: 317–322. 10.1016/0042-6822(89)90249-3 [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304: 76–78. 10.1038/304076a0 [DOI] [PubMed] [Google Scholar]
- Rudrawar S, Dyason JC, Rameix-Welti MA, Rose FJ, Kerry PS, Russell RJ, van der Werf S, Thomson RJ, Naffakh N, von Itzstein M. 2010. Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat Commun 1: 113. 10.1038/ncomms1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443: 45–49. 10.1038/nature05114 [DOI] [PubMed] [Google Scholar]
- Russell CA, Fonville JM, Brown AE, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, et al. 2012. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336: 1541–1547. 10.1126/science.1222526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi WF, Shi Y, Wu Y, Liu D, Gao GF. 2013a. Origin and molecular characterization of the human-infecting H6N1 influenza virus in Taiwan. Protein Cell 4: 846–853. 10.1007/s13238-013-3083-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, et al. 2013b. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342: 243–247. 10.1126/science.1242917 [DOI] [PubMed] [Google Scholar]
- Shi Y, Wu Y, Zhang W, Qi J, Gao GF. 2014. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol 12: 822–831. 10.1038/nrmicro3362 [DOI] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS, Guan Y. 2009a. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci 106: 11709–11712. 10.1073/pnas.0904991106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, et al. 2009b. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Song MS, Pascua PN, Lee JH, Baek YH, Park KJ, Kwon HI, Park SJ, Kim CJ, Kim H, Webby RJ, et al. 2011. Virulence and genetic compatibility of polymerase reassortant viruses derived from the pandemic (H1N1) 2009 influenza virus and circulating influenza A viruses. J Virol 85: 6275–6286. 10.1128/JVI.02125-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Marathe BM, Kumar G, Wong SS, Rubrum A, Zanin M, Choi YK, Webster RG, Govorkova EA, Webby RJ. 2015. Unique determinants of neuraminidase inhibitor resistance among N3, N7, and N9 avian influenza viruses. J Virol 89: 10891–10900. 10.1128/JVI.01514-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Qi J, Xiao H, Bi Y, Zhang W, Xu Y, Wang F, Shi Y, Gao GF. 2017. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep 20: 1201–1214. 10.1016/j.celrep.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Su S, Bi Y, Wong G, Gray GC, Gao GF, Li S. 2015. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol 89: 8671–8676. 10.1128/JVI.01034-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279: 393–396. 10.1126/science.279.5349.393 [DOI] [PubMed] [Google Scholar]
- Sun X, Shi Y, Lu X, He J, Gao F, Yan J, Qi J, Gao GF. 2013. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep 3: 769–778. 10.1016/j.celrep.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, Vavricka CJ, Gao GF. 2014. Structure of influenza virus N7: the last piece of the neuraminidase “jigsaw” puzzle. J Virol 88: 9197–9207. 10.1128/JVI.00805-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E, Meijer A, Lackenby A, Gubareva L, Rebelo-de-Andrade H, Besselaar T, Fry A, Gregory V, Leang SK, Huang W, et al. 2015. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013-2014. Antiviral Res 117: 27–38. 10.1016/j.antiviral.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7: 440–451. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, et al. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci 109: 4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong XC, Weng SS, Xue F, Wu X, Xu TM, Zhang WH. 2018. First human infection by a novel avian influenza A(H7N4) virus. J Infect 77: 255–257. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, et al. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310: 77–80. 10.1126/science.1119392 [DOI] [PubMed] [Google Scholar]
- Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, et al. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 10: 2196–2199. 10.3201/eid1012.040961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vries E, Collins PJ, Vachieri SG, Xiong X, Liu J, Walker PA, Haire LF, Hay AJ, Schutten M, Osterhaus AD, et al. 2012. H1N1 2009 pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog 8: e1002914. 10.1371/journal.ppat.1002914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, McKimm-Breschkin JL, Caldwell JB, Kortt AA, Colman PM. 1992. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 14: 327–332. 10.1002/prot.340140302 [DOI] [PubMed] [Google Scholar]
- Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, et al. 2011. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7: e1002249. 10.1371/journal.ppat.1002249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Qi J, Liu Y, Vavricka CJ, Wu Y, Li Q, Gao GF. 2011. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85: 8431–8435. 10.1128/JVI.00638-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Qi J, Bi Y, Zhang W, Wang M, Zhang B, Wang M, Liu J, Yan J, Shi Y, et al. 2015a. Adaptation of avian influenza A (H6N1) virus from avian to human receptor-binding preference. EMBO J 34: 1661–1673. 10.15252/embj.201590960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhang W, Qi J, Wang F, Zhou J, Bi Y, Wu Y, Sun H, Liu J, Huang C, et al. 2015b. Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus. Nat Commun 6: 5600. 10.1038/ncomms6600 [DOI] [PubMed] [Google Scholar]
- Webster RG, Govorkova EA. 2014. Continuing challenges in influenza. Ann N Y Acad Sci 1323: 115–139. 10.1111/nyas.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Geraci J, Petursson G, Skirnisson K. 1981a. Conjunctivitis in human-beings caused by influenza A virus of seals. N Engl J Med 304: 911. [DOI] [PubMed] [Google Scholar]
- Webster RG, Hinshaw VS, Bean WJ, Vanwyke KL, Geraci JR, Staubin DJ, Petursson G. 1981b. Characterization of an influenza A virus from seals. Virology 113: 712–724. 10.1016/0042-6822(81)90200-2 [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56: 152–179. 10.1128/MMBR.56.1.152-179.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SH, Yang JR, Wu HS. 2013. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Resp Med 1: 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MC, Lowen AC. 2018. Implications of segment mismatch for influenza A virus evolution. J Gen Virol 99: 3–16. 10.1099/jgv.0.000989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2012. Meetings of the WHO working group on surveillance of influenza antiviral susceptibility—Geneva, November 2011 and June 2012. Wkly Epidemiol Rec 87: 369–374. [PubMed] [Google Scholar]
- WHO. 2019a. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2019. https://www.who.int/influenza.
- WHO. 2019b. Human infection with avian influenza A(H7N9) virus—China: update. https://www.who.int/influenza.
- WHO. 2019c. Influenza at the human-animal interface–summary and assessment, from 25 June 2019 to 27 September 2019. https://www.who.int/influenza.
- Wiley DC, Skehel JJ. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann Rev Biochem 56: 365–394. 10.1146/annurev.bi.56.070187.002053 [DOI] [PubMed] [Google Scholar]
- Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, et al. 2013a. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14: 446–452. 10.1016/j.chom.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, et al. 2013b. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 23: 1347–1355. 10.1038/cr.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF. 2013c. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 3: 1551. 10.1038/srep01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. 2014. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22: 183–191. 10.1016/j.tim.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Vavricka CJ, Wu Y, Li Q, Rudrawar S, Thomson RJ, von Itzstein M, Gao GF, Qi J. 2015. Atypical group 1 neuraminidase pH1N1-N1 bound to a group 1 inhibitor. Protein Cell 6: 771–773. 10.1007/s13238-015-0197-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xu D, Huang B, Ma X, Qi W, Shi F, Liu X, Zhang Y, Xu W. 2014. Discovery of N-substituted oseltamivir derivatives as potent and selective inhibitors of H5N1 influenza neuraminidase. J Med Chem 57: 8445–8458. 10.1021/jm500892k [DOI] [PubMed] [Google Scholar]
- Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, et al. 2013a. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497: 392–396. 10.1038/nature12144 [DOI] [PubMed] [Google Scholar]
- Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, et al. 2013b. Receptor binding by an H7N9 influenza virus from humans. Nature 499: 496–499. 10.1038/nature12372 [DOI] [PubMed] [Google Scholar]
- Xiong XL, McCauley JW, Steinhauer DA. 2014. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr Top Microbiol 385: 63–91. [DOI] [PubMed] [Google Scholar]
- Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. 2010. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol 84: 1715–1721. 10.1128/JVI.02162-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. 2012a. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol 86: 982–990. 10.1128/JVI.06322-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012b. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol 86: 9221–9232. 10.1128/JVI.00697-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Peng R, Zhang W, Qi J, Song H, Liu S, Wang H, Wang M, Xiao H, Fu L, et al. 2019. Avian-to-human receptor-binding adaptation of avian H7N9 influenza virus hemagglutinin. Cell Rep 29: 2217–2228.e5. 10.1016/j.celrep.2019.10.047 [DOI] [PubMed] [Google Scholar]
- Yang Y, Wong G, Yang L, Tan S, Li J, Bai B, Xu Z, Li H, Xu W, Zhao X, et al. 2019. Comparison between human infections caused by highly and low pathogenic H7N9 avian influenza viruses in wave five: clinical and virological findings. J Infect 78: 241–248. 10.1016/j.jinf.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Yee KS, Carpenter TE, Cardona CJ. 2009. Epidemiology of H5N1 avian influenza. Comp Immunol Microbiol Infect Dis 32: 325–340. 10.1016/j.cimid.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Zhang W, Shi Y, Lu X, Shu Y, Qi J, Gao GF. 2013a. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science 340: 1463–1467. 10.1126/science.1236787 [DOI] [PubMed] [Google Scholar]
- Zhang W, Shi Y, Qi J, Gao F, Li Q, Fan Z, Yan J, Gao GF. 2013b. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J Virol 87: 5949–5958. 10.1128/JVI.00545-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Bi Y, Tian H, Li X, Liu D, Wu Y. 2014. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis 20: 2076–2079. 10.3201/eid2012.140911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Bi Y, Wang J, Wong G, Shi W, Hu F, Yang Y, Yang L, Deng X, Jiang S, et al. 2017. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect 75: 71–75. 10.1016/j.jinf.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, et al. 2012. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci 109: 18903–18908. 10.1073/pnas.1212579109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Dong J, Zhang Y, Yang L, Li X, Chen T, Zhao X, Wei H, Bo H, Zeng X, et al. 2018. A gene constellation in avian influenza A (H7N9) viruses may have facilitated the fifth wave outbreak in China. Cell Rep 23: 909–917. 10.1016/j.celrep.2018.03.081 [DOI] [PubMed] [Google Scholar]
- Zohari S, Neimanis A, Härkönen T, Moraeus C, Valarcher JF. 2014. Avian influenza A(H10N7) virus involvement in mass mortality of harbour seals (Phoca vitulina) in Sweden, March through October 2014. Euro Surveill 19: 20967. 10.2807/1560-7917.ES2014.19.46.20967 [DOI] [PubMed] [Google Scholar]