Abstract
Robust immunity to intracellular infections is mediated by antigen-specific naive CD8 T cells that become activated and differentiate into phenotypically and functionally diverse subsets of effector cells, some of which terminally differentiate and others that give rise to memory cells that provide long-lived protection. This developmental system is an outstanding model with which to elucidate how regulation of chromatin structure and transcriptional control establish gene expression programs that govern cell fate determination, insights from which are likely to be useful for informing the design of immunotherapeutic approaches to engineer durable immunity to infections and tumors. A unifying framework that describes how naive CD8 T cells develop into memory cells is still outstanding. We propose a model that incorporates a common early linear path followed by divergent paths that slowly lose capacity to interconvert and discuss classical and contemporary observations that support these notions, focusing on insights from transcriptional control and chromatin regulation.
During infections and cancer, the activation of antigen-specific naive CD8 T cells results in the development of a tapestry of “effector” and “memory” CD8 T-cell populations (Box 1) that progressively acquire differences in their life spans, abilities to manifest rapid effector functions, to self-renew, to expand extensively upon rechallenge, and to traffic between or localize within distinct lymphoid and nonlymphoid tissue and microanatomic locales (Table 1; Mueller et al. 2013; Jameson and Masopust 2018). The developmental process through which naive CD8 T cells become cells that terminally differentiate, or that develop into one of many CD8 T-cell subclasses has been described by a number of theoretical models. However, a unifying conceptual framework capturing all aspects of observed results is still outstanding. In this review, we take the perspective that naive CD8 T cells initially differentiate along a common linear pathway, which subsequently diverges along developmental paths that may be reinforced, or reprogrammed, and are actively maintained by DNA-sequence-specific transcription factors (TFs) and chromatin remodeling factors (CRFs). We attempt to synthesize the observations from a wide range of studies by drawing on cell developmental aspects of CD8 T-cell responses, the defined roles of specific TFs and CRFs in CD8 T cells, and the basic principles that underlie chromatin remodeling, transcription, and cell fate determination. A more advanced perspective of how the early differentiation of naive CD8 T cells determines the development of memory cells might provide important guideposts that could be informative when considering the design of vaccination and immunotherapies aimed at engineering durable immunity to infections and tumors.
BOX 1.
THE VERNACULAR OF POSTACTIVATION CD8 T CELLS
Antigen-experienced CD8 T cells are classically defined as “effector” or “memory” cells in the context of acute infections, a scenario in which the pathogen is successfully cleared. In this case, naive cells undergo a prototypical pattern of cell accumulation and differentiation, followed by numerical “contraction” of the population and conversion into a stable population of memory cells (Kaech et al. 2002; Badovinac and Harty 2006). Cells in the effector and contraction phases are referred to as “effector” cells, and those that persist after contraction abates as “memory” cells. Those classified under each umbrella term actually manifest extensive phenotypic and functional heterogeneity (Table 1). In addition, the postactivation cells that arise during chronic infection and tumors do not neatly fall into either umbrella term, because in these scenarios antigen is not cleared, typical effector phase contraction does not occur, and a prototypical memory T-cell compartment that can persist in the absence of antigen does not form (Wherry et al. 2004; Kaech and Wherry 2007). Although the effector and memory parlance are handy for discussion, these generalizations can be misleading or confusing given current appreciation of the phenotypic and functional heterogeneity between cells found in the effector and memory phases of the immune response. Furthermore, because immune responses to chronic infections and tumors, wherein the distinct response phases observed in prototypical acute infections are not evident and instead antigen is not cleared, referring to cells as “effector” or “memory” in either acute or chronic immune reaction scenarios becomes dubious. Furthermore, because the word “activated” T cell is often used interchangeably with “effector” T cell, confusion easily arises because the two are not necessarily equivalent. The former implies a cell in a state of acute stimulation, and the latter a cell that manifests a specialized function, such as the ability to destroy an immunological threat, as in the case of a genuine cytotoxic T lymphocyte (CTL). This distinction is important, because many if not all postactivation CD8 T cells (including memory cells) readily manifest “effector” functions such as cytolytic activity, especially in vivo, even though some might be more proficient on short time scales than others (Barber et al. 2003). Thus, the terms “effector” and “memory” do very little to distinguish a cell that has productively differentiated beyond the naive CD8 T-cell state. A potential, if not imperfect solution is to refer to the precise features used to experimentally enumerate the cells in question, rather than trying to oversimplify with general terms that could be misconstrued.
Table 1.
Effector and memory CD8 T-cell subclasses
| Subset | Abbreviation | Surface phenotype | Time observed | Transcription factor (TF) phenotype | Ancestors | Lineage potential | Location | Key characteristics |
|---|---|---|---|---|---|---|---|---|
| Early effector cell | EEC, EE | KLRG1lo, CD127lo | Effector phase, peak of response | Runx3hi, TCF1hi | MP, TE, DP, central memory, effector memory | Mainly lymphoid tissues, circulating through blood | Able to give rise to MP, TE, and DP effector cells and memory cells | |
| Memory precursor effector cell | MPEC, MP | KLRG1lo, CD127hi, CD27hi, CX3CR1lo | Peak of response | Id3hi, TCF1hi, Blimp1lo, T-betlo | Early effectora | CD62Lhi central memory, CD62Llo effector memory | Mainly lymphoid tissues, circulating through blood | Enhanced survival and persistence into contraction and memory phase |
| Terminal effector cell (short-lived effector cell) | SLEC, TE | KLRG1hi, CD127lo, CX3CR1hi, CD27lo | Peak of response | Tbethi, Id2hi, Zeb2hi, Blimp1hi, Id3lo, TCF1lo | Early effectora | LLE, Tem | Lymphoid tissues, circulating through blood | Less enhanced persistence into contraction and memory phase |
| Double- positive effector cell | DPEC, DP | KLRG1hi, CD127hi, CX3CR1int, CD27hi | Peak of response | Tbethi, Id2hi, Zeb2hi, Blimp1int/hi | Early effectora | LLE, Tem, Tpm | Lymphoid tissues, circulating through blood | Some of these cells are able to down-regulate KLRG1 (exKLRG1) and contribute to Tem, Tpm |
| Long-lived effector cell (effector-like memory) | LLE, ELM | KLRG1hi, CD127lo, CX3CR1hi, CD27lo, CD43lo | Contraction phase, early memory phase | Id2hi, Tbethi, Eomeslo | TE, DP | Not established | Lymphoid tissues, circulating through blood | Weak proliferative recall capacity, sufficient for protection |
| Central memory cell | Tcm, CM | KLRG1lo, CD127hi, CD27hi, CD62Lhi, CD43hi | Memory phase | Eomeshi, Id3hi, TCF1hi, Bcl6hi, Klf2hi | EE, MP | Tem | Lymphoid tissues | Exhibit greatest proliferation capacity upon secondary infection, needed for protective immunity |
| Effector memory cell | Tem, EM | KLRG1int, CD127hi, CD62Llo, CD27hi, CD43lo | Contraction phase, memory phase | Blimp1int, Tbetint, Zeb2hi | TE, DP, EE, MP | Tcm, Tem | Vasculature and intravascular space | Exhibit less enhanced proliferative capacity upon secondary infection, more enhanced effector functions |
| Tissue-resident memory cell | Trm | KLRG1lo, CD127int, CD27lo, CD69+,b CD103+,b | Contraction phase, memory phase | Blimp1int, Runx3hi, Nr4a1hi | Not well established | Some Tcm, Trm | Nonlymphoid tissue (NLT) | Provide protection within NLT, do not recirculate |
| Peripheral memory cell | Tpm | KLRG1int, CD127hi, CD62Llo, CX3CR1int, CD27hi | Contraction phase, memory phase | Blimp1int, Tbetint | EE, MP, DPa | Some Tcm-like, Tem | Circulating through blood, lymphoid, and nonlymphoid tissues | Surveillance of peripheral tissues |
| T stem-like memory cell | Tscm | KLRG1lo, CD127hi | Memory phase | TCF1hi, Bcl2hi, Eomeshi, Id3hi | Not well established | Tcm, Tem, Tpma | Lymphoid tissue | Self-renewal, able to give rise to effector and memory lineagesa |
aNot well established.
bVaries by tissue.
LINEAGE RELATIONSHIPS UNDERLYING MEMORY CD8 T-CELL DEVELOPMENT
CD8 T-Cell Heterogeneity
Cells found in the effector phase exhibit substantial phenotypic heterogeneity, which correlates with different potentials for terminal differentiation or developing into specific memory CD8 T-cell subclasses during infections that resolve acutely. One of the most well-defined phenotypic categorization schemes involves delineating antigen-experienced cells using KLRG1 and CD127 expression (Kaech et al. 2003; Joshi et al. 2007). Additional profiles defined using CD27, CD43, CD62L, CXCR3, and CX3CR1 are also regularly applied (Buchholz et al. 2013; Gerlach et al. 2013, 2016; Olson et al. 2013; Youngblood et al. 2017), and we have summarized many of these definitions (Table 1). Although not all of these CD8 T-cell subsets are necessarily stable cell “lineages” and likely include many differentiation intermediates (Jameson and Masopust 2018; Hudson et al. 2019), they are nevertheless critical for the establishment of immunity, and uncovering the transcriptional mechanisms that accounts for this diversity is of intense and long-standing interest within the field (Kaech and Cui 2012; Chang et al. 2014; Milner and Goldrath 2018).
Classical Models of CD8 T-Cell Memory Formation and Their Conundrums
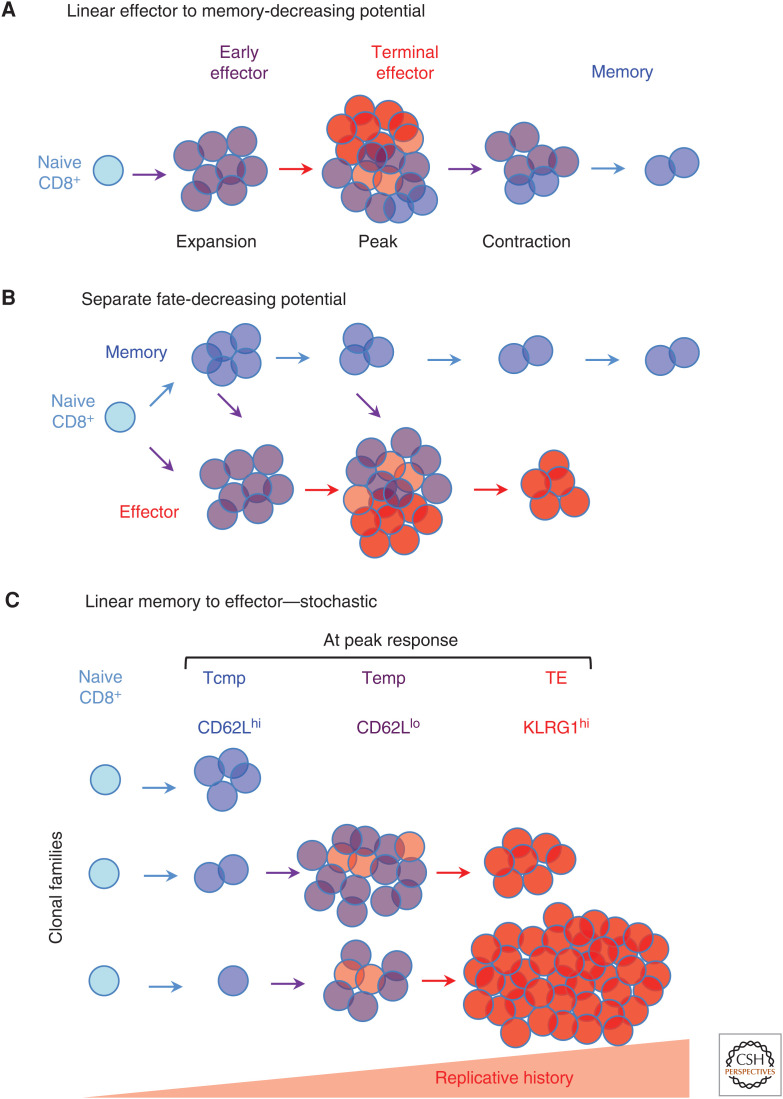
The classical theoretical models that have been used to describe memory CD8 T-cell formation during a prototypical acute infection can be generally grouped into three categories: (1) linear differentiation in which naive cells first develop into “effector” cells before some convert into memory cells (Fig. 1A; Opferman et al. 1999; Kaech and Wherry 2007; Kaech and Cui 2012), (2) separate fate (asymmetric) models in which cells diverge and commit at early times along distinct developmental paths leading to distinct memory or terminally differentiated effector cell “fates” (Fig. 1B; Kaech and Wherry 2007; Kaech and Cui 2012; Reiner and Adams 2014), and (3) linear differentiation in which naive cells initially differentiate into central memory (Tcm) precursor cells that progressively develop into terminally differentiated cells in a manner coupled to the extent of cell division (Fig. 1C; Buchholz et al. 2016).
Figure 1.
Classical models of memory CD8 T-cell formation during acute infection. (A) Linear differentiation decreasing potential model. The responding population differentiates into effector cells, but some cells accumulate more differentiation signals than others, which drives terminal differentiation. (B) Separate fate with progressive differentiation model. Activated cells initially diverge into distinct effector and memory developmental paths based on early signal strength. Cells in the memory pathway that continue to receive stimulation progressively differentiate and ultimately join the effector pathway. (C) Linear differentiation, stochastic model. Individual naive cells unpredictably commit to a proliferative fate that is coupled to the degree of differentiation. Naive cells first develop into slow cycling central memory precursors (Tcmp), then faster dividing effector memory precursors (Temp), and ultimately highly proliferative terminally differentiated progenies.
Each of these models and their derivatives are built on extensive experimental support, suggesting that they all describe important aspects of how the process unfolds. However, considered together they pose some contradictions. Effector cells cannot give rise to memory cells if memory cells actually develop first, and vice versa. In addition, if cells diverge along separate trajectories with actual “fates,” then by definition they do not convert one into the other once they have diverged. The actual biology appears to be more complicated. There is clear evidence that some cells might be on one developmental path before altering or “reversing” course (Youngblood et al. 2017; Herndler-Brandstetter et al. 2018). Furthermore, these models describe development in the context of prototypical acute infections and might not inform what occurs in the context of persistent infections or cancer, even though cells in those settings also originate from naive cells.
HYPOTHESIS
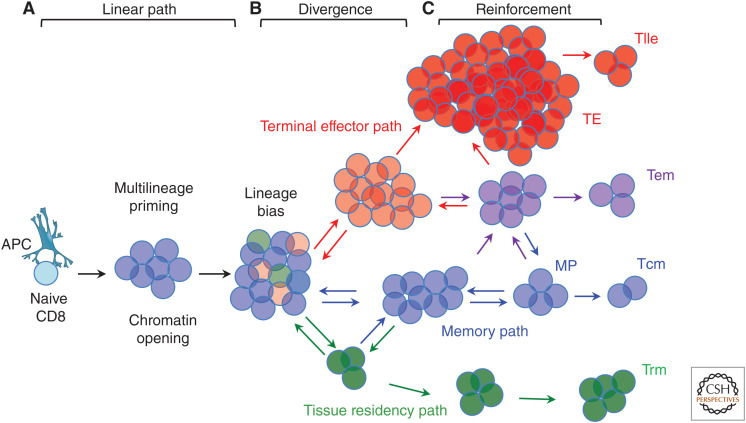
A model composed of an initial linear differentiation path that establishes multilineage potential, followed by divergent developmental trajectories that undergo lineage-restriction but retain potential for interconversion that is gradually reduced might provide a unifying framework (Fig. 2).
Figure 2.
Common linear differentiation, lineage divergence, and progressive restriction. (A) Upon activation, naive cells establish a common linear differentiation program that installs multiple capacities in postactivation cells, such as responsiveness to multiple chemokines, cytokines, and additional costimulatory and coinhibitory receptors, engages migratory capabilities, metabolic capacities, effector functions, and, importantly, comingles expression of regulatory factors that are normally associated with one or another dedicated cell fate. This initial process would facilitate cells to ultimately adopt one of many potential differentiated outcomes later, while being committed to none of them initially, rather serving as progenitors of both terminally differentiated and memory cell progenies. (B) Stochastic transcriptional events result in metastable gene and protein expression states that causes transient lineage bias in some cells among an otherwise homogeneous population (Chang et al. 2008). (C) Cell fate–determining factors reinforce these spontaneous fluctuations, encouraging differentiation along one pathway by enhancing gene expression of the chosen direction, while silencing gene expression that opposes the alternative path at the chromatin level. Cells may interconvert (arrows) until late in their paths.
EVIDENCE FOR UNCOMMITTED COMMON PROGENITORS BEFORE LINEAGE DIVERGENCE
Effector and Memory Cell Potential at the Peak Response
During a model acute intracellular infection, such as infection with the Armstrong strain of lymphocytic choriomeningitis virus (LCMVArm), a spectrum of CD8 T-cell progenies exists near the peak response (Table 1; Kaech and Wherry 2007; Kaech and Cui 2012). For simplicity, the KLRG1/CD127 system suffices to convey the concepts. At this time, cells that are KLRG1hi CD127lo generally predominate numerically depending on the infection type (Obar et al. 2011) and are classified definitively as short-lived effector cells (terminal effector [TE]) because they inefficiently form memory cells (Kaech et al. 2003; Joshi et al. 2007; Obar et al. 2011; Plumlee et al. 2015; Youngblood et al. 2017). Conversely, KLRG1lo CD127hi cells are numerically small and deemed memory precursor effector cells (MPECs [MP]), because they efficiently form memory cells (Kaech et al. 2003; Joshi et al. 2007; Obar et al. 2011; Plumlee et al. 2015; Youngblood et al. 2017). The TE and MP subsets are relatively stable phenotypes that do not efficiently interconvert after transfer into timed, infection-matched hosts (Kaech et al. 2003; Joshi et al. 2007; Obar et al. 2011; Plumlee et al. 2015; Youngblood et al. 2017). However, cells from the KLRG1lo CD127lo population, which are referred to as early effector cells (EECs) are not phenotypically stable, and give rise to all classes of KLRG1/CD127 subsets and circulating memory cells after transfer (Joshi et al. 2007; Obar et al. 2011; Plumlee et al. 2015). Thus, many postactivated CD8 T cells are actually common precursors of both effector and memory CD8 T-cell lineages (Plumlee et al. 2015; Diao and Pipkin 2019).
Multilineage Priming and Lineage Bias Develop before Definitive TE and MP Cells Arise
Cells with both TE and MP potentials are present at early times in the response. Up until approximately day 5 postinfection (pi) with LCMVArm, responding cells manifest substantial flexibility in differentiation outcomes, both in terms of bias toward differentiating into either TE or MP cells at the peak response, as well as the balance in formation of circulating memory CD8 T-cell subsets (Obar et al. 2011; Plumlee et al. 2015; Gerlach et al. 2016), and cells that depart secondary lymphoid organs and generate tissue-resident memory (Trm) cells in nonlymphoid tissues (Masopust et al. 2010). KLRG1 up-regulation indicates commitment toward terminal differentiation in this early time frame (Joshi et al. 2007). However, both KRLG1hi and KLRG1lo cells isolated from day 4.5 post-LCMVArm infection exhibit memory formation potential (albeit KLRG1hi cells less so) (Sarkar et al. 2008) compared to those isolated at the peak response (Joshi et al. 2007). This suggests that regardless of KLRG1 expression, cells at early times are not strongly differentiated or committed. Consistent with this, overall gene expression of KLRG1hi and KLRG1lo cells on day 5 pi is more similar to each other than either subset compared to naive cells, and gene expression in either KLRG1 subset on day 5 is distinct from that found in definitive early effector (EE), TE, and MP phenotypic cells at the peak response on day 8 post-LCMVArm infection (Sarkar et al. 2008; Wang et al. 2018).
Gene expression in responding cells at early times is dynamic and probably unstable at the single-cell level, which suggests cells might undergo substantial gene expression programming before adopting a definitive phenotype. Single-cell RNA sequencing (scRNA-seq) studies have shown that gene expression signatures of either mature TE or Tcm cells are enriched within cells that have undergone their first cell division (Kakaradov et al. 2017). However, this extreme polarity is not evident in their progeny several days later, although expression of effector and memory “fate-classifier” genes indicates lineage bias in some cells (Kakaradov et al. 2017). Moreover, prior to the peak response, additional heterogeneity in single-cell gene expression appears to exist in the spleen and nonlymphoid tissues, which indicates cells begin to diverge toward distinct differentiation outcomes at early times, although the developmental routes to these cell states are not yet defined (Boland et al. 2020; Milner et al. 2020b). These studies argue that gene expression in activated cells early in the response is less differentiated and more flexible than in cells at the peak response, and that individual cells several days after the response has begun are still uncommitted to either TE or memory cell differentiation, but many have become lineage biased.
Early Gene Expression Is Not Reinforced in All Resulting Progenies
Before developing into definitive TE or MP cells, activated CD8 T cells manifest promiscuous expression of key regulatory factors that are otherwise selectively expressed in the definitive subsets. In other settings, this process underlies the propensity of progenitor cells to be able to undergo differentiation along multiple potential paths (Laslo et al. 2006; Chang et al. 2008; Soldatov et al. 2019). Analysis of mice with gene-targeted Id2-YFP and Id3-GFP reporter alleles demonstrated that CD8 T cells coexpress both Id2 and Id3 at early times after infection, before their expression becomes more polarized later (Yang et al. 2011). Likewise, using a targeted Tcf7-GFP allele to trace cells expressing TCF1 (encoded by Tcf7), a TF that is important for Tcm and T-stem-cell-like (Tscm) qualities, shows that its expression is initially sustained for several generations postactivation (Lin et al. 2016), before down-regulation in extensively divided cells that acquire a terminally differentiated phenotype (Zhou et al. 2010; Im et al. 2016; Jadhav et al. 2019; Siddiqui et al. 2019). Notably, considerable fractions of Tcf7-GFPhi cells at early times also express granzyme B or KLRG1 (Lin et al. 2016), which demonstrates coexpression of both effector and memory traits manifest in cells prior to their selective expression in definitive TE or MP cells several days later. These results are consistent with analyses of CD8 T cells from GzmbCre and Klrg1Cre “fate-reporter” mice, which have shown that many memory cells derive from ancestor cells that expressed these effector-cell-associated genes at some point in their history, and subsequently down-regulated them (Bannard et al. 2009; Herndler-Brandstetter et al. 2018). Therefore, many activated CD8 T cells manifest lineage-promiscuous and unstable gene expression that is only reinforced at later times in some fully developed progenies, a process that is a key hallmark of uncommitted progenitors undertaking lineage choice (Laslo et al. 2006; Chang et al. 2008; Soldatov et al. 2019).
LINEAGE DIVERGENCE AND STABILIZATION OF EARLY LINEAGE BIAS
The quality and quantity of signals that recently activated naive CD8 T-cell experience regulate the extent to which the emerging progenies proliferate, which is coupled to terminal differentiation and loss of memory potential (Kaech and Cui 2012; Buchholz et al. 2016). These signals likely function in a cooperative and opposing manner to reinforce or antagonize transcriptional fluctuations that develop in lineage-biased cells early during differentiation to drive lineage commitment (Laslo et al. 2006; Chang et al. 2008; Singer et al. 2008; Pipkin and Rao 2009; Soldatov et al. 2019).
Signals and TF Activities that Reinforce Terminal Differentiation
The collective input of T-cell receptor (TCR) signals, costimulation, and inflammatory cytokine stimulation (e.g., IL-12 and IL-2) predict the extent to which activated CD8 T cells will divide, with greater sums of stimulation increasing proliferative capacity (Marchingo et al. 2014). Cells from progenies with greater cell division history correlate positively with terminally differentiated states because of positive feedback initiated by early inflammatory cytokines. These signals promote proliferation by prolonging expression of IL-2Rα and IL-2 responsiveness, and enhancing transcriptional activities that directly induce effector cell gene expression (Curtsinger et al. 2005; Joshi et al. 2007; Kalia et al. 2010; Pipkin et al. 2010; Starbeck-Miller et al. 2014). For example, inflammatory cytokines and IL-2 each enhance expression of TFs such as T-bet, Zeb2, and Blimp-1 (encoded by Prdm1) that coordinately promote terminal differentiation (Joshi et al. 2007; Rutishauser et al. 2009; Kalia et al. 2010; Pipkin et al. 2010; Xin et al. 2016). Most if not all activated CD8 T cells initially induce T-bet upon initial TCR stimulation, but its enhanced expression in response to inflammatory cues is what drives terminal differentiation (Joshi et al. 2007; Harty and Badovinac 2008). Although T-bet drives terminal differentiation, it requires additional TFs to do so (Kaech and Cui 2012; Chang et al. 2014; Xin et al. 2016). T-bet and Zeb2 positively regulate terminal differentiation through a canonical feedforward pathway (Dominguez et al. 2015; Omilusik et al. 2015). T-bet binds to the Zeb2 locus, induces Zeb2 expression, and both TFs facilitate optimal T-bet binding to cis-regulatory regions in other downstream genes that they both control (Dominguez et al. 2015).
Despite the potency of these regulatory circuits, not all cells that presumably experience early inflammatory signals necessarily commit to terminal differentiation (Buchholz et al. 2013; Gerlach et al. 2013; Buchholz et al. 2016). Many cells show evidence of having activated some degree of gene expression that is normally induced by inflammatory cytokines but nevertheless develop into memory cells. For example, 20%–40% all memory cells develop from cells that previously expressed KLRG1 and then down-regulated it after pathogen clearance (referred to as “ex-KLRG1” cells) (Joshi et al. 2007; Herndler-Brandstetter et al. 2018). Thus, additional factors operating in the face of inflammatory cues arbitrate commitment to terminal differentiation.
Signals and TF Activities that Preserve Memory Potential
The TF Zeb1 is induced in response to TGF-β signals and is necessary for memory CD8 T-cell differentiation. Both Zeb1 and action of mir-200 family microRNAs repress Zeb2 expression (Guan et al. 2018), which provides a mechanism that could interrupt the T-bet-driven program that otherwise drives the terminal differentiation program forward. In addition, the TFs Runx3 and Bach2 both restrain terminal differentiation. Runx3 prevents high expression of T-bet and Zeb2 at early times during infection and also promotes chromatin accessibility to Bach2 motifs during initial TCR stimulation (Wang et al. 2018). Bach2 is a bZIP family TF that can compete with Jun proteins to block TCR-induced AP-1 binding in cis-regulatory regions (Hu and Chen 2013; Roychoudhuri et al. 2016). Bach2 is necessary for the conversion of KLRG1-expressing effector cells into exKLRG1 cells that down-regulate KLRG1 and develop into memory cells (Herndler-Brandstetter et al. 2018). Although Runx3 promotes accessibility to Bach2 motifs during acute LCMV infection, it also represses Bach2 gene expression, and both TFs appear to function antagonistically in the formation of Tscm cells during chronic LMCV infection (Yao et al. 2021). Thus, a complex interplay and perhaps a critical balance between Runx3 and Bach2 activity appears to be important for establishing distinct memory-like T cells during both acute and chronic infections.
Cells that develop into MP cells are insulated from terminal differentiation by the cytokines IL-10 and IL-21 via the TF STAT3. CD8 T cells lacking in any of these factors preferentially manifest a terminally differentiated phenotype and do not normally form Tcm cells (Cui et al. 2011). Stat3 is necessary for expression of SOCS3, which negatively regulated IL-12 responsiveness, and thus counteracts signals that promote terminal differentiation. Stat3 is also necessary for expression of the TFs Bcl6 and Blimp-1, mutual antagonists (Johnston et al. 2009), which regulate formation of distinct memory CD8 T-cell subclasses. Disruption of Bcl6 impairs formation of KLRG1lo CD127hi cells and Tcm cells with self-renewal capacity (Cui et al. 2011). Conversely, disruption of Prdm1 impairs terminal differentiation and results in inflation of Tcm-like cells (Kallies et al. 2009; Rutishauser et al. 2009; Shin et al. 2009). Therefore, signals that govern the early expression of Bcl6 and Prdm1 could be important for differentially regulating terminal differentiation and memory precursor cell formation. However, the fact that Stat3 seems necessary for expression of both of these TFs implies it could contribute to maintaining multilineage potential, in addition to more specifically being required for Tcm formation.
Bcl6 and Blimp1 Regulation Suggests Memory Cell Developmental Paths Are Not Monolithic
Differential regulation of Bcl6 and Blimp1 is complex and might account for early heterogeneity in gene expression that leads to distinct memory developmental potentials that generate Tscm cells, which are embedded within the Tcm phenotype population (Gattinoni et al. 2011, 2012; Graef et al. 2014) and other circulating Tem and Trm subsets. On the one hand, Prdm1 expression is maintained in certain memory cell subsets well after contraction but not those that up-regulate CD62L, which implies that Prdm1-expressing cells cannot develop into Tcm cells or that they down-regulate Prdm1 to do so (Rutishauser et al. 2009; Behr et al. 2019). Prdm1-deficiency impairs formation of Trm and TE cells (Mackay et al. 2016; Milner and Goldrath 2018; Behr et al. 2019). Thus, the amount of Blimp1 activity in cells at early times appears to influence the balance of circulating and noncirculating effector and memory cell lineages. On the other hand, Tcm and Tscm cells that arise during acute infection, and Tscm cells that arise during chronic infection, which exhibit traits found in follicular CD4 T cells that function in germinal centers, are known to depend on the TFs Bcl6, Id3, and TCF1 (Tcf7), which are down-regulated in response to Blimp1 activity (Gattinoni et al. 2012; He et al. 2016b; Im et al. 2016; Crotty 2019). Exactly how early regulation of Bcl6 and Blimp1 contributes to these differential regimes is still unclear.
Naive CD8 T cells express Bcl6 and maintain its expression during TCR stimulation, before up-regulation of Prdm1 (Kalia et al. 2010; Pipkin et al. 2010). Bcl6 is repressed upon cessation of TCR stimulation, and coincides with Prdm1 up-regulation. This process requires IL-2Rα and is governed by IL-2R signal strength (Kalia et al. 2010; Pipkin et al. 2010; Boulet et al. 2014; Xin et al. 2016), suggesting that initial Bcl6 activity could titrate early Blimp1 concentrations (Crotty 2019). In addition, Runx3 participates in this regulation, and is integrated with regulation by both Tcf7 and Id3; Runx3-deficient CD8 T cells overexpress Bcl6, Tcf7, and Id3, and underexpress both Il2ra and Prdm1, and these cells aberrantly acquire follicular T-helper-cell-like characteristics during infection (Kalia et al. 2010; Pipkin et al. 2010; Shan et al. 2017; Wang et al. 2018). Thus, reduced Prdm1 expression in Runx3-deficient cells could result from ectopic expression of Bcl6 and Tcf7 (both repress Prdm1 in CD4s) (Shao et al. 2019), and/or a failure of Runx3 to activate Prdm1 and Il2ra. Runx3 binds to cis-acting regions in all of these genes, suggesting it functions directly in these loci (Shan et al. 2017; Wang et al. 2018), but the order of operations in these regulatory networks is still ill-defined. Elucidating the transcriptional control of Bcl6 and Prdm1 could be important for clarifying the early developmental paths that CD8 T cells take in the context of both acute and chronic infections as well as in cancer (Shin et al. 2009; Chen et al. 2019; Hudson et al. 2019; Miller et al. 2019).
CHROMATIN REMODELING INITIATES TRANSCRIPTIONAL REPROGRAMMING DURING MEMORY CD8 T-CELL FORMATION
Chromatin Accessibility in Naive, Effector, and Memory Cells Suggest a Common History
Chromatin regions that are hypersensitive to cleavage with nucleases such as DNase I (Weintraub and Groudine 1976) and transposase occur where locally disrupted nucleosomes give way to bound TFs, and these accessible regions can be mapped genome-wide by using the assay for transposase-accessible chromatin and high-throughput sequencing (ATAC-seq) (Buenrostro et al. 2013). Compared to naive cells, widespread alterations in chromatin accessibility develop at cis-regulatory regions in mature TE, MP, and KRLG1lo Tmem (day 35) CD8 T-cell populations from mice after acute infection with LCMVArm and in progenitor-like and exhausted CD8 T-cell subsets isolated from mice chronically infected with the clone 13 strain of LCMV (LCMVCl13) (Pauken et al. 2016; Scott-Browne et al. 2016; Sen et al. 2016; Scharer et al. 2017; Yu et al. 2017; Jadhav et al. 2019). Bulk effector (day 8 pi) and memory (day 35 pi) cells after LCMVArm infection each exhibit several thousand regions with altered accessibility compared to naive cells but overlap extensively with each other. Moreover, there are only several hundred regions where accessibility is different when comparing purified TE or MP cells. The remarkable dissimilarity between either Tmem or TE cells and naive cells, in contrast to the relative similarity between TE and MP cells, is consistent with the idea that all cells might initially share a common differentiation history before acquiring more subtle differences along terminal or memory cell paths. Notably, the chromatin accessibility landscape of human effector cells elicited initially by yellow fever vaccine is remarkably similar to that found in the resulting Tmem cells that persist nearly a decade later (Akondy et al. 2017).
Pioneering Chromatin Accessibility upon Naive Cell Activation Establishes Multilineage Potential
Consistent with the notion of a common early differentiation program, chromatin remodeling in naive CD8 T cells that is induced prior to the first cell division upon initial TCR and costimulation alters accessibility of ∼15% of all regions that are accessible later in mature TE, MP, and Tmem subsets (Wang et al. 2018). Many of these de novo accessible regions correspond to those that are stably maintained in mature memory CD8 T cells (Wang et al. 2018; van der Veeken et al. 2019), which demonstrates that chromatin remodeling of memory CD8 T-cell-associated cis-regulatory regions occurs well before “effector” cell differentiation is manifest. DNA sequences underlying de novo accessible regions most frequently encode enriched motifs recognized by RUNX, ETS, bZIP, T-BOX, IRF, RHD, PRDM1, and ZF-KLF TF families, which suggests that TFs in these families account for initial transcriptional reprogramming during naive CD8 T-cell activation (Scott-Browne et al. 2016; Scharer et al. 2017; Wang et al. 2018; van der Veeken et al. 2019). Specific factors from all of the TF families that recognize these motifs have differential requirements for the in vivo development of currently recognized effector and memory CD8 T-cell subsets (Kaech and Wherry 2007; Kaech and Cui 2012; Milner and Goldrath 2018), implying these TF-binding sites function at early times to establish multiple avenues of T-cell differentiation.
Specialized TFs pioneer entry into nucleosome-occluded sites to establish initial chromatin accessibility of previously “inaccessible” cis-regulatory regions (Iwafuchi-Doi and Zaret 2016). In naive CD8 T cells, cooperative activity between multiple TFs whose DNA-binding activity increases in response to TCR stimulation is most likely what initiates pioneering of chromatin accessibility. TCR stimulation transiently activates RHD and bZIP TFs (i.e., NFAT and AP-1) and chromatin accessibility develops transiently at “inducible” regions at NFAT/AP-1-binding sites, adjacent to “primed” regions enriched with RUNX and ETS motifs that remain accessible persistently after cessation of TCR signals (Bevington et al. 2016, 2017a,b). Consistent with this, development of chromatin accessibility at regulatory regions bound by Runx3 or AP-1 during initial TCR stimulation is severely impaired in Runx3-deficient cells (Wang et al. 2018), or cells in which AP-1 TF activity is blocked using a dominant-negative FOS protein (Yukawa et al. 2020). In the absence of Runx3, many fewer cis-regulatory regions encoding RUNX, IRF, bZIP, RHD, PRDM1, and T-BOX motifs become accessible in naive cells upon TCR stimulation (Wang et al. 2018). Notably, Fos and Jun (AP-1) expression is induced during cell activation, whereas Runx3 is expressed prior to stimulation and binds tightly to bulk chromatin in naive cells, wherein it is unable to be extracted fully unless salt concentrations that completely disassemble nucleosomes are used (Wang et al. 2018). Thus, pre-engagement of Runx3 on chromatin prior to stimulation could be an antecedent to chromatin remodeling initiated by TCR stimulation. Direct or indirect cooperativity between Runx proteins and AP-1 (or other bZIP family members) and presumably other TFs, especially those in the ETS-family, which can dimerize with Runx TFs on DNA (Ito 1999), might stabilize the remodeled state.
CHROMATIN REMODELING GOVERNS LINEAGE RESTRICTION OF EFFECTOR AND MEMORY CELL SUBSETS
Important General Features of Chromatin Structure
Differential remodeling of chromatin structure is a key mechanism of cell fate determination that facilitates activating one developmental program and silencing an alternative, opposing program(s) (Pipkin and Rao 2009). However, an easily misconstrued concept is that epigenetic changes to chromatin structure imply developmentally fixed states that are passively inherited through cell generations (Ptashne 2013). In contrast, most available evidence points to the fact that chromatin configurations and transcriptional activity is actively maintained by the function of specific regulatory factors that continuously provide the enzymatic tools to affect transcriptional outputs. More to the point, chromatin structure is known to be extremely dynamic in vitro and in vivo. DNA surrounding nucleosomes spontaneously and rapidly unwraps and rewraps (Li et al. 2005), and histones in nucleosomes at cis-regulatory regions are dynamic in vivo and turn over repeatedly during a single cell generation (Deal et al. 2010). All chromatin modifications can be “erased,” and the overall in vivo topology of chromatin is likely to be a decondensed fiber, as opposed to a hierarchically folded, compacted filament, even at repressed loci (Ou et al. 2017). Thus, chromatin structure can provide both stability and flexibility for dynamically regulating transcriptional programs during cell fate determination.
Differential Activity of Enhancers and Transcription Stabilizes Terminal and Memory Cell Subsets
Definitive TE cells exhibit chromatin structure profiles that reflect more extensive consolidation of transcriptional activity that reinforces terminal differentiation. The overall signal of chromatin accessibility in cis-regulatory regions that are more accessible in TE cells relative to MP cells is substantially greater than the signal in those that are more accessible in MP cells relative to TE cells, which suggests that the TE-specific regions are more strongly occupied by TFs that drive their activity. TE-specific accessible regions are more strongly enriched with T-box and bZIP family TF motifs and more frequently occupied by T-bet and BATF as judged by ChIP-seq compared to regions that are more accessible in MP cells (Kurachi et al. 2014; Scott-Browne et al. 2016). In addition, ChIP-seq mapping of histone modifications (H3K4me1, H3K4me3, H3K27me3, and H3K27Ac) analyzed with algorithms trained to identify enhancer regions and infer their activity indicates that TE cells gain approximately twofold more active enhancers than MP or Tmem cells relative to naive cells; these enhancers are positively correlated with genes that are more highly expressed in TE cells, which implies that development of these cells results from enhanced transcriptional activity (Firpi et al. 2010; Calo and Wysocka 2013; Rajagopal et al. 2013; He et al. 2016a; Yu et al. 2017). Consistent with this, depletion of P-TEFb (positive transcription elongation factor b), which normally induces transcriptional elongation of paused RNA polymerase II and enhances both transcription and maturation of spliced mRNAs (Adelman and Lis 2012), impairs TE cell differentiation and redirects cells into an MP phenotype (Chen et al. 2014). Conversely, cis-regulatory regions that are specifically accessible and active in Tcm cells relative to TE cells encode binding motifs for Tcf1, Lef1, Foxo1, Foxp1, Eomes, Stat5, Gabpa, Gfi1, and Nr3c1 (as well as others) (He et al. 2016a; Yu et al. 2017). Many of these TFs have established roles for activating gene expression that promote T-cell quiescence, lymphoid homing, homeostasis, and the potential for self-renewal (Chandele et al. 2008; Kerdiles et al. 2009; Banerjee et al. 2010; Zhou et al. 2010; Lin et al. 2016). Thus, the activity of cis-regulatory regions as judged at the level of chromatin structure predicts that continuous TF activity maintains the specific transcriptional identities of distinct CD8 T-cell subsets.
Transcription Factors Actively Maintain Lineage Stability in Mature Memory Cells
Runx and T-box motifs frequently co-occur in stably remodeled cis-regulatory regions of memory CD8 T cells, and many are occupied by the T-box TFs, T-bet, and Eomesodermin, and the obligate Runx-TF partner Cbfb (van der Veeken et al. 2019). This suggests that cooperativity between Runx and T-box proteins establishes and maintains the identity of effector and memory CD8 T cells after naive cell stimulation (Intlekofer et al. 2005; Cruz-Guilloty et al. 2009; Hu and Chen 2013; Olesin et al. 2018; Wang et al. 2018; van der Veeken et al. 2019), perhaps by outcompeting nucleosomes that would otherwise form at these sites (Li et al. 2005). Furthermore, binding of Runx3, IRF4, and multiple bZIP family TFs overlaps extensively (Lotem et al. 2013; Kurachi et al. 2014; Wang et al. 2018), which suggests that potential concerted action of these TFs initially establishes chromatin accessibility of cis-regulatory regions during initial CD8 T-cell stimulation, some of which are preserved at later times.
The stability of mature memory CD8 T-cell phenotypes is actively enforced by TFs. Many KLRG1hi cells develop at the peak response that persist into the early aspects of the memory phase (Olson et al. 2013; Milner et al. 2020a). The proteins Id2 and Zeb2 are both important for maintaining the phenotype of these cells, which otherwise decay toward a Tcm-like phenotype if either factor is depleted after memory cells form (Omilusik et al. 2015, 2018). The persistent expression of multiple other TFs in memory cells that are expressed during initial activation or are associated with effector cell differentiation (e.g., Tbx21 and Prdm1) implies that they also are critical for maintaining the differentiated states of various memory CD8 T-cell subsets (Intlekofer et al. 2005; Rutishauser et al. 2009; Yu et al. 2017; Wang et al. 2018). Approaches that can reversibly inactivate these and other factors at different times during cell development will be instrumental for delineating the time-resolved contribution of these factors to specific cell fates.
Chromatin Modifications that Promote Transcriptional Silencing Enforce Terminal Differentiation
Terminal differentiation is enforced by repressing genes that promote lymphoid homing and quiescence and other features of memory cells that promote their maintenance during homeostasis (Best et al. 2013; Gray et al. 2017; Youngblood et al. 2017; Pace et al. 2018). Methylation of histone H3K9 and H3K27 are two well-defined examples of histone modifications that promote gene silencing during cell development (Blackledge et al. 2015). Following TCR stimulation, naive CD8 T cells develop islands of H3K9 trimethylation (H3K9me3) including at the pro-memory genes Il7r and Sell, which correlates with their down-regulation (Pace et al. 2018). The Suppressor Of Variegation 3–9 Homolog 1 (Suv39h1) is one histone methylase that deposits H3K9me3, and Suv39h1-deficient CD8 T cells fail to repress naive and stem-cell-associated genes, and lose the inverse correlation between H3K9me3 density and stem cell gene expression (Pace et al. 2018). These cells accumulate poorly, inefficiently develop a normal TE CD8 T-cell phenotype, and the resulting memory cells are not protective (Pace et al. 2018).
Regions marked by trimethylated histone H3K9 (H3K9me3) can occlude chromatin via multiple Chromobox (Cbx) family proteins, which bind H3K9me3 on adjacent nucleosomes, linking them together, and in some cases promote additional spreading of H3K9me3 deposition (Bannister et al. 2001; Lachner et al. 2001). Extensive diversity among Cbx-family proteins likely provides specificity in how CD8 T cells read H3K9me3-modified genes. For example, CD8 T cells deficient in Cbx3 undergo more effector-like differentiation following activation, and provide stronger antitumor responses in vivo, because Cbx3 naturally represses genes that promote effector cell differentiation and appears to direct H3K9me3 deposition (Sun et al. 2017).
Deposition of H3K27me3 in cis-regulatory regions also appears to be a key step in terminal differentiation. Notably, this occurs after substantial commitment to terminal differentiation is evident, around the peak response, but not earlier (Gray et al. 2017; Kakaradov et al. 2017). Ezh2 catalyzes H3K27me3 deposition, is up-regulated upon stimulation of naive CD8 T cells (Gray et al. 2017), and is more highly expressed in individual “preterminal” effector cells (Kakaradov et al. 2017). Disruption of Ezh2 impairs CD8 T-cell accumulation and effector cell differentiation (Gray et al. 2017; Kakaradov et al. 2017), and correlates with reduced H3K27me3 at the Eomes, Tcf7, and Klf2 genes whose expression increases. These genes encode TFs that promote competitive fitness of Tcm, their maintenance, and their lymphoid retention (Schober et al. 1999; Banerjee et al. 2010; Zhou et al. 2010; Kakaradov et al. 2017). Thus, genes encoding memory-cell-associated determinants are targeted for H3K9 and H3K27 trimethylation in effector cells by distinct histone methyltransferases, which repress memory-cell-specific gene expression and promotes terminal differentiation.
Conversion of cytosine residues in DNA to 5-methylcytosine (5mC) promotes gene repression and contributes to reduced expression of memory-cell-associated genes and terminal differentiation. 5mC leads to gene repression through several mechanisms, including recruitment of chromatin reader proteins containing methyl-DNA-binding domains (Mbd), which bind 5mC-modified DNA and recruit enzymatic corepressor complexes (Li and Zhang 2014), such as the nucleosome remodeling and deacetylase complex (NuRD) (Bowen et al. 2004; Williams et al. 2004). Genome-wide 5mC develops in activated cells as early as day 4.5 after infection, and marks memory-associated genes such as Sell (CD62L) and Tcf7 in both definitive TE and MP cells, correlating with their reduced expression at this time (Youngblood et al. 2017). Disruption of the de novo methyltransferase Dnmt3a in CD8 T cells shows that many of these de novo methylated regions are Dnmt3a dependent in cells responding to infection. Although these cells did not show a major defect in cell accumulation or initial repression of Sell during the effector phase, they more rapidly up-regulated CD127 and CD62L after viral clearance. Thus, de novo DNA methylation appears to promote repression of genes that normally remain silent in terminally differentiated effector cells. Although unexplored, it is possible that targeting of both H3K9me3 and H3K27me3 deposition at certain pro-memory genes could initially involve recruitment to sites of methylated DNA by Mbd-family proteins that interact with Suv39h1 and Ezh2 enzymes (Rose and Klose 2014; Gray et al. 2017; Kakaradov et al. 2017; Youngblood et al. 2017; Pace et al. 2018). Such a model suggests a stepwise mechanism that could restrict memory-cell-lineage gene expression.
Reversible Chromatin Modifications Indicate Lineage Interconversion Is a Facet of Memory Formation
The regulation of histone modifications following naive CD8 T-cell activation and differentiation into distinct effector and memory cell subsets is dynamic. Many genes in naive cells are co-enriched with H3K4me3, a correlate of active genes, as well as H3K27me3, a correlate of inactive genes, and then lose H3K27me3 enrichment after differentiation into effector or memory cells (Russ et al. 2014). Other genes undergo de novo acquisition of H3K4me3 or H3K27me3 deposition during effector and memory cell differentiation. These studies demonstrate that regulation of histone modifications is dynamic during effector and memory cell differentiation. The mammalian genome encodes multiple methylase proteins that can deposit these marks and demethylase proteins that can remove them (Kouzarides 2007; Swigut and Wysocka 2007). Exactly how these different family members function concertedly to control gene activity and stabilize, or reprogram, distinct effector and memory cell lineages is incompletely understood.
There is clear evidence that some effector cells reverse course to develop into memory cells. MP cells at the peak antiviral response are CD62Llo but, after isolation and adoptive transfer, some undergo DNA demethylation in the Sell locus (encodes CD62L) and induce its expression prior to their initial homeostatic cell division (Youngblood et al. 2017). Thus, active removal of 5mC in some MP cells from the effector phase appears to drive their conversion into memory cells. Active DNA demethylation also occurs in the Il2 locus upon TCR stimulation (Bruniquel and Schwartz 2003), but how this occurs is still unknown. Interestingly, deficiency in the protein Mbd2, which binds methylated DNA and could have demethylase activity, impairs normal memory CD8 T-cell formation during LCMV infection (Detich et al. 2002; Kersh 2006). Conversely, DNA hypermethylation and enhanced formation of memory cells occurs in CD8 T cells deficient in ten-eleven translocation 2 (Tet2), a dioxygenase that converts 5mC to 5-hydroxymethylcytosine, and ultimately relieves DNA methylation (Carty et al. 2018). These results generally emphasize the extent to which modifications to chromatin are dynamic, and specifically demonstrate that reversible control of 5mC is pivotal in the formation of memory cells.
Whether and to what extent the developmental potential of memory cells becomes fixed is incompletely understood. Some fully developed Trm cells, after purification from the small intestine epithelium and serial transfer to new hosts subjected to repeated infections, are able to differentiate into Tcm cells (Fonseca et al. 2020). However, these cells retain gene expression and a predilection to take up residence again in their initial tissue of origin, indicating that the original developmental imprint has been, to a degree, maintained over many cell generations. Thus, although cells might initially develop along one path, they retain the ability to change course given the necessary signals but tend to remain fashioned according to their initial developmental origins.
CONCLUDING REMARKS
Antigen-specific responses initially emerge from naive CD8 T cells, whether the inciting stimulus is a pathogen that is successfully cleared, or persists chronically, or is a tumor. One hypothesis is that all responses initially involve an early set of differentiation steps that might be common in all of these settings, and then adopt distinct trajectories in response to external cues that impact their fluctuating cell-intrinsic gene-expression regimes (Fig. 2). The stochasticity in transcriptional events and inherent flexibility of chromatin structure indicates that, in the end, most or all of the developed cell subsets can be effectively reprogrammed if exposed to the required set of regulatory inputs. Somatic cells can be reprogrammed into pluripotent stem cells with ectopic manipulation of a limited number of factors (Rais et al. 2013), and a chromosome encoding human PRF1 isolated from a fibroblast cell can be directly reprogrammed to express physiological amounts of perforin upon transfer into cytotoxic lymphocytes (Pipkin et al. 2007). These observations argue that CD8 T-cell states can be reprogrammed directly. Of the nearly 2000 genes encoding conventional DNA-sequence-specific TFs, and the more than 300 genes encoding chromatin regulatory factors in the mammalian genome, we have only scratched the surface in terms of our knowledge about how the regulatory regimes are configured and implemented in activated T cells (reviewed comprehensively in Kaech and Cui 2012; Chang et al. 2014; Milner and Goldrath 2018). Development and application of new single-cell and computational approaches to link cell genealogies to transcriptomes should facilitate clarifying how transcriptional histories influence cell developmental paths and the trajectories that individual cells chart to their fates. Combining conditional and reversible RNA interference and CRISPR-guided tools to perturb the spatiotemporal functions of genes during different types of responses should help to rapidly elucidate key regulators that can be used to engineer durable immunity to a wide range of challenges.
ACKNOWLEDGMENTS
We acknowledge the important influence that both Dr. Anjana Rao and Dr. Mathias Lichtenheld have had on our view of how transcription is controlled and how cell fates are determined. We apologize to authors whose work we did not directly cite but which has educated us and informed our perspectives. This work was supported by National Institutes of Health (NIH) Grants R01 AI095634, U19 AI109976, and DoD Grant W81XWH-16-1-0006 to M.E.P. and Frenchmen's Creek Women for Cancer Research.
Footnotes
Editors: David Masopust and Rafi Ahmed
Additional Perspectives on T-Cell Memory available at www.cshperspectives.org
REFERENCES
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552: 362–367. 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Harty JT. 2006. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol Rev 211: 67–80. 10.1111/j.0105-2896.2006.00384.x [DOI] [PubMed] [Google Scholar]
- Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. 2010. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185: 4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannard O, Kraman M, Fearon DT. 2009. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science 323: 505–509. 10.1126/science.1166831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Ahmed R. 2003. Cutting edge: rapid in vivo killing by memory CD8 T cells. J Immunol 171: 27–31. 10.4049/jimmunol.171.1.27 [DOI] [PubMed] [Google Scholar]
- Behr FM, Kragten NAM, Wesselink TH, Nota B, van Lier RAW, Amsen D, Stark R, Hombrink P, van Gisbergen K. 2019. Blimp-1 rather than hobit drives the formation of tissue-resident memory CD8+ T cells in the lungs. Front Immunol 10: 400. 10.3389/fimmu.2019.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW, Monach P, Shinton SA, et al. 2013. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat Immunol 14: 404–412. 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, Gilding LN, Ott S, Bonifer C, Cockerill PN. 2016. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J 35: 515–535. 10.15252/embj.201592534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Cockerill PN. 2017a. Chromatin priming elements establish immunological memory in T cells without activating transcription: T cell memory is maintained by DNA elements which stably prime inducible genes without activating steady state transcription. Bioessays 39: 1600184. 10.1002/bies.201600184 [DOI] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Withers DR, Lane PJ, Cockerill PN. 2017b. T cell receptor and cytokine signaling can function at different stages to establish and maintain transcriptional memory and enable T helper cell differentiation. Front Immunol 8: 204. 10.3389/fimmu.2017.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland BS, He Z, Tsai MS, Olvera JG, Omilusik KD, Duong HG, Kim ES, Limary AE, Jin W, Milner JJ, et al. 2020. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol 5: eabb4432. 10.1126/sciimmunol.abb4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet S, Daudelin JF, Labrecque N. 2014. IL-2 induction of Blimp-1 is a key in vivo signal for CD8+ short-lived effector T cell differentiation. J Immunol 193: 1847–1854. 10.4049/jimmunol.1302365 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57. 10.1016/j.bbaexp.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. 2003. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4: 235–240. 10.1038/ni887 [DOI] [PubMed] [Google Scholar]
- Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Graf P, Verschoor A, Schiemann M, Hofer T, Busch DH. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science 340: 630–635. 10.1126/science.1235454 [DOI] [PubMed] [Google Scholar]
- Buchholz VR, Schumacher TN, Busch DH. 2016. T cell fate at the single-cell level. Annu Rev Immunol 34: 65–92. 10.1146/annurev-immunol-032414-112014 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty SA, Gohil M, Banks LB, Cotton RM, Johnson ME, Stelekati E, Wells AD, Wherry EJ, Koretzky GA, Jordan MS. 2018. The loss of TET2 promotes CD8+ T cell memory differentiation. J Immunol 200: 82–91. 10.4049/jimmunol.1700559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandele A, Joshi NS, Zhu J, Paul WE, Leonard WJ, Kaech SM. 2008. Formation of IL-7Rαhi and IL-7Rαlo CD8 T cells during infection is regulated by the opposing functions of GABPα and Gfi-1. J Immunol 180: 5309–5319. 10.4049/jimmunol.180.8.5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. 2008. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453: 544–547. 10.1038/nature06965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Wherry EJ, Goldrath AW. 2014. Molecular regulation of effector and memory T cell differentiation. Nat Immunol 15: 1104–1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Bélanger S, Frederick MA, Li B, Johnston RJ, Xiao N, Liu YC, Sharma S, Peters B, Rao A, et al. 2014. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity 41: 325–338. 10.1016/j.immuni.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, et al. 2019. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51: 840–855.e5. 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2019. T follicular helper cell biology: a decade of discovery and diseases. Immunity 50: 1132–1148. 10.1016/j.immuni.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206: 51–59. 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35: 792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger JM, Valenzuela JO, Agarwal P, Lins D, Mescher MF. 2005. Cutting edge: type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol 174: 4465–4469. 10.4049/jimmunol.174.8.4465 [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164. 10.1126/science.1186777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Theberge J, Szyf M. 2002. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem 277: 35791–35794. 10.1074/jbc.C200408200 [DOI] [PubMed] [Google Scholar]
- Diao H, Pipkin M. 2019. Stability and flexibility in chromatin structure and transcription underlies memory CD8 T-cell differentiation. F1000Res 8: 1278. 10.12688/f1000research.18211.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 212: 2041–2056. 10.1084/jem.20150186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firpi HA, Ucar D, Tan K. 2010. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics 26: 1579–1586. 10.1093/bioinformatics/btq248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, Scott MC, Fares-Frederickson NJ, Wijeyesinghe S, Thompson EA, et al. 2020. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat Immunol 21: 412–421. 10.1038/s41590-020-0607-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. 2011. A human memory T cell subset with stem cell-like properties. Nat Med 17: 1290–1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Klebanoff CA, Restifo NP. 2012. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer 12: 671–684. 10.1038/nrc3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Rohr JC, Perie L, van Rooij N, van Heijst JW, Velds A, Urbanus J, Naik SH, Jacobs H, Beltman JB, et al. 2013. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 340: 635–639. 10.1126/science.1235487 [DOI] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH. 2016. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45: 1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, Drexler I, Höfer T, Riddell SR, Busch DH. 2014. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8+ central memory T cells. Immunity 41: 116–126. 10.1016/j.immuni.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Gray SM, Amezquita RA, Guan T, Kleinstein SH, Kaech SM. 2017. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46: 596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J, Kaech SM. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J Exp Med 215: 1153–1168. 10.1084/jem.20171352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. 2008. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol 8: 107–119. 10.1038/nri2251 [DOI] [PubMed] [Google Scholar]
- He B, Xing S, Chen C, Gao P, Teng L, Shan Q, Gullicksrud JA, Martin MD, Yu S, Harty JT, et al. 2016a. CD8+ T cells utilize highly dynamic enhancer repertoires and regulatory circuitry in response to infections. Immunity 45: 1341–1354. 10.1016/j.immuni.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. 2016b. Follicular CXCR5- expressing CD8+ T cells curtail chronic viral infection. Nature 537: 412–416. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, Lietzenmayer M, Kroehling L, Takumi A, Kometani K, et al. 2018. KLRG1+ effector CD8+ T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity 48: 716–729.e8. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chen J. 2013. A genome-wide regulatory network identifies key transcription factors for memory CD8+ T-cell development. Nat Commun 4: 2830. 10.1038/ncomms3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, et al. 2019. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51: 1043–1058.e4. 10.1016/j.immuni.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6: 1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Ito Y. 1999. Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells 4: 685–696. 10.1046/j.1365-2443.1999.00298.x [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2016. Cell fate control by pioneer transcription factors. Development 143: 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin JX, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ. 2019. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci 116: 14113–14118. 10.1073/pnas.1903520116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. 2018. Understanding subset diversity in T cell memory. Immunity 48: 214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27: 281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12: 749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ. 2007. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 27: 393–405. 10.1016/j.immuni.2007.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, Ahmed R. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111: 837–851. 10.1016/S0092-8674(02)01139-X [DOI] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4: 1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, et al. 2017. Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol 18: 422–432. 10.1038/ni.3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32: 91–103. 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kallies A, Xin A, Belz GT, Nutt SL. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31: 283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol 10: 176–184. 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersh EN. 2006. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. J Immunol 177: 3821–3826. 10.4049/jimmunol.177.6.3821 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15: 373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755–766. 10.1016/j.cell.2006.06.052 [DOI] [PubMed] [Google Scholar]
- Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. 10.1101/cshperspect.a019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12: 46–53. 10.1038/nsmb869 [DOI] [PubMed] [Google Scholar]
- Lin WW, Nish SA, Yen B, Chen YH, Adams WC, Kratchmarov R, Rothman NJ, Bhandoola A, Xue HH, Reiner SL. 2016. CD8+ T lymphocyte self-renewal during effector cell determination. Cell Rep 17: 1773–1782. 10.1016/j.celrep.2016.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y. 2013. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS ONE 8: e80467. 10.1371/journal.pone.0080467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352: 459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Marchingo JM, Kan A, Sutherland RM, Duffy KR, Wellard CJ, Belz GT, Lew AM, Dowling MR, Heinzel S, Hodgkin PD. 2014. T cell signaling, antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science 346: 1123–1127. 10.1126/science.1260044 [DOI] [PubMed] [Google Scholar]
- Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA, Barber DL, Kawamura KS, et al. 2010. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med 207: 553–564. 10.1084/jem.20090858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20: 326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Goldrath AW. 2018. Transcriptional programming of tissue-resident memory CD8+ T cells. Curr Opin Immunol 51: 162–169. 10.1016/j.coi.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Nguyen H, Omilusik K, Reina-Campos M, Tsai M, Toma C, Delpoux A, Boland BS, Hedrick SM, Chang JT, et al. 2020a. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc Natl Acad Sci 117: 25667–25678. 10.1073/pnas.2008571117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, Quezada L, Widjaja CE, Witherden DA, Crowl JT, et al. 2020b. Heterogenous populations of tissue-resident CD8+ T cells are generated in response to infection and malignancy. Immunity 52: 808–824.e7. 10.1016/j.immuni.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Gebhardt T, Carbone FR, Heath WR. 2013. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31: 137–161. 10.1146/annurev-immunol-032712-095954 [DOI] [PubMed] [Google Scholar]
- Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrancois L. 2011. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J Immunol 187: 4967–4978. 10.4049/jimmunol.1102335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesin E, Nayar R, Saikumar-Lakshmi P, Berg LJ. 2018. The transcription factor Runx2 is required for long-term persistence of antiviral CD8+ memory T cells. Immunohorizons 2: 251–261. 10.4049/immunohorizons.1800046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38: 1250–1260. 10.1016/j.immuni.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 212: 2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik KD, Nadjsombati MS, Shaw LA, Yu B, Milner JJ, Goldrath AW. 2018. Sustained Id2 regulation of E proteins is required for terminal differentiation of effector CD8+ T cells. J Exp Med 215: 773–783. 10.1084/jem.20171584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Ober BT, Ashton-Rickardt PG. 1999. Linear differentiation of cytotoxic effectors into memory T lymphocytes. Science 283: 1745–1748. 10.1126/science.283.5408.1745 [DOI] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC. 2017. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025. 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace L, Goudot C, Zueva E, Gueguen P, Burgdorf N, Waterfall JJ, Quivy JP, Almouzni G, Amigorena S. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359: 177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354: 1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Rao A. 2009. Snapshot: effector and memory T cell differentiation. Cell 138: 606.e1–606.e2. 10.1016/j.cell.2009.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Ljutic B, Cruz-Guilloty F, Nouzova M, Rao A, Zúñiga-Pflücker JC, Lichtenheld MG. 2007. Chromosome transfer activates and delineates a locus control region for perforin. Immunity 26: 29–41. 10.1016/j.immuni.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32: 79–90. 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumlee CR, Obar JJ, Colpitts SL, Jellison ER, Haining WN, Lefrancois L, Khanna KM. 2015. Early effector CD8 T cells display plasticity in populating the short-lived effector and memory-precursor pools following bacterial or viral infection. Sci Rep 5: 12264. 10.1038/srep12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. 2013. Epigenetics: core misconcept. Proc Natl Acad Sci 110: 7101–7103. 10.1073/pnas.1305399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. 2013. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502: 65–70. 10.1038/nature12587 [DOI] [PubMed] [Google Scholar]
- Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, Ernst J, Kellis M, Ren B. 2013. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol 9: e1002968. 10.1371/journal.pcbi.1002968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL, Adams WC. 2014. Lymphocyte fate specification as a deterministic but highly plastic process. Nat Rev Immunol 14: 699–704. 10.1038/nri3734 [DOI] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. 2014. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 1839: 1362–1372. 10.1016/j.bbagrm.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, Ji Y, Sukumar M, Eil RL, Yu Z, et al. 2016. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17: 851–860. 10.1038/ni.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, Stock AT, Croom HA, Cullen JG, Nguyen ML, et al. 2014. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 41: 853–865. 10.1016/j.immuni.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. 2009. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31: 296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med 205: 625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer CD, Bally AP, Gandham B, Boss JM. 2017. Cutting edge: chromatin accessibility programs CD8 T cell memory. J Immunol 198: 2238–2243. 10.4049/jimmunol.1602086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. 1999. Expression of the transcription factor lung Krüppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol 163: 3662–3667. [PubMed] [Google Scholar]
- Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. 2016. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45: 1327–1340. 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. 2016. The epigenetic landscape of T cell exhaustion. Science 354: 1165–1169. 10.1126/science.aae0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, et al. 2017. The transcription factor Runx3 guards cytotoxic CD8+ effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol 18: 931–939. 10.1038/ni.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao P, Li F, Wang J, Chen X, Liu C, Xue HH. 2019. Cutting edge: Tcf1 instructs T follicular helper cell differentiation by repressing Blimp1 in response to acute viral infection. J Immunol 203: 801–806. 10.4049/jimmunol.1900581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HM, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. 2009. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31: 309–320. 10.1016/j.immuni.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. 2019. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50: 195–211.e10. 10.1016/j.immuni.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol 8: 788–801. 10.1038/nri2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, et al. 2019. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364: eaas9536. 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Starbeck-Miller GR, Xue HH, Harty JT. 2014. IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med 211: 105–120. 10.1084/jem.20130901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Ha N, Pham DH, Frederick M, Sharma B, Naruse C, Asano M, Pipkin ME, George RE, Thai TH. 2017. Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8+ T cells. Sci Rep 7: 42888. 10.1038/srep42888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Wysocka J. 2007. H3k27 demethylases, at long last. Cell 131: 29–32. 10.1016/j.cell.2007.09.026 [DOI] [PubMed] [Google Scholar]
- van der Veeken J, Zhong Y, Sharma R, Mazutis L, Dao P, Pe'er D, Leslie CS, Rudensky AY. 2019. Natural genetic variation reveals key features of epigenetic and transcriptional memory in virus-specific CD8 T cells. Immunity 50: 1202–1217.e7. 10.1016/j.immuni.2019.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, Pipkin ME. 2018. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48: 659–674.e6. 10.1016/j.immuni.2018.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193: 848–856. 10.1126/science.948749 [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. 2004. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci 101: 16004–16009. 10.1073/pnas.0407192101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. 2004. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20: 719–733. 10.1016/j.immuni.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, Olshansky M, Lin JX, Li P, Speed TP, et al. 2016. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol 17: 422–432. 10.1038/ni.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 12: 1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Lou G, Sun HW, Zhu Z, Sun Y, Chen Z, Chauss D, Moseman EA, Cheng J, D'Antonio MA, et al. 2021. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8+ T cells. Nat Immunol 22: 370–380. 10.1038/s41590-021-00868-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552: 404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, Pereira RM, Crotty S, Chang JT, Pipkin ME, et al. 2017. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol 18: 573–582. 10.1038/ni.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M, Jagannathan S, Vallabh S, Kartashov AV, Chen X, Weirauch MT, Barski A. 2020. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J Exp Med 217: e20182009. 10.1084/jem.20182009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. 2010. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33: 229–240. 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]