Abstract
Accurate transcription start site (TSS) annotations are essential for understanding transcriptional regulation and its role in human disease. Gene collections such as GENCODE contain annotations for tens of thousands of TSSs, but not all of these annotations are experimentally validated nor do they contain information on cell type–specific usage. Therefore, we sought to generate a collection of experimentally validated TSSs by integrating RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression (RAMPAGE) data from 115 cell and tissue types, which resulted in a collection of approximately 50 thousand representative RAMPAGE peaks. These peaks are primarily proximal to GENCODE-annotated TSSs and are concordant with other transcription assays. Because RAMPAGE uses paired-end reads, we were then able to connect peaks to transcripts by analyzing the genomic positions of the 3′ ends of read mates. Using this paired-end information, we classified the vast majority (37 thousand) of our RAMPAGE peaks as verified TSSs, updating TSS annotations for 20% of GENCODE genes. We also found that these updated TSS annotations are supported by epigenomic and other transcriptomic data sets. To show the utility of this RAMPAGE rPeak collection, we intersected it with the NHGRI/EBI genome-wide association study (GWAS) catalog and identified new candidate GWAS genes. Overall, our work shows the importance of integrating experimental data to further refine TSS annotations and provides a valuable resource for the biological community.
Accurate maps of genes and their transcription start sites (TSSs) are essential for studying gene regulation and determining the impact of genetic variation. Although gene and transcript annotations have improved substantially over the years, benefiting from advances in experimental and computational technologies, accurate, cell type–specific annotations are far from complete. Efforts such as the GENCODE project (Frankish et al. 2019) have generated detailed annotations for over 60 thousand genes and 100 thousand transcripts across the human genome. These widely used annotations combine transcriptomic, proteomic, and homology evidence through manual curation and automated computational pipelines. However, these annotations are built in a cell type–agnostic manner; they represent the collective transcriptomic landscape across thousands of unique cell and tissue types. Therefore, it is difficult to know which transcripts are actively transcribed in a particular cell or tissue type and, by extension, which regulatory elements and genetic variants may impact gene expression.
Although public RNA-seq data are accumulating across a wide array of tissues and cells types, many of which are from coordinated efforts such as the Genotype-Tissue Expression (GTEx) (The GTEx Consortium 2020) and Encyclopedia of DNA Elements (ENCODE) (The ENCODE Project Consortium et al. 2020) projects, these experiments are not optimal for annotating specific transcripts and their start sites. Most RNA-seq protocols perform short-read sequencing, which can accurately quantify gene expression levels but cannot delineate transcript isoforms fully nor precisely map the 5′ ends of transcripts. Therefore, assays that target and preserve 5′ ends, such as the cap analysis gene expression (CAGE) assay (Kodzius et al. 2006), are preferred for TSS identification. The FANTOM Consortium generated a TSS catalog across the human genome by integrating thousands of CAGE experiments (The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014). However, CAGE uses short, single-end reads, which have low mappability and cannot connect TSSs to their downstream transcripts. To overcome these limitations, Gingeras and colleagues developed the RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression (RAMPAGE) assay (Batut et al. 2013), which captures the 5′ end of capped RNAs using paired-end reads to enable more accurate genomic mapping and transcript characterization. The Gingeras laboratory generated both RAMPAGE and RNA-seq data for more than 100 human samples during the ENCODE Project (The ENCODE Project Consortium et al. 2020).
Here, we integrated 115 high-quality ENCODE RAMPAGE experiments to identify 52,546 representative RAMPAGE peaks (rPeaks), a curated collection of TSSs, and their activities across the 115 human samples. These rPeaks are supported by other transcription assays including CAGE, long-read RNA-seq using the Pacific Biosciences (PacBio) platform, and high-resolution nuclear run-on of capped transcripts (GRO-cap) (Core et al. 2014). Using paired-end RAMPAGE reads, we linked the majority of rPeaks to annotated genes and identified TSSs of unannotated spliced transcripts. These verified rPeaks were more enriched for transcriptomic and epigenomic features than GENCODE TSSs for the same genes not supported by RAMPAGE. Finally, we used this collection of rPeaks to annotate human variants associated with genome-wide association studies (GWAS) and identify novel phenotype-associated genes. Overall, our TSS collection complements existing gene annotations and shows the utility of cell type–specific TSS annotations in integrative analyses.
Results
Curation of 52,546 representative RAMPAGE peaks
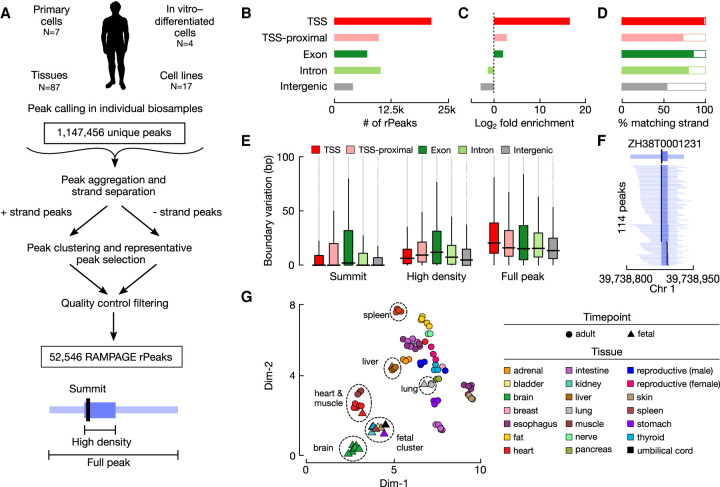
We curated 115 high-quality RAMPAGE experiments (Supplemental Table S1A) from ENCODE to generate our collection of representative rPeaks (Fig. 1A). These RAMPAGE experiments spanned 87 tissues and 28 cell types from a variety of biological contexts. We called peaks using the 5′ ends of RAMPAGE reads in individual experiments as previously described (Zhang et al. 2019), identifying three components for each peak: (1) a full peak; (2) a high-density region in the peak that accounts for 80% of the peak's total RAMPAGE signal; and (3) a summit, which is the genomic position with the highest signal. Given that the RAMPAGE assay enriched for reads at the 5′ ends of transcripts, we filtered out the small subset of RAMPAGE peaks that had higher RNA-seq signals than RAMPAGE signals in the matched biosample, which may be owing to fragmentation, degradation, and cytosolic recapping of the transcripts (see Methods; Supplemental Fig. S1A; Trotman and Schoenberg 2019), retaining about ten thousand peaks per experiment (Supplemental Table S1A). We then clustered overlapping peaks across the 115 experiments for two genomic strands separately and selected a representative peak (rPeak) for each cluster with the highest reads per kilobase per million mapped reads (RPKM) (Fig. 1A). Additional filtering was performed to remove low-signal, single-experiment rPeaks that were likely false positives; in total, we arrived at 52,546 rPeaks (Supplemental Table S1B). The full rPeaks and their high-density regions occupy 0.23% and 0.09% of the human genome, having median widths of 121 and 43 nucleotides (nt), respectively (Supplemental Fig. S1B,C).
Figure 1.
Curating a collection of representative RAMPAGE peaks (rPeaks) across 115 biosamples. (A) Workflow for curating RAMPAGE rPeaks. First, we called peaks in individual RAMPAGE data sets across 115 cell types and tissues. We then pooled these peaks (N = 1,147,456) and separated them by genomic strand. We clustered overlapping peaks on the same strand, selected the peak with the highest RAMPAGE signal (i.e., the rPeak) to represent each cluster, removed all the peaks overlapping the rPeak from the pool, and performed clustering on the remaining peaks. We repeated this process iteratively until all peaks were accounted for by rPeaks. We performed additional filtering using RNA-seq data, removing peaks that had a higher RNA-seq signal than RAMPAGE signal, finally arriving at 52,546 rPeaks. (B) Bar plots showing the number of RAMPAGE rPeaks stratified into distinct sets by genome context: overlapping GENCODE V31 TSSs (red), proximal (±500 bp) to TSSs (pink), overlapping exons (dark green), overlapping introns (light green), and intergenic (gray). (C) Bar plots showing the fold enrichment for the number of genomic positions covered by rPeaks over the footprints of the genomic contexts in B. (D) Bar plots showing the percentage of rPeaks in each genomic context as in B that are on the same strand as their overlapping TSS, gene (exon and intron), or nearest gene (TSS-proximal and intergenic). (E) Box plots displaying the variation in the positions of rPeak summits (left), high-density region boundaries (middle), and full peak boundaries (right), stratified by the genomic contexts as in B. (F) An example TSS-overlapping rPeak ZH38T000123 from K562 cells and the RAMPAGE peaks it represents in 113 other biosamples. For each peak, the full width is denoted in light blue, high-density regions in blue, and summit in black. (G) Scatterplot displaying a two-dimensional Uniform Manifold Approximation and Projection (UMAP) embedding of 87 tissue samples using RAMPAGE signal across all rPeaks as input features. Circles denote adult tissues, and triangles denote fetal tissues. Markers are colored by tissue of origin as defined in the legend.
The majority (59%) of rPeaks either overlapped or were proximal to (±500 bp but did not overlap) a GENCODE-annotated TSS (GENCODE 31 basic TSSs) (Fig. 1B). As an assessment for the extent of divergent transcription at TSSs, we detected an rPeak upstream on the opposite strand within 2 kb for the 6666 TSS-overlapping rPeaks (31%); this percentage is comparable to the FANTOM5 Consortium's CAGE peak collection, for which 8601 (34%) TSS-overlapping peaks have a peak upstream on the opposite strand within 2 kb. The remaining rPeaks overlapped exonic, intronic, or intergenic regions (14%, 18%, and 9% of rPeaks, respectively). We used these genomic contexts (TSS, TSS-proximal, exon, intron, and intergenic) throughout our analyses. As expected, RAMPAGE rPeaks were highly enriched for annotated TSSs and depleted in intergenic regions compared with the genomic footprints of these contexts (Chi-square test, P < 1 × 10−300) (Fig. 1C). Additionally, TSS, TSS-proximal, exonic, and intronic rPeaks had higher strand concordance than intergenic rPeaks, meaning they were more likely to fall on the same strand as their overlapping or closest gene (Fig. 1D). This finding suggests that intergenic rPeaks could result from misannotated or novel TSSs or from transcription at regulatory elements such as enhancer RNAs (eRNAs).
Next, we analyzed the ability of each rPeak to accurately represent their underlying clusters. When we analyzed the range of biosample activities of the RAMPAGE peak clusters, we observed a bimodal distribution (Supplemental Fig. S1D; Supplemental Table S1B), indicating that some rPeaks represent peaks from many RAMPAGE experiments, whereas others represent only a few, reflecting varying levels of cell and tissue type specificity of the TSS usage. The rPeaks in different genomic contexts differ greatly in this regard. TSS rPeaks represent peaks from 26 experiments on average, much higher than other rPeaks (pairwise Fisher's exact test, P < 1 × 10−300) (Supplemental Fig. S1E), indicating that annotated TSSs tend to be active in multiple cell and tissue types. For the vast majority of rPeaks, their summits were at nearly identical positions to the RAMPAGE peaks in individual experiments that they represented (we excluded the peak that was chosen as the rPeak for this analysis), with a difference in median of 0 bp across experiments (Fig. 1E). We note that although the RAMPAGE technique is of single–base pair resolution and annotated TSSs are assigned single–base pair genomic locations, RAMPAGE peaks reflect the firing patterns of RNA polymerases, which can vary across the genome. Polymerase firing originates predominantly from one genomic position at some loci but is more dispersed at other loci, corresponding to narrow, broad, and dispersed TSS shapes (Carninci et al. 2006; Fan et al. 2021). Although the median width of the high-density region of the rPeaks is 43 nt, the high-density region of 3984 rPeaks is a single base pair. Thus, RAMPAGE is capable of determining the resolution of TSSs down to a single base pair. We observed more variability in the boundaries of the high-density regions and full RAMPAGE peaks, with a difference in median of 8 and 18 nt, respectively, which is still relatively small on a genome-wide scale. As an illustrative example, the rPeak (ZH38T0001231) that overlapped a TSS of the PPIE gene represented RAMPAGE peaks from 114 experiments precisely at their summits and high-density regions (Fig. 1F; Supplemental Table S1C).
To assess the biological spectrum of rPeak activity, we performed dimensionality reduction with UMAP for all tissue samples (N = 87), using RAMPAGE signal profiles across the rPeaks in these samples (Fig. 1G). Similar tissues were generally clustered together, for example, brain, heart and leg muscle, and gastrointestinal tissues, respectively. Some fetal tissues were clustered with their corresponding adult tissues, including heart, liver, and lung. However, fetal thyroid and stomach tissues clustered exclusively with other fetal tissue samples, suggesting that these fetal samples share developmental transcriptional patterns at the surveyed life stages. We observed similar patterns using all 115 biosamples, albeit with tissues clustering separately from primary cells and cell lines (Supplemental Fig. S1F). Overall, these results indicate that our RAMPAGE rPeaks are a unified set of transcriptional sites that enables systematic investigations into the transcriptional landscape across multiple biosamples.
RAMPAGE rPeaks are concordant with other TSS annotations
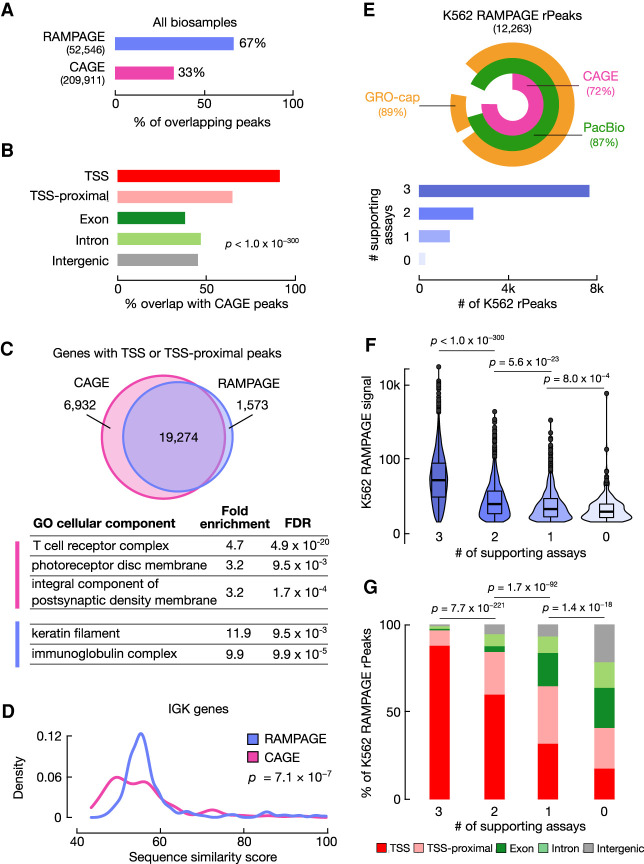
To evaluate the accuracy and comprehensiveness of our RAMPAGE rPeaks collection, we compared it with other collections of TSS annotations. The largest and most biologically diverse of these collections is the atlas of CAGE peaks generated by the FANTOM5 Consortium, which comprises 209,911 peaks annotated across 1816 experiments (The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014; Abugessaisa et al. 2017). Approximately two-thirds of our rPeaks overlapped a CAGE peak, whereas only one-third of the CAGE peaks overlapped an rPeak (Fig. 2A). Stratified by genomic context, the CAGE-overlapping rPeaks were more likely to be at GENCODE TSSs or TSS-proximal loci and less likely to be at exonic, intergenic, or intronic loci (Chi-square test, P < 1.0 × 10−300) (Fig. 2B). Additionally, we intersected our rPeaks with FANTOM enhancers, and only 4% of our rPeaks (N = 2195) and 3% FANTOM enhancers (N = 1977) overlapped. These two results suggest that the RAMPAGE assay preferentially identifies TSSs of genes rather than transcription at regulatory elements.
Figure 2.
RAMPAGE rPeaks are concordant with other transcriptome annotations. (A) Bar graph showing the percentage of RAMPAGE rPeaks that overlap CAGE peaks (purple) and the percentage of CAGE peaks that overlap RAMPAGE rPeaks (pink). (B) Bar graph showing the percentages of CAGE-overlapping RAMPAGE peaks in specific genomic contexts as defined in Figure 1B. (C) Venn diagram depicting the overlap of genes whose TSSs have at least one peak within 500 bp between the sets of CAGE peaks (pink) and RAMPAGE rPeaks (purple). Below are representative Gene Ontology terms (cellular component) enriched in CAGE-only genes (pink) or RAMPAGE-only genes (purple). A full list of enriched terms can be found in Supplemental Table S2. (D) Density plot showing the distributions of the similarity scores for sequences surrounding the TSSs of immunoglobulin kappa (IGK) genes supported by only RAMPAGE peaks (purple) or only CAGE peaks (pink). Sequence similarity was calculated as the maximal score of all pairwise local alignments. P-value corresponds to a two-sided Wilcoxon test. (E, top) VennPie diagram with concentric circles displaying K562 RAMPAGE rPeaks that overlap K562 CAGE peaks (pink) or PacBio 5′ ends (green) or have high GRO-seq signals (orange). The overall percentages are shown in parentheses. (Bottom) Bar plot with the number of K562 rPeaks stratified by the number of supporting transcriptomic assays in the above VennPie. (F) Violin-boxplot showing the distributions of the K562 RAMPAGE signal of rPeaks stratified by the number of supporting assays as defined in E. P-values correspond to two-sided pairwise Wilcoxon tests with FDR correction. (G) Stacked bar graphs showing the percentage of K562 rPeaks belonging to each genomic context (TSS: red, TSS-proximal: pink, exon: dark green, intron: light green, intergenic: gray) stratified by the number of supporting assays as defined in E. P-values correspond to Chi-square tests.
We also observed that although the majority (65%) of RAMPAGE rPeaks overlapped one or no CAGE peaks, some rPeaks overlapped multiple CAGE peaks. We investigated such cases to determine if we were missing alternative TSSs owing to our wider peak calls and found that these were generally sites of dispersed transcription (Supplemental Fig. S2). Using RAMPAGE data, we previously showed that TSS shape is linked with cell type–specific activity because narrow TSSs are more likely to be cell type–specific compared with broad and disperse TSSs, which are more ubiquitously expressed (Fan et al. 2021). Indeed, RAMPAGE rPeaks that overlap multiple CAGE peaks were 2.2 times more likely to overlap ubiquitously active promoters compared with RAMPAGE rPeaks that only overlapped a single CAGE peak (Fisher's exact test, P < 1.0 × 10−300). The structure of our rPeaks—full peak width, high density region, and summit—allows us to detect these types of dispersed transcription events.
Though a majority of TSS and TSS-proximal rPeaks were shared between RAMPAGE and CAGE, we identified sets of genes with TSSs that were exclusively proximal to CAGE peaks (CAGE-only genes) (Supplemental Table S2A) or were exclusively proximal to RAMPAGE rPeaks (RAMPAGE-only genes) (Supplemental Table S2B). Gene Ontology analysis revealed that the 6932 CAGE-only genes were enriched in terms such as T-cell receptor complex and photoreceptor disc membrane (Fig. 2C; Supplemental Table S2C). Enrichment in these terms is not surprising as the corresponding biosamples—T cells and eye tissues—were assayed by CAGE but not RAMPAGE. Additional enrichment for neuronal terms such as integral components of postsynaptic density membranes was unexpected as we integrated RAMPAGE data from seven fetal brain samples and in vitro–differentiated neurons. Alternatively, the 1573 RAMPAGE-only genes were primarily enriched for two terms, immunoglobulin production and keratinization (Fig. 2C; Supplemental Table S2D), owing to an abundance of immunoglobulin kappa (IGK) genes and keratin associated protein (KRTAP) genes, respectively. Although we observed genes from these families in both the RAMPAGE-only and CAGE-only gene sets, the sequences flanking the TSSs of RAMPAGE-only IGK and KRTAP genes shared higher sequence similarity than the corresponding CAGE-only genes (Wilcoxon tests, P = 7.1 × 10−7 and 4.7 × 10−5, respectively) (Fig. 2D; Supplemental Fig. S3A). We hypothesize that the 101-nt, paired-end RAMPAGE reads can uniquely map to genomic regions sharing higher sequence similarity better than the 36-nt, single-end CAGE reads, allowing us to identify rPeaks in more paralogs of these two gene families. In conclusion, the primary differences in the gene coverage by RAMPAGE rPeaks and CAGE peaks largely reflect differences in their biosample collections and assay read length.
To avoid coverage differences owing to biosample composition, we directly compared RAMPAGE rPeaks and CAGE peaks active in K562 and GM12878 cell lines—biosamples used in both peak collections—along with GRO-cap (Core et al. 2014) and PacBio long-read RNA-seq data (Wyman et al. 2020) in the respective cell lines. Generally, the four sets of TSS annotations were highly concordant, with the majority of RAMPAGE rPeaks, CAGE peaks, and PacBio 5′ ends overlapping one another and containing high GRO-cap signals (Supplemental Fig. S3B). In total, 98% of K562 and GM12878 rPeaks were substantiated by overlaps with at least one other transcriptome annotation, and a majority of the rPeaks—65% in K562 and 74% in GM12878—was supported by all of the other assays (Fig. 2E; Supplemental Fig. S3C). These supported rPeaks had higher RAMPAGE signals (pairwise Wilcoxon test, P < 1.0 × 10−300) (Fig. 2F; Supplemental Fig. S3D) and were more likely to overlap TSSs (Chi-square test, P = 2.5 × 10−259) (Fig. 2G; Supplemental Fig. S3E) than rPeaks supported by fewer or no other assays. When we analyzed the CAGE, PacBio, and GRO-cap signals at rPeaks in K562 and GM12878 cells, we found that signal was highly enriched directly at the rPeak summits, showing that our TSS annotations were supported by these other assays at base pair resolution (Supplemental Fig. S3F–K). We also analyzed the number of genes with TSSs supported by RAMPAGE, CAGE, or PacBio data and observed that RAMPAGE rPeaks identified an average of 17% fewer genes than CAGE and PacBio (Supplemental Fig. S3L,M). On the other hand, 96% of RAMPAGE genes were supported by another assay compared with 90% of CAGE genes and 88% of PacBio genes. Thus, our rPeak approach slightly compromises recall for better precision.
Our pairwise comparison between the four transcription assays (Supplemental Fig. S3B) indicates that the majority of RAMPAGE, CAGE, and PacBio annotations overlapped one another (55%–88%) and were supported by high GRO-cap signals (80%–91%). However, when we analyzed GRO-cap peaks, only a small percentage of the over 100,000 peaks overlapped the other assays. This is likely either owing to the ability of GRO-cap to detect transcription of unstable transcripts and/or more lenient calling of GRO-cap peaks.
Stratified by genomic context, TSS rPeaks had the highest levels of GRO-cap signal and overlapped the greatest number of PacBio 5´-ends, followed by TSS-proximal, intronic, and intergenic rPeaks (Supplemental Fig. S3N–P). In contrast, exonic rPeaks consistently had the lowest levels of GRO-cap signal, and although they overlapped a moderate number of PacBio 5´-ends (Supplemental Fig. S3O), these PacBio reads were significantly shorter than those overlapping other rPeak classes (pairwise Wilcoxon test with FDR correction, P < 1.0 × 10−16) (Supplemental Fig. S3P). These results suggest that many of the exonic rPeaks likely are not TSSs and may arise from mRNA recapping (Trotman and Schoenberg 2019).
To evaluate the ability of the RAMPAGE assay to detect low-abundance and unstable transcripts, we compared our rPeak annotations to TSSs classified by stability through the integration of GRO-cap and CAGE data (Core et al. 2014). In K562 and GM12878, 64% and 57%, respectively, of stable TSSs overlapped RAMPAGE rPeaks annotated in those cell types (Supplemental Fig. S3Q). In contrast, <1% of unstable TSSs overlapped rPeaks, showing that the RAMPAGE assay can only detect TSSs of stable transcripts. This was further highlighted when we compared the overlap of our rPeaks collection to enhancers identified by the NET-CAGE assay, which can identify transcription from unstable transcripts (Hirabayashi et al. 2019). Only 1.5% of NET-CAGE-specific enhancers (N = 315) overlapped an rPeak compared with the aforementioned 3% of FANTOM CAGE enhancers (Fisher's exact test, P = 2.0 × 10−33). Therefore, we conclude that our RAMPAGE rPeak collection preferentially contains TSSs for stable, gene-associated transcripts.
Overall, only 2% of K562 and GM12878 RAMPAGE rPeaks were not supported by one of the other assays (N = 301 and 212, respectively). These peaks had the lowest RAMPAGE signals and were more likely to overlap exons, introns, and intergenic regions (Fig. 2F,G; Supplemental Fig. S3D,E). These weaker transcription sites are not as reproducible across assays or may be false positives. Thus, our set of RAMPAGE rPeaks are highly concordant with other transcriptome annotations and likely represent a conservative set of TSSs.
Three-quarters of RAMPAGE rPeaks are assigned to genes via spliced transcripts
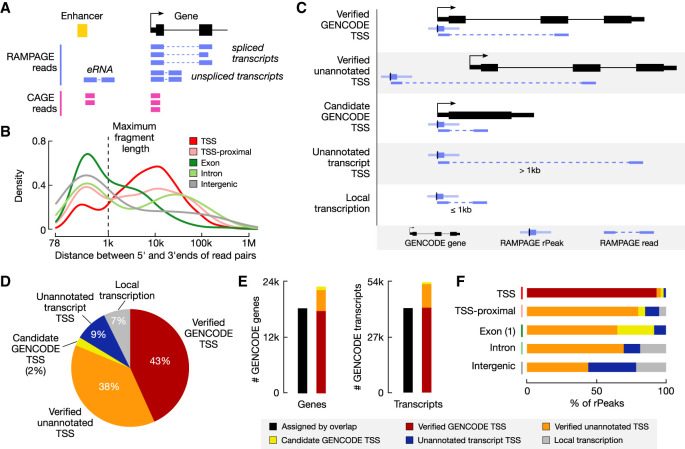
One advantage of the RAMPAGE assay is that it produces paired-end reads, which not only result in more accurately mapped fragments but also have the ability to assign rPeaks to genes. rPeaks are derived from the 5′ ends of RAMPAGE read pairs, and by analyzing the genomic positions of 3′ ends of the read pairs, we can attempt to link rPeaks to downstream transcripts and consequently assign rPeaks to genes. Such a process could go down one of two general paths (Fig. 3A). If the generated transcript is spliced (e.g., mRNAs and lncRNAs), the 3′ end of the read pair would map to an exon that is thousands of base pairs downstream from the rPeak, and we can assign the rPeak to the gene that the exon belongs to. However, if the generated transcript is unspliced (e.g., pre-mRNAs, single-exon transcripts, small RNAs, and most eRNAs), the 3′ end will map <1 kb downstream (the maximum selected fragment size for the RAMPAGE assay; median = 335 nt), and we cannot confidently assign the rPeak to a specific gene.
Figure 3.
Assigning RAMPAGE rPeaks to genes using paired-end reads. (A) Schematic showing how paired-end RAMPAGE reads (purple) can distinguish between spliced and unspliced transcripts, unlike single-end CAGE reads (pink). (B) Density plot of the distances between the 5′ and 3′ ends of RAMPAGE read pairs, stratified by rPeak genomic context. The maximum fragment length (1 kb) is shown by the dashed line. (C) Schematic depicting the paired-end read method for linking RAMPAGE rPeaks with genes and the resulting five categories. (D) Pie chart displaying the percentage of RAMPAGE rPeaks classified as the five categories in C: verified GENCODE TSSs (red), verified unannotated TSSs (orange), candidate GENCODE TSSs (yellow), unannotated transcript TSSs (blue), or local transcription (gray). (E) Bar graphs showing the number of GENCODE genes (left) and transcripts (right) that are accounted for by overlapping RAMPAGE rPeaks (black) versus the paired-end read method illustrated in A and C (colors). Bars for the paired-end method are stratified by TSS class (as defined in C,D). Genes with multiple TSSs were counted only once with the following priority: verified GENCODE TSSs, verified unannotated TSSs, and then candidate GENCODE TSSs. (F) Bar graphs showing the percentage of rPeaks that are classified as verified GENCODE TSSs (red), verified unannotated TSSs (orange), candidate GENCODE TSSs (yellow), unannotated transcript TSSs (blue), or local transcription (gray), stratified by genomic context.
To determine the portions of RAMPAGE reads derived from spliced and unspliced transcripts, we calculated the distance between the 5′ and 3′ ends of the read pairs that support an rPeak (i.e., the 5′ end of the read pair overlaps an rPeak). For TSS-overlapping rPeaks, we observed a bimodal distribution, with 83% of reads from spliced transcripts (distance > 1 kb) and 17% from unspliced transcripts (distance ≤ 1 kb) (Fig. 3B). For other rPeak classes, we also observed substantial percentages of reads deriving from spliced transcripts (43%–68%), suggesting that these rPeaks may correspond to TSSs for misannotated transcripts, novel isoforms, or novel genes. These results indicated that we can use RAMPAGE read pairs from spliced transcripts to assign rPeaks to genes.
We developed a computational pipeline to systematically assign rPeaks to genes (Supplemental Fig. S4A). Because of the aforementioned low GRO-cap signal at exonic rPeaks, we excluded all exonic rPeaks that did not overlap the first exon of an annotated transcript (N = 6709) from this analysis as they likely capture recapping (Trotman and Schoenberg 2019) or degradation events rather than sites of transcription. We further discarded four rPeaks whose supporting reads mapped >500 kb away. We classified the remaining 45,833 rPeaks into five general categories depending on whether an rPeak overlaps a GENCODE-annotated TSS and whether its supporting reads overlapped a GENCODE-annotated exon, with the most prominent scenarios summarized as follows (Fig. 3C) and with details provided in Supplemental Table S3A. First, if an rPeak overlaps a GENCODE-annotated TSS and its supporting reads overlap a downstream exon of the same gene, the rPeak is classified as a verified GENCODE TSS. Second, if an rPeak does not overlap a GENCODE-annotated TSS but its supporting reads overlap a GENCODE-annotated exon, the rPeak is classified as a verified unannotated TSS. Third, if an rPeak overlaps a GENCODE-annotated TSS and its supporting reads overlap the first exon of the gene and if the gene has only one exon or its first exon is >500 nt, then the rPeak is classified as a candidate GENCODE TSS. Fourth, if an rPeak's supporting reads map to >1 kb downstream from the rPeak and do not overlap a GENCODE-annotated exon, the rPeak is deemed to be the TSS of an unannotated transcript. Fifth, if an rPeak's supporting reads are within 1 kb of the rPeak and the rPeak is not a candidate GENCODE TSS (third category above), then it is deemed to originate from local (i.e., unspliced) transcription. Using our pipeline, we assigned 84% of rPeaks to genes (the first three categories) (Fig. 3D), which is 4641 more genes and 12,466 more transcripts than simply using overlaps (Fig. 3E). In total, we curated 19,821 verified GENCODE TSSs, 17,447 verified unannotated TSSs, and 1088 candidate GENCODE TSSs for 22,801 genes and 4129 TSSs for unannotated transcripts (Supplemental Table S3A).
The vast majority of TSS-overlapping rPeaks (19,821 of 21,278, 93%) are verified GENCODE TSSs, indicating that our approach is highly accurate (Fig. 3F; Supplemental Table S3B). These verified GENCODE TSSs amount to 43% of all rPeaks. The next largest category of rPeaks is verified unannotated TSS (38%, N = 17,447) (Supplemental Table S3C), and these rPeaks are potentially novel TSSs of spliced GENCODE-annotated genes revealed by our collection of RAMPAGE data. Three examples from K562 cells are highlighted (Supplemental Fig. S4B–D): exonic rPeak ZH38T0014149, a verified TSS of ING1, (Supplemental Fig. S4B); intronic rPeak ZH38T0050003, a verified TSS of GALNT12 (Supplemental Fig. S4C); and intergenic rPeak ZH38T0049993, a verified TSS of NANS (Supplemental Fig. S4D). These three verified TSSs and their linked transcripts were also supported by PacBio reads in K562 cells.
To determine if the verified unannotated TSSs were from minor or cell type–specific isoforms, we compared the activity levels for the TSSs of 6161 genes that had at least one verified GENCODE TSS and one verified unannotated TSS. We found that most verified TSSs fell into three general classes (Supplemental Fig. S5A): (1) ubiquitously expressed TSSs that belong to ubiquitously expressed genes, (2) cell type–specific TSSs that belong to ubiquitously expressed genes (i.e., TSSs of tissue-specific isoforms), and (3) cell type–specific TSSs that belong to cell type–specific genes. Generally, verified GENCODE TSSs fell into the first and third classes (Supplemental Fig. S5B), whereas verified unannotated TSSs fell into the second and third classes (Supplemental Fig. S5C). Furthermore, verified GENCODE TSSs were more likely to correspond to major isoforms compared with verified unannotated TSSs (Wilcoxon test, P < 1.0 × 10−300) (Supplemental Fig. S5D). When we analyzed the expression profiles of these verified annotated TSSs, we found that they had enriched expression in male reproductive tissues (e.g., testis and prostate) (Supplemental Table S3D), supporting previous findings that alternative transcription is prevalent in these tissues (Naro et al. 2021).
The candidate GENCODE TSS category of rPeaks (N = 452) constitutes 88% of the TSS-overlapping rPeaks that were not supported by reads from spliced transcripts (N = 511). The GENCODE genes that overlap these rPeaks either have only one exon or have a long (>500 nt) first exon (Supplemental Table S3E). We observed a similar pattern for TSS-proximal and exonic rPeaks not supported by reads from spliced transcripts, although at lower percentages. These rPeaks are most likely the TSSs of the overlapping genes, although our paired-end mapping approach is not able to make the assignment definitively; thus, we assigned them the candidate designation. Of the 8298 TSS-proximal and 547 exonic rPeaks that we classified as either candidate GENCODE or verified unannotated TSSs, 1627 overlap the coding DNA sequence (CDS) of an annotated GENCODE gene. These alternative TSSs could potentially affect the open reading frame (ORF) of the annotated gene resulting in a different translated protein.
The unannotated transcript category includes 4129 rPeaks (9% of rPeaks), which are likely TSSs of unannotated spliced transcripts. The rPeaks themselves are primarily intergenic, intronic, or antisense TSS-proximal with respect to GENCODE-annotated genes (Supplemental Table S3F). Although this category of rPeaks shows similar levels of evolutionary conservation to the local transcription category of rPeaks, the former category is active in more biosamples and more likely to overlap PacBio TSSs (Supplemental Fig. S5E–H; Supplemental Table S4). Additionally, the PacBio reads that overlapped the verified unannotated transcript rPeaks had a similar length distribution to the PacBio reads that overlapped rPeaks in the verified GENCODE TSS category, suggesting that the verified unannotated transcript rPeaks may correspond to lncRNAs missed by GENCODE (Supplemental Fig. S5I,J). To test this hypothesis, we intersected these rPeaks with lncRNA TSSs curated by the lncBook database (Ma et al. 2019) and found that 30% of our verified unannotated transcript rPeaks overlapped the lncBook lncRNA TSSs, a significant enrichment over the local transcription rPeaks and random genomic regions (7% and <1% overlap, respectively, Fisher's exact test, P < 8.8 × 10−148) (Supplemental Table S3F). Using overlapping PacBio reads, we also scanned for potential of ORFs in the resulting transcripts and found that the unannotated transcripts had fewer computationally discovered ORFs compared with GENCODE annotated transcripts (pairwise Wilcoxon test with FDR correction, P < 2.6 × 10−77) (Supplemental Fig. S5K,L). These results suggest that many of our verified unannotated transcript rPeaks are likely lncRNAs and further expands the growing list of lncRNAs in the human genome.
We built our catalog of RAMPAGE rPeak TSSs using GENCODE V31 basic annotations. Because GENCODE releases new versions quarterly, we evaluated how our catalog compared to six different GENCODE builds. We ran our pipeline using GENCODE V24, V31, and V38, evaluating both basic and comprehensive annotations (Supplemental Fig. S5M; Supplemental Table S3G). As expected, more rPeaks were classified as verified GENCODE TSSs when using the newer, more comprehensive GENCODE builds. For example, 18,000 rPeaks were classified as verified GENCODE TSSs using GENCODEv24 basic annotations compared to 25,000 rPeaks with the GENCODE V38 comprehensive annotations. Nevertheless, even with the GENCODE V38 comprehensive annotations, we still identified 13,000 novel TSSs for annotated GENCODE transcripts and 3700 TSSs for novel transcripts, showing that our rPeak catalog still identifies novel transcriptional events.
RAMPAGE-verified TSSs are enriched for regulatory signatures
We compared our RAMPAGE-verified TSS annotations with GENCODE-annotated TSSs for enrichment in functional, epigenomic, and additional transcriptomic annotations. For these analyses, we only considered genes with both a RAMPAGE-verified unannotated TSS and a GENCODE TSS that did not overlap each other (4751 genes) and used a uniform 100-bp region centered at an rPeak summit or GENCODE TSS to control for gene expression and provide an unbiased comparison.
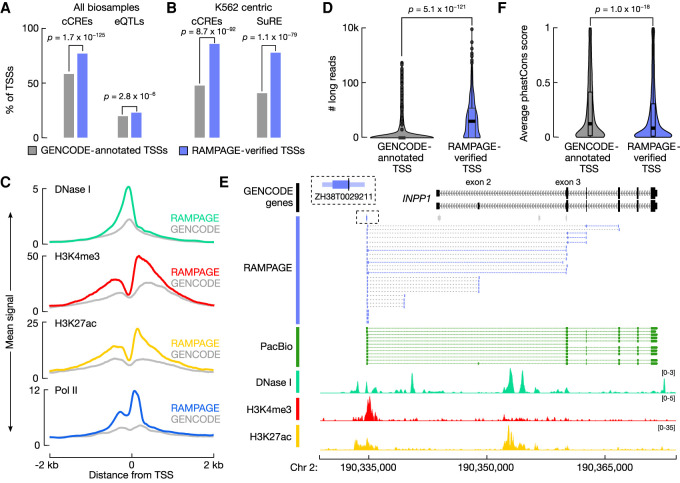
RAMPAGE-verified unannotated TSSs were more likely to overlap ENCODE candidate cis-regulatory elements (cCREs; 1.3-fold enrichment, Fisher's exact test, P = 1.7 × 10−125) (The ENCODE Project Consortium et al. 2020) and GTEx expression quantitative trait loci (eQTLs; 1.2-fold enrichment, Fisher's exact test, P = 2.8 × 10−6) (GTEx Consortium 2017) compared with matched GENOCDE TSSs (Fig. 4A). When we restricted our analysis to RAMPAGE-verified unannotated TSSs active in K562 and their matched GENCODE TSSs (961 genes), we observed that the verified TSSs were more likely to overlap K562 cCREs (1.8-fold enrichment, Fisher's exact test, P = 8.7 × 10−92) (Fig. 4B) and peaks from the Survey of Regulatory Elements (SuRE) assay, a massively parallel reporter assay testing promoter activity (1.9-fold enrichment, Fisher's exact tests, P = 1.1 × 10−79) (Fig. 4A; van Arensbergen et al. 2017). The K562 RAMPAGE-verified unannotated TSSs also had higher H3K4me3 and H3K27ac ChIP-seq signals, which had the canonical asymmetric pattern corresponding to transcriptional direction, chromatin accessibility, and Pol II ChIP-seq signals compared with the matched GENCODE TSSs (Fig. 4C).
Figure 4.
RAMPAGE-verified rPeaks are enriched for regulatory signatures. (A) Bar plots display the percentage of RAMPAGE-verified TSSs (purple) and matched GENCODE-annotated TSSs (gray) that overlap cell type–agnostic cCREs and the full compendium of GTEx eQTLs. P-values are from Fisher's exact test. (B) Bar plots display the percentage of RAMPAGE-verified TSSs expressed in K562 (purple) and matching GENCODE-annotated TSSs (gray) that overlap K562 cCREs and SuRE assay peaks. P-values are from Fisher's exact test. (C) Aggregation plots of epigenomic signals, DNase I (teal), H3K4me3 (red), H3K27ac (yellow), and Pol II (blue), at K562 RAMPAGE-verified TSSs (colors) and matched GENCODE-annotated TSSs across a ±2-kb window centered on the summits and TSSs, respectively. (D) Nested violin boxplots showing the number of PacBio 5′ read ends that overlap K562 RAMPAGE-verified TSSs (purple) and matched GENCODE-annotated TSSs (gray). P-value is from a Wilcoxon rank-sum test. (E) Genome browser view of INPP1 locus in K562. RAMPAGE-verified TSS, ZH38T0029211, is linked to the INPP1 gene by paired-end RAMPAGE reads (purple), whereas the GENCODE-annotated TSSs are not supported by RAMPAGE reads. PacBio reads (green) also support ZH38T0029211 as a verified TSS of INPP1 and epigenomic signals, DNase I (teal), H3K4me3 (red), and H3K27ac (yellow), support promoter activity at ZH38T0029211 and not at the annotated GENCODE TSSs. RAMPAGE rPeaks with RPM > 2 in K562 are shown in purple, and those with RPM ≤ 2 are shown in gray. (F) Nested violin boxplots of average phastCons conservation scores across RAMPAGE-verified TSSs (purple) and matched GENCODE-annotated TSSs (gray). P-value is from a Wilcoxon rank-sum test.
We also compared the verified unannotated TSSs with the K562 and GM12878 PacBio long-read RNA-seq data. RAMPAGE-verified unannotated TSSs were more likely to overlap the 5′ ends of PacBio reads compared with the GENCODE-matched controls (median of three supporting reads vs. zero supporting reads; Wilcoxon test, P = 5.1 × 10−121) (Fig. 4D). One example is highlighted at the INPP1 locus (Fig. 4E). ZH38T0029211 is a RAMPAGE-verified TSS located 8844 bp upstream of two GENCODE-annotated TSSs for the INPP1 gene. The majority of RAMPAGE reads link ZH38T0029211 to the first coding exon (exon 3), whereas a minority links it to exon 2; similarly, PacBio reads support ZH38T0029211 as a TSS of INPP1 with the majority also excluding exon 2. Furthermore, epigenomic signals, such as chromatin accessibility and histone ChIP-seq, also support ZH38T0029211 as a novel TSS of INPP1.
Despite the enrichment for functional, epigenomic, and transcriptomic annotations, the RAMPAGE-verified unannotated TSSs were less evolutionarily conserved than their matched GENCODE TSSs as measured by phastCons (Wilcoxon test, P = 2.5 × 10−19) (Fig. 4F; Supplemental Fig. S5N; Siepel et al. 2005) and liftOver (Hinrichs et al. 2006) to the mm10 genome (Fisher's exact test, P = 1.5 × 10−15) (Supplemental Fig. S5O). However, the RAMPAGE-verified TSSs were still more conserved than distal enhancer cCREs (cCREs-dELS; Wilcoxon test, P = 7.4 × 10−81; Fisher's exact test, P = 1.2 × 10−80) (Supplemental Fig. S5N,O) and much more conserved than random genomic regions (Wilcoxon test, P < 1.0 × 10−300; Fisher's exact test, P < 1.0 × 10−300) (Supplemental Fig. S5N,O). These findings suggest that although the RAMPAGE-verified unannotated TSSs are more biochemically and transcriptionally active in the evaluated cell types, GENCODE TSSs correspond to transcripts expressed in other cell types that have not been surveyed by the RAMPAGE assay. Therefore, for cell type–agnostic data analyses, we suggest users supplement GENCODE TSS annotations with RAMPAGE-annotated TSSs, whereas for cell type–specific analyses, our results show that RAMPAGE TSSs are a more precise and accurate set of TSSs than the using the entire set of GENCODE annotations.
RAMPAGE rPeaks identify novel genes that are associated with GWAS phenotypes
We intersected our RAMPAGE rPeaks with variants reported in the NHGRI-EBI GWAS catalog to evaluate the utility of our collection of experimentally derived TSSs (Buniello et al. 2019). Accounting for population-specific linkage disequilibrium (LD; r2 > 0.7), our rPeaks overlapped 1345 variants associated with 208 phenotypes (Supplemental Table S5A). These GWAS SNPs were slightly more likely to overlap the TSS of a major isoform compared with matched controls (70.5% vs. 66.5%; Fisher's exact test, P = 0.01) and were also more likely to be eQTLs (89% vs. 65% of controls; Fisher's exact test, P = 6.6 × 10−119) with 86% of the eQTLs overlapping the rPeak TSSs of their eGenes. To identify disease-associated cell and tissue types, we performed biosample enrichment analysis using our previously published pipeline (The ENCODE Project Consortium et al. 2020). However, unlike our previous work, which used nearly one million cCREs, covering ∼8% of the human genome, our rPeaks had a much smaller genomic footprint; therefore, we only observed enrichments passing our FDR thresholds for three phenotypes: (1) obesity-related traits, (2) intelligence, and (3) general cognitive ability (see Methods; Supplemental Table S5B). Generally, enriched cell types were related to disease etiology. For example, intelligence and cognitive ability variants were enriched at rPeaks active in the neuroblastoma cell line SK-N-DZ, whereas obesity variants were enriched in rPeaks active in a variety of gastrointestinal and thyroid tissues. This result suggests that although we do not have the power to determine phenotype-relevant cell types for most studies using only RAMPAGE rPeaks, they can still capture biologically relevant enrichments to aid downstream variant interpretation.
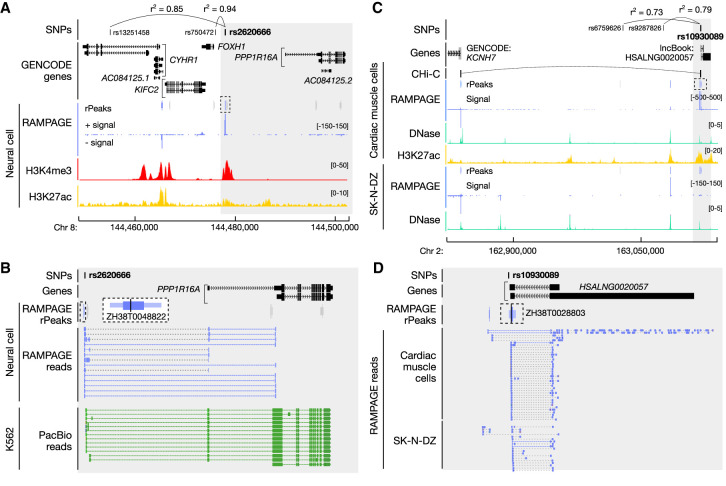
Among the 1345 variants that overlapped RAMPAGE rPeaks, 76% overlapped verified TSSs (50% GENCODE-annotated TSSs and 26% unannotated TSSs) and were therefore linked with an annotated gene by paired-end reads. Of these verified TSS-overlapping variants, 52% were linked with a gene that was not previously reported by the original GWAS and 37% were linked with a gene that was not reported by any GWAS, giving new insights into disease risk (Supplemental Table S5C). Of particular interest were RAMPAGE-verified unannotated transcript TSSs that were originally classified as intergenic using GENCODE annotations; these novel TSSs enabled us to assign 41 intergenic SNPs, which were associated with 68 phenotypes, to genes. Figure 5A highlights rs2620666, which is in high LD with two lead SNPs, rs750472 and rs13251458, reported to be associated with several cognitive traits (Supplemental Table S5D). The original studies reported FOXH1 and CYHR1 as possible candidate genes owing to their close proximity to the lead SNPs. Although rs2620666 lies only 1694 bp upstream of a GENCODE-annotated FOXH1 TSS, it overlaps a RAMPAGE-verified unannotated TSS of PPP1R16A (ZH38T0048822) (Fig. 5B), which encodes a protein phosphatase regulatory subunit. This novel TSS is 11,915 bp upstream of the nearest GENCODE-annotated TSS for PPP1R16A, and this gene assignment is also supported by PacBio reads (Fig. 5B; Supplemental Table S5E). The novel TSS has high RAMPAGE signal in neural cells, brain tissues, and blood cells; moreover, the GTEx Consortium identified rs2620666 as an eQTL for several genes (Supplemental Table S5F), the most significant of which is PPP1R16A in whole-blood samples, suggesting that this variant may influence PPP1R16A expression. This PPP1R16A TSS has been reported by other gene annotation collections and, recently (May 2021), was included as part of the GENCODE V38 basic annotations. This example highlights the importance of having a comprehensive collection of annotated TSSs so that variants are assigned correctly to the linked genes.
Figure 5.
Disease-associated SNPs are linked with new candidate genes using the RAMPAGE rPeak catalog. (A) Genome browser view of the CYHR1–PPP1R16A locus. Rs2620666 is in high LD (shown as r2 values) with GWAS SNPs rs13251458 and rs750472, and overlaps RAMPAGE rPeak ZH38T0048822 (dashed box). RAMPAGE rPeaks with RPM > 2 in neural cells are shown in purple, and those with RPM ≤ 2 are shown in gray; RAMPAGE signal is shown in purple. Supporting epigenomic signals from neural cells, H3K4me and H3K27ac, are shown in red and yellow, respectively. The region shaded in gray is magnified in B. (B) Zoomed-in genome browser view (gray highlight in A) displaying RAMPAGE reads (purple) and PacBio reads (green) supporting RAMPAGE peak ZH38T0048822 (dashed box), which is a verified unannotated TSS of PPP1R16A and overlaps GWAS SNP rs2620666. RAMPAGE peaks are colored as in A, and a magnified image of ZH38T0048822 is shown in a larger dashed box with white background. (C) Genome browser view of the KCNH7 locus. Rs10930089 is in high LD with GWAS SNPs rs6759626 and rs9287826 and overlaps RAMPAGE rPeak ZH38T0028803 (dashed box). RAMPAGE peaks are colored as described in A for cardiac muscle and SK-N-DZ cells. Supporting epigenomic signals from cardiac muscle cells and SK-N-DZ are shown with DNase I in teal and H3K27ac in yellow. CHi-C links for cardiac cells are shown in black. The region shaded in gray is magnified in D. (D) Zoomed-in genome browser view (gray highlight in C) displaying RAMPAGE reads (purple) from cardiac muscle and SK-N-DZ cells supporting RAMPAGE peak ZH38T0028803 (in dashed box), which overlaps two transcripts of the lncBook lncRNA HSALNG0020057 and GWAS SNP rs10930089. RAMPAGE peaks shown in purple have RPM > 2 in both cardiac muscle and SK-N-DZ cells.
Finally, we investigated the 34 GWAS variants that overlapped TSSs of RAMPAGE-verified unannotated transcripts (Supplemental Table S5A). Of particular interest was rs10930089, an intergenic SNP in high LD with rs6759626 and rs9287826, two lead SNPs associated with general cognitive ability (Davies et al. 2018). Rs10930089 overlaps ZH38T0028803, the TSS of a RAMPAGE-verified unannotated transcript that has high RAMPAGE signal in SK-N-DZ (a neuronal cell line), cardiac tissues, and male reproductive tissues (Fig. 5C; Supplemental Table S5G). ZH38T0028803 overlaps the TSSs of two lncRNA transcripts annotated in lncBook, both of which are consistent with the RAMPAGE reads pairs (Fig. 5D). In the other direction of the genome, ZH38T0028803 lies 282,766 bp upstream of KCNH7, which encodes a potassium voltage channel that has known roles in neurons and the heart (https://www.genecards.org/cgi-bin/carddisp.pl?gene=KCNH7; accessed September 4, 2020). Variants in KCNH7 have also been previously associated with bipolar disorder (Strauss et al. 2014) and treatment response in schizophrenia (Wang et al. 2019), suggesting it may play an important role in neuronal pathways. We found that 3D chromatin contact data linked ZH38T0028803 with KCNH7 in cardiac myocytes (Fig. 5C; Montefiori et al. 2018) but not in iPSC-derived neurons (Rajarajan et al. 2018; Song et al. 2019). Furthermore, ZH38T0028803 has high chromatin accessibility in SK-N-DZ, cardiac cells, and heart tissues, but low chromatin accessibility in fetal brain and iPSC-derived neurons (Supplemental Table S5H). Taken together, these results suggest that rs10930089 may modulate the function of ZH38T0028803, the TSS of a lncRNA expressed in neuronal and cardiac cells, and this TSS may also acts as an enhancer for KCNH7 in both of these two types of cells, with the caveat that the 3D connection is in neuronal cell types other than iPSC-derived neurons.
Discussion
We annotated 52,546 RAMPAGE rPeaks by integrating 115 RAMPAGE experiments, uniformly curating sites of transcription in hundreds of human cell and tissue types. Using paired-end RAMPAGE reads, we assigned the majority of these rPeaks as TSSs of annotated genes and additionally identified TSSs of over 4000 novel transcripts. We then showed that the TSSs in our catalog were enriched for various regulatory signatures defined using epigenetic and functional data and that our catalog complements existing TSS annotations such as those by GENCODE. Through systematic comparisons with CAGE, GRO-cap, and PacBio long-read data, we also determined that our catalog of RAMPAGE rPeaks was highly precise and accurate. In particular, PacBio and RAMPAGE had the highest overlap in both GM1878 and K562 cells. PacBio long reads not only supported our RAMPAGE TSS annotations but also supported our assignments of these TSSs to genes (Figs. 4E, 5B; Supplemental Fig. S3B–D). PacBio long-read data are particularly advantageous as they allow us to identify novel isoforms and annotate the 3′ ends of transcripts in addition to annotating TSSs. As these data continue to be produced for a wide variety of biosamples by the ENCODE Consortium, they will be very useful for further expanding our TSS catalog and enriching transcript annotations.
In both K562 and GM12878 cells, CAGE peaks tended to be the least concordant with the RAMPAGE rPeaks and PacBio 5′ ends (Supplemental Fig. S3B). We also noted that CAGE-specific peaks were much more likely to be intronic and intergenic than RAMPAGE rPeaks. However, CAGE peaks were supported by GRO-cap signals at a comparable level as RAMPAGE rPeaks, suggesting that CAGE-specific peaks contain true TSSs (Supplemental Fig. S3B). We hypothesize that the CAGE assay can identify a subclass of intergenic and intronic transcription sites, likely eRNAs, that are not detected by RAMPAGE or PacBio long-read RNA-seq. This ability can be used to annotate TSS-distal regulatory elements. Thus, additional comparisons need to be performed with transcription assays that have high rates of eRNA detection, such as BruUV-seq (Magnuson et al. 2016) and PRO-seq/cap (Kwak et al. 2013).
When we compared our catalog of RAMPAGE rPeaks to the FANTOM Consortium's CAGE peak collection, we found that loci missed by RAMPAGE were primarily owing to differences in surveyed biosamples (Fig. 2C). This result indicates that there is high variability in the transcriptional landscapes among different cell types, and a more comprehensive TSS collection can be achieved by surveying a larger collection of biosamples; however, there are additional considerations regarding the composition of a sample collection. Although we currently include over 100 biosamples in our RAMPAGE rPeak catalog, the majority of these biosamples are bulk tissue samples that comprise many different cell types. We found that tissue samples generally clustered separately from primary and in vitro–differentiated cell samples despite some sharing similar biological profiles (Supplemental Fig. S1F), possibly owing to the technical differences in assaying tissues versus cells. The impact of biosample composition on TSS annotation was also apparent when we observed an enrichment of neuron-related Gene Ontology terms for CAGE-only genes despite the presence of fetal brain tissues and iPSC-derived neurons in our RAMPAGE sample collection. This result suggests that these early developmental brain tissues may be dominated by precursor cells such as immature neuronal progenitors or radial glia and that the iPSC-derived neurons may represent alternative cell states from mature neurons. On a genome-wide scale, SK-N-DZ has a transcriptional profile that is more similar to iPSC-derived neurons than to mature neurons, as is evident from UMAP embedding (Supplemental Fig. S1F). The discrepancy among the different types of neuronal cells was further highlighted by our GWAS analysis in which we observed that the cognitive phenotype-related SNPs overlapped a novel TSS active in SK-N-DZ cells but not in iPSC-derived neurons. Therefore, although SK-N-DZ cells overall share similar transcriptomic signatures to iPSC-derived neurons, there are subtle differences in cellular state that may have important impacts on variant and disease interpretation. With further developments of single-cell transcriptomic technologies to capture the 5′ end of transcripts, it will be important to expand our TSS identification methods to build a comprehensive catalog by cell type, particularly in heterogeneous tissues such as the brain.
Even though we observed enrichments in some tissues for GWAS variants associated with three phenotypes, our comparisons were underpowered compared with our previous work (The ENCODE Project Consortium et al. 2020) owing to the small genomic footprint of RAMPAGE rPeaks. Despite this, we showed that accurate TSS annotations, particularly those TSSs linked with known transcripts, are important for interpreting variants reported by GWAS. Additionally, we anticipate that such collections will also be important for the detection and interpretation of rare and de novo variants uncovered by whole-genome sequencing efforts, as these variants have larger effect sizes and may be more likely to fall within promoter regions than in distal regulatory elements. For example, a recent study found an enrichment of de novo variants associated with autism spectrum disorder in promoters (An et al. 2018). Therefore, accurate, cell type–specific TSS annotations can improve our power for interpreting the impact of de novo genetic variation across cell types.
Finally, we identified 4129 TSSs for unannotated transcripts, many of which we hypothesize to be lncRNAs although we could not test this hypothesis with only the beginning portion of these transcripts. It is also unclear if these transcripts carry out any cellular functions. A wide range of functional mechanisms have been reported for lncRNAs, varying from transcriptional regulation of other genes via epigenetic or antisense means to simply being the byproducts of strong enhancers (Fang and Fullwood 2016; Quinn and Chang 2016). With development of antisense oligonucleotide (ASO) and CRISPR perturbation technologies, it is now possible to perform screens to identify functional lncRNAs in a high-throughput manner (Joung et al. 2017; Liu et al. 2017; Ramilowski et al. 2020). As these collections of functionally validated lncRNAs become available across diverse cellular contexts, we plan to further refine our TSS catalog to include such functional information.
There are some limitations to our catalog of RAMPAGE rPeaks, which should be considered as they may bias results toward highly expressed, stable transcripts. One caveat to our catalog is that it primarily contains TSSs for stable transcripts, as comparisons with NET-CAGE and GRO-cap data showed that the RAMPAGE assay is generally unable to detect TSSs of unstable transcripts such as eRNAs. Additionally, using RAMPAGE rPeaks, we identified 17% fewer genes compared with CAGE and PacBio data in the same cell types. Thus, although we showed that our collection likely has a lower false-discovery rate, we may be underreporting transcriptional events. Finally, the 5′ ends of some RAMPAGE read pairs may be imprecise owing to fragmentation and degradation. However, aggregation analysis of other transcription assays revealed a sharp peak of signal centered on RAMPAGE rPeaks (Supplemental Fig. S3F–K), suggesting that only a very small percentage of sites may be impacted by such technical artifacts. Overall, our rPeak catalog is highly concordant with other assays even at the base pair level. In the future, we hope to expand this catalog using transcription annotations from more assays as they become available in a wider variety of cell and tissue types.
In summary, our catalog of RAMPAGE rPeaks expands the human transcriptional landscape across over 100 cell and tissue types. The catalog provides a valuable resource to the biological community by improving annotations for studying gene regulation and aiding in the interpretation of genetic variants associated with human diseases.
Methods
Detailed methods can be found in the Supplemental Methods.
Generating a collection of RAMPAGE rPeaks
We downloaded RAMPAGE BAM alignment files that contained reads mapped to the GRCh38/hg38 reference genome. We then removed redundant reads as previously described (Zhang et al. 2019) and pooled read pairs from biological replicates. We created signal files of the 5′ ends of R1 reads that we used for all subsequent signal quantifications. Finally, we excluded all experiments with a nonredundancy fraction of less than 0.25, which resulted in a final collection of 115 high-quality RAMPAGE experiments (Supplemental Table S1). We then called RAMPAGE peaks as previously described (Zhang et al. 2019). For each peak, we identified a high-density region, which contained 80% of the reads in each original peak, and a summit, which was the genomic position with the highest number of 5′ read ends. For each RAMPAGE experiment, the Gingeras laboratory also performed a matching total RNA-seq experiment on the same biosample, which we used to filter RAMPAGE peaks. We excluded peaks whose RNA-seq signals were greater than their RAMPAGE signals (i.e., peaks that fell below the x = y line) (Supplemental Fig. S1). Finally, to further select for high-quality annotations, we only retained peaks with reads per million (RPM) > 2 (Supplemental Table S1).
To generate RAMPAGE rPeaks, we adapted the representative DNase I hypersensitivity site (rDHS) pipeline as previousy described (The ENCODE Project Consortium et al. 2020). First, to retain strand-specific information, we separated peaks based on DNA strand and then clustered the strand-specific peaks across all 115 experiments. For each cluster, we selected the peak with the highest reads per kilobase per million (RPKM) signal as the rPeak. All peaks that overlapped this rPeak were then removed. We iteratively repeated this process until all 1.1 M RAMPAGE peaks were represented by a collection of 80,157 nonoverlapping rPeaks. To reduce false positives, we discarded all singleton rPeaks (i.e., rPeaks that represented only one experiment) unless they had an RPM > 5, resulting in a final set of 52,546 rPeaks.
Genomic context and enrichment
We used the following hierarchical approach to assign genomic context to annotations (including RAMPAGE rPeaks and FANTOM CAGE peaks).
TSS-overlapping: rPeak overlapped an annotated TSS from GENCODEv31 basic annotations.
TSS-Proximal: rPeak fell within ±500 bp of an annotated TSS from GENCODEv31 basic; required at least 50% of the RAMPAGE rPeak to overlap region.
Exon: rPeak overlapped “exon” annotation from GENCODEv31 basic that include coding exons (CDSs), exons of noncoding genes, and untranslated regions (UTRs); required at least 50% of the RAMPAGE rPeak to overlap exon.
Intron: rPeak overlapped an annotated gene from GENCODEv31 basic but not an exon; required at least 50% of the RAMPAGE rPeak to overlap gene.
Intergenic: all remaining rPeaks.
We annotated each rPeak with strand information by assigning the strand of the overlapping transcript for TSS-overlapping, exon, and intron rPeaks or closest gene for TSS-proximal and intergenic rPeaks. To determine the genomic background, we calculated the percentage of the GRCh38 genome comprising each of the annotations. We then determined the percentage of total rPeaks falling in each annotation and calculated fold enrichment.
Boundary and summit analysis
For each rPeak, we calculated the median peak boundary, high-density boundary, and summit variation for each peak that was represented. We did not include peaks that were selected as the rPeaks in this analysis.
UMAP
We performed two separate UMAP analyses: one using all 115 biosamples (Supplemental Fig. S1F) and one using the subset of all 87 tissue samples (Fig. 1G). For each biosample, we calculated the RPKM at each rPeak. We then took the Log10 and normalized these values before implementing the UMAP algorithm.
Comparisons with other transcription annotations
Comparison with CAGE peaks
We downloaded CAGE peaks and quantifications from the FANTOM Consortium (Abugessaisa et al. 2017). To compare the overall concordance of peak collections, we intersected the entire collection of CAGE peaks with the entire collection of RAMPAGE peaks, requiring at least 25% of the CAGE peak to overlap the RAMPAGE peak and the peaks to fall on the same strand. To extract peaks active in K562 and GM12878, we selected all peaks with an average transcripts per million (TPM) > 2 across the three surveyed replicates. We intersected these peaks with RAMPAGE rPeaks with RPM > 2 in K562 and GM12878, respectively, requiring overlapping peaks to be on the same strand and an overlap of a minimum of 25% of the CAGE peak.
Comparison with CAGE and NET-CAGE enhancers
We downloaded CAGE & NET-CAGE enhancers from Hirabayashi et al. (2019). We lifted the enhancers to the hg38 genome and intersected them with RAMPAGE rPeaks. We then stratified the enhancer annotations as to whether they were detected by CAGE (N = 65,423) or only NET-CAGE (N = 20,363) and calculated the total percent overlap with the RAMPAGE rPeaks.
Comparison with PacBio long-read RNA-seq data
We downloaded the PacBio BAM files from the ENCODE project data portal and merged replicates. We then intersected PacBio 5′ read ends with RAMPAGE and CAGE peaks and only considered strand matching intersections.
Comparison with GRO-cap signal
We downloaded GRO-cap signal files from Core et al. (2014). To calculate average signal at RAMPAGE rPeaks, CAGE peaks, and PacBio 5′ ends, we lifted down the 1-bp summits or read ends to the hg19 genome. We then set region width to a uniform 50 bp centered on the peak summits or 5′ ends and calculated the average signal across each region. To determine a signal threshold for high GRO-cap signal, we first randomly selected 500,000 50-bp genomic regions and calculated their average GRO-cap signal. We then selected the 99.5th percentile as the threshold for high signal, which was 0.06 in K562 and 0.08 in GM12878.
Comparison with GRO-cap peaks
We downloaded GRO-cap peak calls from Core et al. (2014). We intersected these GRO-cap peaks with RAMPAGE rPeaks, CAGE peaks, and RAMPAGE PacBio reads, requiring annotations to be on the same strand. From the same study, we also obtained sets of paired GRO-cap peaks in GM12878 and K562 that were classified by stability. We lifted these peaks to the hg38 genome and intersected them with RAMPAGE rPeaks, requiring annotations to be on the same strand. We then calculated the overall percentage of each category that overlapped the rPeaks.
Comparison of GENCODE-covered genes
We first set peak width to a uniform 100 bp centered around each peak summit or 5′ read end and then intersected these regions with annotated TSSs of GENCODE V31 genes, requiring annotations to be on the same strand. We performed Gene Ontology analysis using PantherDB's online database (Mi et al. 2017). We first performed this analysis for the entire sets of RAMPAGE and CAGE peaks and then for peaks and PacBio 5′ read ends in K562 and GM12878 cells.
Aggregate transcriptomic signals at RAMPAGE rPeaks
Using 1-bp bins, we calculated the average CAGE, PacBio, and GRO-cap signals along a 4-kb window centered across the summits of RAMPAGE rPeaks active in either K562 or GM12878 cells. In all three assays, we calculated strand-specific signal for each rPeak.
Assigning RAMPAGE rPeaks to Genes
Curating verified GENCODE TSSs, verified unannotated TSSs, unannotated transcript TSSs, and local transcription rPeaks
We developed the following computational workflow to link RAMPAGE rPeaks with genes, which is detailed in Supplemental Figure S3A. Briefly, based on the genomic context of the rPeak and the location of its supporting 3′ reads, we assigned the rPeak into one of six categories.
Verified GENCODE TSS: rPeak overlaps an annotated GENCODE TSS and its 3′ read ends overlap a downstream exon.
Verified unannotated TSS: rPeak does not overlap an annotated GENCODE TSS (i.e., rPeak is either TSS-proximal, exonic, intronic, or intergenic), and its 3′ read ends overlap a downstream exon.
Candidate GENCODE TSS: rPeak overlaps a TSS or first exon or is TSS-proximal to either a single exon transcript or to a transcript with a first exon >500 nt.
Unannotated transcript TSS: rPeak is supported by reads with 3′ ends that do not overlap an annotated GENCODE exon.
Local transcription: rPeak is supported by reads that span <1 kb or map to the first exon of the transcript.
Discard: We discarded all rPeaks that overlapped exons that were not the first exon of a transcript or only supported by reads that spanned >500 kb.
Overlap of novel transcripts with lncRNAs
We downloaded lncRNA annotations from lncBook (Ma et al. 2019) and extracted annotated TSSs. Then, we intersected RAMPAGE rPeaks, requiring annotations to be on the same strand. We also calculated the overlap of lncBook TSSs with 500,000 100-bp random genomic regions.
Scanning transcripts for open reading frames
We intersected our RAMPAGE rPeaks with PacBio reads to delineate produced transcripts and then scanned these transcripts using NCBI's ORFfinder tool (Wheeler et al. 2003). Stratifying by our rPeak TSS assignment, we calculated the number of uniquely identified ORFs for each rPeak.
Characterizing biosample profiles of RAMPAGE TSSs
We selected all GENCODE genes with at least one linked RAMPAGE rPeak (either verified GENCODE or verified unannotated). For each gene, we calculated two metrics:
The total number of biosamples in which the gene was expressed.
The total biosample space, which was a concatenated list of all biosamples for which any linked RAMPAGE rPeak was expressed.
To evaluate the cell type specificity of gene and transcript expression, we compared the number of active biosamples (RPM > 2) for each RAMPAGE rPeak and its linked gene. To determine whether the transcripts resulting from rPeak TSSs correspond to major or minor isoforms, we calculated the total number of biosamples for which the rPeak has an RPM > 2 and then divided this by the total biosample space of its linked gene.
Comparison of GENCODE and verified TSSs
Generating sets of matched GENCODE TSSs
We first selected all GENCODE genes that did not have a single annotated TSS overlapping a RAMPAGE rPeak. Of these, we then selected all genes with a RAMPAGE-verified TSS. Because of the no overlapping requirement, these RAMPAGE-verified TSSs were either TSS-proximal, exonic, intronic, or intergenic. The GENCODE-annotated TSSs of these genes served as the matched GENCODE TSS set. We also curated K562-specific annotations by selecting all RAMPAGE-verified TSSs with an RPM > 2 in K562 and their matched GENCODE TSSs. Unlike the RAMPAGE-verified TSSs, GENCODE TSSs were only 1 bp in width; therefore, to eliminate biases owing to region width, we generated uniform 100-bp regions centered on either RAMPAGE-verified TSS summits or GENCODE TSSs, respectively.
Overlap of RAMPAGE-verified and matched GENCODE TSSs with ENCODE cCREs, GTEx eQTLs, and SuRE peaks
We intersected the uniform 100-bp-sized TSS regions with genomic annotations as follows. We downloaded cell type–agnostic cCREs and K562-specific cCREs from the ENCODE SCREEN database (https://screen.encodeproject.org). For the K562 cCREs, we filtered out “low-DNase” cCREs, which are regulatory regions deemed inactive in the cell type. We downloaded version 8 eQTLs from the GTEx database and reformatted them into BED format. We downloaded SuRE peaks from van Arensbergen et al. (2017) and lifted the regions to the hg38 genome.
Aggregate epigenomic signals at RAMPAGE-verified and matched GENCODE TSSs
We calculated the average DNase-seq and H3K4me3, H3K27ac, and Pol II ChIP-seq signals along a 4-kb window centered across the RAMPAGE-verified rPeak summit or matched GENCODE TSS, respectively, accounting for strand orientation. We used the following signal files from the ENCODE portal: ENCFF971AHO, ENCFF847JMY, ENCFF779QTH, and ENCFF321FZQ.
Conservation of RAMPAGE-verified and -matched GENCODE TSSs
We calculated the average 100-way vertebrate phastCons conservation across the uniform 100-bp TSS regions. We also lifted the uniform 100-bp-sized TSS regions to the mm10 genome and calculated the percentage of total regions that successfully lifted over. We also compared the lift over rates of ENCODE cCREs-dELS—extracted from the cell type–agnostic set of cCREs—and 500,000 random regions of the genome. For comparison, both these sets of regions were resized to 100 bp around the region center.
Interpreting GWAS variants with the RAMPAGE rPeak catalog
Overlap of GWAS variants
We curated SNPs reported by the NHGRI-EBI GWAS catalog as of January 2019 and using population-specific LD, incorporating all SNPs in high LD (r2 > 0.7) with this collection, as previously described (The ENCODE Project Consortium et al. 2020). We intersected this collection with our RAMPAGE rPeak catalog. To compare gene assignments, we extracted reported and mapped genes from the original studies and determined if our rPeak linked genes (from read pair analysis) were represented in the list.
Comparison with eQTLs
As previously mentioned, we downloaded eQTLs from the GTEx database. We then compared the overlap between GWAS SNPs and matched controls as previously defined (The ENCODE Project Consortium et al. 2020) and calculated the number of SNPs in each group that was linked to the same gene by both RAMPAGE reads and expression changes (eQTL).
Cell type enrichment
We tested whether sets of GWAS SNPs were enriched in RAMPAGE rPeaks activity in specific biosamples using the same GWAS enrichment pipeline as previously described (The ENCODE Project Consortium et al. 2020). Because RAMPAGE rPeaks have a much smaller genomic footprint than other collections of genomic regions (e.g., cCREs), we only included studies for which at least 15 LD blocks contained a SNP that overlapped a RAMPAGE rPeak. We reported all enrichments with an FDR corrected P-value less than 0.05 (Supplemental Table S5B).
3D chromatin interactions between ZH38T0028803 and KCNH7
We downloaded the cardiomyocyte promoter capture Hi-C data from Montefiori et al. (2018) and iPSC neuron promoter capture Hi-C data from Song et al. (2019). We also requested iPSC neuron Hi-C loop calls directly from Rajarajan et al. (2018), who generously provided these annotations. We intersected links with the KCNH7 locus, requiring one of the KCNH7 GENCODE TSSs to overlap one anchor and ZH38T0028803 to overlap the other anchor.
Data access
All raw and processed sequencing data generated in this study have been submitted to the ENCODE Project Data Portal (https://www.encodeproject.org/) under the data collection https://www.encodeproject.org/carts/2ac8b407-bee2-4ed3-ac2e-d284cdc48e41/. A UCSC Genome Browser track hub for the hg38 genome build is available at http://users.wenglab.org/moorej3/RAMPAGE/hub.txt. Code for computational analysis is available at GitHub (https://github.com/weng-lab/RAMPAGE-Analysis) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Gabriela Balderrama-Gutierrez, Diane Trout, and Julien Lagarde for discussions on how to best analyze TSSs from long-read PacBio data. This work was supported by grants from the National Institutes of Health, National Human Genome Research Institute under U24HG009446 to Z.W. and UM1HG009443 to A.M.
Author contributions: J.E.M. and Z.W. conceived and designed the project. A.M. led the production of PacBio long-read data. Z.W. supervised the project. J.E.M. led the bioinformatics analysis with contributions from X.-O.Z., S.I.E., K.F., H.E.P., and F.R.; J.E.M. and Z.W. analyzed the data and wrote the paper with contributions from X.-O.Z., S.I.E., K.F., H.E.P., F.R., and A.M.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275723.121.
Competing interest statement
Z.W. is a cofounder of Rgenta Therapeutics, and she serves on its scientific advisory board.
References
- Abugessaisa I, Noguchi S, Hasegawa A, Harshbarger J, Kondo A, Lizio M, Severin J, Carninci P, Kawaji H, Kasukawa T. 2017. FANTOM5 CAGE profiles of human and mouse reprocessed for GRCh38 and GRCm38 genome assemblies. Sci Data 4: 170107. 10.1038/sdata.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J-Y, Lin K, Zhu L, Werling DM, Dong S, Brand H, Wang HZ, Zhao X, Schwartz GB, Collins RL, et al. 2018. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362: eaat6576. 10.1126/science.aat6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. 2013. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res 23: 169–180. 10.1101/gr.139618.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. 2019. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47: D1005–D1012. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. 10.1038/ng1789 [DOI] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46: 1311–1320. 10.1038/ng.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, Hagenaars SP, Ritchie SJ, Marioni RE, Fawns-Ritchie C, et al. 2018. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun 9: 2098. 10.1038/s41467-018-04362-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K, Moore JE, Zhang X-O, Weng Z. 2021. Genetic and epigenetic features of promoters with ubiquitous chromatin accessibility support ubiquitous transcription of cell-essential genes. Nucleic Acids Res 49: 5705–5725. 10.1093/nar/gkab345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fullwood MJ. 2016. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 14: 42–54. 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2020. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369: 1318–1330. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Bhagat S, Matsuki Y, Takegami Y, Uehata T, Kanemaru A, Itoh M, Shirakawa K, Takaori-Kondo A, Takeuchi O, et al. 2019. NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements. Nat Genet 51: 1369–1379. 10.1038/s41588-019-0485-9 [DOI] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, et al. 2017. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548: 343–346. 10.1038/nature23451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. 2006. CAGE: cap analysis of gene expression. Nat Methods 3: 211–222. 10.1038/nmeth0306-211 [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: eaah7111. 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, Bajic VB, Zhang Z. 2019. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res 47: 2699. 10.1093/nar/gkz073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Veloso A, Kirkconnell KS, de Andrade Lima LC, Paulsen MT, Ljungman EA, Bedi K, Prasad J, Wilson TE, Ljungman M. 2016. Identifying transcription start sites and active enhancer elements using BruUV-seq. Sci Rep 5: 17978. 10.1038/srep17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, Bozek G, Moskowitz IP, McNally EM, Nóbrega MA. 2018. A promoter interaction map for cardiovascular disease genetics. eLife 7: e35788. 10.7554/eLife.35788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro C, Cesari E, Sette C. 2021. Splicing regulation in brain and testis: common themes for highly specialized organs. Cell Cycle 20: 480–489. 10.1080/15384101.2021.1889187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, Powell S, Yashaswini C, LaMarca EA, Kassim B, et al. 2018. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362: eaat4311. 10.1126/science.aat4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Yip CW, Agrawal S, Chang J-C, Ciani Y, Kulakovskiy IV, Mendez M, Ooi JLC, Ouyang JF, Parkinson N, et al. 2020. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res 30: 1060–1072. 10.1101/gr.254219.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Yang X, Ren X, Maliskova L, Li B, Jones IR, Wang C, Jacob F, Wu K, Traglia M, et al. 2019. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat Genet 51: 1252–1262. 10.1038/s41588-019-0472-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Markx S, Georgi B, Paul SM, Jinks RN, Hoshi T, McDonald A, First MB, Liu W, Benkert AR, et al. 2014. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Hum Mol Genet 23: 6395–6406. 10.1093/hmg/ddu335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman JB, Schoenberg DR. 2019. A recap of RNA recapping. Wiley Interdiscip Rev RNA 10: e1504. 10.1002/wrna.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen J, FitzPatrick VD, de Haas M, Pagie L, Sluimer J, Bussemaker HJ, van Steensel B. 2017. Genome-wide mapping of autonomous promoter activity in human cells. Nat Biotechnol 35: 145–153. 10.1038/nbt.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su Y, Yan H, Huang Z, Huang Y, Yue W. 2019. Association study of KCNH7 polymorphisms and individual responses to risperidone treatment in schizophrenia. Front Psychiatry 10: 633. 10.3389/fpsyt.2019.00633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al. 2003. Database resources of the national center for biotechnology. Nucleic Acids Res 31: 28–33. 10.1093/nar/gkg033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman D, Balderrama-Gutierrez G, Reese F, Jiang S, Rahmanian S, Forner S, Matheos D, Zeng W, Williams B, Trout D, et al. 2020. A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. bioRxiv 10.1101/672931 [DOI]
- Zhang XO, Gingeras TR, Weng Z. 2019. Genome-wide analysis of polymerase III–transcribed Alu elements suggests cell-type–specific enhancer function. Genome Res 29: 1402–1414. 10.1101/gr.249789.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.