Abstract
Structural hippocampal abnormalities are common in many neurological and psychiatric disorders, and variation in hippocampal measures is related to cognitive performance and other complex phenotypes such as stress sensitivity. Hippocampal subregions are increasingly studied, as automated algorithms have become available for mapping and volume quantification. In the context of the Enhancing Neuro Imaging Genetics through Meta Analysis Consortium, several Disease Working Groups are using the FreeSurfer software to analyze hippocampal subregion (subfield) volumes in patients with neurological and psychiatric conditions along with data from matched controls. In this overview, we explain the algorithm's principles, summarize measurement reliability studies, and demonstrate two additional aspects (subfield autocorrelation and volume/reliability correlation) with illustrative data. We then explain the rationale for a standardized hippocampal subfield segmentation quality control (QC) procedure for improved pipeline harmonization. To guide researchers to make optimal use of the algorithm, we discuss how global size and age effects can be modeled, how QC steps can be incorporated and how subfields may be aggregated into composite volumes. This discussion is based on a synopsis of 162 published neuroimaging studies (01/2013–12/2019) that applied the FreeSurfer hippocampal subfield segmentation in a broad range of domains including cognition and healthy aging, brain development and neurodegeneration, affective disorders, psychosis, stress regulation, neurotoxicity, epilepsy, inflammatory disease, childhood adversity and posttraumatic stress disorder, and candidate and whole genome (epi‐)genetics. Finally, we highlight points where FreeSurfer‐based hippocampal subfield studies may be optimized.
Keywords: ENIGMA, FreeSurfer, hippocampal subfields, hippocampal subregions, hippocampus, MRI, quality control, segmentation
Hippocampal subfield analysis is increasingly performed with the availability of automated magnetic resonance imaging segmentation methods. We give a synopsis of the FreeSurfer hippocampal subfield segmentation algorithm, measurement reliability studies and application domains. We discuss how global size and age effects can be modeled and suggest a standardized hippocampal subfield segmentation quality control procedure for improved pipeline harmonization.
1. INTRODUCTION
The hippocampus is an intensely studied brain region in preclinical and clinical neuroscience (Andersen, Morris, Amaral, Bliss, & O'Keefe, 2007). It is involved in multiple aspects of cognition, including spatial navigation (O'Keefe, 1990), learning (Morris, 2006), and episodic memory (Burgess, Maguire, & O'Keefe, 2002), affective processing (Koelsch et al., 2015), and stress response regulation in humans (Herman et al., 2016; McEwen & Akil, 2020). Given its many functions—and its circumscribed, exposed position in the medial temporal lobe—it has been the target of volumetric studies in neurology and psychiatry. Early hippocampal studies with computed tomography used indirect metrics such as widened hippocampal fissure, increased hippocampal lucency, or temporal horn diameter (de Leon, George, Stylopoulos, Smith, & Miller, 1989; George et al., 1990; Sandor, Albert, Stafford, & Harpley, 1988; Scheltens, Weinstein, & Leys, 1992). Its contrast‐rich boundaries on magnetic resonance imaging (MRI) against the ventricular space may even have led to an “investigation bias” toward this structure compared to other less circumscribed brain structures. Meanwhile, MRI‐based hippocampal morphometry has advanced through improved contrast, spatial resolution, and computational progress. Images with 0.5–0.9 mm isotropic spatial resolution are now regularly collected with standard anatomical sequences on 3 Tesla platforms (e.g., Human Connectome Project, http://protocols.humanconnectome.org). The spread of high‐field MRI further accelerated this development. Recent work validating in vivo 7‐Tesla human MRI with histology has pushed MRI toward “in vivo neuropathology” (DeKraker, Ferko, Lau, Köhler, & Khan, 2018) and allows subfield segmentation techniques to take advantage of the resolution and contrast available at 7 Tesla (Giuliano et al., 2017).
Attempts to subdivide the hippocampal formation into substructures from MRI have been made for several years. Based on sublayers and boundaries with neighboring structures, subdividing the hippocampus has been the objective of several manual segmentation protocols (Jeukens et al., 2009; La Joie et al., 2010; Mueller et al., 2007; Mueller et al., 2018; Mueller & Weiner, 2009; Shing et al., 2011; Wisse et al., 2012). Detecting and following boundaries in the complex, “swiss roll‐”like structure and recognizing landmarks when the internal contrast is insufficient, is a complex visual task that requires expert anatomical knowledge and practice. The procedure may take up to 40 hours per case (Winterburn et al., 2013), exhausting realistically available resources, particularly in larger imaging genetics studies, some of which now exceed 50,000 individuals (Satizabal et al., 2019, Grasby, Jahanshad et al., 2020). Manual segmentations are still represent the gold standard and have been carefully interwoven into fully automated algorithms (Iglesias et al., 2015; Van Leemput et al., 2008, 2009; Yushkevich et al., 2009; Yushkevich et al., 2015).
The Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) consortium conducts imaging and imaging genetics analyses in population cohorts and clinical samples using an international multisite framework, bringing together imaging and genetics groups for large‐scale collaborations. The principle of distributed analyses—in which local sites perform image postprocessing and association statistics on their own authority—makes harmonization an important element to enhance reliability. Collaborative studies allow for detection of small effects sizes, but, the resulting summary statistics rely heavily on the quality of the incoming local association results and extracted data. As disease working groups are increasingly engaged (Thompson et al., 2020) in cross‐diagnostic comparisons or genuine transdiagnostic (“denosologization”) studies, these efforts might benefit from pipeline harmonization (Figure 1).
FIGURE 1.
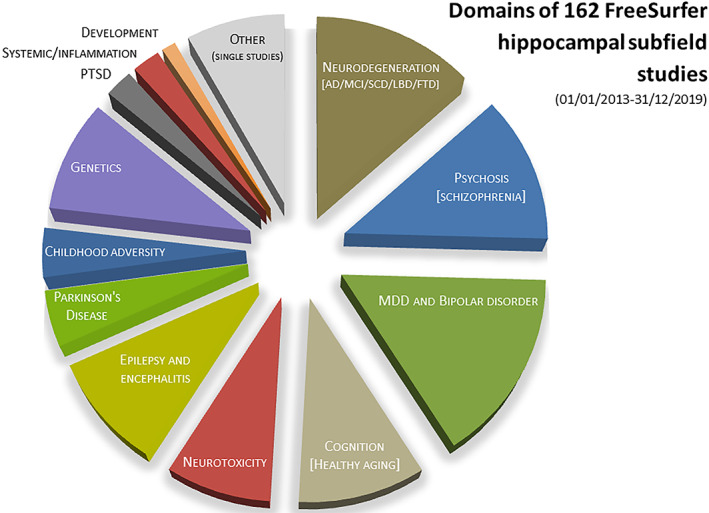
Domains covered by 162 FreeSurfer subfields studies published between 2013 and 2019. Ten studies performed on patient groups with a genetic analysis included (see Supplemental Table 1, domain/subdomain “genetics”) were counted double to avoid under‐representation of genetic studies, that is, the pie chart contains a total of 172 entries. The following additional aggregations were made: major depressive disorder (MDD) and bipolar disorder (21 and 6 studies) were pooled; single neurological (8), single psychiatric studies (5) and one study on perceived stress were pooled to a category “other (single studies)”; studies on epilepsy (10) and encephalitis (5) were pooled. AD, Alzheimer's disease; FTD, frontotemporal dementia; LBD, Lewy body disease; MCI, mild cognitive impairment; PTSD, posttraumatic stress disorders; SCD, subjective cognitive deficits
The purpose of this report is to give an overview of the hippocampal subfield segmentation algorithm implemented in FreeSurfer (FS) and present a novel protocol for standardizing quality control (QC). To put the pipeline into context and to guide researchers to optimally use the algorithm, we review reports that have employed this tool, pointing out typical methodological challenges and pitfalls. In this article, we follow the recently established practice to refer to the resulting substructures either as “subregion(s)” or “subfield(s)”, in an interchangeable way despite the erstwhile anatomical nomenclatures that reserved “subfields” for the cornu ammonis (CA). Section 1 provided background and rationale for this study. In Section 2, we describe the core algorithm of the FS‐based hippocampus segmentation, summarize findings of reliability studies, with examples of the correlation between subfield volumes, and discuss the relationship between size and reliability. Next, Section 3 describes the QC procedure developed in the context of the ENIGMA consortium to improve pipeline harmonization. In Section 4, we give an overview on FS‐based hippocampal subfield studies published between January 2013 and December 2019, focusing on (a) the clinical or neuroscience domains being studied, (b) statistical methods, and (c) QC procedures. Finally, Section 5 concludes with a summary, discussion, and outlook.
2. THE FS HIPPOCAMPAL SUBFIELD SEGMENTATION ALGORITHM
2.1. Principle and output
The hippocampal segmentation tool examined in this paper is part of the widely used and freely available neuroimaging analysis package, FS (Fischl, 2012). The current version of this tool is FreeSurfer 7.1, and this version has no significant changes to the hippocampal subfield tool compared to version 6.0 [FS6.0]. FreeSurfer 7.1 includes a segmentation of the amygdala (Saygin et al., 2017) that does not affect the hippocampal subregion segmentation that was introduced with FS6.0. The automated segmentation of hippocampal subfields is driven by a probabilistic atlas, built from two manually delineated datasets: one comprised of 15 post mortem subjects, including four with Alzheimer's disease, from whom ex vivo MRI (7 Tesla, FLASH sequence) was obtained at very high resolution (0.10–0.20 mm isotropic); and a second that comprised 39 T1‐weighted in vivo images (1 mm isotropic voxels) from 29 controls and 10 mildly demented individuals. The key developmental leap from FS5.3 to FS6.0 was the inclusion of manually delineated high resolution ex vivo MRI data (Iglesias et al., 2015). Both labeled datasets were combined by a Bayesian algorithm into a probabilistic atlas that is encoded in a tetrahedral mesh, where each node carries probabilistic information on its assignment to subregions (van Leemput, 2009). 1
The challenge of segmenting a new, unseen hippocampus was then posed as a Bayesian inference problem of maximizing the probability of the segmentation—given the atlas and the input image. Robustness to changes in MRI contrast was achieved by modeling voxel intensities as samples from a Gaussian mixture model conditioned on the hidden segmentation. Parameters of this model were informed by the individual input scan more specifically from five intensity compartments: gray matter, white matter, cerebrospinal fluid, alveus, and the molecular layer). Although the alveus and molecular layer resemble white matter, they are modeled separately, given that partial volume effects influence their voxel intensities. The outputs of the hippocampal segmentation are left and right hemisphere images with label assignments for voxels in the hippocampal area (at resolutions of 1‐mm isotropic and 0.333‐mm isotropic) to either background or one of twelve subregions. These are (sorted by average size): CA1, molecular layer (ML), hippocampal tail, subiculum, presubiculum, granule cell layer of dentate gyrus (GC‐ML‐DG), CA4, CA3, hippocampal fissure, hippocampus‐amygdala‐transition‐area (HATA), and fimbria (Figure 2a,b). The manual delineation protocol and the grouping of its subregions into 12 output labels are detailed in Iglesias et al. (2015). Alternative aggregations, for example, ones that consider head, body and tail subdivisions, are available online (https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfieldsAndNucleiOfAmygdala). In this report, we use reaggregation to refer to aggregations deviating from the three standard aggregations.
FIGURE 2.
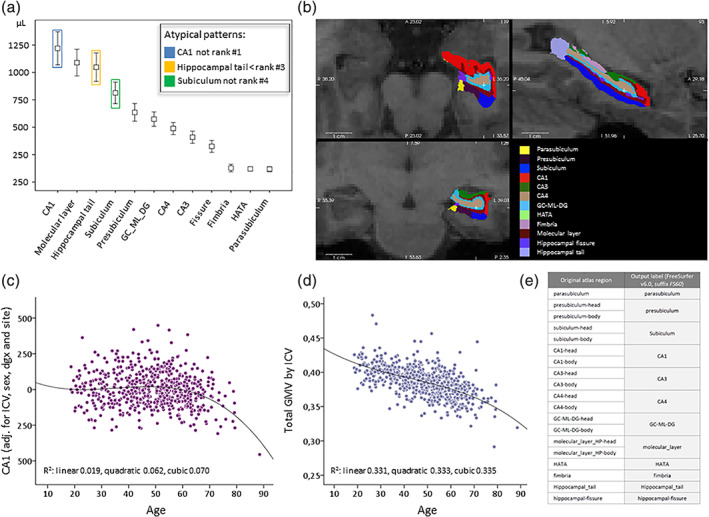
Typical subfield size ranking, overlay in FreeSurfer viewer and exemplary age‐volume relationships. (a) Typically raw subfield volumes ranked according to their average size are depicted from a local Max Planck Institute of Psychiatry (MPIP)‐based sample of N = 614 subjects (T1WI, FS6.0, mean values and 1 SD). The volume ranking order is extremely robust across other samples including 3 Tesla samples (data not shown). The colored frames point to subregions that underlie ranking violation rules (see Section 2.3 for details). (b) A 3 Tesla example viewed in FS in three corresponding planes (see white cross‐hair) with the FS inherent color scheme. The same scheme for the 12 subregions was adopted in the ENIGMA quality control (QC) algorithm. (c) An example of a tendency for nonlinear age effects for the bilateral CA1 region, adjusted for intracranial volume (ICV), sex, diagnosis (major depressive disorder [MDD]/healthy) and site (here coding for coil upgrade related raw image differences). Quadratic or cubic polynomial fits are superior to a linear correlation. (d) The same principle is plotted for the ratio between total gray matter volume and ICV; a less strong nonlinear influence can be read from the fit values. (e) One of three aggregation schemes available in recent development versions of FS, referred to as “FS60,” explaining the mapping between the 12 output labels (which are the labels in the FS6.0 atlas used in this study) and the underlying regions
2.2. Measurement reliability and validity
The output of the automated FS subfield segmentation consists of both the spatial map of a subfield and its absolute volume, and reliability measures have been reported for both, as detailed in Sections 2.2.1 and 2.2.2. For volumes, the difference between measurements (e.g., test–retest measures) can be expressed as percentage error or as intraclass coefficient (ICC). The latter quantifies the consistency of repeated measurements of the same quantity. Different subtypes of the ICC exist, and some are sensitive also to systematic biases of one measurement, a feature that may remain undetected by the Pearson correlation. The spatial reliability of subfield segmentations is often expressed as the Sørensen–Dice index often abbreviated to Dice index; range 0–1; 1 represents perfect overlap of both measurements). The Dice index can be calculated for a single pair of repeatedly measured subregions and is relevant for using the subfield segmentation as regions of interest (ROIs) for other (e.g., functional imaging) experiments.
2.2.1. Test–retest reliability (same scanner)
Several studies have examined test–retest reliability of FS6.0 subfield volumes, all reporting ICC values larger than 0.5 and most around 0.9 (Brown et al., 2020; Elvsåshagen et al., 2016; Quattrini et al., 2020; Whelan et al., 2016; Worker et al., 2018). More specifically, Whelan et al. (2016) included data from four independent cohorts (total N > 1,740), reporting high ICC values (0.70–0.97) for a 3 Tesla sample, and moderate to high for a 4 Tesla sample (0.50–0.89). Another 3‐Tesla study applied FS6.0 to data from 22 healthy subjects scanned three times, and 40 patients with AD scanned twice, again reporting high ICC values (>0.9) for 20 of 24 investigated hippocampal subregions, and significantly lower ICC for the hippocampal fissure and fimbria (Worker et al., 2018). In addition, high sensitivity toward AD progression at both 2‐year and 6‐week follow‐up times was detected, using intraindividually optimized co‐registration of repeated scans (Worker et al., 2018). In their report on bipolar disorder, Elvsåshagen et al. (2016) reported test–retest ICC over 0.94 for 3‐Tesla T1‐weighted images selectively for the dentate gyrus/CA4, for within‐session repeats (N = 53) and two separate scanning sessions (N = 21). Quattrini et al. (2020) also reported good volume reliability (mean error <5%, ICC > 0.92) and excellent spatial agreement (mean Dice index >0.92) for different 3 Tesla scanner platforms. Another recent report on FS6.0 (Brown et al., 2020) confirmed high test–retest reliability (measured by volume differences, ICC, and Dice overlap) with a similar regional ranking, that is, high reliability for all subregions except for the parasubiculum, hippocampal fissure, and HATA.
The reliability of FS5.3 hippocampal subfield segmentations have also been investigated in 65 subjects scanned at 13 sites. Marizzoni et al. (2015) reported good or very good volumetric (reproducibility errors 2–5%) and spatial reproducibility (Dice indices ~0.9 or >0.9) for all subregions except for the hippocampal fissure and fimbria.
Of note, since FS7.1 (released 04/2020), hippocampal subfield segmentation can be combined with longitudinal input data to optimally assess within‐subject changes. This approach reduces measurement noise compared with two separate segmentations, increases test–retest reliability and thus leads to higher sensitivity to longitudinal changes (Iglesias et al., 2016). The two more recent reports strongly confirmed this finding both for within‐ and across‐scanner comparisons, particularly for older subjects (Brown et al., 2020; Quattrini et al., 2020). For repeated measures, this approach is thus clearly recommended.
2.2.2. Reliability across vendor platforms and field‐strengths
Whelan et al. (2016) reported on a FS6.0 study comparing T1WI of 1.5 Tesla with 3 Tesla and found high ICC for 11 subregions (0.721–0.915) and a low value for the hippocampal fissure (0.575). Another small trans‐platform analysis (1.5 and 3 Tesla, FS5.3, performed on seven children) was included in the report of Tamnes et al. (2014), showing high linear correlations (r values .80–.97) for all subfields except the fimbria. More recently, reliability across vendors (or before and after typical hardware upgrades) has been reported in great detail (Brown et al., 2020; Quattrini et al., 2020). As a general pattern, these studies show that there is higher sensitivity of the spatial reproducibility to MR scanner effects, compared to the subfield volumes. Within‐session T1‐averages are suggested as one possibility to improve test–retest reliability.
2.2.3. Relationship between average subregion size and measurement reliability
For FS5.3, Marizzoni et al. (2015) reported that specific test–retest measures (reproducibility errors and Dice index) were positively associated with regional volumes. In the interest of this report, based on published results (Whelan et al., 2016), Table 2), we correlated the rank of the average subregion size with corresponding ICC values for two FS6.0 samples (unweighted averages across both samples and both hemispheres), and found a high positive rank correlation (Spearman's rho 0.85). This pattern was further confirmed and extended to the Dice overlap index for FS6.0 by Quattrini et al. (2020) who also concluded that the best test–retest reproducibility was achieved for hippocampal subfields >300 mm3 (see typical size ranking in Figure 2a). This could imply that the power to detect group differences or other clinical correlations may be biased toward larger subregions, and this should be considered when interpreting results.
TABLE 2.
Overview of reported FS‐based hippocampal subfield studies, 01/2013‐12/2019. Studies from the first category (heritability studies) are summarized in Section 4.2. The clinical/behavioral/biomarker field was categorized into 15 domains, and the genetic field into 4 domains. Minimum keywords are given for single studies or groups of studies to characterize subdomains
| Domain | Studies |
|---|---|
| Heritability | |
| Heritability | Heritability (Greenspan, Arakelian, & van Erp, 2016; van der Meer et al., 2018; Whelan et al., 2016), heritability and genetic correlation (Elman et al., 2019) |
| Clinical diagnoses, behavioral and cognitive phenotypes, or other biomarkers | |
| Neurodevelopmental studies | Cognitive aspects during early childhood until early adulthood (Krogsrud et al., 2014; Riggins et al., 2018; Tamnes et al., 2014) |
| Healthy aging |
Cognitive performance (Aslaksen, Bystad, Ørbo, & Vangberg, 2018; Delazer et al., 2019; Engvig et al., 2012; Ezzati, Katz, Lipton, Lipton, & Verghese, 2015; Foster, Kennedy, Hoagey, & Rodrigue, 2019; Pereira et al., 2014; Zammit et al., 2017; Zheng, Cui, et al., 2018; Zheng, Liu, et al., 2018), hormonal influences (Pintzka & Håberg, 2015), protective effects of education (Jiang, Cao, et al., 2019) Other intermediate aging phenotypes: Beta‐amyloid and tau pathology (Caldwell et al., 2018; Hsu et al., 2015; Lindberg et al., 2017; Parker, Cash, et al., 2019; Parker, Slattery, et al., 2019; Rahayel et al., 2019) Memory functions in homeless and marginally housed persons (Gicas et al., 2019) Recollection processes in older age (Hartopp et al., 2018) Subfields as ROI for fMRI (memory formation) (Thavabalasingam, O'Neil, Tay, Nestor, & Lee, 2019) |
| Pathological aging |
Amnestic or vascular MCI, established AD or LBD (de Flores et al., 2015; Evans et al., 2018; Gomar et al., 2017; Györfi et al., 2017; Hirjak, Wolf, et al., 2017; Hirjak, Sambataro, et al., 2019; Kälin et al., 2017; Kang, Lim, Joo, Lee, & Lee, 2018; Khan et al., 2015; Li et al., 2016; Li, Dong, Xie, & Zhang, 2013; Liang et al., 2018; Lim et al., 2012; Lim et al., 2013; Mak et al., 2016; Mak et al., 2017; Parker, Slattery, et al., 2019; Sarica et al., 2018; Shim et al., 2017; Wang, Yu, et al., 2018) Vitamin D in MCI (Al‐Amin et al., 2019), IL‐4 levels in MCI and AD (Boccardi et al., 2019) Spectral analyses (including healthy elderly, subjective memory complaints, MCI and AD; risk of conversion from MCI to AD) (DeVivo et al., 2019; Grajski & Bressler, 2019; Izzo, Andreassen, Westlye, & van der Meer, 2019; Marizzoni et al., 2019; Zhao et al., 2019) |
| Other neurodegenerative conditions |
PD (including dementia) (Foo et al., 2017; Lenka et al., 2018; Low, Foo, Yong, Tan, & Kandiah, 2019; Park et al., 2019; Pereira et al., 2013; Stav et al., 2016; Uribe et al., 2018) Frontotemporal dementia (Bocchetta et al., 2018), LBD, and beta amyloid (Mak et al., 2019) Multisystem atrophy and PD (Wang, Zhang, Yang, Luo, & Fan, 2019) Amyotrophic lateral sclerosis (Christidi et al., 2019), primary lateral sclerosis (Finegan et al., 2019) |
| Epilepsy |
Mesial temporal lobe epilepsy (Costa et al., 2019; Donos et al., 2018; Duarte et al., 2018; Kim, Suh, & Kim, 2015; Kreilkamp, Weber, Elkommos, Richardson, & Keller, 2018; Lee, Seo, & Park, 2019; Long, Feng, Liao, Zhou, & Urbin, 2018; Peixoto‐Santos et al., 2018; Schoene‐Bake et al., 2014; Sone et al., 2016) Subfields as ROI for fiber tracking (Rutland et al., 2018) |
| CNS (autoimmune) inflammatory disorder | Multiple sclerosis (González Torre et al., 2017; Zuppichini & Sandry, 2018), clinically isolated syndrome (Cacciaguerra et al., 2019), neuromyelitis optica spectrum disorder (Chen et al., 2019), anti‐LGI1 encephalitis (Finke et al., 2017), anti‐NMDA receptor encephalitis (Finke et al., 2016), subtypes of limbic encephalitis (Ernst et al., 2019) |
| Neurotoxicity | Toxic agents, radiotherapy or hypoxia (Decker et al., 2017; Lv et al., 2018; Ørbo, Vangberg, Tande, Anke, & Aslaksen, 2018; Phillips et al., 2020; Stamenova et al., 2018), alcoholism (Lee et al., 2016; Mole, Mak, Chien, & Voon, 2016; Zahr, Pohl, Saranathan, Sullivan, & Pfefferbaum, 2019), alcohol withdrawal (Kühn et al., 2014), familial risk for alcohol use disorder (adolescents) (Maksimovskiy et al., 2019), cannabis use (Beale et al., 2018), cigarette smoking (Durazzo, Meyerhoff, & Nixon, 2013), preterm birth in school‐aged children (Aanes et al., 2019) |
| Stress response |
Plasma markers of oxidative stress (van Velzen et al., 2017), peripheral inflammatory markers in HIV (Fleischman et al., 2018) expression of glucocorticoid inducible genes in MDD (Frodl, Carballedo, et al., 2014) Socioeconomic status and chronic physiological stress (hair cortisol) (Merz et al., 2019). Perceived stress (Zimmerman et al., 2016) |
| MDD |
Female MDD patients (Han, Won, Sim, & Tae, 2016; Kühn et al., 2012), ECT and treatment response to ECT (Cao et al., 2018; Gryglewski et al., 2019) or antidepressants (Hu et al., 2019; Maller et al., 2017) Acute and remitted depression (Kraus et al., 2019), depression symptom severity (Brown et al., 2019) Neurovascular disease in late onset MDD (Choi et al., 2017), MDD and aging (Szymkowicz et al., 2017), MDD and interleukin‐6 (Kakeda et al., 2018), MDD and other inflammatory markers (Lindqvist et al., 2014), MDD and tryptophan (Doolin et al., 2018) Subfields as ROI for fiber tracking (Rutland et al., 2019) |
| Bipolar disease |
Bipolar disease (Elvsåshagen et al., 2013, 2016), comparison with MDD in adults (Cao et al., 2017; Han et al., 2019) and in children and adolescents (Tannous et al., 2018), lithium effects (Giakoumatos et al., 2015; Hartberg et al., 2015; Simonetti et al., 2016) Predominant polarity (Janiri, Simonetti, et al., 2019) |
| Schizophrenia |
Schizophrenia (Zheng et al., 2019), first episode and chronic disease (Kawano et al., 2015; McHugo et al., 2018), symptom correlates (Han et al., 2016; Kühn et al., 2012), first episode psychosis (Baglivo et al., 2018; Buchy et al., 2016; Li et al., 2018), response to electroconvulsive therapy (Jiang, Xu, et al., 2019), conversion of high risk patients (Provenzano et al., 2020), young relatives at risk (Francis et al., 2013; Ho, Iglesias, et al., 2017; Ho, Holt, et al., 2017), progression patterns (Ho, Holt, et al., 2017; Ho, Iglesias, et al., 2017), genetic and cognitive correlates of schizophrenia (Nakahara et al., 2019) Metacognition and insight deficits (Alkan, Davies, Greenwood, & Evans, 2019; Hýža, Kuhn, Češková, Ustohal, & Kašpárek, 2016; Orfei et al., 2017) |
| PTSD and early life adversity | PTSD (Averill et al., 2017; Bøen et al., 2014; Hayes et al., 2017; L. Chen et al., 2016), childhood maltreatment (Chalavi et al., 2015; Teicher, Anderson, & Polcari, 2012), verbal abuse (Lee et al., 2018), childhood trauma in schizophrenia and healthy controls (du Plessis et al., 2019), in bipolar disorders and healthy controls (Janiri, Sani, et al., 2019) and in adolescence (Malhi et al., 2019) |
| Other neuropsychiatric conditions |
Pain symptoms (Ezzati et al., 2014), ADHD (Al‐Amin, Zinchenko, & Geyer, 2018), anorexia nervosa (Myrvang et al., 2018), obsessive compulsive disorder (Zhang et al., 2019), and transient global amnesia (Wang, Zhang, et al., 2018), essential tremor (Prasad et al., 2019), rapid eye movement sleep behavior disorder (Campabadal et al., 2019), trigeminal neuralgia (Vaculik, Noorani, Hung, & Hodaie, 2019) Thalamic infarction (Chen et al., 2016), sequelae of microsurgical aneurysm clipping (Hedderich et al., 2019) |
| Systemic disease | Prediabetes (Dong et al., 2019) primary biliary cholangitis (Mosher et al., 2018), systemic lupus erythematosus (Bódi et al., 2017) |
| Transdiagnostic approach |
Psychosis spectrum (Francis et al., 2013; Haukvik et al., 2015; Mathew et al., 2014; Vargas et al., 2017), unipolar–bipolar spectrum in adults (Cao et al., 2017; Han et al., 2019) or children and adolescents (Tannous et al., 2018) Bipolar disease and schizophrenia (pooling different subfield methods) (Haukvik, Tamnes, Söderman, & Agartz, 2018) Social anxiety disorder, childhood trauma and PTSD (Ahmed‐Leitao et al., 2019) |
| Genetic or epigenetic candidate analyses and exploratory imaging genetics analyses | |
| Candidate SNPs/genes | BDNF val66met variants (Aas et al., 2014; Frodl, Skokauskas, et al., 2014), TESC gene and MDD (Han et al., 2017), COMT and first‐episode MDD (Otsuka et al., 2019), oxytocin receptor gene and MDD (Na et al., 2018), 22q11.2 deletion syndrome (Mancini et al., 2019) |
| Epigenetics and gene–environment effects | Epigenetic modifications of glucocorticoid receptor in MDD and controls (Na et al., 2014), environmental adversity and COMT, BDNF, and 5‐HTTLPR (Rabl et al., 2014) |
| Polygenic risk | Aerobic exercise and polygenic risk for schizophrenia (Papiol et al., 2017; Papiol et al., 2019) |
| GWAS | GWAS of all subfields (van der Meer et al., 2018), GWAS of the dentate gyrus (Nakahara et al., 2019), GWAS with a methodological focus (Couvy‐Duchesne et al., 2019) |
Abbreviations: AD, Alzheimer's disease; ECT, electroconvulsive therapy; FS, FreeSurfer; GWAS, Genome wide association analysis; LBD, Lewy body dementia; MCI, mild cognitive impairment; MDD, major depressive disorder; PD, Parkinson's disease; PTSD, posttraumatic stress disorder; ROI, region of interest.
2.2.4. Other reliability aspects
T2WI as input
It is known that a T2‐weighted image (mostly in the coronal plane, perpendicular to the main axis of the hippocampus), facilitates the identification of the molecular layer and of its boundary with the CA1, which are usually not clearly visible on standard T1‐weighted data (Iglesias et al., 2015; Wisse et al., 2014). In this context, Mueller et al. (2018) compared four automated segmentation protocols, both T1WI‐ and T2WI‐based and manual segmentation, particularly with regard to increasing the sensitivity to disease effects. Indeed, effect sizes were higher for methods that included a high resolution T2WI compared to solely T1WI‐based methods. Yet, no direct comparison between a single T1WI and combined T1‐/T2WI input to version 6.0 was reported. Such a direct comparison is reported in Iglesias et al. (2015), who demonstrated increased sensitivity to MCI/controls differences with the combined input.
FS subfield tool to measure total HV
As the total hippocampal volume (HV) is included in some analyses of subfield data (as a dependent variable, or as a correction variable), a recent study (N = 664) validated manually traced HVs against several FS versions (5.2, 5.3, and 6.0) (Schmidt et al., 2018). In brief, automatically measured HV values were systematically larger than manually traced HVs, with a small influence of age and HV itself on this bias. Still, ICC values were high as were measures of spatial overlap. The study also reported detailed failure rates for FS6.0, estimated from 708 cases: The general segmentation failed in 25 cases (3.5%), with six cases (0.8%) showing incorrect orientation and two cases (0.3%) showing insufficient quality or contrast, leaving 17 cases (or 2.4%) without clearly explained failure. Several other reports focused on the reliability of automated total HV measurement: as concluded from eight reliability measures, FS6.0 subfield‐based total HV proved more reliable than other automated algorithms (Khlif, Egorova, et al., 2019; Khlif, Werden, et al., 2019), with acceptable biases. When FS6.0 is applied to children and adolescents, estimation biases compared to manual tracing may appear (Herten, Konrad, Krinzinger, Seitz, & von Polier, 2019; Schoemaker et al., 2016). For the analysis of brains with marked atrophy, the false segmentation of dura in FS has been identified as a problem for medial temporal lobe studies (Xie et al., 2019).
2.2.5. Measurement validity
Studies of test–retest reliability properties cannot account for deviation from the ground truth labeling, so, regardless of the study design, these studies are not informative on the actual validity of the subfield measurements. General external validity of the FS‐based hippocampal subfield volume measures is rooted in the cytoarchitectonic principle of the ex vivo sample labeling. Further validity aspects are still investigated with a latent assumption that subfield measures hold indirect information on functional or circuit differences with (patho‐)physiological relevance. Here, studies on subregional functional connectivity (FC), including intrahippocampal connectivity, or morphological covariance analysis provide relevant contributions (Dalton, McCormick, de Luca, Clark, & Maguire, 2019; Dalton, McCormick, & Maguire, 2019; Ge et al., 2019). An intriguing approach to derive hippocampal subregions is to search for subfield‐specific connectivity patterns, instead of hard histological ground truth: purely data‐driven, FC‐based hippocampal parcellations impressively match manual and—to a lesser degree—automated segmented subfields (Wu et al., 2018). Further, combined anatomical and functional features from 3 Tesla delivered a similar accuracy to 7 Tesla data. So far, no FC studies have been presented that directly incorporate FS6.0‐based subfield volumes, or that relate subfield volumes to connectivity measures.
One report critically addressed different aspects of the FS5.3 hippocampal subfield protocol (Wisse et al., 2014):
The effect of using low resolution T1WI as exclusive input to the algorithm, as these images might not provide sufficient contrast, for example, of the white matter bands between the dentate gyrus and the CA.
A bias in the original parcellation scheme that extrapolated one coronal section to the entire longitudinal axis of the hippocampus (Van Leemput et al., 2008, 2009), leading to overestimations of CA1 and atypical result patterns.
The mismatch between high‐resolution 3 Tesla T2WI (0.19 mm isometric) on which the algorithm has been developed, and its application to low resolution (1 mm isometric) T1WI.
The second concern, also expressed by de Flores et al. (2015), is directed toward a core element of the algorithm and was addressed by integrating ex vivo data into the atlas. This improved the consistency with manual segmentations (i.e., validity), in terms of the boundaries of CA2/3, presubiculum and the transition between the subiculum and CA1 (Iglesias et al., 2015).
The first and third concerns that have been re‐emphasized in a recent comment (Wisse et al., 2020) boil down to the question of whether low resolution T1WI as inputs can lead to sufficiently valid subfield results. For optimal correspondence with the gold standard, good GM/WM contrast is required to detect the white matter bands and the molecular layer. There is currently no clear demonstration that this can be only achieved by adding a T2WI, rather than by a T1WI with high resolution at 0.4–0.5 mm isotropic or 0.4 × 0.4 × 1.0 mm3. At an empirical level, also when this feature is suboptimally present, lower resolution T1WI have detected expected biological effects, for example, with good discrimination between MCI and controls (Iglesias et al., 2015). Still, a methodological rather than clinical validation study is missing that compares manual tracings according to the ex vivo protocol on very high resolution MRI data (e.g., 0.2 mm isotropic) to atlas based segmentations of downsampled images or newly acquired lower resolution images of the same subjects.
2.3. Cross‐correlations among subfield volumes and multiple testing considerations
Due to the explorative character of many hippocampal subfield studies, multiple test correction needs to be performed to control for false positive results. In the following exemplary analyses, we demonstrate how the correlation of (bilaterally added) subfield volumes with each other and with total HV in two samples. The purpose of these analyses is to demonstrate how the correlation structure is influenced by the global correction variable (intracranial volume [ICV] vs. total HV), and how subfield volumes relate to the total HV on a 1.5 Tesla compared to a 3 Tesla platform. Sample A was a 1.5 Tesla major depressive disorder (MDD)/controls sample acquired at the Max Planck Institute of Psychiatry (MPIP) and included in previous ENIGMA studies (Schmaal et al., 2017) (General Electric, Signa/Signa Excite, T1WI, SPGR‐3D or spin‐echo, voxel size 0.9 × 0.9 × 1.2 mm3, N = 597 [209 controls, 388 MDD patients], age 19–89 years); and Sample B, a 3 Tesla sample of healthy subjects (Münster Cohort, 3T Philips Gyroscan Intera, T1WI, 3D fast gradient echo sequence, voxel size 0.5 mm isometric; N = 211, age 17–61 years). The data were processed with FS5.3 (general segmentation) and FS6.0 (subfield segmentation).
In Sample A, Pearson correlations between raw subregional volumes ranged between r = −0.130 (fimbria vs. fissure, p = 0.756) and r = 0.988 (CA4 vs. GC‐ML‐DG, p < 1e‐20) (median 0.531). Correcting for ICV, age, age‐squared, sex, and site led to a slight left‐shift (r‐values lowered by ~0.113 on average, resulting median 0.379) within a similar total range (r = [−0.112; 0.982]). Correcting for total HV instead of ICV led to a marked left‐shift with r between −0.661 (molecular layer vs. hippocampal tail) and 0.942 (CA4 vs. GC‐ML‐DG) (median −0.108).—This “global correction” effect is best explained by the strong collinearity between subfield volumes and the total HV, even after correction for ICV, age, age‐squared, sex, and site (r = [0.327; 0.974]) (see Supplemental Figure 1a).
In Sample B, correlations between raw volumes were similar (r between 0.095 [fimbria vs. hippocampal tail, p = .170] and 0.994 [CA4 vs. GC‐ML‐DG, p < 1e‐20], median 0.575). Partial correlation coefficients between subregional volumes corrected for the same covariates as in Sample A were again mildly left‐shifted (range r = [−0.085; 0.992], median 0.475). For comparability with published cross‐correlations of residualized subregional volumes (van der Meer et al., 2018), the age‐squared term was omitted which changed the distribution only marginally (r = [−0.074; 0.992], median 0.471) and basically matched the mentioned report. Correcting for total HV again led to a strong left shift of the distribution (r = [−0.663; 0.964], median −0.024) (see Supplemental Figure 1b).
2.3.1. Comparing the subregional volumes versus total HV correlations (referred to as “autocorrelations”) between Samples A and B
For this analysis, Sample A was restricted to control subjects only for better comparability (N = 209, age range 19–79 years). Similar autocorrelation ranges were detected in both samples after correcting for age, age‐squared, sex, ICV, and site (Sample A: [0.201; 0.967] vs. Sample B: [0.390; 0.979]). After Fisher's z transformation, no difference was detected between the samples (two‐sided two‐sample t test, unequal variances assumed, p = .385). Six of twelve subregions (three with the lowest, and three with the highest correlations) were equally ranked between the samples, whereas for the remaining subregions the ranking positions were different (Table 1). Still, the correlation between the corresponding Fisher's z scores of the two samples was high (r = 0.967).
TABLE 1.
Correlation between total HV and subregions, corrected for ICV, age, squared age, sex, and site, in healthy subjects at 1.5 and 3 Tesla. Sorting is according to the partial correlation values (r p ) in descending order per sample. The last two columns denote relative shifts of the volume rank and the correlation rank of the 3 Tesla compared with the 1.5 Tesla sample. Negligible volume rank shifts for the two smallest regions (parasubiculum, HATA) were noted. For the correlation rank, identical ranking was found for 6 subregions, and a minor perturbation—by a maximum of two ranks—for the remaining subregions
| Partial correlations between total HV and subregions, adjusted for ICV, age, age‐squared, sex, site | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1.5 Tesla (N = 209) | 3 Tesla (N = 211) | Volume rank shift | Correlation rank shift | ||||||
| Region | Volume rank | r p | Fisher's z | Region | r p | Fisher's z | Volume rank | ||
| Molecular layer | 2 | 0.967 | 2.044 | Molecular layer | 0.997 | 2.276 | 2 | 0 | 0 |
| CA1 | 1 | 0.863 | 1.305 | CA1 | 0.929 | 1.648 | 1 | 0 | 0 |
| GC‐ML‐DG | 6 | 0.841 | 1.225 | GC‐ML‐DG | 0.909 | 1.519 | 6 | 0 | 0 |
| Subiculum | 4 | 0.815 | 1.142 | CA4 | 0.889 | 1.416 | 7 | 0 | −1 |
| CA4 | 7 | 0.806 | 1.116 | Subiculum | 0.856 | 1.279 | 4 | 0 | +1 |
| Presubiculum | 5 | 0.662 | 0.796 | CA3 | 0.685 | 0.838 | 8 | 0 | −2 |
| Hippocampal tail | 3 | 0.654 | 0.782 | Presubiculum | 0.664 | 0.800 | 5 | 0 | +1 |
| CA3 | 8 | 0.588 | 0.675 | HATA | 0.648 | 0.771 | 12 | +1 | −1 |
| HATA | 11 | 0.557 | 0.628 | Hippocampal tail | 0.627 | 0.737 | 3 | 0 | +2 |
| Hippocampal fissure | 9 | 0.386 | 0.407 | Hippocampal fissure | 0.612 | 0.712 | 9 | 0 | 0 |
| Parasubiculum | 12 | 0.359 | 0.376 | Parasubiculum | 0.528 | 0.588 | 11 | −1 | 0 |
| Fimbria | 10 | 0.201 | 0.204 | Fimbria | 0.390 | 0.412 | 10 | 0 | 0 |
Abbreviation: ICV, intracranial volume.
2.3.2. Autocorrelations and subfield size
The degree of autocorrelation of a subfield was linearly correlated with the average raw volume (r = 0.766 [Sample A], and r = 0.690 [Sample B]).
2.3.3. Interpretation
The concordance between the autocorrelation of subfields with their size likely reflects the above (Section 2.2.3) summarized lower measurement reliability of smaller subfields. Although imperfect in design, as measurements were not repeated in the same subjects, these cross‐sectional results point out that subregion volumes at 1.5 Tesla do not show a systematically stronger influence of the probability atlas over the contrast in the input data compared to 3 Tesla. Still, ranking shifts were observed for half of the subfields between platforms, highlighting that pooling results from different field strengths (or highly different acquisition schemes) is not recommended; rather, these should be treated as distinct samples.
2.3.4. Implications for multiple test correction
For multiple testing corrections, given the demonstrated and reported high correlation among subfield volumes (Elman et al., 2019; van der Meer et al., 2018), Bonferroni correction might be too strict. More recently, it was estimated that the 12 subfield volumes, after correction for covariates including ICV, represent seven effectively independent tests (software available at https://neurogenetics.qimrberghofer.edu.au/matSpDlite) (Couvy‐Duchesne et al., 2019; Li & Ji, 2005; Li, Yeung, Cherny, & Sham, 2012). False discovery rate based correction methods as used in several studies so far seem a good option. More complex methods that take into account the actual correlation pattern like approximations to permutation analysis (Conneely & Boehnke, 2007; Zugman et al., 2020) or spectral decomposition (Nyholt, 2004) can be considered.
3. QC ASPECTS
3.1. Reported QC procedures
QC procedures were mentioned in about 44% of 162 reviewed reports (Supplemental Table 1, see Section 4 for details on the review procedure), with heterogeneous elaborateness and sometimes involving up to three independent raters. This does not necessarily indicate that no QC was performed in the remaining studies, as for most imaging laboratories, plausibility checks are expected and may thus be underreported. The overall technical success rate of the subfield segmentation is influenced by installation preconditions and rarely explicitly stated (e.g., 96.5% for FS6.0 in Schmidt et al. (2018)), so cannot be reliably quantified due to the heterogeneity of reports but only estimated to be over 95%. Most QC descriptions point to a visual and mostly qualitative, holistic assessment. Ten studies (~6%) excluded cases only based on outlier features (e.g., > 5 SDs), the largest being the study of van der Meer et al. (2018) (see Supplemental Table 1, column QC details). In turn, most studies used outlier features to guide visual inspection but did not exclude outliers without identifying a technical segmentation failure. Five studies (~3%) report manual editing with no further details (see Supplemental Table 1, column QC details). Only exceptionally, cases were excluded due to complete failure of the segmentation, some of them related to gross brain pathology. Several studies mentioned FS viewing tools to inspect segmentations, or mention the ENIGMA protocol, or leave this unspecified. There appears to be a consensus that in the case of peculiarities or segmentation failure, the whole case is excluded (at least the affected hemisphere), and not single subfields.
3.2. Theoretical considerations for QC
The FS6.0‐based hippocampal segmentation algorithm is fully automated and manual corrections are technically feasible, but not designated. Still, the algorithm should not be used in a completely “unsupervised way,” as segmentation failures in the complex, multistep procedure may occur for several reasons. The FS algorithm as such is very robust, and depending on basic checks before starting the segmentation, a map with hippocampal subfield volumes is generally obtained. The FS tool package allows one to overlay the hippocampal segmentation output on a background image with a simple command, usually in native space, for visual QC (Figure 2b), which is useful in smaller studies.
Still, there is no standardized QC procedure for the hippocampal segmentation tool in FS. While requiring a certain amount of time, a visual QC step combined with an automated recommendation of critical cases, appears to be a good compromise to control the risk of technical or algorithmic problems that—if undetected—may lead to distorted results and conclusions. Based on the Bayesian approach of the probabilistic atlas mesh being matched with the actual data, the output represents a data‐informed modification of this mesh, similar to other deformation‐based procedures, as in tensor‐based morphometry, for example. As they are explicitly penalized, extreme deviations of one subregion (in terms of 3D shape/surface and eventually volume) are thus unlikely, and will not occur independently of other subregions. The QC rater, in our opinion, needs no formal neuroradiological training, yet should be sufficiently informed to distinguish between:
normal anatomical variants that occur in the hippocampal area and might adversely affect the automated algorithm;
minor, visually conspicuous, yet volumetrically negligible phenomena;
incomplete segmentations or otherwise atypical segmentation results.
Category (1) includes, for example, cystic formations with CSF intensity within or close to the hippocampal formation, such as sulcus remnant cysts (also referred to as the “hippocampal sulcus residual cavity”) (Li et al., 2006; Sasaki, Sone, Ehara, & Tamakawa, 1993) or choroidal fissure cysts (that can, histologically, represent arachnoid, neuroglial, or neuroepithelial cysts) (Osborn & Preece, 2006). These might stand out on the raw image, the fissure overlay or the full segmentation overlay, yet with little effect on the validity of the actual adjacent subfield volume measures. Following this logic, most subjects in this category do not need to be excluded. Research on the effect of known normal variants is ongoing; more recently, a correlation between CA1 volumes and incomplete hippocampal inversion (IHI), a very common normal variant found in about 20% of healthy subjects (Colenutt, McCann, Knight, Coulthard, & Kauppinen, 2018), has been reported
Category (2) represents phenomena that could be referred to as “small artifacts” with no critical effect on the resulting volumes. Yet, it seems useful to identify examples in this category, particularly for raters assessing a smaller study that may only contain very few examples of such phenomena (see Figure 3b–e and legends for specific examples).
FIGURE 4.
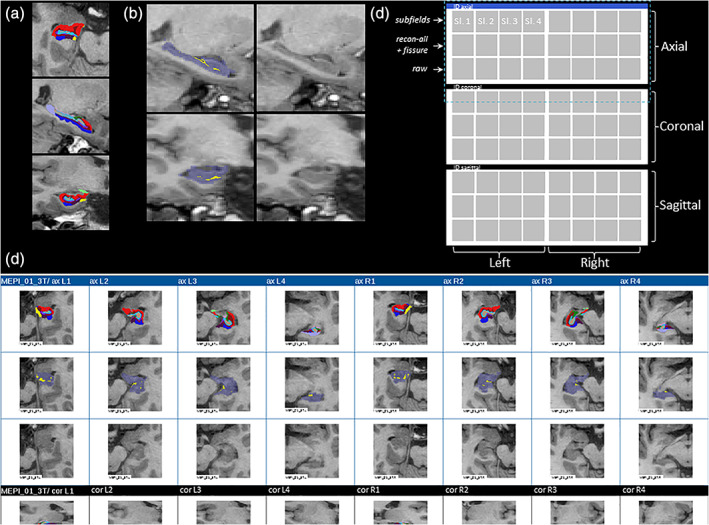
Exemplary overlays of full segmentation results, combined hippocampal starting mask and fissure, and full visual quality control (QC) output. (a) 3 Tesla examples of (bias field corrected) T1‐weighted input image and exemplary axial and sagittal colored subfield segmentation. (b) Principle of combined background image, and hippocampal starting mask (from general subcortical segmentation [recon‐all]), and hippocampal fissure, allowing a check of basic orientation/rotation, successful basic hippocampal segmentation (purple transparent overlay) and correct placement of hippocampal fissure (yellow) within the hippocampal starting mask in one glance. (c) Layout structure of HTML output of one case. One 8 × 3 set of images usually fits on a large screen, so a maximum of one page flip is needed to finalize one case. Dark blue horizontal bars separate cases from each other. Dashed purple box marks the area for which a real example is depicted in (d). (d) Exemplary HTML‐output of one case (limited to axial and part of the coronal output images). Sl., slice number
Category (3) is potentially complex and comprises heterogeneous underlying causes, including technical failures (e.g., algorithm failure, or incorrect orientation of the input image), low input image quality, or extreme pathology with gross atrophy or intrahippocampal signal intensity changes. Particularly the latter category—extreme pathology—may lead to a decision to keep the subject in the sample, or exceptionally exclude only selected subfields. Such a scenario may for example be marked postoperative or posttraumatic changes of the medial temporal lobe that prevent proper detection of certain subfields, strong intrahippocampal intensity changes caused by hippocampal sclerosis, ischemia or encephalitis, or strongly progressed AD or other forms of neurodegeneration. For this reason, the underlying T1WI is displayed in parallel.
Figure 3 covers examples of Categories (1) and (2). Additional examples of deviations may be found in Figure 12 and supplemental Figure 22 of the original report on the ex vivo atlas‐based method (Iglesias et al., 2015).
Two further points deserve attention: First, circularity is problematic—in the sense of (visually) rating those features that will be used as the dependent variable of the analysis. In addition, as the hippocampal segmentation is based on—though not fully dependent on—a proper general segmentation of the hippocampus (as part of the recon‐all command in FS), such a control step should be incorporated in the QC (see below). For our QC pipeline, we thus developed a triple overlay of (a) the resulting hippocampal fissure (an output of the subfield segmentation) on (b) the starting hippocampus mask of the general segmentation, both overlaid on (c) the T1‐weighted background image as the basis for the visual QC. In addition to this, yet not in isolation, the full set of hippocampal subregions is shown (Figure 2). Second, the general cortical and subcortical segmentation should also undergo a QC, for example, by the packages supported online, as of April 2017 (http://enigma.ini.usc.edu/protocols/imaging-protocols), for two main reasons: (a) the generation of intensity distributions is based on defined white matter areas and successful cortical/subcortical segmentation and (b) global measures such as ICV, total brain volume or total gray matter (TGM) are usually needed for the statistical model and rely on a proper general registration or segmentation.
As both the general QC and the subfield QC show example slices of the original input image, this should help in detecting very low‐quality input images (e.g., blurred margins due to motion artifacts). Such images increase the risk of general segmentation failure (e.g., parts of lobes missing, or considerable underestimates of cortical thickness) and/or incomplete progression of the hippocampal segmentation to the borders of the hippocampus. The latter may be found in case of an extremely degenerated hippocampus where internal CSF signals may mimic a true margin. In our experience, such global low‐quality cases often lead to strongly underestimated TGM values or TGM/ICV ratios that appear as outlier values, particularly when plotted against age.
3.3. A proposed QC pipeline
Within the ENIGMA consortium, we have developed a standardized set of MATLAB, Linux shell, and R scripts for QC of hippocampal subfields derived from the FS6.0 algorithm that provide several functions, including (a) starting subfield segmentations on a group of subjects that have undergone the general recon‐all segmentation, (b) gaining a standardized readout table of subregional volumes and global volumes from the general segmentation, (c) a list of cases that should be inspected individually, and (d) a browsable hypertext markup language (HTML) file that allows to perform this visual QC also in larger samples (Figure 4).
FIGURE 3.
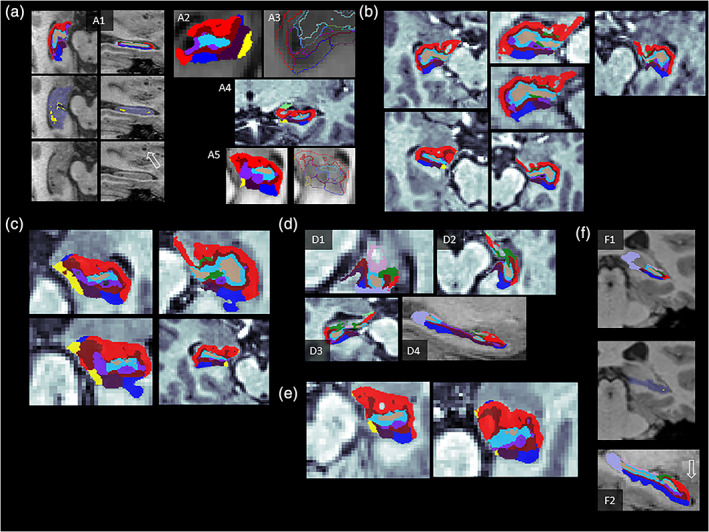
Examples of normal anatomical variants, peculiarities and phenomena, and discrepancy between the binary hippocampal mask and segmentation results. Images are displayed at (unsmoothed) 1‐mm isometric resolution, and subfield overlays at 0.333‐mm isometric resolution. Anonymized examples are differently contrasted as they stem from several sites with different display settings. All shown examples would not necessarily need to be excluded, yet see specific comments on a2 and a5. (a1) Example of sulcus remnant cysts that are found in ~25% of adults between the dentate gyrus and the cornu ammonis; considered an incidental finding with no pathological implication; classified as hippocampal fissure (yellow on mid row image) which represents CSF intensity. Similar peri‐ or intrahippocampal cysts exist, such as choroidal fissure cysts (etiologically arachnoid or neuroglial or neuroepithelial cysts) may similarly be classified as hippocampal fissure, or extra‐hippocampal (“background”) by the algorithm, depending on location details. Certain types of cysts may co‐occur with enlarged perivascular spaces (Virchow Robin spaces), for example, in the lower basal ganglia or midbrain (see open arrows in a1). (a2) A cystic area not classified as fissure, but as extra‐hippocampal background, and a part of CA1 seems neglected. Depending on the amount of such truncated hippocampal tissue, extreme cases of this type should be excluded. (a3–a5) Examples of cysts either classified clearly as fissure, or as background (i.e., no hippocampal subregion). a5 likely classified the fissure correctly that, however, appears brighter than CSF on the raw image due to partial volume effects. More extreme cases (hippocampal fissure intensity being too bright on T1WI or not represented on T2WI) should be excluded, and the hippocampal fissure volume may contain cysts. (b) Very frequent (90%) yet practically negligible observed discontinuous appearance of CA1 and/or smaller spared regions (“holes”) within CA1. (c) Subfields extending into another subfield or forming small extensions/islands; though visually conspicuous the volume effect of such extensions is negligible. (d) “Holes” localizable to fimbria (light pink, d1), or CA3 (green) or CA1 (red) as in d2 or d3 are caused by the removal of the alveus (white matter layer on the superior rim of the hippocampus) area in the final binary classification step, which may cause the impression of an abrupt ending of the segmentation. (e) “Bulky” appearance of CA1 in the sense of this subfield strongly dominating the appearance on one slice. This may occur due to strict orthogonal slicing in native space. (f1) Example of standard hippocampal segmentation (recon‐all) missing parts of the posterior hippocampus, while the subfield segmentation v6.0 correctly detects these parts. Incompleteness of the standard binary hippocampal starting mask should therefore not be an exclusion criterion per se. (f2) Anterior parts seem truncated, yet lie in the amygdala complex; this appearance is normal and does not indicate a failed segmentation
Steps (a) and (b) are based on Linux shell scripts. Step (c) is an R script based on two principles: First, an outlier calculation scheme (+/− 2.98 SDs, each representing the roughly 1% of cases in the tails of a normal distribution) directed to all subfield volumes, yet also total brain volume, total GM volume, ICV, and GM/ICV ratio. Second, as the rank order of the subregional volumes is very robust (Figure 2a) across MR platforms and samples, a set of three violations of this ranking order has been defined from an analysis of N = 626 subjects in which the deviation pattern of the actual rank from the group mean rank was calculated: (a) CA1 not being the largest subregion (i.e., not rank #1; frequency ~ 11%), (b) hippocampal tail being below rank #3 (frequency ~2%), and (c) subiculum not being ranked #4 (frequency ~ 2%). The first rule is optional (switched off by default) as it may be oversensitive and too unspecific in larger samples.
For Step (d), the number of cases summarized per HTML‐file can be defined by the user. In this way, several separate files are generated, for example, for distribution to several raters. Further, a random order mode (to avoid QC order biases) can be requested, or a sparse version of the HTML output with fewer example slices that allows one to accelerate the throughput in very large samples (e.g., N > 1,000).
Overall, when using our ENIGMA Hippocampal Subfields QC protocol, we recommend the following steps, as also shown in Figure 5, before statistical analyses of subregional volumes:
FIGURE 5.

Overall quality control (QC) flow scheme for a FreeSurfer‐based hippocampal subfield study. Depending on local pipelines, general QC steps regarding the raw data quality, motion artifacts and complete coverage may be performed directly on the picture archiving system and be study‐independent. A general inspection of the cortical/subcortical segmentation result of FS is recommended before subfield specific operations. Steps 2–7 are supported by a script package written in MATLAB, R, and Shell. “freeview + subfields” refers to an individual check using the interactive FS viewing tool (https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfields)
Perform general FS segmentation and append the hippocampal segmentation module, ideally using the same version (currently 6.0 or 7.0) (Note: Combining FS.5.3 general segmentation with FS6.0 subfield version is feasible and produces highly similar results, compared to FS6.0 only; also see supplemental material in van der Meer et al. (2018).
Perform a QC of the general cortical and subcortical segmentation, using the respective ENIGMA scripts and instructions (http://enigma.ini.usc.edu/protocols/imaging-protocols).
Read out hippocampal subregional volume and general segmentation results using the script from the SUBFIELDS package; run the R script outliers_hippo_and_QC_support.R on this output to generate a list of cases that could harbor problems (see Figure 5 for an example).
Run the three QC preparation scripts to eventually generate the QC‐HTML files; adapt graphical and random order or sparse output settings in the scripts as required.
-
Inspect all datasets in the QC‐HTML files with a standard browser, with particular attention to the flagged datasets. Additional recommendations are:
To become familiar with the study specific variance of segmentations and to avoid an order bias, it may be efficient to view a random selection of cases at a higher pace, collecting “global impressions” first.
Before browsing the static HTML‐file and if unfamiliar with the FS hippocampal segmentation output, it may be beneficial to explore a handful of cases using the FS viewer Freeview that allows scrolling in all three dimensions. As FS output is in native space, this step is useful to become acquainted with the variance caused by imperfectly corresponding anatomical slice positions between subjects.
4. FS‐BASED HIPPOCAMPAL SUBFIELD ANALYSIS: STATUS OF CLINICAL NEUROSCIENCE PUBLICATIONS
The studies referred to in this section were retrieved from a PubMed literature search performed on December 14, 2019. The search combination was “hippocampal subfields” OR “hippocampal subregions” OR “hippocampal subfield” OR “hippocampal subregion” AND “MRI” (limited [filter] to human studies and a time frame January 1, 2013–December 14, 2019), resulting in 401 reports. All 401 abstracts were reviewed (P. G. S.) and those in which the segmentation technique was not the automated FS tool (i.e., other automated tools or manual segmentation protocols) were excluded. Reviews and retrospective meta‐analyses were excluded to avoid double inclusion of original studies. After excluding reliability and purely methodological studies that we refer to in Section 4.2, 162 reports were available for further analyses.
From these studies, we aimed to identify domains, topic, and selected methodological information: field strength, MRI sequence (T1WI, T2WI, or both), FS version, global volume correction (if used, and the specific volume used), statistical approach for global correction (regression method, proportion method, covariate method), presence of age and sex as covariates, modeling of nonlinear age effects, and additional covariates. We extracted if a QC of the subfield segmentation was reported, if outliers were excluded and if manual corrections were performed. Finally, we noted if and how hippocampal subfields were (re‐)aggregated to create composite markers. Three studies used hippocampal subfield segmentation merely as a tool to define ROIs, leaving 159 studies for the analysis of statistical aspects.
As one goal of ENIGMA is to advance the endophenotype concept (Gottesman & Gould, 2003)for genetic association studies, the returned hippocampal subfield studies were organized into studies of (a) heritability; (b) clinical diagnoses, behavioral and cognitive phenotypes, or other biomarkers; and (c) studies of candidate genes or exploratory imaging genetics analyses. Results of (a) are summarized as text in Section 4.1. Results of all Categories (a) and (c) are presented at three different levels of granularity: first, in an iterative process based on the full papers, studies were aggregated into 19 domains (15 clinical/ behavioral domains, 4 (epi‐)genetic domains) that are depicted in a pie chart (Figure 1, see legend for details on category fusions). Second, in Table 2, subcharacterizations and topics have been added to single studies or groups of studies for a quick overview and references. Third, Supplemental Table 1 lists all 162 studies in more detail focusing on the relevant technical features extracted for the methodological and QC review.
4.1. Heritability analyses
Hippocampal subfield volumes from FS6.0 have been analyzed with regard to their heritability—mainly to corroborate their utility as endophenotype—based on monozygotic/dizygotic twin studies (Elman et al., 2019; Greenspan et al., 2016; Whelan et al., 2016) or on genotype information (van der Meer et al., 2018).
Whelan et al. (2016) reported heritability between 56 and 88% for hippocampal subvolumes and total HV, after controlling for age, sex, and age‐by‐sex effects. Heritability estimates h 2 as measured by the proportion of variance in volume attributable to genetics via twin studies were larger than 70% for regions with relatively larger volumes (whole hippocampus, molecular layer, CA1, CA3, CA4, hippocampal tail, granule cell layer, subiculum, and presubiculum) and moderate to high (55% < h 2 < 70%) for smaller subregions (HATA, fimbria, parasubiculum, hippocampal fissure). Projection to a hippocampal surface model demonstrated that heritability is larger for posterior subregions (including the hippocampal tail), and smaller for anteromedial subregions (parasubiculum, presubiculum, fimbria), suggesting a gradient of genetic influences. Even so, the relationship between subregional volume and measurement reliability may also have influenced estimates of heritability. Elman et al. (2019) reported heritability values between 37% (HATA) and 89% (molecular layer) from values controlled for age and sex. Genetic correlations as a measure of shared genetic influences were high between subfields and the total HV, indicating that for standard resolution T1WI, no substantial additional information on the genetic underpinning is gained through the subfield volumes (Elman et al., 2019). Despite finding significant genetic covariance of the subfields, no stable latent genetic traits (composed from combinations of subfields) were found in a factor analysis (Elman et al., 2019).
Greenspan et al. (2016) reported heritability estimates for FS6.0 subfield volumes between 20 and 87% along with high shared genetic variance with total HV (mean 0.79, range 0.50–0.98). After regressing out total HV—or when percentages based on the ratio of the subregional volume/total HV) were considered—heritabilities ranged between 4 and 86% (7 and 84%, respectively). Here, heritability values were not significant for the fimbria, hippocampal fissure and HATA.
From a recent GWAS on hippocampal subregional volumes residualized for ICV, age, sex and site (van der Meer et al., 2018), significant SNP‐based heritability was reported from a very large sample (N > 20,000), ranging between 14% (for the parasubiculum) and 27% (for the hippocampal tail).
4.2. Application to clinical diagnoses, behavioral, and cognitive phenotypes, (epi‐)genetic markers or other biomarkers
We found that hippocampal subfield analyses using the FS tool so far have been performed in a broad range of domains that are quantified in Figure 1 and grouped into subdomains/topics in Table 2. Beyond activity in expected major fields such as hippocampus‐dependent cognitive functions, healthy and pathological aging, major psychiatric diseases of the affective/psychosis spectrum and different neurological disorders, a growing interest in psychotraumatology, and neurotoxic influences including systemic inflammation is noted. Subfield volumes are now also probed as endophenotypes, in studies analyzing candidate genes, epigenetic methylation patterns polygenic risk scores, but also exploring the whole genome for associations with single subfields (Nakahara et al., 2019) or all subfields (van der Meer et al., 2018), or for methodological comparisons (Couvy‐Duchesne et al., 2019).
4.3. Statistical aspects
4.3.1. Modeling global volume effects
One important source of interindividual variance in regional brain volumes is the variation in overall head size (Mathalon, Sullivan, Rawles, & Pfefferbaum, 1993). To increase the specificity of regional results, ICV is often included in the statistical model—of the 159 reports in which subfield volumes were the outcome variable, 78% reported results corrected for ICV. Technically, in FS, ICV is estimated from the matrix created during a linear registration of the individual MRI scan to Talairach space, and is thus also referred to as “estimated total intracranial volume” (Buckner et al., 2004). FS‐based studies thus tend to employ this measure as an ICV surrogate. Alternatively, after segmentation (e.g., using Statistical Parametric Mapping [SPM] software), TGM, white matter and CSF volumes can be summed up. Both methods correlate well with the gold standard of manual delineation, yet overestimations have been reported for both SPM8 and FS (version 5.1.0) (Nordenskjöld et al., 2013). Excellent agreement with the gold standard, superior to FS5.3, was recently achieved with SPM12 based segmentation augmented by a manually edited intracranial mask (in MNI space) (Malone et al., 2015). A smaller proportion of studies considered total brain volume (4.4%), supratentorial gray matter volume (2.5%) or total HV (5.7%) as a correction variable. Choosing these variables gradually increases the regional specificity which is particularly relevant if effects in extra‐hippocampal brain areas are expected (e.g., neurodegenerative disorders). Correction for total HV allows to study subregional effects that exceed total HV effects, maximizing the regional specificity.
ICV (or another global volume) can be modeled in various ways (O'Brien et al., 2011): first, it can be included as a nuisance covariate in analyses of covariance, linear mixed models or regression analyses, along with other confounding variables (analysis of covariance approach). This approach was used in most (79%) of the 136 studies that included a global volume. Second, volumes can be adjusted by regression analysis (residual approach), so volcorr = vol ‐ b*(ICV‐ICVmean), where b is the slope of the regression of vol on ICV (Hayes et al., 2017; Mathalon et al., 1993). About 13% of 136 studies applied this approach. Third, ICV may be used for a direct individual standardization of regional volumes. This approach, applied in 25% of the reports, is referred to as the proportion (or ratio) approach. In its simplest form, volcorr = vol/ICV. In an extension of this, the sample mean ICV can be multiplied with this term. The advantages and disadvantages of each of these methods are a long discussed topic (Arndt, Cohen, Alliger, Swayze 2nd, & Andreasen, 1991; Buckner et al., 2004; Jack Jr et al., 1989; O'Brien et al., 2011; Sanfilipo, Benedict, Zivadinov, & Bakshi, 2004; Voevodskaya et al., 2014). No generally valid recommendation can be given that applies to all study types and questions and each approach requires certain assumptions (O'Brien et al., 2011). For example, the covariate allows for a clear attribution of different sources of variance; the regression approach may be unreliable in small samples, but yields a more Gaussian distribution compared to the proportion approach.
An increasingly recognized problem is the possibility of nonlinear (allometric) rather than isometric relationships between total and local cerebral measures (Jäncke, Liem, & Merillat, 2019; O'Brien et al., 2011; Reardon et al., 2016). Toro et al. (2009) demonstrated that assumptions on brain allometry made a critical difference in a genetic study on HV. More recently, van Eijk et al. (2020) reported that allometric scaling may also be found for hippocampal subfield volumes.
4.3.2. Modeling of age effects
Modeling of age effects is another critical point for studies on hippocampal morphology. For TGM volume, when corrected for ICV, the gain in explained variance through nonlinear terms is rather weak (Figure 2d), whereas hippocampal subregion volumes (and other subcortical structures) show nonlinear effects, with a plateau phase in mid adulthood and a decrease starting around age 65 (de Flores et al., 2015) (also see Figure 2c). Similarly, nonlinear developmental trajectories have been reported for the age range between 8 and 30 years for subcortical brain structures including the total hippocampus (Ostby et al., 2009), and for hippocampal subregions between age 4 and 22 years (Krogsrud et al., 2014). This implies that in hippocampal subregional studies of larger age ranges, either guided by theoretical considerations or by data exploration, models with nonlinear terms should be used. In our collection of reports, age effects were modeled in most studies (~89%); four (2.5%) of these studies modeled nonlinear age effects.
In addition, aging processes may be abnormal in clinical conditions, and disease‐specific aging processes may falsely translate into group main effects. Respective age‐by‐sex and age‐by‐diagnosis interaction terms, and their higher order versions, can help to refine the model and reduce interpretational ambiguity. Here, pooling of multisite data (e.g., mega‐analysis) is useful, and methods to remove batch effects under preservation of covariate effects have been suggested for age harmonization (Fortin et al., 2018; Pomponio et al., 2020).
4.3.3. Skipping or reaggregation of hippocampal subfields
Skipping or, in turn, reaggregating hippocampal subfields to create composite measures (differing from those in the original FS6.0 or FS5.3 scheme) was reported in a handful of studies (~4%). Subfields excluded due to their relatively low reliability were mostly the fimbria and the hippocampal fissure (as the latter does not represent hippocampal tissue, but a CSF cleft). Some studies define the subicular complex from the subiculum, the parasubiculum and presubiculum (see Supplemental Table 1, last column, and table legend for details). In FS6.0 and FS7.1, three variants of how the algorithms' hidden fine‐grained subfield parcellation is aggregated to the final labels are provided: (a) a head‐body‐tail aggregation, useful for comparisons with earlier landmark‐based segmentation work or calculation of more coarse ROIs regions, for example, for functional MRI, (b) the aggregation to 12 subregions presented here, and (c) a version called “CA,” in which the internal labels (GC‐ML‐DG and molecular layer) are absorbed by the CA subfields. More specifically, GC‐ML‐DG is assigned to CA4, and voxels in the molecular layer are assigned to the closest voxel that is neither in the molecular layer nor background.
McHugo et al. (2018) in their work on psychosis suggested two aggregation schemes for easier comparison with data generated from other commonly used segmentation algorithms (Mueller et al., 2018; Yushkevich et al., 2015) and to account for imperfect boundary definitions for smaller subregions (CA3, GC‐DG, molecular layer) in the absence of multispectral input data. These schemes are interesting variants, particularly when volumetric effects along the longitudinal axis of the hippocampus are investigated.
4.3.4. Multivariate analyses and structural covariance analysis
Given the good measurement reliability of most subfield volumes, multivariate analysis including machine learning approaches are particularly attractive, allowing for the detection of complex morphological patterns. For example, hippocampal subfield information predicted treatment outcome in MDD with an accuracy of 83% (Cao et al., 2018). Similarly, linear discriminant analysis using hippocampal subfield information (from both FS5.3 and FS6.0) proved superior to total HV for discriminating AD patients from healthy controls (Iglesias et al., 2015). Both studies add indirect clinical validity to the subfield segmentation. Structural covariance analysis that is based on the coherence of subfield volumetric patterns across subjects is another interesting approach: For example, Wang, Yu, et al. (2018) and Wang, Zhang, et al. (2018) reported higher covariance between hippocampal subfields and extra‐hippocampal gray matter volume in amnestic and vascular MCI patients compared with control subjects. Unsupervised learning tasks such as clustering have revealed groupings within the subfields and with other brain areas (van der Meer et al., 2018). To our knowledge, no clustering of patient samples into subgroups, based on hippocampal subfield patterns, has been reported so far.
5. SUMMARY DISCUSSION AND OUTLOOK
The FS‐based hippocampal subfield segmentation algorithm is a widely applied tool that can be used to study physiological hippocampus‐dependent processes and pathological conditions. Healthy and pathological aging studies, including the MCI/AD spectrum, together with studies on the psychosis and the affective disorders spectrum make up about 50% of the studies. Early environmental, and genetic influences and studies of neurotoxic influences on hippocampal integrity make up the majority of the remaining studies. The tool provides an opportunity to improve upon the long‐known nonspecificity of (total) hippocampal volumetry findings in many neuropsychiatric diseases (Geuze, Vermetten, & Bremner, 2005). Due to its easy, automated use, it naturally fosters exploratory studies, and some reports now exist performing GWAS on hippocampal subfield volumes (Morey et al., 2020; Nakahara et al., 2019; van der Meer et al., 2018). Here, recent methodological work has highlighted that for hippocampal subfield volumes multivariate GWAS could be a useful addition to the classical multiple univariate approach (Couvy‐Duchesne et al., 2019).
Most clinical/behavioral studies used analysis of covariance or linear mixed models, covarying for age, sex, and ICV. Still, considerable variability exists regarding the modeling of age and global volumes, which hampers retrospective meta‐analyses. A similar degree of heterogeneity was noted for QC steps. Prospective harmonized analyses of existing datasets, as performed in ENIGMA, and also the analysis of prospectively acquired subfield data could benefit from our QC protocol as a guideline. Multivariate analyses of hippocampal subfield data are still rarely used but have begun to show promising results, for example, for predicting treatment response in MDD (Cao et al., 2018). In conjunction with good across‐site and across‐vendor reliability (Brown et al., 2020; Quattrini et al., 2020), this strengthens the potential of subfield volumes as a biomarker for clinical trials. One possible advantage of subregional volumes over voxel‐wise or vertex‐wise features is a higher degree of interpretability, as effect weights can be translated back to histology or other levels of validation.
Only very few studies (3.1%) made use of two MRI input channels despite evidence that a second, high resolution T2WI—by detecting critical internal hippocampal boundaries—can improve subfield measurement reliability and validity (Iglesias et al., 2015; Mueller et al., 2018; Winterburn et al., 2013). We suggest that in the currently available FS hippocampal subfield studies the potential of the algorithm is not fully exploited and optimized MRI protocols are considered in future prospective studies.
At least two other automated hippocampal subfield segmentation algorithms exist, both developed and evaluated on 3 Tesla platforms and both based on manual tracings on high resolution MRI images as a reference: MaGET‐Brain (multiple automatically generated templates) (Pipitone et al., 2014) and automatic segmentation of hippocampal subfields (ASHS) (Yushkevich et al., 2015). A complex challenge in this methodological field is the definition of an agreed manual segmentation protocol: here, the hippocampal subfields group of the EU Joint Programme–Neurodegenerative Disease Research is aiming at harmonizing a reliable and valid protocol for postmortem data, also fostering its translation to MRI data. A protocol for 7 Tesla MRI data with good agreement between manual and ASHS performance has been published (Wisse et al., 2016), and comparing ex vivo to high‐field MRI data (9.4 Tesla) has resulted in further progress in protocol definitions (de Flores et al., 2019). This work is ongoing and will serve as the basis for new validation experiments within the MRI domain (Olsen et al., 2019). The influence of IHI—a common, atypical rotation of the hippocampal formation (Cury et al., 2015) that impacts subfield volumes (Colenutt et al., 2018)—presents another challenge.
As an outlook, hippocampal shape analysis is worthwhile as it can be combined with subfield information to localize shape effects. The shape analysis pipeline of ENIGMA (http://enigma.usc.edu/ongoing/enigma-shape-analysis) is based on the FS subcortical segmentation, followed by topological correction steps and smoothing based on the topology‐preserving level set algorithm (Gutman et al., 2015; Xiao Han, Xu, & Prince, 2003). After registration to standard templates (Gutman et al., 2015; Gutman, Wang, Rajagopalan, Toga, & Thompson, 2012), two metrics are calculated for each vertex point: a radial distance that is comparable to a “shape thickness” measure, and the Jacobian determinant (Gutman et al., 2015; Wang et al., 2011), a metric of localized tissue reduction or enlargement of surface area relative to the template shape. Using a subfield surface projection (first presented for FS5.3 by Cong et al. (2014)), shape effects can be assigned to specific subfields. Such a combined approach has recently helped to localize hippocampal shape changes in MDD (Ho et al., 2020) and to identify volume changes and a complex bending and displacement of parts of the hippocampus in obsessive–compulsive disorder (Zhang et al., 2019). Beyond this, using a surface‐based atlas of hippocampal subvolume projections built on the medial core modeling (Gutman et al., 2012), proportional contributions of volume subfields to a surface effect can be quantified (Figure 6).
FIGURE 6.
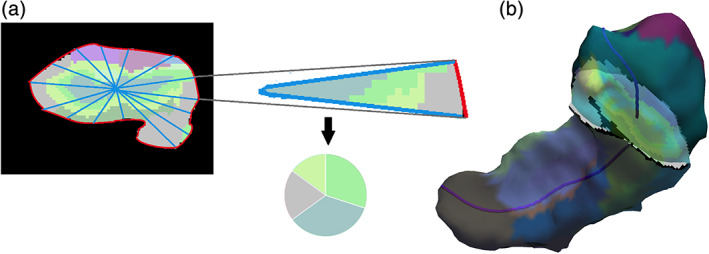
Relationship between surface and volumetric subfield composition. (a) Surface subfields are defined by the volume subfield nearest to the surface. For each surface element, we integrate the 3D subfields between the surface and the medial curve. The pie chart represents the proportion of volume subfields underlying the surface line (red). The figure is drawn in 2D for illustration purposes only, yet, the principle can be equally applied to a 2D patch at the surface. (b) An individual hippocampal model with surface subfields and a coronal slice of the volumetric subfields from FS6.0. The dark purple line is the medial curve
In conclusion, the FS‐based hippocampal subfield segmentation tool provides a valuable, fine‐morphological phenotyping instrument with good measurement reliability and validity. Use of a standardized QC procedure can help to harmonize large‐scale, collaborative studies and reduce methodological heterogeneity. Acquisition protocols specifically designed for subfield studies, as well as further external validation and multivariate analysis techniques, could further improve its usefulness in clinical neuroscience.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplemental Figure 1 Effect of covariate structure on subfield correlations. (A) depicts distribution of 66 Pearson (partial) correlation coefficients of 12 raw (bilateral) subfield volumes correlated with each other in sample A (1.5 Tesla, 597 subjects) (top row). Note mild leftward shift of (partial) correlation coefficients after correction for ICV, age, age‐squared and sex (middle row), and strong leftward shift after replacing ICV by the total HV (bottom row). (B) shows equivalent distributions obtained from sample B (3 Tesla, 211 subjects).
Supplemental Table 1 Brief characterization of 162 FreeSurfer hippocampal subfield studies. Numbers in the last column refer to subfield reaggregation/skipping details: 1Fimbria, fissure, hippocampal tail excluded. 2CA3 and CA4 aggregated, subicular complex aggregated (pre‐, para‐ and subiculum), DG aggregated (granule cell layer and molecular layer). 3Fimbria and fissure excluded. 4Fimbria and alveus combined. 5CA4 excluded, subicular complex aggregated (pre‐, para‐ and subiculum), DG aggregated (granule cell layer and molecular layer). 6Parasubiculum, presubiculum, alveus, fissure, fimbria and HATA omitted. 7Fimbria and fissure excluded. 8Three aggregations: classical FS6.0, anterior/posterior composite regions, composite subfields of body/head. ADHS, attention deficit hyperactivity syndrome; AD, Alzheimer's Disease; aMCI amnestic mild cognitive impairment; Cov, Covariate; CSF, cerebrospinal fluid; ECT, electroconvulsive therapy; FTD, frontotemporal dementia; GWAS, genome wide association study; HV, hippocampal volume; ICV, intracranial volume; LBD, Lewy Body Disease; MCI, mild cognitive impairment; MDD, major depressive disorder; MTLE, mesio‐temporal lobe epilepsy; PTSD, posttraumatic stress disorder; QC, quality control; ROI, region of interest; Regr, Regression; sGMV, supratentorial gray matter volume; SAD, social anxiety disorder; SCD, subjective cognitive deficits; UHR, ultra‐high risk; TBV, total brain volume
ACKNOWLEDGMENTS
The authors thank Ida Kippersund and Stener Nerland of the ENIGMA‐Bipolar Disorders Working Group for providing QC examples from their local samples. The authors are grateful to Filip Jukic for supporting literature search and QC work on the MPIP sample. This work was supported by the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17 to U. D.). C. W., N. J., and P. M. T. were supported in part by NIH grant U54 EB020403, from the NIH Big Data to Knowledge (BD2K) program, which is supported by multiple NIH institutes. J. E. I. is supported by European Research Council (Starting Grant 677697, project BUNGEE‐TOOLS). R.A.M. and E.K.C. were supported by the VA Mid‐Atlantic Mental Illness Research and Clinical Center and NIH RO1 MH111671.
Sämann PG, Iglesias JE, Gutman B, et al. FreeSurfer‐based segmentation of hippocampal subfields: A review of methods and applications, with a novel quality control procedure for ENIGMA studies and other collaborative efforts. Hum Brain Mapp. 2022;43:207–233. 10.1002/hbm.25326
[Corrections added in affiliations on 02nd January, 2021, after first online publication]
Funding information European Research Council; National Institutes of Health, Grant/Award Number: U54 EB020403; Interdisciplinary Center for Clinical Research, Münster, Grant/Award Numbers: 677697, Dan3/012/17, SFB‐TRR58; German Research Foundation, Grant/Award Numbers: Z02, C09, DA1151/5‐2, FOR2107 DA1151/5‐1
Endnote
The modification from FS5.3 to FS6.0 incorporated anatomical and theoretical refinements, as well as a new subregional parcellation that correlated more strongly with gold standards and was derived from higher resolution ex vivo scans. The atlas in FS5.3 had been built from in vivo scans at 0.38 × 0.38 × 0.80 mm3 resolution, which yielded voxels approximately 100 times larger than the ex vivo protocol and required the human labeller to rely on geometric criteria in their delineations. This had led to underestimation of CA1 at the expense of the subiculum and CA2/CA3 (de Flores et al., 2015; Iglesias et al., 2015; Wisse, Biessels, & Geerlings, 2014). Thus, while analyses of between version (FS5.3/FS6.0) agreement have been performed (Whelan et al., 2016), their interpretation is difficult and they are therefore not reported in detail here.
DATA AVAILABILITY STATEMENT
The data used for the analysis in Section 2.3 are made available on request (saemann@psych.mpg.de).
REFERENCES
- Aanes, S. , Bjuland, K. J. , Sripada, K. , Sølsnes, A. E. , Grunewaldt, K. H. , Håberg, A. , … Skranes, J. (2019). Reduced hippocampal subfield volumes and memory function in school‐aged children born preterm with very low birthweight (VLBW). NeuroImage: Clinical, 23, 101857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas, M. , Haukvik, U. K. , Djurovic, S. , Tesli, M. , Athanasiu, L. , Bjella, T. , … Melle, I. (2014). Interplay between childhood trauma and BDNF val66met variants on blood BDNF mRNA levels and on hippocampus subfields volumes in schizophrenia spectrum and bipolar disorders. Journal of Psychiatric Research, 59, 14–21. [DOI] [PubMed] [Google Scholar]
- Ahmed‐Leitao, F. , Rosenstein, D. , Marx, M. , Young, S. , Korte, K. , & Seedat, S. (2019). Posttraumatic stress disorder, social anxiety disorder and childhood trauma: Differences in hippocampal subfield volume. Psychiatry Research: Neuroimaging, 284, 45–52. [DOI] [PubMed] [Google Scholar]
- Al‐Amin, M. , Bradford, D. , Sullivan, R. K. P. , Kurniawan, N. D. , Moon, Y. , Han, S.‐H. , … Burne, T. H. J. (2019). Vitamin D deficiency is associated with reduced hippocampal volume and disrupted structural connectivity in patients with mild cognitive impairment. Human Brain Mapping, 40, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Amin, M. , Zinchenko, A. , & Geyer, T. (2018). Hippocampal subfield volume changes in subtypes of attention deficit hyperactivity disorder. Brain Research, 1685, 1–8. [DOI] [PubMed] [Google Scholar]
- Alkan, E. , Davies, G. , Greenwood, K. , & Evans, S. L. H. (2019). Brain structural correlates of metacognition in first‐episode psychosis. Schizophrenia Bulletin, 46, 552–561. 10.1093/schbul/sbz116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, P. , Morris, R. , Amaral, D. , Bliss, T. , & O'Keefe, J. (2007). Division of neurophysiology. In The hippocampus book, Oxford, England: Oxford University Press. [Google Scholar]
- Arndt, S. , Cohen, G. , Alliger, R. J. , Swayze, V. W., 2nd , & Andreasen, N. C. (1991). Problems with ratio and proportion measures of imaged cerebral structures. Psychiatry Research, 40, 79–89. [DOI] [PubMed] [Google Scholar]
- Aslaksen, P. M. , Bystad, M. K. , Ørbo, M. C. , & Vangberg, T. R. (2018). The relation of hippocampal subfield volumes to verbal episodic memory measured by the California verbal learning test II in healthy adults. Behavioural Brain Research, 351, 131–137. [DOI] [PubMed] [Google Scholar]
- Averill, C. L. , Satodiya, R. M. , Scott, J. C. , Wrocklage, K. M. , Schweinsburg, B. , Averill, L. A. , … Abdallah, C. G. (2017). Posttraumatic stress disorder and depression symptom severities are differentially associated with hippocampal subfield volume loss in combat veterans. Chronic Stress, 1. 10.1177/2470547017744538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglivo, V. , Cao, B. , Mwangi, B. , Bellani, M. , Perlini, C. , Lasalvia, A. , … GET UP Group . (2018). Hippocampal subfield volumes in patients with first‐episode psychosis. Schizophrenia Bulletin, 44, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale, C. , Broyd, S. J. , Chye, Y. , Suo, C. , Schira, M. , Galettis, P. , … Solowij, N. (2018). Prolonged cannabidiol treatment effects on hippocampal subfield volumes in current cannabis users. Cannabis Cannabinoid Res, 3, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccardi, V. , Westman, E. , Pelini, L. , Lindberg, O. , Muehlboeck, J.‐S. , Simmons, A. , … Mecocci, P. (2019). Differential associations of IL‐4 with hippocampal subfields in mild cognitive impairment and Alzheimer's disease. Frontiers in Aging Neuroscience, 10. 10.3389/fnagi.2018.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchetta, M. , Iglesias, J. E. , Scelsi, M. A. , Cash, D. M. , Cardoso, M. J. , Modat, M. , … Rohrer, J. D. (2018). Hippocampal subfield volumetry: Differential pattern of atrophy in different forms of genetic frontotemporal dementia. Journal of Alzheimer's Disease, 64, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bódi, N. , Polgár, A. , Kiss, E. , Mester, Á. , Poór, G. , & Kéri, S. (2017). Reduced volumes of the CA1 and CA4‐dentate gyrus hippocampal subfields in systemic lupus erythematosus. Lupus, 26, 1378–1382. [DOI] [PubMed] [Google Scholar]
- Bøen, E. , Westlye, L. T. , Elvsåshagen, T. , Hummelen, B. , Hol, P. K. , Boye, B. , … Malt, U. F. (2014). Smaller stress‐sensitive hippocampal subfields in women with borderline personality disorder without posttraumatic stress disorder. Journal of Psychiatry & Neuroscience, 39, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E. M. , Pierce, M. E. , Clark, D. C. , Fischl, B. R. , Iglesias, J. E. , Milberg, W. P. , … Salat, D. H. (2020). Test‐retest reliability of FreeSurfer automated hippocampal subfield segmentation within and across scanners. NeuroImage, 210, 116563. [DOI] [PubMed] [Google Scholar]
- Brown, S. S. G. , Rutland, J. W. , Verma, G. , Feldman, R. E. , Alper, J. , Schneider, M. , … Balchandani, P. (2019). Structural MRI at 7T reveals amygdala nuclei and hippocampal subfield volumetric association with major depressive disorder symptom severity. Scientific Reports, 9, 10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchy, L. , Barbato, M. , MacMaster, F. P. , Bray, S. , Clark, D. , Deighton, S. , & Addington, J. (2016). Cognitive insight is associated with cortical thickness in first‐episode psychosis. Schizophrenia Research, 172, 16–22. [DOI] [PubMed] [Google Scholar]
- Buckner, R. L. , Head, D. , Parker, J. , Fotenos, A. F. , Marcus, D. , Morris, J. C. , & Snyder, A. Z. (2004). A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas‐based head size normalization: Reliability and validation against manual measurement of total intracranial volume. NeuroImage, 23, 724–738. [DOI] [PubMed] [Google Scholar]
- Burgess, N. , Maguire, E. A. , & O'Keefe, J. (2002). The human Hippocampus and spatial and episodic memory. Neuron, 35, 625–641. 10.1016/s0896-6273(02)00830-9 [DOI] [PubMed] [Google Scholar]
- Cacciaguerra, L. , Pagani, E. , Mesaros, S. , Dackovic, J. , Dujmovic‐Basuroski, I. , Drulovic, J. , … Rocca, M. A. (2019). Dynamic volumetric changes of hippocampal subfields in clinically isolated syndrome patients: A 2‐year MRI study. Multiple Sclerosis, 25, 1232–1242. [DOI] [PubMed] [Google Scholar]
- Caldwell, J. Z. K. , Berg, J.‐L. , Shan, G. , Cummings, J. L. , Banks, S. J. , & Alzheimer's Disease Neuroimaging Initiative . (2018). Sex moderates the impact of diagnosis and amyloid PET positivity on hippocampal subfield volume. Journal of Alzheimer's Disease, 64, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campabadal, A. , Segura, B. , Junque, C. , Serradell, M. , Abos, A. , Uribe, C. , … Iranzo, A. (2019). Cortical gray matter and hippocampal atrophy in idiopathic rapid eye movement sleep behavior disorder. Frontiers in Neurology, 10, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, B. , Luo, Q. , Fu, Y. , Du, L. , Qiu, T. , Yang, X. , … Qiu, H. (2018). Predicting individual responses to the electroconvulsive therapy with hippocampal subfield volumes in major depression disorder. Scientific Reports, 8, 5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, B. , Passos, I. C. , Mwangi, B. , Amaral‐Silva, H. , Tannous, J. , Wu, M.‐J. , … Soares, J. C. (2017). Hippocampal subfield volumes in mood disorders. Molecular Psychiatry, 22, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalavi, S. , Vissia, E. M. , Giesen, M. E. , Nijenhuis, E. R. S. , Draijer, N. , Cole, J. H. , … Reinders, A. A. T. S. (2015). Abnormal hippocampal morphology in dissociative identity disorder and post‐traumatic stress disorder correlates with childhood trauma and dissociative symptoms. Human Brain Mapping, 36, 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Luo, T. , Lv, F. , Shi, D. , Qiu, J. , Li, Q. , … Li, Y. (2016). Relationship between hippocampal subfield volumes and memory deficits in patients with thalamus infarction. European Archives of Psychiatry and Clinical Neuroscience, 266, 543–555. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Fu, J. , Luo, Q. , Han, Y. , Zheng, Q. , Xie, M. , & Li, Y. (2019). Altered volume and microstructural integrity of hippocampus in NMOSD. Multiple Sclerosis and Related Disorders, 28, 132–137. [DOI] [PubMed] [Google Scholar]
- Choi, W. H. , Jung, W. S. , Um, Y. H. , Lee, C. U. , Park, Y. H. , & Lim, H. K. (2017). Cerebral vascular burden on hippocampal subfields in first‐onset drug‐naïve subjects with late‐onset depression. Journal of Affective Disorders, 208, 47–53. [DOI] [PubMed] [Google Scholar]
- Christidi, F. , Karavasilis, E. , Rentzos, M. , Velonakis, G. , Zouvelou, V. , Xirou, S. , … Bede, P. (2019). Hippocampal pathology in amyotrophic lateral sclerosis: Selective vulnerability of subfields and their associated projections. Neurobiology of Aging, 84, 178–188. [DOI] [PubMed] [Google Scholar]
- Colenutt, J. , McCann, B. , Knight, M. J. , Coulthard, E. , & Kauppinen, R. A. (2018). Incomplete hippocampal inversion and its relationship to hippocampal subfield volumes and aging. Journal of Neuroimaging, 28, 422–428. [DOI] [PubMed] [Google Scholar]
- Cong, S. , Rizkalla, M. , Du, E. Y. , West, J. , Risacher, S. , Saykin, A. , & Shen, L. (2014). For the Alzheimer's Disease Neuroimaging Initiative (2014): Building a Surface Atlas of Hippocampal Subfields from MRI Scans using FreeSurfer, FIRST and SPHARM. Conf Proc:813–816. [DOI] [PMC free article] [PubMed]
- Conneely, K. N. , & Boehnke, M. (2007). So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. American Journal of Human Genetics, 81, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, B. S. , Santos, M. C. V. , Rosa, D. V. , Schutze, M. , Miranda, D. M. , & Romano‐Silva, M. A. (2019). Automated evaluation of hippocampal subfields volumes in mesial temporal lobe epilepsy and its relationship to the surgical outcome. Epilepsy Research, 154, 152–156. [DOI] [PubMed] [Google Scholar]
- Couvy‐Duchesne, B. , Strike, L. T. , McMahon, K. L. , de Zubicaray, G. I. , Thompson, P. M. , Martin, N. G. , … Wright, M. J. (2019). A fast method for estimating statistical power of multivariate GWAS in real case scenarios: Examples from the field of imaging genetics. Behavior Genetics, 49, 112–121. [DOI] [PubMed] [Google Scholar]
- Cury, C. , Toro, R. , Cohen, F. , Fischer, C. , Mhaya, A. , Samper‐González, J. , … IMAGEN Consortium . (2015). Incomplete hippocampal inversion: A comprehensive MRI study of over 2000 subjects. Frontiers in Neuroanatomy, 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, M. A. , McCormick, C. , de Luca, F. , Clark, I. A. , & Maguire, E. A. (2019). Functional connectivity along the anterior‐posterior axis of hippocampal subfields in the ageing human brain. Hippocampus, 29, 1049–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, M. A. , McCormick, C. , & Maguire, E. A. (2019). Differences in functional connectivity along the anterior‐posterior axis of human hippocampal subfields. NeuroImage, 192, 38–51. 10.1016/j.neuroimage.2019.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores, R. , Berron, D. , Ding, S.‐L. , Ittyerah, R. , Pluta, J. B. , Xie, L. , … Wisse, L. E. M. (2019). Characterization of hippocampal subfields using ex vivo MRI and histology data: Lessons for in vivo segmentation. In Hippocampus (Vol. 30, pp. 545–564). 10.1002/hipo.23172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores, R. , la Joie, R. , Landeau, B. , Perrotin, A. , Mézenge, F. , de La Sayette, V. , … Chételat, G. (2015). Effects of age and Alzheimer's disease on hippocampal subfields: Comparison between manual and FreeSurfer volumetry. Human Brain Mapping, 36, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon, M. , George, A. , Stylopoulos, L. , Smith, G. , & Miller, D. (1989). Early markers for Alzheimer's disease: The atrophic hippocampus. Lancet, 334, 672–673. [DOI] [PubMed] [Google Scholar]
- Decker, A. L. , Szulc, K. U. , Bouffet, E. , Laughlin, S. , Chakravarty, M. M. , Skocic, J. , … Mabbott, D. J. (2017). Smaller hippocampal subfield volumes predict verbal associative memory in pediatric brain tumor survivors. Hippocampus, 27, 1140–1154. [DOI] [PubMed] [Google Scholar]
- DeKraker, J. , Ferko, K. M. , Lau, J. C. , Köhler, S. , & Khan, A. R. (2018). Unfolding the hippocampus: An intrinsic coordinate system for subfield segmentations and quantitative mapping. NeuroImage, 167, 408–418. [DOI] [PubMed] [Google Scholar]
- Delazer, M. , Zamarian, L. , Benke, T. , Wagner, M. , Gizewski, E. R. , & Scherfler, C. (2019). Is an intact hippocampus necessary for answering 3 × 3?—Evidence from Alzheimer's disease. Brain and Cognition, 134, 1–8. [DOI] [PubMed] [Google Scholar]
- DeVivo, R. , Zajac, L. , Mian, A. , Cervantes‐Arslanian, A. , Steinberg, E. , Alosco, M. L. , … Alzheimer's Disease Neuroimaging Initiative . (2019). Differentiating between healthy control participants and those with mild cognitive impairment using volumetric MRI data. Journal of the International Neuropsychological Society, 25, 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Dongwei, L. , Zhang, J. , Liang, J. , Sun, Z. , & Fang, J. (2019). Individuals in the prediabetes stage exhibit reduced hippocampal tail volume and executive dysfunction. Brain and Behavior: A Cognitive Neuroscience Perspective, 9, e01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donos, C. , Breier, J. , Friedman, E. , Rollo, P. , Johnson, J. , Moss, L. , … Tandon, N. (2018). Laser ablation for mesial temporal lobe epilepsy: Surgical and cognitive outcomes with and without mesial temporal sclerosis. Epilepsia, 59, 1421–1432. [DOI] [PubMed] [Google Scholar]
- Doolin, K. , Allers, K. A. , Pleiner, S. , Liesener, A. , Farrell, C. , Tozzi, L. , … O'Keane, V. (2018). Altered tryptophan catabolite concentrations in major depressive disorder and associated changes in hippocampal subfield volumes. Psychoneuroendocrinology, 95, 8–17. [DOI] [PubMed] [Google Scholar]
- du Plessis, S. , Scheffler, F. , Luckhoff, H. , Asmal, L. , Kilian, S. , Phahladira, L. , & Emsley, R. (2019). Childhood trauma and hippocampal subfield volumes in first‐episode schizophrenia and healthy controls. Schizophrenia Research, 215, 308–313. 10.1016/j.schres.2019.10.009 [DOI] [PubMed] [Google Scholar]
- Duarte, J. T. C. , Jardim, A. P. , Comper, S. M. , de Marchi, L. R. , Gaça, L. B. , Fernandes, M. T. , … Yacubian, E. M. T. (2018). The impact of epilepsy duration in a series of patients with mesial temporal lobe epilepsy due to unilateral hippocampal sclerosis. Epilepsy Research, 147, 51–57. 10.1016/j.eplepsyres.2018.08.009 [DOI] [PubMed] [Google Scholar]
- Durazzo, T. C. , Meyerhoff, D. J. , & Nixon, S. J. (2013). Interactive effects of chronic cigarette smoking and age on hippocampal volumes. Drug and Alcohol Dependence, 133, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elman, J. A. , Panizzon, M. S. , Gillespie, N. A. , Hagler, D. J., Jr. , Fennema‐Notestine, C. , Eyler, L. T. , … Kremen, W. S. (2019). Genetic architecture of hippocampal subfields on standard resolution MRI: How the parts relate to the whole. Human Brain Mapping, 40, 1528–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvsåshagen, T. , Westlye, L. T. , Bøen, E. , Hol, P. K. , Andersson, S. , Andreassen, O. A. , … Malt, U. F. (2013). Evidence for reduced dentate gyrus and fimbria volume in bipolar II disorder. Bipolar Disorders, 15, 167–176. [DOI] [PubMed] [Google Scholar]
- Elvsåshagen, T. , Zuzarte, P. , Westlye, L. T. , Bøen, E. , Josefsen, D. , Boye, B. , … Andreazza, A. C. (2016). Dentate gyrus‐cornu ammonis (CA) 4 volume is decreased and associated with depressive episodes and lipid peroxidation in bipolar II disorder: Longitudinal and cross‐sectional analyses. Bipolar Disorders, 18, 657–668. [DOI] [PubMed] [Google Scholar]
- Engvig, A. , Fjell, A. M. , Westlye, L. T. , Skaane, N. V. , Sundseth, Ø. , & Walhovd, K. B. (2012). Hippocampal subfield volumes correlate with memory training benefit in subjective memory impairment. NeuroImage, 61, 188–194. [DOI] [PubMed] [Google Scholar]
- Ernst, L. , David, B. , Gaubatz, J. , Domínguez‐Narciso, I. , Lüchters, G. , Becker, A. J. , … Rüber, T. (2019). Volumetry of mesiotemporal structures reflects serostatus in patients with limbic encephalitis. AJNR. American Journal of Neuroradiology, 40, 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. E. , Adams, H. H. H. , Licher, S. , Wolters, F. J. , van der Lugt, A. , Ikram, M. K. , … Ikram, M. A. (2018). Subregional volumes of the hippocampus in relation to cognitive function and risk of dementia. NeuroImage, 178, 129–135. [DOI] [PubMed] [Google Scholar]
- Ezzati, A. , Katz, M. J. , Lipton, M. L. , Lipton, R. B. , & Verghese, J. (2015). The association of brain structure with gait velocity in older adults: A quantitative volumetric analysis of brain MRI. Neuroradiology, 57, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzati, A. , Zimmerman, M. E. , Katz, M. J. , Sundermann, E. E. , Smith, J. L. , Lipton, M. L. , & Lipton, R. B. (2014). Hippocampal subfields differentially correlate with chronic pain in older adults. Brain Research, 1573, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan, E. , Li Hi Shing, S. , Chipika, R. H. , Doherty, M. A. , Hengeveld, J. C. , Vajda, A. , … Bede, P. (2019). Widespread subcortical grey matter degeneration in primary lateral sclerosis: A multimodal imaging study with genetic profiling. NeuroImage: Clinical, 24, 102089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke, C. , Kopp, U. A. , Pajkert, A. , Behrens, J. R. , Leypoldt, F. , Wuerfel, J. T. , … Paul, F. (2016). Structural hippocampal damage following anti‐N‐methyl‐D‐aspartate receptor encephalitis. Biological Psychiatry, 79, 727–734. [DOI] [PubMed] [Google Scholar]
- Finke, C. , Prüss, H. , Heine, J. , Reuter, S. , Kopp, U. A. , Wegner, F. , … Bartsch, T. (2017). Evaluation of cognitive deficits and structural hippocampal damage in encephalitis with leucine‐rich, glioma‐inactivated 1 antibodies. JAMA Neurology, 74, 50–59. [DOI] [PubMed] [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman, D. A. , Arfanakis, K. , Leurgans, S. , Keating, S. M. , Lamar, M. , Bennett, D. A. , … Barnes, L. L. (2018). Neopterin is associated with hippocampal subfield volumes and cognition in HIV. Neurology Neuroimmunology & Neuroinflammation, 5, e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, H. , Mak, E. , Chander, R. J. , Ng, A. , Au, W. L. , Sitoh, Y. Y. , … Kandiah, N. (2017). Associations of hippocampal subfields in the progression of cognitive decline related to Parkinson's disease. NeuroImage: Clinical, 14, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin, J.‐P. , Cullen, N. , Sheline, Y. I. , Taylor, W. D. , Aselcioglu, I. , Cook, P. A. , … Shinohara, R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, C. M. , Kennedy, K. M. , Hoagey, D. A. , & Rodrigue, K. M. (2019). The role of hippocampal subfield volume and fornix microstructure in episodic memory across the lifespan. Hippocampus, 29, 1206–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, A. N. , Seidman, L. J. , Tandon, N. , Shenton, M. E. , Thermenos, H. W. , Mesholam‐Gately, R. I. , … Keshavan, M. S. (2013). Reduced subicular subdivisions of the hippocampal formation and verbal declarative memory impairments in young relatives at risk for schizophrenia. Schizophrenia Research, 151, 154–157. [DOI] [PubMed] [Google Scholar]
- Frodl, T. , Carballedo, A. , Frey, E.‐M. , O'Keane, V. , Skokauskas, N. , Morris, D. , … Connor, T. (2014). Expression of glucocorticoid inducible genes is associated with reductions in cornu ammonis and dentate gyrus volumes in patients with major depressive disorder. Development and Psychopathology, 26, 1209–1217. [DOI] [PubMed] [Google Scholar]
- Frodl, T. , Skokauskas, N. , Frey, E.‐M. , Morris, D. , Gill, M. , & Carballedo, A. (2014). BDNF Val66Met genotype interacts with childhood adversity and influences the formation of hippocampal subfields. Human Brain Mapping, 35, 5776–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, R. , Kot, P. , Liu, X. , Lang, D. J. , Wang, J. Z. , Honer, W. G. , & Vila‐Rodriguez, F. (2019). Parcellation of the human hippocampus based on gray matter volume covariance: Replicable results on healthy young adults. Human Brain Mapping, 40, 3738–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, A. E. , de Leon, M. J. , Stylopoulos, L. A. , Miller, J. , Kluger, A. , Smith, G. , & Miller, D. C. (1990). CT diagnostic features of Alzheimer disease: Importance of the choroidal/hippocampal fissure complex. AJNR. American Journal of Neuroradiology, 11, 101–107. [PMC free article] [PubMed] [Google Scholar]
- Geuze, E. , Vermetten, E. , & Bremner, J. D. (2005). MR‐based in vivo hippocampal volumetrics: 2. Findings in neuropsychiatric disorders. Molecular Psychiatry, 10, 160–184. [DOI] [PubMed] [Google Scholar]
- Giakoumatos, C. I. , Nanda, P. , Mathew, I. T. , Tandon, N. , Shah, J. , Bishop, J. R. , … Keshavan, M. S. (2015). Effects of lithium on cortical thickness and hippocampal subfield volumes in psychotic bipolar disorder. Journal of Psychiatric Research, 61, 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicas, K. M. , Thornton, A. E. , Waclawik, K. , Wang, N. , Jones, A. A. , Panenka, W. J. , … Honer, W. G. (2019). Volumes of the hippocampal formation differentiate component processes of memory in a community sample of homeless and marginally housed persons. Archives of Clinical Neuropsychology, 34, 548–562. 10.1093/arclin/acy066 [DOI] [PubMed] [Google Scholar]
- Giuliano, A. , Donatelli, G. , Cosottini, M. , Tosetti, M. , Retico, A. , & Fantacci, M. E. (2017). Hippocampal subfields at ultra high field MRI: An overview of segmentation and measurement methods. Hippocampus, 27, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomar, J. J. , Ragland, J. D. , Uluğ, A. M. , Sousa, A. , Huey, E. D. , Conejero‐Goldberg, C. , … Goldberg, T. E. (2017). Differential medial temporal lobe morphometric predictors of item‐ and relational‐encoded memories in healthy individuals and in individuals with mild cognitive impairment and Alzheimer's disease. Alzheimers Dement, 3, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Torre, J. A. , Cruz‐Gómez, Á. J. , Belenguer, A. , Sanchis‐Segura, C. , Ávila, C. , & Forn, C. (2017). Hippocampal dysfunction is associated with memory impairment in multiple sclerosis: A volumetric and functional connectivity study. Multiple Sclerosis, 23, 1854–1863. [DOI] [PubMed] [Google Scholar]
- Gottesman, I. I. , & Gould, T. D. (2003). The endophenotype concept in psychiatry: Etymology and strategic intentions. The American Journal of Psychiatry, 160, 636–645. [DOI] [PubMed] [Google Scholar]
- Grajski, K. A. , & Bressler, S. L. (2019). Differential medial temporal lobe and default‐mode network functional connectivity and morphometric changes in Alzheimer's disease. NeuroImage: Clinical, 23. 10.1016/j.nicl.2019.101860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby, K. L. , Jahanshad, N. , Painter, J. N. , Colodro‐Conde, L. , Bralten, J. , Hibar, D. P. , & (ENIGMA)—Genetics working group (2020). The genetic architecture of the human cerebral cortex. Science, 367(6484), eaay6690 10.1126/science.aay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, K. S. , Arakelian, C. R. , & van Erp, T. G. M. (2016). Heritability of hippocampal formation sub‐region volumes. Journal of Neurology and Neuroscience, 7, 159. 10.21767/2171-6625.1000159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryglewski, G. , Baldinger‐Melich, P. , Seiger, R. , Godbersen, G. M. , Michenthaler, P. , Klöbl, M. , … Lanzenberger, R. (2019). Structural changes in amygdala nuclei, hippocampal subfields and cortical thickness following electroconvulsive therapy in treatment‐resistant depression: Longitudinal analysis. The British Journal of Psychiatry, 214, 159–167. 10.1192/bjp.2018.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman, B. A. , Jahanshad, N. , Ching, C. R. K. , Wang, Y. , Kochunov, P. V. , Nichols, T. E. , & Thompson, P. M. (2015). Medial demons registration localizes the degree of genetic influence over subcortical shape variability: An N= 1480 meta‐analysis. Proceedings IEEE International Symposium on Biomedical Imaging, 2015, 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman, B. A. , Wang, Y. , Rajagopalan, P. , Toga, A. W. , & Thompson, P. M. (2012). Shape matching with medial curves and 1‐D group‐wise registration. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI) . 10.1109/isbi.2012.6235648 [DOI]
- Györfi, O. , Nagy, H. , Bokor, M. , Moustafa, A. A. , Rosenzweig, I. , Kelemen, O. , & Kéri, S. (2017). Reduced CA2‐CA3 hippocampal subfield volume is related to depression and normalized by l‐DOPA in newly diagnosed Parkinson's disease. Frontiers in Neurology, 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, K.‐M. , Kim, A. , Kang, W. , Kang, Y. , Kang, J. , Won, E. , … Ham, B.‐J. (2019). Hippocampal subfield volumes in major depressive disorder and bipolar disorder. European Psychiatry, 57, 70–77. [DOI] [PubMed] [Google Scholar]
- Han, K.‐M. , Won, E. , Kang, J. , Choi, S. , Kim, A. , Lee, M.‐S. , … Ham, B.‐J. (2017). TESC gene‐regulating genetic variant (rs7294919) affects hippocampal subfield volumes and parahippocampal cingulum white matter integrity in major depressive disorder. Journal of Psychiatric Research, 93, 20–29. [DOI] [PubMed] [Google Scholar]
- Han, K.‐M. , Won, E. , Sim, Y. , & Tae, W.‐S. (2016). Hippocampal subfield analysis in medication‐naïve female patients with major depressive disorder. Journal of Affective Disorders, 194, 21–29. [DOI] [PubMed] [Google Scholar]
- Hartberg, C. B. , Jørgensen, K. N. , Haukvik, U. K. , Westlye, L. T. , Melle, I. , Andreassen, O. A. , & Agartz, I. (2015). Lithium treatment and hippocampal subfields and amygdala volumes in bipolar disorder. Bipolar Disorders, 17, 496–506. [DOI] [PubMed] [Google Scholar]
- Hartopp, N. , Wright, P. , Ray, N. J. , Evans, T. E. , Metzler‐Baddeley, C. , Aggleton, J. P. , & O'Sullivan, M. J. (2018). A key role for subiculum‐fornix connectivity in recollection in older age. Frontiers in Systems Neuroscience, 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukvik, U. K. , Tamnes, C. K. , Söderman, E. , & Agartz, I. (2018). Neuroimaging hippocampal subfields in schizophrenia and bipolar disorder: A systematic review and meta‐analysis. Journal of Psychiatric Research, 104, 217–226. [DOI] [PubMed] [Google Scholar]
- Haukvik, U. K. , Westlye, L. T. , Mørch‐Johnsen, L. , Jørgensen, K. N. , Lange, E. H. , Dale, A. M. , … Agartz, I. (2015). In vivo hippocampal subfield volumes in schizophrenia and bipolar disorder. Biological Psychiatry, 77, 581–588. [DOI] [PubMed] [Google Scholar]
- Hayes, J. P. , Hayes, S. , Miller, D. R. , Lafleche, G. , Logue, M. W. , & Verfaellie, M. (2017). Automated measurement of hippocampal subfields in PTSD: Evidence for smaller dentate gyrus volume. Journal of Psychiatric Research, 95, 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedderich, D. M. , Reess, T. J. , Thaler, M. , Berndt, M. T. , Moench, S. , Lehm, M. , … Friedrich, B. (2019). Hippocampus subfield volumetry after microsurgical or endovascular treatment of intracranial aneurysms—An explorative study. European Radiology Experimental, 3, 13. 10.1186/s41747-019-0092-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, J. P. , McKlveen, J. M. , Ghosal, S. , Kopp, B. , Wulsin, A. , Makinson, R. , … Myers, B. (2016). Regulation of the hypothalamic‐pituitary‐adrenocortical stress response. Comprehensive Physiology, 6, 603–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herten, A. , Konrad, K. , Krinzinger, H. , Seitz, J. , & von Polier, G. G. (2019). Accuracy and bias of automatic hippocampal segmentation in children and adolescents. Brain Structure & Function, 224, 795–810. [DOI] [PubMed] [Google Scholar]
- Hirjak, D. , Sambataro, F. , Remmele, B. , Kubera, K. M. , Schröder, J. , Seidl, U. , … Thomann, P. A. (2019). The relevance of hippocampal subfield integrity and clock drawing test performance for the diagnosis of Alzheimer's disease and mild cognitive impairment. The World Journal of Biological Psychiatry, 20, 197–208. [DOI] [PubMed] [Google Scholar]
- Hirjak, D. , Wolf, R. C. , Remmele, B. , Seidl, U. , Thomann, A. K. , Kubera, K. M. , … Thomann, P. A. (2017). Hippocampal formation alterations differently contribute to autobiographic memory deficits in mild cognitive impairment and Alzheimer's disease. Hippocampus, 27, 702–715. [DOI] [PubMed] [Google Scholar]
- Ho, N. F. , Holt, D. J. , Cheung, M. , Iglesias, J. E. , Goh, A. , Wang, M. , … Zhou, J. (2017). Progressive decline in hippocampal CA1 volume in individuals at ultra‐high‐risk for psychosis who do not remit: Findings from the longitudinal youth at risk study. Neuropsychopharmacology, 42, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, N. F. , Iglesias, J. E. , Sum, M. Y. , Kuswanto, C. N. , Sitoh, Y. Y. , de Souza, J. , … Holt, D. J. (2017). Progression from selective to general involvement of hippocampal subfields in schizophrenia. Molecular Psychiatry, 22, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T. C. , Gutman, B. , Pozzi, E. , Grabe, H. J. , Hosten, N. , Wittfeld, K. , … Schmaal, L. (2020). Subcortical shape alterations in major depressive disorder: Findings from the ENIGMA major depressive disorder working group. Human Brain Mapping. 10.1002/hbm.24988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. J. , Shou, H. , Benzinger, T. , Marcus, D. , Durbin, T. , Morris, J. C. , & Sheline, Y. I. (2015). Amyloid burden in cognitively normal elderly is associated with preferential hippocampal subfield volume loss. Journal of Alzheimer's Disease, 45, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Zhang, L. , Hu, X. , Lu, L. , Tang, S. , Li, H. , … Huang, X. (2019). Abnormal hippocampal subfields may be potential predictors of worse early response to antidepressant treatment in drug‐Naïve patients with major depressive disorder. Journal of Magnetic Resonance Imaging, 49, 1760–1768. [DOI] [PubMed] [Google Scholar]
- Hýža, M. , Kuhn, M. , Češková, E. , Ustohal, L. , & Kašpárek, T. (2016). Hippocampal volume in first‐episode schizophrenia and longitudinal course of the illness. The World Journal of Biological Psychiatry, 17, 429–438. [DOI] [PubMed] [Google Scholar]
- Iglesias, J. E. , Augustinack, J. C. , Nguyen, K. , Player, C. M. , Player, A. , Wright, M. , … Alzheimer's Disease Neuroimaging Initiative . (2015). A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage, 115, 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, J. E. , van Leemput, K. , Augustinack, J. , Insausti, R. , Fischl, B. , Reuter, M. , & Alzheimer's Disease Neuroimaging Initiative . (2016). Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject‐specific atlases. NeuroImage, 141, 542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo, J. , Andreassen, O. A. , Westlye, L. T. , & van der Meer, D. (2019). The association between hippocampal subfield volumes in mild cognitive impairment and conversion to Alzheimer's disease. Brain Research, 1728, 146591. 10.1016/j.brainres.2019.146591 [DOI] [PubMed] [Google Scholar]
- Jack, C. R., Jr. , Twomey, C. K. , Zinsmeister, A. R. , Sharbrough, F. W. , Petersen, R. C. , & Cascino, G. D. (1989). Anterior temporal lobes and hippocampal formations: Normative volumetric measurements from MR images in young adults. Radiology, 172, 549–554. [DOI] [PubMed] [Google Scholar]
- Jäncke, L. , Liem, F. , & Merillat, S. (2019). Scaling of brain compartments to brain size. Neuroreport, 30, 573–579. [DOI] [PubMed] [Google Scholar]
- Janiri, D. , Sani, G. , de Rossi, P. , Piras, F. , Banaj, N. , Ciullo, V. , … Spalletta, G. (2019). Hippocampal subfield volumes and childhood trauma in bipolar disorders. Journal of Affective Disorders, 253, 35–43. [DOI] [PubMed] [Google Scholar]
- Janiri, D. , Simonetti, A. , Piras, F. , Ciullo, V. , Spalletta, G. , & Sani, G. (2019). Predominant polarity and hippocampal subfield volumes in bipolar disorders. Bipolar Disorders, 22, 490–497. 10.1111/bdi.12857 [DOI] [PubMed] [Google Scholar]
- Jeukens, C. R. L. P. N. , Vlooswijk, M. C. G. , Majoie, H. J. M. , de Krom, M. C. T. F. M. , Aldenkamp, A. P. , Hofman, P. A. M. , … Backes, W. H. (2009). Hippocampal MRI volumetry at 3 Tesla: Reliability and practical guidance. Investigative Radiology, 44, 509–517. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Cao, X. , Jiang, J. , Li, T. , Wang, J. , Yang, Z. , & Li, C. (2019). Atrophy of hippocampal subfield CA2/3 in healthy elderly men is related to educational attainment. Neurobiology of Aging, 80, 21–28. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Xu, L. , Li, X. , Tang, Y. , Wang, P. , Li, C. , … Luo, C. (2019). Common increased hippocampal volume but specific changes in functional connectivity in schizophrenia patients in remission and non‐remission following electroconvulsive therapy: A preliminary study. NeuroImage: Clinical, 24, 102081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakeda, S. , Watanabe, K. , Katsuki, A. , Sugimoto, K. , Igata, N. , Ueda, I. , … Korogi, Y. (2018). Relationship between interleukin (IL)‐6 and brain morphology in drug‐naïve, first‐episode major depressive disorder using surface‐based morphometry. Scientific Reports, 8, 10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kälin, A. M. , Park, M. T. M. , Chakravarty, M. M. , Lerch, J. P. , Michels, L. , Schroeder, C. , … Leh, S. E. (2017). Subcortical shape changes, hippocampal atrophy and cortical thinning in future Alzheimer's disease patients. Frontiers in Aging Neuroscience, 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D. W. , Lim, H. K. , Joo, S.‐H. , Lee, N. R. , & Lee, C. U. (2018). The association between hippocampal subfield volumes and education in cognitively normal older adults and amnestic mild cognitive impairment patients. Neuropsychiatric Disease and Treatment, 14, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, M. , Sawada, K. , Shimodera, S. , Ogawa, Y. , Kariya, S. , Lang, D. J. , … Honer, W. G. (2015). Hippocampal subfield volumes in first episode and chronic schizophrenia. PLoS One, 10, e0117785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, W. , Westman, E. , Jones, N. , Wahlund, L.‐O. , Mecocci, P. , Vellas, B. , … AddNeuroMed Consortium and for the Alzheimer's Disease Neuroimaging Initiative . (2015). Automated hippocampal subfield measures as predictors of conversion from mild cognitive impairment to Alzheimer's disease in two independent cohorts. Brain Topography, 28, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlif, M. S. , Egorova, N. , Werden, E. , Redolfi, A. , Boccardi, M. , DeCarli, C. S. , … Brodtmann, A. (2019). A comparison of automated segmentation and manual tracing in estimating hippocampal volume in ischemic stroke and healthy control participants. NeuroImage: Clinical, 21, 101581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlif, M. S. , Werden, E. , Egorova, N. , Boccardi, M. , Redolfi, A. , Bird, L. , & Brodtmann, A. (2019). Assessment of longitudinal hippocampal atrophy in the first year after ischemic stroke using automatic segmentation techniques. NeuroImage: Clinical, 24, 102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. B. , Suh, S.‐I. , & Kim, J. H. (2015). Volumetric and shape analysis of hippocampal subfields in unilateral mesial temporal lobe epilepsy with hippocampal atrophy. Epilepsy Research, 117, 74–81. [DOI] [PubMed] [Google Scholar]
- Koelsch, S. , Jacobs, A. M. , Menninghaus, W. , Liebal, K. , Klann‐Delius, G. , von Scheve, C. , & Gebauer, G. (2015). The quartet theory of human emotions: An integrative and neurofunctional model. Physics of Life Reviews, 13, 1–27. 10.1016/j.plrev.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Kraus, C. , Seiger, R. , Pfabigan, D. M. , Sladky, R. , Tik, M. , Paul, K. , … Lanzenberger, R. (2019). Hippocampal subfields in acute and remitted depression‐an ultra‐high field magnetic resonance imaging study. The International Journal of Neuropsychopharmacology, 22, 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreilkamp, B. A. K. , Weber, B. , Elkommos, S. B. , Richardson, M. P. , & Keller, S. S. (2018). Hippocampal subfield segmentation in temporal lobe epilepsy: Relation to outcomes. Acta Neurologica Scandinavica, 137, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsrud, S. K. , Tamnes, C. K. , Fjell, A. M. , Amlien, I. , Grydeland, H. , Sulutvedt, U. , … Walhovd, K. B. (2014). Development of hippocampal subfield volumes from 4 to 22 years. Human Brain Mapping, 35, 5646–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn, S. , Charlet, K. , Schubert, F. , Kiefer, F. , Zimmermann, P. , Heinz, A. , & Gallinat, J. (2014). Plasticity of hippocampal subfield volume cornu ammonis 2+3 over the course of withdrawal in patients with alcohol dependence. JAMA Psychiatry, 71, 806–811. [DOI] [PubMed] [Google Scholar]
- Kühn, S. , Musso, F. , Mobascher, A. , Warbrick, T. , Winterer, G. , & Gallinat, J. (2012). Hippocampal subfields predict positive symptoms in schizophrenia: First evidence from brain morphometry. Translational Psychiatry, 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie, R. , Fouquet, M. , Mézenge, F. , Landeau, B. , Villain, N. , Mevel, K. , … Chételat, G. (2010). Differential effect of age on hippocampal subfields assessed using a new high‐resolution 3T MR sequence. NeuroImage, 53, 506–514. [DOI] [PubMed] [Google Scholar]
- Lee, H.‐J. , Seo, S. A. , & Park, K. M. (2019). Quantification of thalamic nuclei in patients diagnosed with temporal lobe epilepsy and hippocampal sclerosis. Neuroradiology, 62, 185–195. 10.1007/s00234-019-02299-6 [DOI] [PubMed] [Google Scholar]
- Lee, J. , Im, S.‐J. , Lee, S.‐G. , Stadlin, A. , Son, J.‐W. , Shin, C.‐J. , … Kim, S. (2016). Volume of hippocampal subfields in patients with alcohol dependence. Psychiatry Research: Neuroimaging, 258, 16–22. [DOI] [PubMed] [Google Scholar]
- Lee, S. W. , Yoo, J. H. , Kim, K. W. , Kim, D. , Park, H. , Choi, J. , … Jeong, B. (2018). Hippocampal subfields volume reduction in high schoolers with previous verbal abuse experiences. Clinical Psychopharmacology and Neuroscience, 16, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka, A. , Ingalhalikar, M. , Shah, A. , Saini, J. , Arumugham, S. S. , Hegde, S. , … Pal, P. K. (2018). Hippocampal subfield atrophy in patients with Parkinson's disease and psychosis. Journal of Neural Transmission, 125, 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , & Ji, L. (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity, 95, 221–227. [DOI] [PubMed] [Google Scholar]
- Li, M.‐X. , Yeung, J. M. Y. , Cherny, S. S. , & Sham, P. C. (2012). Evaluating the effective numbers of independent tests and significant p‐value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics, 131, 747–756. 10.1007/s00439-011-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Li, K. , Guan, P. , Chen, Y. , Xiao, Y. , Lui, S. , … Gong, Q. (2018). Volume alteration of hippocampal subfields in first‐episode antipsychotic‐naïve schizophrenia patients before and after acute antipsychotic treatment. NeuroImage: Clinical, 20, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Li, D. , Li, Q. , Li, Y. , Li, K. , Li, S. , & Han, Y. (2016). Hippocampal subfield volumetry in patients with subcortical vascular mild cognitive impairment. Scientific Reports, 6, 20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Li, J. , Segal, S. , Wegiel, J. , de Santi, S. , Zhan, J. , & de Leon, M. J. (2006). Hippocampal cerebrospinal fluid spaces on MR imaging: Relationship to aging and Alzheimer disease. AJNR. American Journal of Neuroradiology, 27, 912–918. [PMC free article] [PubMed] [Google Scholar]
- Li, Y.‐D. , Dong, H.‐B. , Xie, G.‐M. , & Zhang, L.‐J. (2013). Discriminative analysis of mild Alzheimer's disease and normal aging using volume of hippocampal subfields and hippocampal mean diffusivity: An in vivo magnetic resonance imaging study. American Journal of Alzheimer's Disease and Other Dementias, 28, 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Yin, Z. , Liu, R. , Zhao, H. , Wu, S. , Lu, J. , … Zhang, B. (2018). The role of MRI biomarkers and their interactions with cognitive status and APOE ε4 in nondemented elderly subjects. Neurodegenerative Diseases, 18, 270–280. [DOI] [PubMed] [Google Scholar]
- Lim, H. K. , Hong, S. C. , Jung, W. S. , Ahn, K. J. , Won, W. Y. , Hahn, C. , … Lee, C. U. (2012). Automated hippocampal subfield segmentation in amnestic mild cognitive impairments. Dementia and Geriatric Cognitive Disorders, 33, 327–333. [DOI] [PubMed] [Google Scholar]
- Lim, H. K. , Hong, S. C. , Jung, W. S. , Ahn, K. J. , Won, W. Y. , Hahn, C. , … Lee, C. U. (2013). Automated segmentation of hippocampal subfields in drug‐naïve patients with Alzheimer disease. AJNR. American Journal of Neuroradiology, 34, 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, O. , Mårtensson, G. , Stomrud, E. , Palmqvist, S. , Wahlund, L.‐O. , Westman, E. , & Hansson, O. (2017). Atrophy of the posterior subiculum is associated with memory impairment, tau‐ and Aβ pathology in non‐demented individuals. Frontiers in Aging Neuroscience, 9, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist, D. , Mueller, S. , Mellon, S. H. , Su, Y. , Epel, E. S. , Reus, V. I. , … Wolkowitz, O. M. (2014). Peripheral antioxidant markers are associated with total hippocampal and CA3/dentate gyrus volume in MDD and healthy controls‐preliminary findings. Psychiatry Research, 224, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J. , Feng, Y. , Liao, H. , Zhou, Q. , & Urbin, M. A. (2018). Motor sequence learning is associated with hippocampal subfield volume in humans with medial temporal lobe epilepsy. Frontiers in Human Neuroscience, 12, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, A. , Foo, H. , Yong, T. T. , Tan, L. C. S. , & Kandiah, N. (2019). Hippocampal subfield atrophy of CA1 and subicular structures predict progression to dementia in idiopathic Parkinson's disease. Journal of Neurology, Neurosurgery, and Psychiatry, 90, 681–687. [DOI] [PubMed] [Google Scholar]
- Lv, X. , He, H. , Yang, Y. , Han, L. , Guo, Z. , Chen, H. , … Xie, C. (2018). Radiation‐induced hippocampal atrophy in patients with nasopharyngeal carcinoma early after radiotherapy: A longitudinal MR‐based hippocampal subfield analysis. Brain Imaging and Behavior, 13, 1160–1171. 10.1007/s11682-018-9931-z [DOI] [PubMed] [Google Scholar]
- Mak, E. , Donaghy, P. C. , McKiernan, E. , Firbank, M. J. , Lloyd, J. , Petrides, G. S. , … O'Brien, J. T. (2019). Beta amyloid deposition maps onto hippocampal and subiculum atrophy in dementia with Lewy bodies. Neurobiology of Aging, 73, 74–81. [DOI] [PubMed] [Google Scholar]
- Mak, E. , Gabel, S. , Su, L. , Williams, G. B. , Arnold, R. , Passamonti, L. , … O'Brien, J. T. (2017). Multi‐modal MRI investigation of volumetric and microstructural changes in the hippocampus and its subfields in mild cognitive impairment, Alzheimer's disease, and dementia with Lewy bodies. International Psychogeriatrics, 29, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, E. , Su, L. , Williams, G. B. , Watson, R. , Firbank, M. , Blamire, A. , & O'Brien, J. (2016). Differential atrophy of hippocampal subfields: A comparative study of dementia with Lewy bodies and Alzheimer disease. The American Journal of Geriatric Psychiatry, 24, 136–143. [DOI] [PubMed] [Google Scholar]
- Maksimovskiy, A. L. , Oot, E. N. , Seraikas, A. M. , Rieselbach, M. , Caine, C. , Sneider, J. T. , … Silveri, M. M. (2019). Morphometric biomarkers of adolescents with familial risk for alcohol use disorder. Alcoholism, Clinical and Experimental Research, 43, 2354–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi, G. S. , Das, P. , Outhred, T. , Irwin, L. , Gessler, D. , Bwabi, Z. , … Mannie, Z. (2019). The effects of childhood trauma on adolescent hippocampal subfields. The Australian and New Zealand Journal of Psychiatry, 53, 447–457. [DOI] [PubMed] [Google Scholar]
- Maller, J. J. , Broadhouse, K. , Rush, A. J. , Gordon, E. , Koslow, S. , & Grieve, S. M. (2017). Increased hippocampal tail volume predicts depression status and remission to anti‐depressant medications in major depression. Molecular Psychiatry, 23, 1737–1744. 10.1038/mp.2017.224 [DOI] [PubMed] [Google Scholar]
- Malone, I. B. , Leung, K. K. , Clegg, S. , Barnes, J. , Whitwell, J. L. , Ashburner, J. , … Ridgway, G. R. (2015). Accurate automatic estimation of total intracranial volume: A nuisance variable with less nuisance. NeuroImage, 104, 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini, V. , Sandini, C. , Padula, M. C. , Zöller, D. , Schneider, M. , Schaer, M. , & Eliez, S. (2019). Positive psychotic symptoms are associated with divergent developmental trajectories of hippocampal volume during late adolescence in patients with 22q11DS. Molecular Psychiatry, 25, 2844–2859. 10.1038/s41380-019-0443-z [DOI] [PubMed] [Google Scholar]
- Marizzoni, M. , Antelmi, L. , Bosch, B. , Bartrés‐Faz, D. , Müller, B. W. , Wiltfang, J. , … PharmaCog Consortium . (2015). Longitudinal reproducibility of automatically segmented hippocampal subfields: A multisite European 3T study on healthy elderly. Human Brain Mapping, 36, 3516–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marizzoni, M. , Ferrari, C. , Jovicich, J. , Albani, D. , Babiloni, C. , Cavaliere, L. , … PharmaCog Consortium . (2019). Predicting and tracking short term disease progression in amnestic mild cognitive impairment patients with prodromal Alzheimer's disease: Structural brain biomarkers. Journal of Alzheimer's Disease, 69, 3–14. [DOI] [PubMed] [Google Scholar]
- Mathalon, D. H. , Sullivan, E. V. , Rawles, J. M. , & Pfefferbaum, A. (1993). Correction for head size in brain‐imaging measurements. Psychiatry Research, 50, 121–139. [DOI] [PubMed] [Google Scholar]
- Mathew, I. , Gardin, T. M. , Tandon, N. , Eack, S. , Francis, A. N. , Seidman, L. J. , … Keshavan, M. S. (2014). Medial temporal lobe structures and hippocampal subfields in psychotic disorders: Findings from the bipolar‐schizophrenia network on intermediate phenotypes (B‐SNIP) study. JAMA Psychiatry, 71, 769–777. [DOI] [PubMed] [Google Scholar]
- McEwen, B. S. , & Akil, H. (2020). Revisiting the stress concept: Implications for affective disorders. The Journal of Neuroscience, 40, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugo, M. , Talati, P. , Woodward, N. D. , Armstrong, K. , Blackford, J. U. , & Heckers, S. (2018). Regionally specific volume deficits along the hippocampal long axis in early and chronic psychosis. NeuroImage: Clinical, 20, 1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz, E. C. , Desai, P. M. , Maskus, E. A. , Melvin, S. A. , Rehman, R. , Torres, S. D. , … Noble, K. G. (2019). Socioeconomic disparities in chronic physiologic stress are associated with brain structure in children. Biological Psychiatry, 86, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole, T. B. , Mak, E. , Chien, Y. , & Voon, V. (2016). Dissociated accumbens and hippocampal structural abnormalities across obesity and alcohol dependence. The International Journal of Neuropsychopharmacology, 19, pyw039. 10.1093/ijnp/pyw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, R. A. , Garrett, M. E. , Stevens, J. S. , Clarke, E. K. , Haswell, C. C. , van Rooij, S. J. H. , … Ashley‐Koch, A. E. (2020). Genetic predictors of hippocampal subfield volume in PTSD cases and trauma‐exposed controls. European Journal of Psychotraumatology. 10.1080/20008198.2020.1785994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R. G. M. (2006). Elements of a neurobiological theory of hippocampal function: The role of synaptic plasticity, synaptic tagging and schemas. The European Journal of Neuroscience, 23, 2829–2846. [DOI] [PubMed] [Google Scholar]
- Mosher, V. A. L. , Swain, M. G. , Pang, J. X. Q. , Kaplan, G. G. , Sharkey, K. A. , MacQueen, G. M. , & Goodyear, B. G. (2018). Magnetic resonance imaging evidence of hippocampal structural changes in patients with primary biliary cholangitis. Clinical and Translational Gastroenterology, 9, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S. G. , Stables, L. , Du, A. T. , Schuff, N. , Truran, D. , Cashdollar, N. , & Weiner, M. W. (2007). Measurement of hippocampal subfields and age‐related changes with high resolution MRI at 4T. Neurobiology of Aging, 28, 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S. G. , & Weiner, M. W. (2009). Selective effect of age, Apo e4, and Alzheimer's disease on hippocampal subfields. Hippocampus, 19, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, SG , Yushkevich, PA , Das, S , Wang, L , Van Leemput, K , Iglesias, JE , Alpert, K , Mezher, A , Ng, P , Paz, K , Weiner, MW , Alzheimer's Disease Neuroimaging Initiative (2018): Systematic comparison of different techniques to measure hippocampal subfield volumes in ADNI2. NeuroImage: Clinical 17:1006–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrvang, A. D. , Vangberg, T. R. , Stedal, K. , Rø, Ø. , Endestad, T. , Rosenvinge, J. H. , & Aslaksen, P. M. (2018). Hippocampal subfields in adolescent anorexia nervosa. Psychiatry Research: Neuroimaging, 282, 24–30. [DOI] [PubMed] [Google Scholar]
- Na, K.‐S. , Chang, H. S. , Won, E. , Han, K.‐M. , Choi, S. , Tae, W. S. , … Ham, B.‐J. (2014). Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One, 9, e85425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, K.‐S. , Won, E. , Kang, J. , Kim, A. , Choi, S. , Kim, Y.‐K. , … Ham, B.‐J. (2018). Interaction effects of oxytocin receptor gene polymorphism and depression on hippocampal volume. Psychiatry Research: Neuroimaging, 282, 18–23. [DOI] [PubMed] [Google Scholar]
- Nakahara, S. , Turner, J. A. , Calhoun, V. D. , Lim, K. O. , Mueller, B. , Bustillo, J. R. , … van Erp, T. G. M. (2019). Dentate gyrus volume deficit in schizophrenia. Psychological Medicine, 50, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenskjöld, R. , Malmberg, F. , Larsson, E.‐M. , Simmons, A. , Brooks, S. J. , Lind, L. , … Kullberg, J. (2013). Intracranial volume estimated with commonly used methods could introduce bias in studies including brain volume measurements. NeuroImage, 83, 355–360. [DOI] [PubMed] [Google Scholar]
- Nyholt, D. R. (2004). A simple correction for multiple testing for single‐nucleotide polymorphisms in linkage disequilibrium with each other. American Journal of Human Genetics, 74, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, L. M. , Ziegler, D. A. , Deutsch, C. K. , Frazier, J. A. , Herbert, M. R. , & Locascio, J. J. (2011). Statistical adjustments for brain size in volumetric neuroimaging studies: Some practical implications in methods. Psychiatry Research, 193, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, J. (1990). A computational theory of the hippocampal cognitive map. Progress in Brain Research, 83, 301–312. [DOI] [PubMed] [Google Scholar]
- Olsen, R. K. , Carr, V. A. , Daugherty, A. M. , la Joie, R. , Amaral, R. S. C. , Amunts, K. , … Hippocampal Subfields Group . (2019). Progress update from the hippocampal subfields group. Alzheimer's and Dementia, 11, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørbo, M. C. , Vangberg, T. R. , Tande, P. M. , Anke, A. , & Aslaksen, P. M. (2018). Memory performance, global cerebral volumes and hippocampal subfield volumes in long‐term survivors of out‐of‐hospital cardiac arrest. Resuscitation, 126, 21–28. [DOI] [PubMed] [Google Scholar]
- Orfei, M. D. , Piras, F. , Banaj, N. , di Lorenzo, G. , Siracusano, A. , Caltagirone, C. , … Spalletta, G. (2017). Unrealistic self‐overconfidence in schizophrenia is associated with left presubiculum atrophy and impaired episodic memory. Cortex, 86, 132–139. [DOI] [PubMed] [Google Scholar]
- Osborn, A. G. , & Preece, M. T. (2006). Intracranial cysts: Radiologic‐pathologic correlation and imaging approach. Radiology, 239, 650–664. [DOI] [PubMed] [Google Scholar]
- Ostby, Y. , Tamnes, C. K. , Fjell, A. M. , Westlye, L. T. , Due‐Tønnessen, P. , & Walhovd, K. B. (2009). Heterogeneity in subcortical brain development: A structural magnetic resonance imaging study of brain maturation from 8 to 30 years. The Journal of Neuroscience, 29, 11772–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, Y. , Kakeda, S. , Sugimoto, K. , Katsuki, A. , Nguyen, L. H. , Igata, R. , … Yoshimura, R. (2019). COMT polymorphism regulates the hippocampal subfield volumes in first‐episode, drug‐naive patients with major depressive disorder. Neuropsychiatric Disease and Treatment, 15, 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiol, S. , Keeser, D. , Hasan, A. , Schneider‐Axmann, T. , Raabe, F. , Degenhardt, F. , … Falkai, P. (2019). Polygenic burden associated to oligodendrocyte precursor cells and radial glia influences the hippocampal volume changes induced by aerobic exercise in schizophrenia patients. Translational Psychiatry, 9, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papiol, S. , Popovic, D. , Keeser, D. , Hasan, A. , Schneider‐Axmann, T. , Degenhardt, F. , … Malchow, B. (2017). Polygenic risk has an impact on the structural plasticity of hippocampal subfields during aerobic exercise combined with cognitive remediation in multi‐episode schizophrenia. Translational Psychiatry, 7, e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.‐W. , Lee, C.‐N. , Sim, Y. , Ham, H.‐K. , Tae, W.‐S. , & Kim, B.‐J. (2019). Automated subfield volumetric analysis of hippocampus in patients with drug‐Naïve nondementia Parkinson's disease. Parkinson's Disease, 2019, 8254263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, T. D. , Cash, D. M. , Lane, C. A. S. , Lu, K. , Malone, I. B. , Nicholas, J. M. , … Schott, J. M. (2019). Hippocampal subfield volumes and pre‐clinical Alzheimer's disease in 408 cognitively normal adults born in 1946. PLoS One, 14(10), e0224030. 10.1371/journal.pone.0224030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, T. D. , Slattery, C. F. , Yong, K. X. X. , Nicholas, J. M. , Paterson, R. W. , Foulkes, A. J. M. , … Schott, J. M. (2019). Differences in hippocampal subfield volume are seen in phenotypic variants of early onset Alzheimer's disease. NeuroImage: Clinical, 21, 101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto‐Santos, J. E. , de Carvalho, L. E. D. , Kandratavicius, L. , Diniz, P. R. B. , Scandiuzzi, R. C. , Coras, R. , … Leite, J. P. (2018). Manual hippocampal subfield segmentation using high‐field MRI: Impact of different subfields in hippocampal volume loss of temporal lobe epilepsy patients. Frontiers in Neurology, 9, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J. B. , Junqué, C. , Bartrés‐Faz, D. , Ramírez‐Ruiz, B. , Marti, M.‐J. , & Tolosa, E. (2013). Regional vulnerability of hippocampal subfields and memory deficits in Parkinson's disease. Hippocampus, 23, 720–728. [DOI] [PubMed] [Google Scholar]
- Pereira, J. B. , Valls‐Pedret, C. , Ros, E. , Palacios, E. , Falcón, C. , Bargalló, N. , … Junque, C. (2014). Regional vulnerability of hippocampal subfields to aging measured by structural and diffusion MRI. Hippocampus, 24, 403–414. [DOI] [PubMed] [Google Scholar]
- Phillips, N. S. , Cheung, Y. T. , Glass, J. O. , Scoggins, M. A. , Liu, W. , Ogg, R. J. , … Krull, K. R. (2020). Neuroanatomical abnormalities related to dexamethasone exposure in survivors of childhood acute lymphoblastic leukemia. Pediatric Blood & Cancer, 67, e27968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintzka, C. W. S. , & Håberg, A. K. (2015). Perimenopausal hormone therapy is associated with regional sparing of the CA1 subfield: A HUNT MRI study. Neurobiology of Aging, 36, 2555–2562. [DOI] [PubMed] [Google Scholar]
- Pipitone, J. , Park, M. T. M. , Winterburn, J. , Lett, T. A. , Lerch, J. P. , Pruessner, J. C. , … Alzheimer's Disease Neuroimaging Initiative . (2014). Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. NeuroImage, 101, 494–512. [DOI] [PubMed] [Google Scholar]
- Pomponio, R. , Erus, G. , Habes, M. , Doshi, J. , Srinivasan, D. , Mamourian, E. , … Davatzikos, C. (2020). Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage, 208, 116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, S. , Shah, A. , Bhalsing, K. S. , Kumar, K. J. , Saini, J. , Ingalhalikar, M. , & Pal, P. K. (2019). Abnormal hippocampal subfields are associated with cognitive impairment in essential tremor. Journal of Neural Transmission, 126, 597–606. [DOI] [PubMed] [Google Scholar]
- Provenzano, F. A. , Guo, J. , Wall, M. M. , Feng, X. , Sigmon, H. C. , Brucato, G. , … Small, S. A. (2020). Hippocampal pathology in clinical high‐risk patients and the onset of schizophrenia. Biological Psychiatry, 87, 234–242. [DOI] [PubMed] [Google Scholar]
- Quattrini, G. , Pievani, M. , Jovicich, J. , Aiello, M. , Bargalló, N. , Barkhof, F. , … PharmaCog Consortium . (2020). Amygdalar nuclei and hippocampal subfields on MRI: Test‐retest reliability of automated volumetry across different MRI sites and vendors. NeuroImage, 218, 116932. [DOI] [PubMed] [Google Scholar]
- Rabl, U. , Meyer, B. M. , Diers, K. , Bartova, L. , Berger, A. , Mandorfer, D. , … Pezawas, L. (2014). Additive gene‐environment effects on hippocampal structure in healthy humans. Journal of Neuroscience, 34, 9917–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayel, S. , Bocti, C. , Sévigny Dupont, P. , Joannette, M. , Lavallée, M. M. , Nikelski, J. , … Joubert, S. (2019). Subcortical amyloid load is associated with shape and volume in cognitively normal individuals. Human Brain Mapping, 40, 3951–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon, P. K. , Clasen, L. , Giedd, J. N. , Blumenthal, J. , Lerch, J. P. , Chakravarty, M. M. , & Raznahan, A. (2016). An allometric analysis of sex and sex chromosome dosage effects on subcortical anatomy in humans. The Journal of Neuroscience, 36, 2438–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins, T. , Geng, F. , Botdorf, M. , Canada, K. , Cox, L. , & Hancock, G. R. (2018). Protracted hippocampal development is associated with age‐related improvements in memory during early childhood. NeuroImage, 174, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutland, J. W. , Brown, S. , Verma, G. , Feldman, R. E. , Sharma, H. , Markowitz, M. , … Balchandani, P. (2019). Hippocampal subfield‐specific connectivity findings in major depressive disorder: A 7 Tesla diffusion MRI study. Journal of Psychiatric Research, 111, 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutland, J. W. , Feldman, R. E. , Delman, B. N. , Panov, F. , Fields, M. C. , Marcuse, L. V. , … Balchandani, P. (2018). Subfield‐specific tractography of the hippocampus in epilepsy patients at 7 Tesla. Seizure, 62, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor, T. , Albert, M. , Stafford, J. , & Harpley, S. (1988). Use of computerized CT analysis to discriminate between Alzheimer patients and normal control subjects. AJNR. American Journal of Neuroradiology, 9, 1181–1187. [PMC free article] [PubMed] [Google Scholar]
- Sanfilipo, M. P. , Benedict, R. H. B. , Zivadinov, R. , & Bakshi, R. (2004). Correction for intracranial volume in analysis of whole brain atrophy in multiple sclerosis: The proportion vs. residual method. NeuroImage, 22, 1732–1743. [DOI] [PubMed] [Google Scholar]
- Sarica, A. , Vasta, R. , Novellino, F. , Vaccaro, M. G. , Cerasa, A. , Quattrone, A. , & Alzheimer's Disease Neuroimaging Initiative . (2018). MRI asymmetry index of hippocampal subfields increases through the continuum from the mild cognitive impairment to the Alzheimer's disease. Frontiers in Neuroscience, 12, 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satizabal, C. L. , Adams, H. , Hibar, D. P. , White, C. C. , Knol, M. J. , Stein, J.L. , … Ikram, M. A. (2019). Genetic architecture of subcortical brain structures in 38,851 individuals. Nature genetics, 51(11), 1624–1636. 10.1038/s41588-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M. , Sone, M. , Ehara, S. , & Tamakawa, Y. (1993). Hippocampal sulcus remnant: Potential cause of change in signal intensity in the hippocampus. Radiology, 188, 743–746. [DOI] [PubMed] [Google Scholar]
- Saygin, Z. M. , Kliemann, D. , Iglesias, J. E. , van der Kouwe, A. J. W. , Boyd, E. , Reuter, M. , … Alzheimer's Disease Neuroimaging Initiative . (2017). High‐resolution magnetic resonance imaging reveals nuclei of the human amygdala: Manual segmentation to automatic atlas. NeuroImage, 155, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens, P. , Weinstein, H. C. , & Leys, D. (1992). Neuro‐imaging in the diagnosis of Alzheimer's disease I. Computer tomography and magnetic resonance imaging. Clinical Neurology and Neurosurgery, 94, 277–289. [DOI] [PubMed] [Google Scholar]
- Schmaal, L. , Hibar, D. P. , Sämann, P. G. , Hall, G. B. , Baune, B. T. , Jahanshad, N. , … Veltman, D. J. (2017). Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Molecular Psychiatry, 22, 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M. F. , Storrs, J. M. , Freeman, K. B. , Jack, C. R., Jr. , Turner, S. T. , Griswold, M. E. , & Mosley, T. H., Jr. (2018). A comparison of manual tracing and FreeSurfer for estimating hippocampal volume over the adult lifespan. Human Brain Mapping, 39, 2500–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker, D. , Buss, C. , Head, K. , Sandman, C. A. , Davis, E. P. , Chakravarty, M. M. , … Pruessner, J. C. (2016). Hippocampus and amygdala volumes from magnetic resonance images in children: Assessing accuracy of FreeSurfer and FSL against manual segmentation. NeuroImage, 129, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoene‐Bake, J.‐C. , Keller, S. S. , Niehusmann, P. , Volmering, E. , Elger, C. , Deppe, M. , & Weber, B. (2014). In vivo mapping of hippocampal subfields in mesial temporal lobe epilepsy: Relation to histopathology. Human Brain Mapping, 35, 4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, G. , Choi, K.‐Y. , Kim, D. , Suh, S.‐I. , Lee, S. , Jeong, H.‐G. , & Jeong, B. (2017). Predicting neurocognitive function with hippocampal volumes and DTI metrics in patients with Alzheimer's dementia and mild cognitive impairment. Brain and Behavior: A Cognitive Neuroscience Perspective, 7, e00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing, Y. L. , Rodrigue, K. M. , Kennedy, K. M. , Fandakova, Y. , Bodammer, N. , Werkle‐Bergner, M. , … Raz, N. (2011). Hippocampal subfield volumes: Age, vascular risk, and correlation with associative memory. Frontiers in Aging Neuroscience, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, A. , Sani, G. , Dacquino, C. , Piras, F. , De Rossi, P. , Caltagirone, C. , … Spalletta, G. (2016). Hippocampal subfield volumes in short‐ and long‐term lithium‐treated patients with bipolar I disorder. Bipolar Disorders, 18, 352–362. [DOI] [PubMed] [Google Scholar]
- Sone, D. , Sato, N. , Maikusa, N. , Ota, M. , Sumida, K. , Yokoyama, K. , … Matsuda, H. (2016). Automated subfield volumetric analysis of hippocampus in temporal lobe epilepsy using high‐resolution T2‐weighed MR imaging. NeuroImage: Clinical, 12, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenova, V. , Nicola, R. , Aharon‐Peretz, J. , Goldsher, D. , Kapeliovich, M. , & Gilboa, A. (2018). Long‐term effects of brief hypoxia due to cardiac arrest: Hippocampal reductions and memory deficits. Resuscitation, 126, 65–71. [DOI] [PubMed] [Google Scholar]
- Stav, A. L. , Johansen, K. K. , Auning, E. , Kalheim, L. F. , Selnes, P. , Bjørnerud, A. , … Fladby, T. (2016). Hippocampal subfield atrophy in relation to cerebrospinal fluid biomarkers and cognition in early Parkinson's disease: A cross‐sectional study. NPJ Parkinson's Disease, 2, 15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymkowicz, S. M. , McLaren, M. E. , O'Shea, A. , Woods, A. J. , Anton, S. D. , & Dotson, V. M. (2017). Depressive symptoms modify age effects on hippocampal subfields in older adults. Geriatrics & Gerontology International, 17, 1494–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes, C. K. , Walhovd, K. B. , Engvig, A. , Grydeland, H. , Krogsrud, S. K. , Østby, Y. , … Fjell, A. M. (2014). Regional hippocampal volumes and development predict learning and memory. Developmental Neuroscience, 36, 161–174. [DOI] [PubMed] [Google Scholar]
- Tannous, J. , Amaral‐Silva, H. , Cao, B. , Wu, M.‐J. , Zunta‐Soares, G. B. , Kazimi, I. , … Soares, J. C. (2018). Hippocampal subfield volumes in children and adolescents with mood disorders. Journal of Psychiatric Research, 101, 57–62. [DOI] [PubMed] [Google Scholar]
- Teicher, M. H. , Anderson, C. M. , & Polcari, A. (2012). Childhood maltreatment is associated with reduced volume in the hippocampal subfields CA3, dentate gyrus, and subiculum. Proceedings of the National Academy of Sciences of the United States of America, 109, E563–E572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavabalasingam, S. , O'Neil, E. B. , Tay, J. , Nestor, A. , & Lee, A. C. H. (2019). Evidence for the incorporation of temporal duration information in human hippocampal long‐term memory sequence representations. Proceedings of the National Academy of Sciences of the United States of America, 116, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Jahanshad, N. , Ching, C. R. K. , Salminen, L. E. , Thomopoulos, S. I. , Bright, J. , … ENIGMA Consortium . (2020). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, R. , Chupin, M. , Garnero, L. , Leonard, G. , Perron, M. , Pike, B. , … Paus, T. (2009). Brain volumes and Val66Met polymorphism of the BDNF gene: local or global effects? Brain Structure and Function, 213, 501–509. 10.1007/s00429-009-0203-y [DOI] [PubMed] [Google Scholar]
- Uribe, C. , Segura, B. , Baggio, H. C. , Campabadal, A. , Abos, A. , Compta, Y. , … Junque, C. (2018). Differential progression of regional hippocampal atrophy in aging and Parkinson's disease. Frontiers in Aging Neuroscience, 10, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaculik, M. F. , Noorani, A. , Hung, P. S.‐P. , & Hodaie, M. (2019). Selective hippocampal subfield volume reductions in classic trigeminal neuralgia. NeuroImage: Clinical, 23, 101911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, D. , Rokicki, J. , Kaufmann, T. , Palomera, A. C. , Moberget, T. , Alnaes, D. , … Westlye, L. (2018): Brain scans from 21297 individuals reveal the genetic architecture of hippocampal subfield volumes. bioRxiv. Retrieved from https://www.biorxiv.org/content/early/2018/04/11/299578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk, L. , Hansell, N. K. , Strike, L. T. , Couvy‐Duchesne, B. , de Zubicaray, G. I. , Thompson, P. M. , … Wright, M. J. (2020). Region‐specific sex differences in the hippocampus. NeuroImage, 215, 116781. [DOI] [PubMed] [Google Scholar]
- van Leemput, K. (2009). Encoding probabilistic brain atlases using Bayesian inference. IEEE Transactions on Medical Imaging, 28, 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leemput, K. , Bakkour, A. , Benner, T. , Wiggins, G. , Wald, L. L. , Augustinack, J. , … Fischl, B. (2008). Model‐based segmentation of hippocampal subfields in ultra‐high resolution in vivo MRI. Medical Image Computing and Computer‐Assisted Intervention, 11, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leemput, K. , Bakkour, A. , Benner, T. , Wiggins, G. , Wald, L. L. , Augustinack, J. , … Fischl, B. (2009). Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus, 19, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen, L. S. , Wijdeveld, M. , Black, C. N. , van Tol, M.‐J. , van der Wee, N. J. A. , Veltman, D. J. , … Schmaal, L. (2017). Oxidative stress and brain morphology in individuals with depression, anxiety and healthy controls. Progress in Neuro‐Psychopharmacology & Biological Psychiatry, 76, 140–144. [DOI] [PubMed] [Google Scholar]
- Vargas, T. , Dean, D. J. , Osborne, K. J. , Gupta, T. , Ristanovic, I. , Ozturk, S. , … Mittal, V. A. (2017). Hippocampal subregions across the psychosis spectrum. Schizophrenia Bulletin, 44, 1091–1099. 10.1093/schbul/sbx160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voevodskaya, O. , Simmons, A. , Nordenskjöld, R. , Kullberg, J. , Ahlström, H. , Lind, L. , … Alzheimer's Disease Neuroimaging Initiative . (2014). The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer's disease. Frontiers in Aging Neuroscience, 6, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Zhang, L. , Yang, H. , Luo, X. , & Fan, G. (2019). Do multiple system atrophy and Parkinson's disease show distinct patterns of volumetric alterations across hippocampal subfields? An exploratory study. European Radiology, 29, 4948–4956. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Yu, Y. , Zhao, W. , Li, Q. , Li, X. , Li, S. , … Han, Y. (2018). Altered whole‐brain structural covariance of the hippocampal subfields in subcortical vascular mild cognitive impairment and amnestic mild cognitive impairment patients. Frontiers in Neurology, 9, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhang, R. , Wei, W. , Jiang, H. , Gao, Z. , Lin, J. , & Zhang, J. (2018). Long‐term sequelae of hippocampal lesions in patients with transient global amnesia: A multiparametric MRI study. Journal of Magnetic Resonance Imaging, 47, 1350–1358. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Song, Y. , Rajagopalan, P. , An, T. , Liu, K. , Chou, Y.‐Y. , … Alzheimer's Disease Neuroimaging Initiative . (2011). Surface‐based TBM boosts power to detect disease effects on the brain: An N=804 ADNI study. NeuroImage, 56, 1993–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, C. D. , Hibar, D. P. , van Velzen, L. S. , Zannas, A. S. , Carrillo‐Roa, T. , McMahon, K. , … Alzheimer's Disease Neuroimaging Initiative . (2016). Heritability and reliability of automatically segmented human hippocampal formation subregions. NeuroImage, 128, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn, J. L. , Pruessner, J. C. , Chavez, S. , Schira, M. M. , Lobaugh, N. J. , Voineskos, A. N. , & Mallar Chakravarty, M. (2013). A novel in vivo atlas of human hippocampal subfields using high‐resolution 3T magnetic resonance imaging. NeuroImage, 74, 254–265. 10.1016/j.neuroimage.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Wisse, L. , Daugherty, A. M. , la Joie, R. , Insausti, R. , Yassa, M. A. , Carr, V. A. , … Hippocampal Subfields Group . (2016). Automated Hippocampal Subfield Segmentation at 7T MRI. AJNR. American Journal of Neuroradiology, 37(6), 1050–1057. 10.3174/ajnr.A4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Biessels, G. J. , & Geerlings, M. I. (2014). A critical appraisal of the hippocampal subfield segmentation package in FreeSurfer. Frontiers in Aging Neuroscience, 6, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Chételat, G. , Daugherty, A. M. , Flores, R. , Joie, R. , Mueller, S. G. , … Carr, V. A. (2020). Hippocampal subfield volumetry from structural isotropic 1 mm 3 MRI scans: A note of caution. Human Brain Mapping, 1–12. 10.1002/hbm.25234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Gerritsen, L. , Zwanenburg, J. J. M. , Kuijf, H. J. , Luijten, P. R. , Biessels, G. J. , & Geerlings, M. I. (2012). Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. NeuroImage, 61, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Worker, A. , Dima, D. , Combes, A. , Crum, W. R. , Streffer, J. , Einstein, S. , … O'daly, O. (2018). Test‐retest reliability and longitudinal analysis of automated hippocampal subregion volumes in healthy ageing and Alzheimer's disease populations. Human Brain Mapping, 39, 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z. , Gao, Y. , Shi, F. , Ma, G. , Jewells, V. , & Shen, D. (2018). Segmenting hippocampal subfields from 3T MRI with multi‐modality images. Medical Image Analysis, 43, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Han, H. X. , Xu, C. , & Prince, J. L. (2003). Topology preserving level set method for geometric deformable models. IEEE Transactions on Pattern Analysis and Machine Intelligence, 25, 755–768. [Google Scholar]
- Xie, L. , Wisse, L. E. M. , Pluta, J. , de Flores, R. , Piskin, V. , Manjón, J. V. , … Alzheimer's Disease Neuroimaging Initiative . (2019). Automated segmentation of medial temporal lobe subregions on in vivo T1‐weighted MRI in early stages of Alzheimer's disease. Human Brain Mapping, 40, 3431–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Avants, B. B. , Pluta, J. , Das, S. , Minkoff, D. , Mechanic‐Hamilton, D. , … Detre, J. A. (2009). A high‐resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage, 44, 385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Pluta, J. B. , Wang, H. , Xie, L. , Ding, S.‐L. , Gertje, E. C. , … Wolk, D. A. (2015). Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Human Brain Mapping, 36, 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr, N. M. , Pohl, K. M. , Saranathan, M. , Sullivan, E. V. , & Pfefferbaum, A. (2019). Hippocampal subfield CA2+3 exhibits accelerated aging in alcohol use disorder: A preliminary study. NeuroImage: Clinical, 22, 101764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit, A. R. , Ezzati, A. , Zimmerman, M. E. , Lipton, R. B. , Lipton, M. L. , & Katz, M. J. (2017). Roles of hippocampal subfields in verbal and visual episodic memory. Behavioural Brain Research, 317, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Hu, X. , Lu, L. , Li, B. , Hu, X. , Bu, X. , … Huang, X. (2019). Abnormalities of hippocampal shape and subfield volumes in medication‐free patients with obsessive‐compulsive disorder. Human Brain Mapping, 40, 4105–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. , Wang, X. , Yin, C. , He, M. , Li, S. , & Han, Y. (2019). Trajectories of the hippocampal subfields atrophy in the Alzheimer's disease: A structural imaging study. Frontiers in Neuroinformatics, 13. 10.3389/fninf.2019.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, F. , Cui, D. , Zhang, L. , Zhang, S. , Zhao, Y. , Liu, X. , … Qiu, J. (2018). The volume of hippocampal subfields in relation to decline of memory recall across the adult lifespan. Frontiers in Aging Neuroscience, 10, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, F. , Li, C. , Zhang, D. , Cui, D. , Wang, Z. , & Qiu, J. (2019). Study on the sub‐regions volume of hippocampus and amygdala in schizophrenia. Quantitative Imaging in Medicine and Surgery, 9, 1025–1036. 10.21037/qims.2019.05.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, F. , Liu, Y. , Yuan, Z. , Gao, X. , He, Y. , Liu, X. , … Qiu, J. (2018). Age‐related changes in cortical and subcortical structures of healthy adult brains: A surface‐based morphometry study. Journal of Magnetic Resonance Imaging, 49, 152–163. 10.1002/jmri.26037 [DOI] [PubMed] [Google Scholar]
- Zimmerman, M. E. , Ezzati, A. , Katz, M. J. , Lipton, M. L. , Brickman, A. M. , Sliwinski, M. J. , & Lipton, R. B. (2016). Perceived stress is differentially related to hippocampal subfield volumes among older adults. PLoS One, 11, e0154530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugman, A. , Harrewijn, A. , Cardinale, E. M. , Zwiebel, H. , Freitag, G. F. , Werwath, K. E. , … Winkler, A. M. (2020). Mega‐analysis methods in ENIGMA: The experience of the generalized anxiety disorder working group. Human Brain Mapping, 1–23. Retrieved from https://pubmed.ncbi.nlm.nih.gov/32596977/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppichini, M. D. , & Sandry, J. (2018). Pilot investigation of the relationship between hippocampal volume and pattern separation deficits in multiple sclerosis. Multiple Sclerosis and Related Disorders, 26, 157–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Effect of covariate structure on subfield correlations. (A) depicts distribution of 66 Pearson (partial) correlation coefficients of 12 raw (bilateral) subfield volumes correlated with each other in sample A (1.5 Tesla, 597 subjects) (top row). Note mild leftward shift of (partial) correlation coefficients after correction for ICV, age, age‐squared and sex (middle row), and strong leftward shift after replacing ICV by the total HV (bottom row). (B) shows equivalent distributions obtained from sample B (3 Tesla, 211 subjects).
Supplemental Table 1 Brief characterization of 162 FreeSurfer hippocampal subfield studies. Numbers in the last column refer to subfield reaggregation/skipping details: 1Fimbria, fissure, hippocampal tail excluded. 2CA3 and CA4 aggregated, subicular complex aggregated (pre‐, para‐ and subiculum), DG aggregated (granule cell layer and molecular layer). 3Fimbria and fissure excluded. 4Fimbria and alveus combined. 5CA4 excluded, subicular complex aggregated (pre‐, para‐ and subiculum), DG aggregated (granule cell layer and molecular layer). 6Parasubiculum, presubiculum, alveus, fissure, fimbria and HATA omitted. 7Fimbria and fissure excluded. 8Three aggregations: classical FS6.0, anterior/posterior composite regions, composite subfields of body/head. ADHS, attention deficit hyperactivity syndrome; AD, Alzheimer's Disease; aMCI amnestic mild cognitive impairment; Cov, Covariate; CSF, cerebrospinal fluid; ECT, electroconvulsive therapy; FTD, frontotemporal dementia; GWAS, genome wide association study; HV, hippocampal volume; ICV, intracranial volume; LBD, Lewy Body Disease; MCI, mild cognitive impairment; MDD, major depressive disorder; MTLE, mesio‐temporal lobe epilepsy; PTSD, posttraumatic stress disorder; QC, quality control; ROI, region of interest; Regr, Regression; sGMV, supratentorial gray matter volume; SAD, social anxiety disorder; SCD, subjective cognitive deficits; UHR, ultra‐high risk; TBV, total brain volume
Data Availability Statement
The data used for the analysis in Section 2.3 are made available on request (saemann@psych.mpg.de).


