Abstract
Auxin signaling and patterning is an inherently complex process, involving polarized auxin transport, metabolism, and signaling, its effect on developmental zones, as well as growth rates, and the feedback between all these different aspects. This complexity has led to an important role for computational modeling in unraveling the multifactorial roles of auxin in plant developmental and adaptive processes. Here we discuss the basic ingredients of auxin signaling and patterning models for root development as well as a series of key modeling studies in this area. These modeling studies have helped elucidate how plants use auxin signaling to compute the size of their root meristem, the direction in which to grow, and when and where to form lateral roots. Importantly, these models highlight how auxin, through patterning of and collaborating with other factors, can fulfill all these roles simultaneously.
Auxin signaling plays a key role in diverse processes governing the growth, development, and environmental adaptation of plant roots. Auxin maxima signal the presence of stem cell niches fueling the growth of individual root organs, with the auxin gradients originating at these maxima impacting the size and activity of the corresponding meristems (Sabatini et al. 1999; Petersson et al. 2009). Environmental factors such as nitrate presence or soil salinity impact these auxin patterns, either directly or via other hormones, resulting in the adaptation of root growth rates to environmental conditions (Krouk et al. 2010; Fu et al. 2019). At the same time, transient asymmetries in auxin signaling underlie asymmetries in elongation growth that enable tropic responses of roots toward gravity (Swarup et al. 2005) and away from salt (Galvan-Ampudia et al. 2013; van den Berg et al. 2016), while no such asymmetries are detected during hydrotropism and phototropism (Shkolnik et al. 2016; Kimura et al. 2018) and a potential role in nutrient tropisms remains to be investigated. In lateral roots (LRs), additional auxin-based processes influence gravitropism, resulting in nonvertical gravitropic set-point angles for LRs (Roychoudhry et al. 2013) to ensure the widening of the root system necessary to explore a larger soil surface area (Lynch and Brown 2001). Again, environmental conditions impact these auxin patterns and hence LR growth angles, with, for example, phosphate starvation inducing the shallow angles characteristic of topsoil foraging (Lynch and Brown 2001) and nitrate deprivation instead inducing steep root growth angles (Roychoudhry et al. 2017). Additionally, temporal changes in auxin signaling confer on subsets of protoxylem-neighboring pericycle cells the competence for future LR formation (De Smet et al. 2007; Moreno-Risueno et al. 2010; Xuan et al. 2016). Whether or not these competent cells subsequently develop into LRs, and at which rate, is again heavily dependent on auxin signaling (Lavenus et al. 2013; Du and Scheres 2018; Santos Teixeira and ten Tusscher 2019). Finally, auxin signaling specifies the timing and location of cell differentiation dictating, for example, protophloem differentiation rate (Marhava et al. 2018) and the patterning of xylem and phloem poles (Mähönen et al. 2006; Bishopp et al. 2011). Auxin signaling thus enables plant roots to compute where to grow and at what rate, when and where, and at what angle to form LRs, and when and where to form which tissue types. Additionally, it allows plant roots to finely tune these decisions to the environmental conditions of individual root tips, the root system, and overall plant status. Despite the tremendous progress that has been and is being made in identifying the molecular players and networks involved in these processes, a complete mechanistic understanding often does not come easily.
Two major factors are complicating our understanding. First, all of these processes have an inherently multiscale and feedback-regulated nature, involving processes ranging from the rapid subcellular relocalization of auxin-exporting PIN proteins (Geldner et al. 2001; Kleine-Vehn and Friml 2008; Kleine-Vehn et al. 2009), cellular gene-expression changes (Mähönen et al. 2014; Santuari et al. 2016), tissue level growth, and hormone patterning (Sabatini et al. 1999; Dello Ioio et al. 2007; Mähönen et al. 2014; Salvi et al. 2020) to slow overall root system growth and architecture development (Nacry et al. 2005). While at widely different spatial and temporal scales, these processes are all influenced and being influenced by one another, making their mechanistic underpinnings hard to disentangle. As a further complication, the major role of cell division, cell elongation, and tissue growth in these processes gives rise to additional feedback and complicates interpretation (Rutten and ten Tusscher 2019). As an example, rapid cell growth may dilute cellular transcription factor or hormone levels (Band et al. 2012), while the combination of cell displacement with stability of a transcription factor may at the same time cause the presence of that factor well outside of its domain of transcription (Mähönen et al. 2014). Also, a phenotype in which cell differentiation occurs at a longer distance from the root tip may be interpreted as a slowdown in differentiation rate (Pacifici et al. 2018) but could potentially also arise from higher division rates enhancing the cumulative displacement of cells out of the meristem.
A second major issue revolves around the specificity of auxin signaling. Surely, tissue-specific differences in the expression of the auxin-response machinery enable the differential interpretation of similar auxin signals in different tissue contexts (Weijers et al. 2005; Rademacher et al. 2011). Likewise, simultaneously expressed response machinery components with different auxin affinity parameters enable the differential interpretation of quantitatively different auxin signals (Villalobos et al. 2012; Shimizu-Mitao and Kakimoto 2014). Still, this does not fully explain how auxin at similar times and tissues and within similar concentration ranges can control stable root developmental zonation, transient directional growth responses, and temporally periodic priming of LRs (van den Berg and ten Tusscher 2018). Additionally, it does not easily explain how auxin may control cell division, cell expansion, and cell differentiation rates (Evans et al. 1994) as well as control the sizes and locations of zones at which these different cellular processes occur (Ishida et al. 2010; Perrot-Rechenmann 2010). A major question is how, under these conditions, additional contextual information is generated and used for a cell to compute and decide what to do in response to the auxin signals it receives.
In this review, we discuss how computational models, through their capacity to integrate processes at various spatiotemporal scales as well as the feedback between them, have enabled us to gain insight in how auxin signaling controls root development, growth, and adaptation. Additionally, we show how in doing so, models have helped shed light on the issue of auxin signaling specificity.
A BRIEF PRIMER ON MULTISCALE ROOT MODELS
In this section, we provide a brief introduction to the different aspects involved in multiscale models of root growth, development, and adaptation.
Root Anatomy
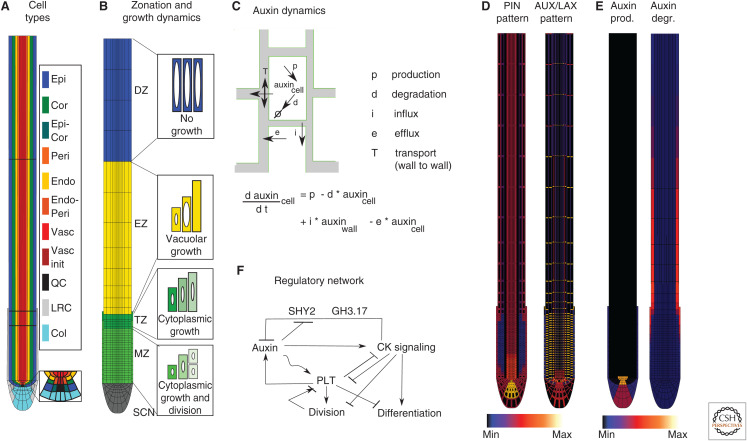
As a first step, one needs to specify the root tissue for which one aims to model the dynamical behavior. Because of both computational efficiency and lack of full three-dimensional data to base the models on, most root models focus either on a longitudinal or transverse cross section of the root. Existing longitudinal root models range from one-dimensional models of a single strand of cells representing a single tissue type (Band et al. 2012; Muraro et al. 2013; Moret et al. 2020) to stylized rectangular models of the entire root tip (Grieneisen et al. 2007; Mironova et al. 2010, 2012; Mähönen et al. 2014; Tian et al. 2014; Moore et al. 2015; Hong et al. 2017; Retzer et al. 2019), to realistically shaped root tip models either idealized from (Cruz-Ramírez et al. 2012; van den Berg et al. 2016; Di Mambro et al. 2017; van den Berg and ten Tusscher 2018; Salvi et al. 2020), or based on, anatomical data (Fig. 1A; Band et al. 2014; Mellor et al. 2016a,b, 2020; Muraro et al. 2016; Liu et al. 2017). One-dimensional models represent a suitable approach if the research question focuses on a single tissue type. In other situations, based on the finding that the absence of a root cap and wedge-shaped root tip has a significant impact on root tip auxin patterning (Cruz-Ramírez et al. 2012; van den Berg et al. 2016), most researchers have moved from using rectangular simplifications to applying realistic root tip anatomies. In addition to incorporating overall root tip shape, two-dimensional models typically incorporate cell-type-specific differences in cell width, as well as the increase in cell length as cells mature from their birth in the meristem, through elongation and subsequent differentiation, creating the typical zoned patterns at the root tip (Fig. 1A,B). While model layouts directly taken from anatomical data may seem the most realistic and therefore preferable, idealized anatomies have the advantage of not introducing any asymmetries, which is important when, for example, studying the origin of auxin asymmetries during tropisms. Most transverse root models are two-dimensional (Péret et al. 2013; De Rybel et al. 2014; Muraro et al. 2014, 2016; el-Showk et al. 2015; Mellor et al. 2017), again either idealized from or directly taken from anatomical data, yet occasionally also one-dimensional, ring-shaped representations (Fàbregas et al. 2015; Mellor et al. 2019) or one-dimensional line sections (Miyashima et al. 2019) have been used.
Figure 1.
Overview of components of root tip models. (A) Root tip architecture indicating the different incorporated cell types. (Epi) Epidermis, (Cor) cortex, (Epi-Cor) epidermis-cortical initials, (Peri) pericycle, (Endo) endodermis, (Endo-Peri) endodermis-pericycle initials, (Vasc) vasculature, (QC) quiescent center, (Col) columella, (LRC) lateral root cap. (B) Layout of the root tip with the locations of the different developmental zones and the growth dynamics occurring inside these zones. (SCN) Stem cell niche, (COL) columella, (MZ) meristematic zone, (TZ) transition zone, (EZ) elongation zone, (DZ) differentiation zone. (C) Schematic depiction of auxin dynamics, with intracellular production and degradation, transmembrane influx into and efflux out of cells, and wall-to-wall transport, and the (simplified) auxin dynamics model equation this results in. (D) Imposed pattern of auxin-exporting PIN and auxin-importing AUX/LAX proteins. (E) Imposed pattern of elevated auxin production and (CK-dependent) degradation. (F) Example network of regulatory interactions (based on data in Salvi et al. 2020).
Auxin Dynamics
To simulate auxin patterning, models incorporate auxin production and degradation, active PIN and ABCB/PGP-mediated auxin export, and passive as well as active AUX/LAX-mediated auxin import (Fig. 1C). Additionally, models incorporate auxin inflow into the vasculature from the (not explicitly modeled) shoot. Typically, cell-type and root-zone-specific levels and polarity of PIN proteins are superimposed on the root anatomy (for an exception, see Fig. 1D; Mironova et al. 2012). In most early models, auxin import was modeled as a constant, cell-type-, and developmental zone-type-independent process (Grieneisen et al. 2007; Mironova et al. 2010, 2012; Mähönen et al. 2014). In Swarup et al. (2005) as well as later models, cell-type- and zone-specific AUX/LAX expression levels were incorporated (Fig. 1D; Band et al. 2014; Di Mambro et al. 2017; Moore et al. 2017; Mellor et al. 2020). Recently, a modeling study also incorporated plasmodesmata, enabling gradient-dependent passive diffusion between neighboring cells (Mellor et al. 2020). On a similar note, early models assumed similar auxin production and degradation across all cells. More recent models incorporate the elevated production of auxin or auxin precursors in the region around the quiescent center and stem cell niche, the columella, and the LR cap (Fig. 1E; Di Mambro et al. 2017; van den Berg and ten Tusscher 2018; Mellor et al. 2020; Salvi et al. 2020). Additionally, some recent models incorporate the GRETCHENHAGEN3.17 (GH3.17)-mediated elevated degradation of auxin in the LR cap and elongation zone epidermis (Fig. 1F; Di Mambro et al. 2017; Salvi et al. 2020).
Although similar in the types of processes considered, models can be divided into two groups based on whether individual cells and cell walls are assumed to have a single homogeneous auxin concentration (Band et al. 2014; Mellor et al. 2020) or whether subcellular and subwall compartment concentration differences are considered (Grieneisen et al. 2007; Moore et al. 2015; van den Berg et al. 2016; Di Mambro et al. 2017; van den Berg and ten Tusscher 2018; Salvi et al. 2020). In the latter type of models, the intracell and intrawall diffusion of auxin is also taken into account. It has thus far remained unclear to what extent such subcompartment auxin gradients exist and whether they are relevant for auxin patterning.
Regulatory Networks
To decipher how auxin signaling impacts growth, developmental, and adaptive decisions in plant roots, root models typically incorporate a regulatory network consisting of the genes and hormone signals considered relevant for the phenomenon under investigation (Fig. 1F). Models thus often incorporate regulation of the auxin-exporting PIN proteins, auxin-importing AUX/LAX proteins, auxin-biosynthesizing TAA and YUCCA enzymes, and auxin-degrading GH3 enzymes that together govern auxin patterns. Additionally, models may incorporate the regulation of the AUX/IAA and ARF factors involved in auxin signaling sensitivity; and downstream, cell-fate- and cell-behavior-controlling factors such as WOX5 and PLETHORAs are modeled (Mähönen et al. 2014; Tian et al. 2014). In addition to auxin dynamics, root models frequently incorporate the dynamics of other hormones such as gibberellic acid (GA) (Band et al. 2012; Muraro et al. 2016), ethylene (Moore et al. 2015), and cytokinin (CK) (signaling) (Muraro et al. 2013, 2016; Moore et al. 2015; Di Mambro et al. 2017; Salvi et al. 2020), which, by affecting auxin transporters, auxin signaling, and downstream factors modulate, antagonize, or synergize with auxin-dependent control of root growth, development, and adaptation.
Cell and Tissue Growth Dynamics
Only a subset of root models explicitly incorporate cell growth, division and expansion, and the resulting tissue growth dynamics (Grieneisen et al. 2007; Mironova et al. 2010; Band et al. 2012; Mähönen et al. 2014; Lebovka et al. 2020; Salvi et al. 2020). Importantly, auxin and gene-expression levels and patterns not only control these growth dynamics, but are subsequently also affected by them, giving rise to additional feedback that is ignored in static root models (Mähönen et al. 2014; Salvi et al. 2020).
So far, models combining hormone, gene expression, and growth dynamics have applied relatively simplistic descriptions of cell and tissue growth, and several aspects of growth dynamics can still be significantly improved. As an example, while mechanically more realistic, root growth models taking into account the symplastic and highly anisotropic growth of root tissue have been developed (Fozard et al. 2013, 2016; Vos et al. 2014; Weise and ten Tusscher 2019), and these have not yet been fully integrated with detailed models of auxin and gene-expression dynamics.
THE FRUITS OF MULTISCALE ROOT MODELING
Computing Zones and Rates of Growth
Mature Root Growth Control
It has long been known that the location of the quiescent center and stem cell niche coincide with an auxin maximum (Sabatini et al. 1999), with the auxin gradient emanating from it impacting root meristem size. Earlier modeling studies focused mostly on the mechanism underlying the formation of the auxin maximum and gradient in the root tip. These studies revealed the important role of the PIN exporter reflux loop (Grieneisen et al. 2007; Mironova et al. 2010) in generating an auxin maximum and gradient and the important modulatory functions of AUX/LAX importers (Band et al. 2014), ethylene, CK cross talk with auxin (Moore et al. 2015; Liu et al. 2017) and, more recently, plasmodesmatal connections (Mellor et al. 2020) for precise auxin patterning. They have been extensively reviewed in Rutten and ten Tusscher (2019).
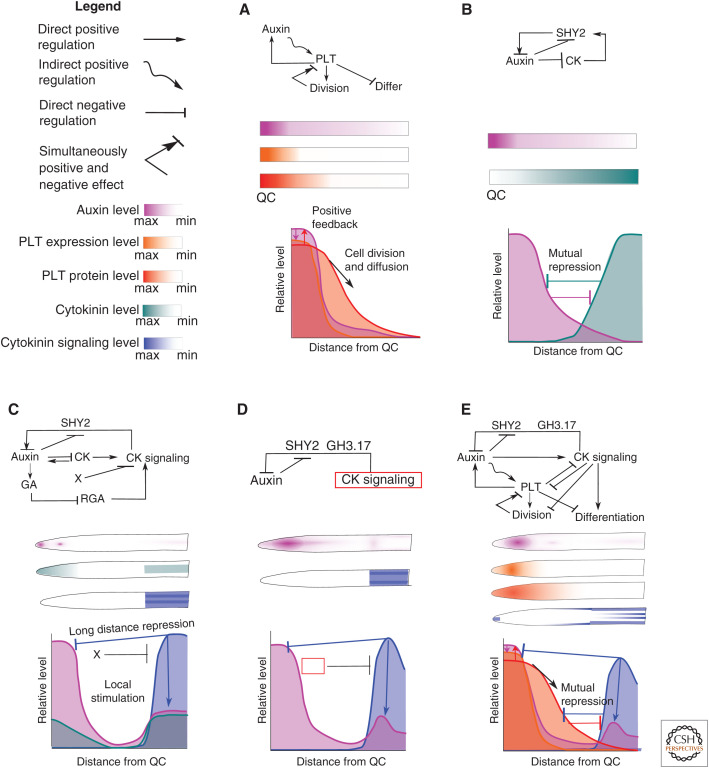
More recent studies have focused on the interplay of auxin with other hormones, particularly its antagonist CK, and how this contributes to the control of root meristem size and root growth (Fig. 2). Because of the discrepancy in sites of CK production (root cap and meristem) (Antoniadi et al. 2015) and CK signaling (transition and elongation zone) (Mason et al. 2004) as well as the intricate feedback between auxin, CK, and other hormones (ethylene and gibberellin among others), reconstructing the auxin-CK patterning network has proven to be highly nontrivial (Moore et al. 2015). Using a model incorporating auxin, CK and GA signaling (Muraro et al. 2016) demonstrated that to correctly simulate CK signaling as occurring outside the meristem, an additional, not yet identified factor operating in the meristem and repressing CK signaling was necessary. Additionally, they demonstrated that CK signaling–dependent repression of PINs resulted in a secondary increase of auxin levels in the vasculature. Using a somewhat tangential approach in which CK signaling was superimposed to only occur in the correct location outside the meristem, Di Mambro and coworkers further investigated the impact of this domain on auxin patterning. They demonstrated that CK-mediated repression of PINs combined with the induction of the GH3.17 auxin-degrading enzyme generates an auxin minimum at the transition zone. In a recent study, Salvi et al. (2020) identify the PLETHORA transcription factors, together with auxin, major determinants of stem cell and meristem status (Galinha et al. 2007), as the factors responsible for repressing CK signaling inside the meristem. Importantly, auxin induces PLT expression (Mähönen et al. 2014), while PLTs through affecting auxin production, degradation, and transport positively affect auxin levels (Santuari et al. 2016), giving rise to a positive feedback loop. Salvi et al. (2020) showed that this positive feedback loop enables the activation and expansion of the root meristem after germination. Additionally, they demonstrated that meristem activation itself, through division-induced dilution of PLETHORA proteins in the distalmost meristem cells, induces the formation of the elongation zone and activation of CK signaling. This CK signaling eventually halts meristem size expansion. Finally, Salvi and coworkers demonstrate that CK not only antagonizes auxin, but also PLTs, either directly (ARR12) or through repressing divisions that contribute to the expansion of the PLT domain (ARR1). By performing hypothetical what-if experiments in which CK antagonizes only auxin or only PLETHORAs, the authors demonstrate that while PLETHORA antagonism enables CK to rapidly exert an effect on meristem size, auxin antagonism is responsible for stronger and longer-term effects on final meristem size.
Figure 2.
Models for auxin, PLT, and cytokinin (CK)-mediated root meristem patterning. (Top to bottom) Simplified cell-level regulatory network, resulting root level patterning of key factors, schematic depiction of tissue-level interactions underlying the tissue patterning. (A) Model for auxin-PLT-based meristem size patterning. (Based on data in Mähonen et al. 2014.) (B) Model for auxin-CK antagonism (additional ethylene interactions not shown). (Figure created from data in Moore et al. 2015.) (C) Model for auxin, gibberellic acid (GA), CK signaling-mediated meristem patterning. (Based on data in Muraro et al. 2016.) (D) Model for auxin and CK signaling–mediated meristem patterning. Red box indicates the superimposed patterning of CK signaling. Auxin-CK: auxin patterning in absence of the superimposed CK signaling. (Figure based on data in Di Mambro et al. 2017.) (E) Model for auxin-PLT-CK signaling–mediated meristem patterning. (Based on data in Salvi et al. 2020.) (QC) Quiescent center.
Thus, the auxin-PLETHORA-CK regulatory network results in two distinct zones: the meristem in which auxin co-occurs with PLETHORAs, and the elongation zone in which auxin co-occurs with CK signaling. Thereby the necessary additional information is generated that enables auxin to control division rates in one domain and elongation, and differentiation rates in the other. Still, this leaves open the question of how auxin can control both the rates of cellular division, expansion, and differentiation processes, as well as—through its interaction with PLTs and CK—the location and size of the domains in which these processes occur. Rapid, transient changes in auxin patterns necessary to, for example, adjust elongation rates during gravitropism, may conflict with the need for long-term, robust control of developmental zonation processes that are also dependent on auxin. A combination of experiments and modeling (Mähönen et al. 2014) revealed critical PLT properties essential to resolve this conflict. The authors used their model to demonstrate that, because of the high auxin levels required for PLETHORA induction, combined with the long timescale for induction, PLETHORA transcription only occurs near the meristem. Additionally, they showed that the stable nature of the PLETHORA protein allows for its maintenance in cells that, as a result of division, are beyond this transcriptional domain. Combined with its cell-to-cell movement through plasmodesmata, this allows for the production of a PLETHORA protein gradient. The result is a slow and stable PLETHORA gradient immune to rapid, transient changes in auxin patterning and capable of maintaining stable root domains. Indeed, using a what-if simulation in which auxin directly controls both elongation rates and root zonation, the authors demonstrated that gravitropism-induced auxin asymmetries would result in a loss of radially coordinated development.
Computing Where to Grow
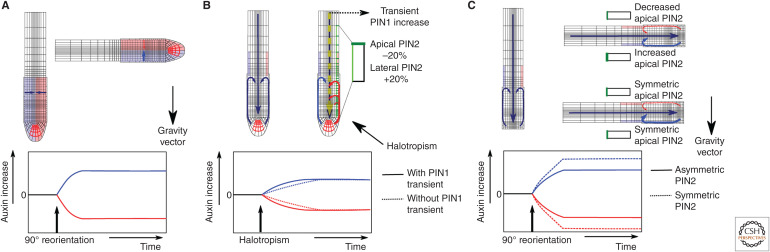
While it has long been known that many directional root growth responses arise from an asymmetry in auxin patterning, the complexities underlying the generation, maintenance, and modulation of these auxin asymmetries are still being unraveled (Fig. 3). A first study combining experiments with modeling to study root tropisms was conducted by Band et al. (2012). Band et al. used a modeling approach to derive auxin levels from DIIVenus measurements during gravitropism, enabling them to quantify gravitropic auxin asymmetry, demonstrating a twofold increase in auxin levels at the lower side.
Figure 3.
Models for root tropism-mediated auxin asymmetry dynamics. Blue = bottom (gravitropism) or nonsalt exposed (halotropism) root side. Red = top (gravitropism) or salt exposed (halotropism) root side. Yellow = change in PIN1 (halotropism only). Green = change in PIN2 membrane patterning. (A) Schematic depiction of the Band et al. (2012) model for root gravitropism. (Top) The two modeled root half compartments and the change in intercompartment auxin exchange upon gravitropic stimulation. (Bottom) Resulting buildup of auxin asymmetry. (B) Schematic depiction of the van den Berg et al. (2016) model for root halotropism. (Top) Change in PIN2 membrane patterning, PIN1-expression level, and the resulting changes in auxin reflux at the salt-exposed and non-exposed root sides. (Bottom) Resulting buildup of auxin asymmetry in presence and absence of transient increase in PIN1 levels. (C) Schematic depiction of the Retzer et al. (2019) model for root gravitropism. (Top) Changes in auxin reflux at the top and bottom root sides in the presence and absence of asymmetric changes in PIN2 membrane levels. (Bottom) Resulting buildup of auxin asymmetry in presence and absence of asymmetric changes in PIN2 membrane levels.
In recent years, roots were also found to be capable of detecting small 5%–10% salt gradients and respond by growing away from the direction of highest salt levels (Galvan-Ampudia et al. 2013). A major question is how roots can translate such a weak and noisy directional signal into a robust directional response. Using a longitudinal root tip model (van den Berg et al. 2016) demonstrated that the originally reported asymmetry in PIN2 can initiate the process of auxin asymmetry formation yet is insufficient to generate a significant auxin asymmetry. Instead, the auxin-dependent expression of the auxin importer AUX1 was shown to be critical in spatially propagating and locally amplifying the auxin asymmetry, while a transient increase in PIN1 was shown to enhance the speed of asymmetry generation (van den Berg et al. 2016). In a more recent study, salt-induced polarization of AUX1 was shown to further contribute to the auxin asymmetry underlying halotropism (Korver et al. 2020).
Intriguingly, while during halotropism PIN2 asymmetry promotes auxin asymmetry, a recent study reported a dampening effect of PIN2 asymmetry on gravitropic auxin asymmetry (Retzer et al. 2019). Previous studies reported that during gravitropism a transient asymmetry in PIN2 levels occurs, with PIN2 levels increasing at the lower, high auxin side and decreasing at the upper, low auxin side (Baster et al. 2013). This PIN2 asymmetry was originally thought to contribute to gravitropic auxin asymmetry. Retzer et al. convincingly show that while a functional PIN2 is required to transport auxin to the elongation zone, the PIN2 asymmetry enhances auxin loss at the lower side while reducing it at the upper side, thereby damping the amplitude of the auxin asymmetry.
An at least partial explanation for the different roles of PIN2 asymmetry reported in the halotropism and gravitropism studies may lie in whether PIN2 changes occur on one or both sides of the root, and whether or not lateral PIN2 changes occur. In the gravitropism study, the combined increase of apical PIN2 on the lower side, reduces auxin accumulation, and decrease of apical PIN2 on the upper side, reduces auxin loss and dampens auxin asymmetry (Retzer et al. 2019). In contrast, in the halotropism study, PIN2 changes only occur on the salt-exposed side and involve the combined decrease of apical and increase of inward lateral PIN2 levels that result in the reshuffling of auxin to the nonsalt exposed side.
Computing Where to Branch
Lateral Root Priming
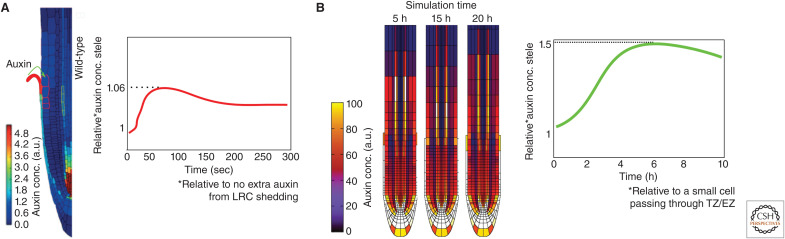
LR formation starts with the priming of subsets of pericycle cells that gain competence for the future formation of LRs. Priming involves periodic oscillations in auxin and/or auxin signaling at the end of the meristem and beginning of the elongation zone. Through growth, this temporal periodicity is transformed into a spatially repetitive pattern of prebranch sites. Several modeling studies have attempted to explain periodic auxin (signaling) oscillations (Lucas et al. 2008; Middleton et al. 2010; Mironova et al. 2010; Mellor et al. 2016b). Here we focus on the two models that take into account root tip morphology and cellular resolution (Fig. 4; Xuan et al. 2016; van den Berg and ten Tusscher 2018). In the paper by Xuan et al. (2016), the authors investigated to what extent periodic apoptosis of the topmost LR cap cells, found to strongly correlate in space and time with pericycle priming, could drive auxin oscillations in the root vasculature. The authors modeled a nongrowing root tip with realistic anatomy and cellular layout, and assumed that, upon apoptosis, all auxin present in the root cap cells was passed on to the apoplast of directly neighboring epidermal cells. Additionally, the authors assumed apoptosis to occur synchronously across the root cap and performed three-dimensional axisymmetric simulations. The authors reported a 2- to 5-minute increase of 6%–12% in vascular auxin levels upon root cap apoptosis (Xuan et al. 2016). Although these auxin increases are lower and shorter than experimentally observed, the model highlights that significant amounts of auxin released from the root cap can be transported to the inner vascular tissues. Additionally, the model demonstrated that, as a result of PIN patterns, the highest auxin accumulation naturally occurs in the outermost vasculature cell file, the protoxylem, in which priming is known to start (De Smet et al. 2007).
Figure 4.
Models for the periodic auxin elevations underlying lateral root priming. (A) Panel created from data in the Xuan et al. (2016) model. (Top) Apoptosis of the uppermost lateral root cap (LRC) cell results in the auxin content of that cell being exported to the cell walls of the neighboring epidermal tissue. (Bottom) Resulting vascular auxin dynamics. (B) (Top) Synergy between root tip reflux loop and growth dynamics lead to emergent periodic auxin elevations. (Panel created from data in the Van den Berg et al. 2018 model.) (Bottom) Resulting vascular auxin dynamics. (TZ) Transition zone, (EZ) elongation zone.
A strong correlation of priming with root cap apoptosis has been observed, yet apoptosis is likely not driving priming given the persistence of priming in smb mutants. van den Berg and ten Tusscher (2018) investigated whether the combination of root growth with root tip auxin patterning could underlie auxin oscillations. They developed a two-dimensional root tip model in which cellular growth, division, expansion, and differentiation dynamics are incorporated. This dynamic model generated oscillations in auxin levels as an emergent outcome of root tip auxin reflux and root growth dynamics. van den Berg and ten Tusscher subsequently took apart their model, removing or modifying different aspects of auxin transport and growth dynamics in an effort to pin down the oscillatory mechanism. First, they demonstrated that the reflux of auxin in the root tip results in an auxin-loading zone at the transition and early elongation zone. Next, they showed that the potential for cells to load auxin increases with cell size, and largely arises from increased passive uptake of auxin through the membrane. Additionally, cellular auxin loading was shown to increase if cells are followed by a smaller cell below, resulting in reduced growth-dependent cumulative displacement and hence more time to grow and load auxin in the relevant domain. Furthermore, since the surface-to-volume ratio increase is largest for vascular cells, these cells load mostly auxin, explaining the preferential loading of auxin in the vasculature. Consistent with the results of Xuan et al. (2015), maximum auxin increase occurred in the vasculature cell file neighboring the pericycle. Finally, the authors demonstrate how the periodic formation of large- and small-cell pairs, generating peak auxin loading in the large cell of the pair, naturally arises from root growth dynamics. The model generated peak oscillation levels resulting in 50%–80% increases in vascular auxin levels that lasted 2–3 hours, which is more consistent with experimental data.
Computing When to Branch
Lateral Root Initiation
After priming, the initial steps in the formation of an actual LR involve the polarization and migration of nuclei in selected prebranch site cells, resulting in the asymmetric divisions with which LR primordium (LRP) development starts (De Smet 2012). Essential for these initial events to occur is the swelling of the pericycle prebranch site cells, for which the shrinkage of overlaying endodermal cells is required (Vermeer et al. 2014). Both this spatial accommodation process and the ensuing asymmetric divisions are auxin dependent (Vermeer et al. 2014; Moret et al. 2020), requiring a highly local elevation of auxin signaling in prebranch site cells (Dubrovsky et al. 2008; De Smet 2010). To investigate potential mechanisms for localized pericycle auxin accumulation, the Grieneisen group first developed a longitudinal root model (Laskowski et al. 2008) and, more recently, a transverse root model (el-Showk et al. 2015). The initial longitudinal model was used to investigate the mechanisms through which mechanical bending may induce LR initiation. Laskowski et al. (2008) demonstrated that root bending, through local expansion of cell lengths in the outer curve of the bend, could initiate auxin accumulation. This potential for auxin accumulation upon bending was shown to be specific for the differentiation zone, which, due to its more apolar patterning of PIN exporters, enables a lateral reflux of auxin essential for auxin accumulation. Additionally, the authors demonstrated that auxin accumulation could be enhanced when incorporating the auxin-dependent induction of AUX1, giving rise to a local positive feedback loop. Finally, Laskowski et al. demonstrated that the decrease in auxin rootward of the bend, through decreased expression and polarity of auxin-dependent PIN3/PIN7, limits rootward while promoting lateral auxin transport, thereby further enhancing the auxin maximum. In their more recent study, Grieneisen and coworkers predominantly focused on vascular patterning. With regard to LR initiation, they demonstrated in this study that the polarity patterns of PIN1 and PIN7 that generate a transverse auxin flux circuit directed toward the xylem axis are critical for auxin-dependent AUX1 expression to result in auxin accumulation specifically in the pericycle (el-Showk et al. 2015).
Nodule development in legumes shares many parallels with LR development, both in terms of involved genes, as well as in the requirement for localized auxin accumulation to start the nodulation process (Franssen et al. 2015; Schiessl et al. 2019), although auxin accumulation occurs in the cortex rather than pericycle. Models of nodule initiation demonstrated that relative to an increase in AUX/LAX-mediated auxin uptake, reduction of PIN-mediated efflux may be even more effective in generating a robust auxin maximum (Deinum et al. 2012, 2016).
Lateral Root Emergence
A next critical step in the development of a new LR is its emergence from within the main root, requiring the buildup of turgor pressure of the LRP relative to its overlaying tissue as well as the separation of cells directly overlaying the LRP (Stoeckle et al. 2018). For this, a tightly localized elevation of auxin signaling resulting in the auxin-dependent modification of water transport and cell wall properties is essential. To investigate the mechanism underlying this localized auxin patterning (Péret et al. 2013), a model of a cross-section of the Arabidopsis root containing a stage 0/I primordium was used, and their modeling was combined with dedicated experiments. The authors first demonstrated that cortical, auxin-dependent expression of the auxin importer LAX3-enabled LRP auxin efflux results in cortical auxin accumulation. However, this auxin accumulation was not tightly constrained to cells overlaying the LRP. If also incorporating that auxin more rapidly induces the PIN3 exporter as compared to LAX3, the more focused PIN3-mediated LRP auxin efflux produced a much more localized auxin and LAX3 activity pattern, consistent with experimental results. Thus, this study again underlines the importance of auxin dependence of auxin importers in generating and amplifying auxin patterns. Additionally, it shows the importance of timing. In an earlier study, using a highly simplified model of water fluxes between an LRP and its surroundings, the same authors had already demonstrated that through an auxin-dependent patterning of aquaporins, the necessary downstream changes in turgor pressure are achieved (Péret et al. 2012).
CONCLUDING REMARKS
In this article, we show how computational models are an invaluable tool in deciphering the complex, multiscale processes through which plant roots use auxin signaling to compute their developmental patterning, growth rate, and direction. Iterating between models and experiments has proven to be a powerful approach to delineate the necessary and sufficient ingredients underlying a particular developmental patterning or adaptation mechanism. This, for example, taught us the importance of PIN exporter polarity patterns, AUX/LAX importer tissue-specific patterns, plasmodesmatal connectivity, and the complex auxin-CK-PLT interaction network in determining root tip auxin pattern, root meristem size, and hence root growth rate (Grieneisen et al. 2007; Band et al. 2014; Mähönen et al. 2014; Di Mambro et al. 2017; Mellor et al. 2020; Salvi et al. 2020).
Through simulating what-if scenarios that are experimentally out of reach, these models additionally provide us with plausible explanations for the selective advantage of particular mechanisms and regulatory network architectures. As an example, what-if simulations assuming a direct control of both gravitropism and developmental zonation by auxin demonstrated that this would cause decorrelated differentiation (Mähönen et al. 2014). Similarly, what-if simulations for CK signaling only antagonizing auxin or PLETHORAs instead of both shows the importance of PLETHORA antagonism for early and auxin antagonism for later stages of meristem development (Salvi et al. 2020). These computational studies also underline that not all explanations should be sought for in genes or hormones, instead highlighting the importance of cell size differences and growth- or bending-induced changes therein for bootstrapping auxin elevations (Laskowski et al. 2008; van den Berg and ten Tusscher 2018). Additionally, by explicitly incorporating cellular and tissue growth processes, models have helped reveal the importance of growth and division-induced dilution for the resetting of cellular hormone and gene-expression patterns that is necessary for cells to switch to the next developmental stage (Salvi et al. 2020).
Taking a computational perspective, modeling studies have also enabled us to decipher exactly how plants use auxin signaling to compute their decision-making processes. In the case of halotropism, it was found that the auxin dependence of the auxin importer AUX1 plays an essential role in amplifying initially small auxin asymmetries, enabling plant roots to translate weak directional signals into a clear decision on the direction of growth (van den Berg et al. 2016). On a similar note, for root vascular patterning, a series of modeling studies have elucidated how the simultaneous auxin-dependent production of CK and AHP6-mediated antagonism of CK signaling modulate PIN-mediated auxin export such that initial small auxin differences are amplified into robustly patterned xylem and phloem poles (De Rybel et al. 2014; Muraro et al. 2014; el-Showk et al. 2015; Mellor et al. 2017). For gravitropism, modeling identified the auxin exporter PIN2 and, for LRs, CK signaling, both antagonizing auxin asymmetry as tuning devices for gravitropic response strength decisions (Retzer et al. 2019; Waidmann et al. 2019). In the case of LR emergence, the sequential auxin-dependent induction of auxin exporters and importers was shown to enable the computation of exactly which overlaying cells should elevate their auxin levels and give way to the growing LRP (Péret et al. 2013).
Finally, taking an information theoretic perspective, modeling studies have helped us understand auxin signaling specificity, by disentangling how auxin may simultaneously convey multiple messages. In root meristems, auxin (signaling) is known to control both meristem size, and hence where cells divide and where they stop dividing and start elongating and differentiating, as well as at which rates cells divide, elongate, and differentiate. Logically, a single auxin signal conveys insufficient information to decide both what processes to perform and at what rate to do this. First, modeling has helped to demonstrate that through directly controlling root elongation rate and, indirectly, via the more stable PLETHORA transcription factors, meristem patterning, auxin can control both rapid root tropisms and robust developmental zonation (Mähönen et al. 2014). Second, modeling highlights how auxin signaling, through interacting with both the PLETHORAs and CK signaling, self-organizes a meristematic PLETHORA and elongation/differentiation CK signaling domain (Salvi et al. 2020). Auxin signaling thus controls what happens where (division vs. expansion/differentiation). Additionally, by teaming up with either PLETHORAs or CK signaling, auxin signaling provides itself with the additional contextual information necessary to also control at which rates these processes take place.
A thus far mostly unexplored territory in the field of plant computational developmental biology is modeling beyond the scale of individual organs. Models simulating overall root system architecture (RSA) traditionally focus on the physiological processes of water and nutrient uptake (Ho et al. 2004). In these models, typically rates of LR formation, main and LR growth, and LR angles or simply final RSA are varied to investigate the consequences for foraging efficiency under different conditions (Rangarajan et al. 2018). Expanding these models to the realm of mechanistic, developmental modeling would enable us to decipher thus far undiscovered, potential feedback between processes such as LR angle and main root growth rate. However, such an expansion not only requires incorporating hormonal–genetic networks controlling different aspects of RSA that may not yet have been fully elucidated, but also plant level fluxes of auxin and other relevant systemic and local signals. A major factor limiting the development of these models is our current lack of detailed knowledge on auxin transport outside the well-studied root tips and forming LRP. In addition to active, PIN-mediated auxin transport (Blilou et al. 2005; Grieneisen et al. 2007), transport of auxin also occurs through the differentiated phloem and is unloaded in meristems (Goldsmith et al. 1974; Ross-Elliott et al. 2017). The relative importance of these two processes, and how this may differ in different regions of the root system and for different developmental stages is currently unclear. Another major limitation lies on the modeling side. While current RSA models are spatiotemporally too coarse grained to successfully incorporate developmental patterning processes, the fine-grained, cell-based models used to simulate individual root tips and LRP are computationally prohibitively complex. Development of root system models at an intermediate spatial resolution and their integration with experiments will likely be the next frontier in deciphering the role of auxin signaling in plant root development and adaptation.
ACKNOWLEDGMENTS
J.R., T.v.d.B., and K.t.T. were supported by Grant No. 864.14.003 of the Netherlands Scientific Organization (NWO).
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, Novák O. 2015. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 27: 1955–1967. 10.1105/tpc.15.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Úbeda-Tomás S, Dyson RJ, Middleton AM, Hodgman TC, Owen MR, Jensen OE, Bennett MJ, King JR. 2012. Growth-induced hormone dilution can explain the dynamics of plant root cell elongation. Proc Natl Acad Sci 109: 7577–7582. 10.1073/pnas.1113632109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, et al. 2014. Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875. 10.1105/tpc.113.119495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baster P, Robert S, Kleine-Vehn J, Vanneste S, Kania U, Grunewald W, De Rybel B, Beeckman T, Friml J. 2013. SCFTIR1/AFB-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J 32: 260–274. 10.1038/emboj.2012.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. 2011. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926. 10.1016/j.cub.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. 2012. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015. 10.1016/j.cell.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum EE, Geurts R, Bisseling T, Mulder BM. 2012. Modeling a cortical auxin maximum for nodulation: different signatures of potential strategies. Front Plant Sci 3: 96. 10.3389/fpls.2012.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum EE, Kohlen W, Geurts R. 2016. Quantitative modelling of legume root nodule primordium induction by a diffusive signal of epidermal origin that inhibits auxin efflux. BMC Plant Biol 16: 254. 10.1186/s12870-016-0935-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682. 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, et al. 2014. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345: 1255215. 10.1126/science.1255215 [DOI] [PubMed] [Google Scholar]
- De Smet I. 2010. Multimodular auxin response controls lateral root development in Arabidopsis. Plant Signal Behav 5: 580–582. 10.4161/psb.11495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I. 2012. Lateral root initiation: one step at a time. New Phytol 193: 867–873. 10.1111/j.1469-8137.2011.03996.x [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, Rybel BD, dit Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci 114: E7641–E7649. 10.1073/pnas.1705833114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2018. Lateral root formation and the multiple roles of auxin. J Exp Bot 69: 155–167. 10.1093/jxb/erx223 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105: 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Showk S, Help-Rinta-Rahko H, Blomster T, Siligato R, Marée AFM, Mähönen AP, Grieneisen VA. 2015. Parsimonious model of vascular patterning links transverse hormone fluxes to lateral root initiation: auxin leads the way, while cytokinin levels out. PLoS Comput Biol 11: e1004450. 10.1371/journal.pcbi.1004450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle MA. 1994. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194: 215–222. 10.1007/BF01101680 [DOI] [Google Scholar]
- Fàbregas N, Formosa-Jordan P, Confraria A, Siligato R, Alonso JM, Swarup R, Bennett MJ, Mähönen AP, Caño-Delgado AI, Ibañes M. 2015. Auxin influx carriers control vascular patterning and xylem differentiation in Arabidopsis thaliana. PLoS Genet 11: e1005183. 10.1371/journal.pgen.1005183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozard JA, Lucas M, King JR, Jensen OE. 2013. Vertex-element models for anisotropic growth of elongated plant organs. Front Plant Sci 4: 233. 10.3389/fpls.2013.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozard JA, Bennett MJ, King JR, Jensen OE. 2016. Hybrid vertex-midline modelling of elongated plant organs. Interface Focus 6: 20160043. 10.1098/rsfs.2016.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen HJ, Xiao TT, Kulikova O, Wan X, Bisseling T, Scheres B, Heidstra R. 2015. Root developmental programs shape the Medicago truncatula nodule meristem. Development 142: 2941–2950. 10.1242/dev.120774 [DOI] [PubMed] [Google Scholar]
- Fu Y, Yang Y, Chen S, Ning N, Hu H. 2019. Arabidopsis IAR4 modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front Plant Sci 10: 522. 10.3389/fpls.2019.00522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, Haring MA, Munnik T, Vernoux T, Testerink C. 2013. Halotropism is a response of plant roots to avoid a saline environment. Curr Biol 23: 2044–2050. 10.1016/j.cub.2013.08.042 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. 2001. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Goldsmith MH, Cataldo DA, Karn J, Brenneman T, Trip P. 1974. The rapid non-polar transport of auxin in the phloem of intact coleus plants. Planta 116: 301–317. 10.1007/BF00390855 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. 10.1038/nature06215 [DOI] [PubMed] [Google Scholar]
- Ho MD, McCannon BC, Lynch JP. 2004. Optimization modeling of plant root architecture for water and phosphorus acquisition. J Theor Biol 226: 331–340. 10.1016/j.jtbi.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Hong JH, Savina M, Du J, Devendran A, Ramakanth KK, Tian X, Sim WS, Mironova VV, Xu J. 2017. A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell 170: 102–113.e14. 10.1016/j.cell.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. 2010. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71. 10.1242/dev.035840 [DOI] [PubMed] [Google Scholar]
- Kimura T, Haga K, Shimizu-Mitao Y, Takebayashi Y, Kasahara H, Hayashi K, Kakimoto T, Sakai T. 2018. Asymmetric auxin distribution is not required to establish root phototropism in Arabidopsis. Plant Cell Physiol 59: 828–840. 10.1093/pcp/pcy018 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J. 2008. Polar targeting and endocytic recycling in auxin-dependent plant development. Ann Rev Cell Dev Biol 24: 447–473. 10.1146/annurev.cellbio.24.110707.175254 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J. 2009. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849. 10.1105/tpc.109.071639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver RA, van den Berg T, Meyer AJ, Galvan-Ampudia CS, ten Tusscher KHWJ, Testerink C. 2020. Halotropism requires phospholipase Dζ1-mediated modulation of cellular polarity of auxin transport carriers. Plant Cell Environment 43: 143–158. 10.1111/pce.13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937. 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CAT, Hogeweg P, Marée AFM, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. 10.1371/journal.pbio.0060307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458. 10.1016/j.tplants.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Lebovka I, Mele BH, Zakieva A, Gursanscky N, Merks R, Greb T. 2020. Computational modelling of cambium activity provides a regulatory framework for simulating radial plant growth. bioRxiv 10.1101/2020.01.16.908715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K. 2017. Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Mol Plant 10: 1480–1496. 10.1016/j.molp.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Lucas M, Guédon Y, Jay-Allemand C, Godin C, Laplaze L. 2008. An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. 10.1371/journal.pone.0003673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. 2001. Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237. 10.1023/A:1013324727040 [DOI] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. 2006. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98. 10.1126/science.1118875 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Bassukas AEL, Zourelidou M, Kolb M, Moret B, Fastner A, Schulze WX, Cattaneo P, Hammes UZ, Schwechheimer C, et al. 2018. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. 10.1038/s41586-018-0186-z [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. 2004. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937. 10.1104/pp.103.038109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor N, Band LR, Pěnčík A, Novák O, Rashed A, Holman T, Wilson MH, Voß U, Bishopp A, King JR, et al. 2016a. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc Natl Acad Sci 113: 11022–11027. 10.1073/pnas.1604458113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor N, Bennett MJ, King JR. 2016b. GH3-mediated auxin conjugation can result in either transient or oscillatory transcriptional auxin responses. Bull Math Biol 78: 210–234. 10.1007/s11538-015-0137-x [DOI] [PubMed] [Google Scholar]
- Mellor N, Adibi M, El-Showk S, De Rybel B, King J, Mähönen AP, Weijers D, Bishopp A. 2017. Theoretical approaches to understanding root vascular patterning: a consensus between recent models. J Exp Bot 68: 5–16. 10.1093/jxb/erw410 [DOI] [PubMed] [Google Scholar]
- Mellor N, Vaughan-Hirsch J, Kümpers BMC, Help-Rinta-Rahko H, Miyashima S, Mähönen AP, Campilho A, King JR, Bishopp A. 2019. A core mechanism for specifying root vascular patterning can replicate the anatomical variation seen in diverse plant species. Development 146: dev172411. 10.1242/dev.172411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor NL, Voß U, Janes G, Bennett MJ, Wells DM, Band LR. 2020. Auxin fluxes through plasmodesmata modify root-tip auxin distribution. Development 147: dev181669. 10.1242/dev.181669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton AM, King JR, Bennett MJ, Owen MR. 2010. Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol 72: 1383–1407. 10.1007/s11538-009-9497-4 [DOI] [PubMed] [Google Scholar]
- Mironova VV, Omelyanchuk NA, Yosiphon G, Fadeev SI, Kolchanov NA, Mjolsness E, Likhoshvai VA. 2010. A plausible mechanism for auxin patterning along the developing root. BMC Syst Biol 4: 98. 10.1186/1752-0509-4-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova VV, Omelyanchuk NA, Novoselova ES, Doroshkov AV, Kazantsev FV, Kochetov AV, Kolchanov NA, Mjolsness E, Likhoshvai VA. 2012. Combined in silico/in vivo analysis of mechanisms providing for root apical meristem self-organization and maintenance. Ann Bot 110: 349–360. 10.1093/aob/mcs069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, Toyokura K, Blob B, Heo J, Mellor N, Help-Rinta-Rahko H, Otero S, Smet W, et al. 2019. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565: 490–494. 10.1038/s41586-018-0839-y [DOI] [PubMed] [Google Scholar]
- Moore S, Zhang X, Mudge A, Rowe JH, Topping JF, Liu J, Lindsey K. 2015. Spatiotemporal modelling of hormonal crosstalk explains the level and patterning of hormones and gene expression in Arabidopsis thaliana wild-type and mutant roots. New Phytol 207: 1110–1122. 10.1111/nph.13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Liu J, Zhang X, Lindsey K. 2017. A recovery principle provides insight into auxin pattern control in the Arabidopsis root. Sci Rep 7: 43004. 10.1038/srep43004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Norman JMV, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311. 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret B, Marhava P, Aliaga Fandino AC, Hardtke CS, ten Tusscher KHW. 2020. Local auxin competition explains fragmented differentiation patterns. Nat Commun 11: 2965. 10.1038/s41467-020-16803-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D, Byrne H, King J, Bennett M. 2013. The role of auxin and cytokinin signalling in specifying the root architecture of Arabidopsis thaliana. J Theor Biol 317: 71–86. 10.1016/j.jtbi.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Muraro D, Mellor N, Pound MP, Help H, Lucas M, Chopard J, Byrne HM, Godin C, Hodgman TC, King JR, et al. 2014. Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc Natl Acad Sci 111: 857–862. 10.1073/pnas.1221766111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D, Larrieu A, Lucas M, Chopard J, Byrne H, Godin C, King J. 2016. A multi-scale model of the interplay between cell signalling and hormone transport in specifying the root meristem of Arabidopsis thaliana. J Theor Biol 404: 182–205. 10.1016/j.jtbi.2016.04.036 [DOI] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Onckelen HV, Rossignol M, Doumas P. 2005. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074. 10.1104/pp.105.060061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E, Di Mambro R, Dello Ioio R, Costantino P, Sabatini S. 2018. Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J 37: e99134. 10.15252/embj.201899134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. 2012. Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998. 10.1038/ncb2573 [DOI] [PubMed] [Google Scholar]
- Péret B, Middleton AM, French AP, Larrieu A, Bishopp A, Njo M, Wells DM, Porco S, Mellor N, Band LR, et al. 2013. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol Syst Biol 9: 699. 10.1038/msb.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. 2010. Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol 2: a001446. 10.1101/cshperspect.a001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K. 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668. 10.1105/tpc.109.066480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. 2011. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606. 10.1111/j.1365-313X.2011.04710.x [DOI] [PubMed] [Google Scholar]
- Rangarajan H, Postma JA, Lynch JP. 2018. Co-optimization of axial root phenotypes for nitrogen and phosphorus acquisition in common bean. Ann Bot 122: 485–499. 10.1093/aob/mcy092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzer K, Akhmanova M, Konstantinova N, Malínská K, Leitner J, Petrášek J, Luschnig C. 2019. Brassinosteroid signaling delimits root gravitropism via sorting of the Arabidopsis PIN2 auxin transporter. Nat Commun 10: 5516. 10.1038/s41467-019-13543-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Elliott TJ, Jensen KH, Haaning KS, Wager BM, Knoblauch J, Howell AH, Mullendore DL, Monteith AG, Paultre D, Yan D, et al. 2017. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6: e24125. 10.7554/eLife.24125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhry S, Del Bianco M, Kieffer M, Kepinski S. 2013. Auxin controls gravitropic setpoint angle in higher plant lateral branches. Curr Biol 23: 1497–1504. 10.1016/j.cub.2013.06.034 [DOI] [PubMed] [Google Scholar]
- Roychoudhry S, Kieffer M, Del Bianco M, Liao CY, Weijers D, Kepinski S. 2017. The developmental and environmental regulation of gravitropic setpoint angle in Arabidopsis and bean. Sci Rep 7: 42664. 10.1038/srep42664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten JP, ten Tusscher K. 2019. In silico roots: room for growth. Trends Plant Sci 24: 250–262. 10.1016/j.tplants.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. 10.1016/S0092-8674(00)81535-4 [DOI] [PubMed] [Google Scholar]
- Salvi E, Rutten JP, Di Mambro R, Polverari L, Licursi V, Negri R, Dello Ioio R, Sabatini S, ten Tusscher K. 2020. A self-organized PLT/auxin/ARR-B network controls the dynamics of root zonation development in Arabidopsis thaliana. Dev Cell 53: 431–443.e23. 10.1016/j.devcel.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Santos Teixeira JA, ten Tusscher KH. 2019. The systems biology of lateral root formation: connecting the dots. Mol Plant 12: 784–803. 10.1016/j.molp.2019.03.015 [DOI] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JLPM, et al. 2016. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell Online 28: 2937–2951. 10.1105/tpc.16.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD, et al. 2019. Nodule inception recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr Biol 29: 3657–3668.e5. 10.1016/j.cub.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Mitao Y, Kakimoto T. 2014. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol 55: 1450–1459. 10.1093/pcp/pcu077 [DOI] [PubMed] [Google Scholar]
- Shkolnik D, Krieger G, Nuriel R, Fromm H. 2016. Hydrotropism: root bending does not require auxin redistribution. Mol Plant 9: 757–759. 10.1016/j.molp.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Stoeckle D, Thellmann M, Vermeer JE. 2018. Breakout-lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol 41: 67–72. 10.1016/j.pbi.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster GTS, Bhalerao R, Bennett MJ. 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065. 10.1038/ncb1316 [DOI] [PubMed] [Google Scholar]
- Tian H, Wabnik K, Niu T, Li H, Yu Q, Pollmann S, Vanneste S, Govaerts W, Rolcík J, Geisler M, et al. 2014. WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol Plant 7: 277–289. 10.1093/mp/sst118 [DOI] [PubMed] [Google Scholar]
- van den Berg T, Korver RA, Testerink C, ten Tusscher KHWJ. 2016. Modeling halotropism: a key role for root tip architecture and reflux loop remodeling in redistributing auxin. Development 143: 3350–3362. 10.1242/dev.135111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg T, ten Tusscher KH. 2018. Lateral root priming synergystically arises from root growth and auxin transport dynamics. bioRxiv 10.1101/361709 [DOI] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N. 2014. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183. 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- Villalobos LIAC, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos DD, Vissenberg K, Broeckhove J, Beemster GTS. 2014. Putting theory to the test: which regulatory mechanisms can drive realistic growth of a root? PLoS Comput Biol 10: e1003910. 10.1371/journal.pcbi.1003910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann S, Ruiz Rosquete M, Schöller M, Sarkel E, Lindner H, LaRue T, Petřík I, Dünser K, Martopawiro S, Sasidharan R, et al. 2019. Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat Commun 10: 3540. 10.1038/s41467-019-11483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885. 10.1038/sj.emboj.7600659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise LD, ten Tusscher KHWJ. 2019. Discrete mechanical growth model for plant tissue. PLoS ONE 14: e0221059. 10.1371/journal.pone.0221059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. 2015. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388. 10.1016/j.cub.2015.03.046 [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Damme DV, Parizot B, Rop GD, Opdenacker D, Möller BK, Skorzinski N, Njo MF, et al. 2016. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387. 10.1126/science.aad2776 [DOI] [PubMed] [Google Scholar]