Abstract
The zygote, a totipotent stem cell, is crucial to the life cycle of sexually reproducing organisms. It is produced by the fusion of two differentiated cells—the egg and sperm, which in plants have radically different siRNA transcriptomes from each other and from multicellular embryos. Owing to technical challenges, the epigenetic changes that accompany the transition from differentiated gametes to totipotent zygote are poorly understood. Because siRNAs serve as both regulators and outputs of the epigenome, we characterized small RNA transcriptomes of zygotes from rice. Zygote small RNAs exhibit extensive maternal carryover and an apparent lack of paternal contribution, indicated by absence of sperm signature siRNAs. Zygote formation is accompanied by widespread redistribution of 24-nt siRNAs relative to gametes, such that ∼70% of the zygote siRNA loci do not overlap any egg cell siRNA loci. Newly detected siRNA loci in zygote are gene-proximal and not associated with centromeric heterochromatin, similar to canonical siRNAs, in sharp contrast to gametic siRNA loci that are gene-distal and heterochromatic. In addition, zygote but not egg siRNA loci are associated with high DNA methylation in the mature embryo. Thus, the zygote begins transitioning before the first embryonic division to an siRNA profile that is associated with future RdDM in embryogenesis. These findings indicate that, in addition to changes in gene expression, the transition to totipotency in the plant zygote is accompanied by resetting of the epigenetic reprogramming that occurred during gamete formation.
Gametes and zygotes constitute critical developmental stages in the life cycle of all sexually reproducing organisms. During fertilization, the egg cell fuses with a sperm cell to form the zygote, which is an undifferentiated and totipotent stem cell that initiates embryogenesis. Flowering plants undergo double fertilization, in which a second sperm cell fuses with the central cell and gives rise to the endosperm, a nutritive tissue that nurtures the developing embryo or germinating seedling (for review, see Lord and Russell 2002). In animals, early embryogenesis is controlled by maternal gene products predeposited in the egg cell. Depending on the organism, the zygotic genome does not become transcriptionally active until a number of cell divisions have occurred (Tadros and Lipshitz 2009). Recent studies show that maternal-to-zygote transition in flowering plants differs markedly from most animals (for review, see Armenta-Medina and Gillmor 2019). In rice zygotes, thousands of genes are up-regulated in zygotes, many of which are undetected in the egg cell, consistent with similar observations in zygotes of maize and Arabidopsis (Chen et al. 2017; Zhao et al. 2019). Furthermore, zygotic transcription was shown to be required for early embryogenesis (Kao and Nodine 2019; Zhao et al. 2019). These observations suggest that in angiosperms, unlike most animals, zygotes are transcriptionally active, and zygotic genome activation (ZGA) occurs in the zygote. However, similar to animals, ZGA in plants is gradual. The initial transcriptome of flowering plant zygotes is thus dominated by egg cell RNA carryover and, although newly expressed genes in the zygote are widespread and represent a significant fraction of the zygote transcriptome, their expression levels are relatively low (Anderson et al. 2017; Chen et al. 2017; Zhao et al. 2019).
Along with dynamic changes in gene expression, epigenomic reprogramming has been observed during flowering plant reproduction. In rice and maize, the egg cell is ∼10 times larger than sperm in diameter and thus ∼1000 times larger than the sperm cell in volume (Kranz et al. 1991; Anderson et al. 2013), and its chromatin is diffused (Scholten et al. 2002). In contrast, the sperm cell chromatin undergoes global condensation, paralleling animal sperm chromatin in which protamines replace histones (Kimmins and Sassone-Corsi 2005). A male-germline-specific histone H3 variant MGH3 (also termed H3.10) is present in the sperm cell (Okada et al. 2005; Borg and Berger 2015), following the removal of H3.1 (Borg et al. 2020). H3.10 is resistant to trimethylation at H3K27 (H3K27me3), thus priming the activation of key genes for sperm differentiation and embryogenesis (Borg et al. 2020). Upon karyogamy, H3.10 is removed from the paternal chromatin via a replication-independent process (Ingouff et al. 2007). Other histone H3 variants, such as H3.3, are also removed from egg cell chromatin upon karyogamy, followed by loading of newly synthesized histones, again via a replication-independent mechanism (Ingouff et al. 2010). In addition, other cells of both male and female gametophytes in Arabidopsis experience global chromatin changes as well. Heterochromatin is decondensed in the central cell, the cell that gives rise to endosperm (Pillot et al. 2010). A similar phenomenon occurs in the pollen vegetative cell, the cell that encapsulates the sperm cells and enables their migration through the style to the ovule (Schoft et al. 2009; Mérai et al. 2014; Hsieh et al. 2016). Relaxation of heterochromatin in the pollen vegetative cell has been reported to produce short interfering RNAs (siRNAs) that traffic into the sperm cells and reinforce transposon silencing in the gametes (Slotkin et al. 2009; Calarco et al. 2012; Martínez et al. 2016; Park et al. 2016; Kim et al. 2019). Similarly, it has been proposed that siRNAs traffic from the central cell to the egg cell, as well as from the endosperm into the developing embryo (Hsieh et al. 2009; Ibarra et al. 2012; Martinez and Köhler 2017).
Concomitant with chromatin reprogramming, there is also evidence for changes in DNA methylation during plant reproduction, especially in the context of RNA-directed DNA methylation (RdDM) (for review, see Gehring 2019). In plants, RdDM can function in both de novo and maintenance DNA methylation (for review, see Cuerda-Gil and Slotkin 2016). Briefly, 24-nt siRNAs are loaded onto an argonaute protein (AGO), which recruits the DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2). DRM2 leads to methylation in all sequence contexts, but methylation in the CHH context (mCHH), where H is A, C, or T, is a strong indicator of RdDM in both rice and maize (Gent et al. 2013; Tan et al. 2016, 2018), but not in all plants (Zemach et al. 2013). Multiple studies reported that disruption of RdDM leads to a variety of reproductive phenotypes, including aborted embryos (Autran et al. 2011; Grover et al. 2018), arrested pollen (Wang et al. 2020), defective triploid block when the seeds were produced from a 2n maternal × 4n paternal cross (Erdmann et al. 2017; Borges et al. 2018; Martinez et al. 2018; Satyaki and Gehring 2019), and defective floral development (Dorweiler et al. 2000; Moritoh et al. 2012). These observations suggest siRNAs and RdDM are important for normal plant reproduction.
In mammals, it has long been proposed that fusion of two epigenetically distinct gametes presents a challenge in reproduction, and resetting of the epigenome is required for the pluripotent state of the early embryo (for review, see Messerschmidt et al. 2014). Epigenome reprogramming in mammals includes large-scale erasure of somatic chromatin signatures in germ cell precursors, establishment of sex-specific signatures in the germline, and postfertilization resetting toward pluripotency (for reviews, see Saitou et al. 2012; Messerschmidt et al. 2014; Tang et al. 2016). The functional consequences of epigenomic changes in gametic fate acquisition and subsequent zygotic totipotency in plants are unclear. It is clear, however, that in plants the majority of DNA methylation is stably transmitted both maternally and paternally (for review, see Gehring 2019). In Caenorhabditis elegans, siRNAs can serve as carriers of transgenerational epigenetic information, in which siRNAs can be inherited across a few generations (for review, see Houri-Zeevi and Rechavi 2017). Whereas multiple changes in siRNA profiles have been observed during plant reproduction (Schoft et al. 2009; Slotkin et al. 2009; Calarco et al. 2012; Ibarra et al. 2012; Grover et al. 2020; Li et al. 2020; Papareddy et al. 2020), transgenerational inheritance of siRNAs, or the lack thereof, has yet to be rigorously demonstrated in plants.
In vegetative tissues such as seedlings, 24-nt siRNAs coincide with mCHH islands—short regions with high CHH methylation—that are enriched around genes and mark the ends of TEs and euchromatin-heterochromatin boundaries (Gent et al. 2013; Li et al. 2015). Hereafter, we refer to such a 24-nt siRNA profile as the canonical siRNA profile, because 24-nt siRNAs are the most abundant length class in most plants (for review, see Cuerda-Gil and Slotkin 2016), including rice gametes (Li et al. 2020). We have previously shown that the siRNA transcriptome is reprogrammed in rice gametes (Li et al. 2020) where siRNA transcriptomes of egg and sperm were distinct from each other in genome-wide distribution, as well as distinct from that of the seedling (Li et al. 2020). The relative magnitude of the egg-borne and sperm-borne contribution of siRNAs to the zygote is unknown. A recent study in Arabidopsis revealed that siRNAs from heterochromatic TEs are transiently up-regulated during embryogenesis, whereas siRNAs from euchromatic TEs peak at mature green embryos (Papareddy et al. 2020). However, currently there are no data available for siRNA transcriptomes before the preglobular embryo stage, and consequently very little is known about the siRNA landscape in plant zygotes. Because siRNA production is influenced by histone modifications and DNA methylation, and siRNAs in turn can direct histone modifications and DNA methylation (Law and Jacobsen 2010; Matzke and Mosher 2014; Parent et al. 2021), the siRNA transcriptome is an output and indicator of the epigenome. Owing to the extreme difficulties associated with plant zygote isolation and corresponding low-input sequencing, epigenome profiles of zygotes have remained poorly characterized in plants. Given the importance of genome-wide epigenetic changes associated with the acquisition of totipotency, we undertook a detailed characterization of the small RNA transcriptome of rice zygotes to investigate the changes that occur soon after fertilization.
Results
We collected rice zygotes ∼9 h after pollination (hap), which corresponds to the completion of S phase, just prior to the first zygotic division (Ding et al. 2009; Anderson et al. 2017). We generated small RNA transcriptomes from six replicates, with ∼50 zygotes in each replicate. As a maternal sporophytic control, we also collected postfertilization ovary of the same developmental stage as zygote (9 hap) and prepared small RNA transcriptomes from three replicates, with 10 ovaries in each replicate. For our analyses, we also included small RNA transcriptome data from rice gametes, pre-anthesis ovary (0-h ovary) and seedlings (Li et al. 2020). Except where indicated otherwise, siRNAs used for analyses were small RNA reads (20- to 25-nt) not overlapping 90% or more of their lengths with known miRNAs (miRBase v22) (Kozomara et al. 2019), 5S rRNA, tRNA, NOR, or phasiRNA loci (as detected in Li et al. 2020), and multimapped reads were included in all analyses unless indicated otherwise (Supplemental Fig. S1A).
Small RNA transcript carryover from egg cell into zygote
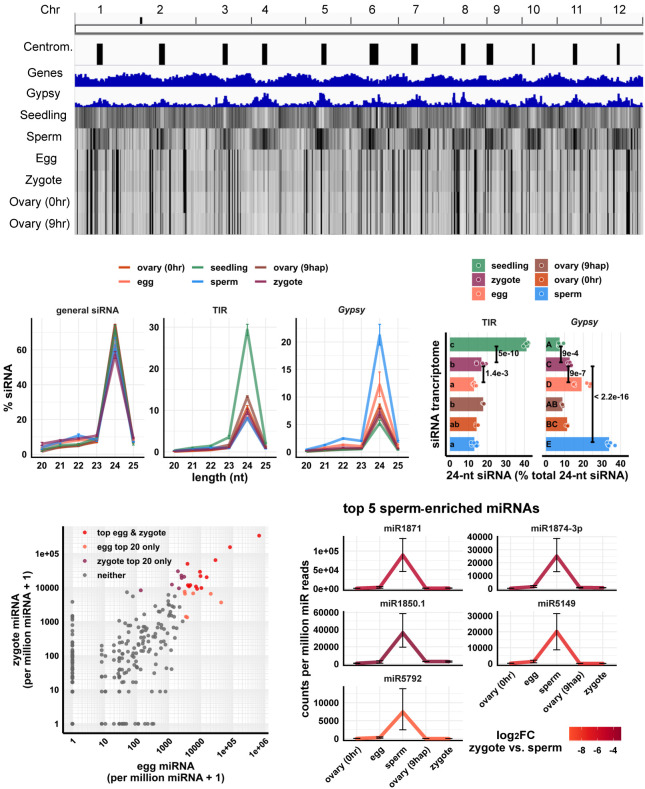
As we previously reported, the sperm cell has an siRNA pattern complementary to the canonical pattern of vegetative tissues, in which its 24-nt siRNAs are spread out across wide heterochromatic regions, including centromeric tandem repeats. The egg cell and ovary have a pattern different from both sperm and vegetative tissues, in which 24-nt siRNAs are concentrated at discrete loci (Fig. 1A). We found that, in a whole-genome view, the zygote had a similar pattern to the egg cell (Fig. 1A, zygote vs. egg track). To confirm that the similarity between zygote and egg cell was not because of large numbers of residual unfertilized egg cells in the zygote samples, we performed a control pollination experiment under similar conditions, and we determined that 98 out of 101 pollinated rice florets produced mature seeds, implying that 3% or less of the rice florets were unfertilized (Supplemental Table S2; see Methods for additional details). Thus, in our zygote samples, unfertilized egg cells might represent at most 3% of the total. We also performed differential expression analyses for miRNAs and detected 14 miRNAs that were lowly expressed in all six replicates of zygote but highly expressed in ovaries of the corresponding developmental stage, that is, 9 hap (Supplemental Fig. S1B). Thus, the similarity between zygote and ovary (Fig. 1A) is unlikely to be caused by small RNA contamination from ovary. A similar analysis was previously used to show that the egg cell samples were also free of prefertilization ovary contamination (Li et al. 2020).
Figure 1.
Genome-wide distribution of zygote small RNAs and comparisons of zygote, egg, and sperm small RNAs. (A) Heat map showing abundance of 24-nt siRNA across genome at 50-kb resolution. The first three tracks are centromeres (as defined by Mizuno et al. 2018), genes, and Gypsy retrotransposons. (B) Length profiles of siRNAs. y-axis values are relative to total siRNA reads (20- to 25-nt siRNAs). (TIR) Terminal inverted repeat transposons, CACTA element excluded; (Gypsy) Gypsy retrotransposons. Error bars are 95% confidence intervals for each cell type. miRNA and phasiRNA are not included in this analysis (Supplemental Fig. S1A). (C) Quantification of TIR and Gypsy panels in B. Each data point is an siRNA transcriptome. Bar heights are averages. x-axis values are relative to total 24-nt siRNAs. Letter grouping (α = 0.05) and P-values are based on Tukey tests. (D) Scatter plot showing miRNA relative abundances in egg and zygote. Each data point is a miRNA. Axes are relative to per million miRNA reads and log10 transformed. “top egg & zygote” refers to intersection of the 20 highest abundant miRNAs in both egg and zygote. (E) Top five sperm-enriched miRNAs. Sperm-enriched is classified as >1000 reads per million miRNA reads in sperm and <500 reads per million miRNA reads in egg. y-axis values are relative to per million miRNA reads. Color code reflects log2FC values for zygote versus sperm, and negative values indicate higher in sperm. Error bars are 95% confidence intervals for each cell type. See Supplemental Figure S1D for additional examples. Zygote and 9-hap ovary data are from this study; all other data from Li et al. (2020).
We next looked at the length profile of siRNAs in zygotes and compared that with published data from other cell and tissue types (Li et al. 2020). We found that in zygotes, as in all other tissues, 24-nt siRNAs predominated (Fig. 1B). Because the abundance of siRNAs of other length classes were all relatively low, we focused on 24-nt siRNAs for further analysis. Based on relative abundance patterns, the zygote siRNAs appeared to resemble egg cell siRNAs. Like the egg cell and unlike seedling tissues, the zygote had a lower abundance of siRNAs overlapping terminal inverted repeat (TIR) transposons (PIF/Harbinger, Tc1/Mariner, Mutator, or hAT superfamilies) than seedling (seedling vs. zygote P = 5 × 10−10, Tukey tests) (Fig. 1B,C). Like the egg cell and unlike the sperm cell, the zygote had a low abundance of siRNAs overlapping Gypsy retrotransposons (sperm vs. zygote P < 2.2 × 10−16, Tukey tests) (Fig. 1B,C). However, whereas the zygote and egg cell were similar, there were some clear differences. Zygote had significantly more siRNAs overlapping TIR elements and significantly less siRNAs overlapping Gypsy retrotransposons than the egg cell (P = 1.4 × 10−3 and P = 9 × 10−7, respectively, Tukey tests) (Fig. 1C).
The similarity between egg and zygote siRNA profiles can be explained by carryover from the egg cell, because the egg cell is ∼1000-fold larger than the sperm cell by volume (Kranz Bautor and Lörz 1991; Anderson et al. 2013; Li et al. 2019a). Although 24-nt siRNAs function in the nucleus, 24-nt siRNAs were found primarily in the cytoplasm of whole-plant homogenates (Ye et al. 2012). Thus, we predict that small RNAs already present in the egg cell before fertilization would contribute to much of the siRNAs present in the zygote. This is consistent with previous observations that the 50 most highly expressed genes in egg cell remained as most highly expressed in zygote, whereas the 50 most highly expressed genes in the sperm cell were lowly expressed in the zygote (Anderson et al. 2013, 2017). Indeed, 13 out of the 20 most abundant miRNAs in egg cells remained among the 20 most abundant miRNAs in zygote (P = 3 × 10−14, Fisher's exact test) (Fig. 1D). However, the miRNA accumulation patterns were not identical between zygote and egg. Thirty-two miRNAs were detected in the zygote but not in the egg cell (>50 reads per million miRNA reads in zygote and undetected in egg cell), and seven miRNAs were detected in the egg cell but not in the zygote (>50 reads per million miRNA reads in egg cell and undetected in zygote). The presence of 32 miRNAs detected in zygote but not egg cell suggests that ZGA is initiated at miRNA loci at this stage, which would be consistent with the known ZGA of other RNA polymerase II transcripts. Meanwhile, top sperm-enriched miRNAs were very much down-regulated in the zygote, consistent with dilution after fertilization (Fig. 1E; Supplemental Fig. S1C). Note that the expression values in the zygote were not used to define these sperm-enriched miRNAs, as we classified sperm-enriched miRNAs relative to egg alone. Specifically, we required >1000 reads per million miRNA reads in sperm, and <500 reads per million miRNA reads in the egg cell for this classification. The expression values of the full set of expressed miRNA genes (miRBase v22) (Kozomara et al. 2019) are provided as a complementary transcriptomics resource (Supplemental Data Set 1). Taken together, these results imply that sperm small RNAs were diluted by the egg cell cytoplasm and that much of the siRNAs detected in the zygote were caused by carryover from the egg cell.
Unusual siRNA loci with abundant siRNAs in egg, ovary, zygote, and endosperm
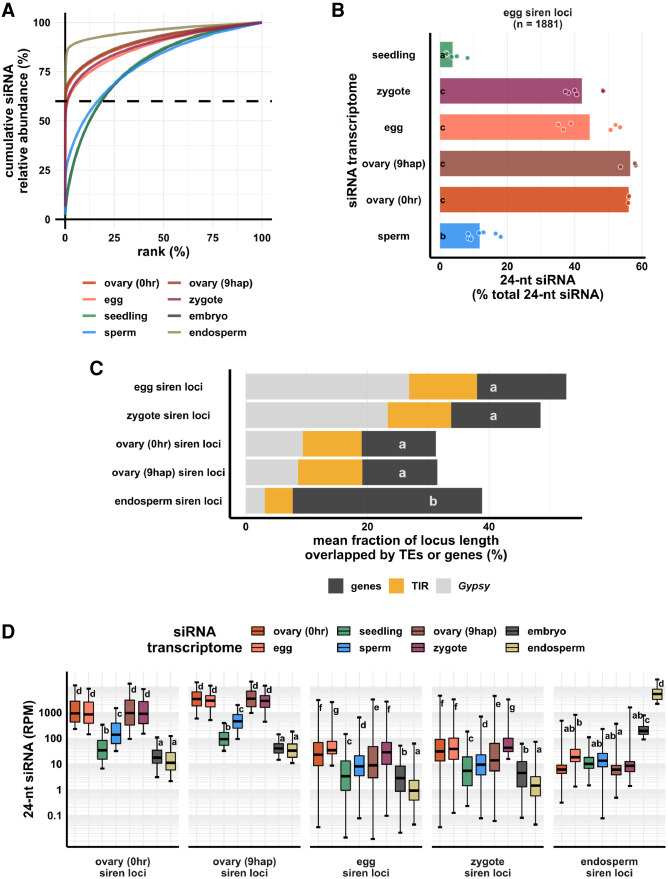
The zygote, egg cell, and ovary displayed an unusual set of highly abundant siRNAs that appeared to be concentrated at discrete sites across the genome, without a clear relationship to gene density (Fig. 1A). It has been previously reported that rice developing endosperm (7–8 d after fertilization) has a unique siRNA profile in which a small number of loci accounts for the majority of siRNAs (Rodrigues et al. 2013). These siRNA loci were termed siren loci (siRNA in the endosperm). A similar phenomenon was recently reported in Brassica rapa and Arabidopsis ovules and endosperm (Grover et al. 2020). The term “siren loci” was also used by Grover et al. to describe these loci. To further investigate this phenomenon in zygote as well as egg, ovary, and endosperm, we defined siRNA loci from these cell types/tissues using Shortstack (Axtell 2013). We ranked siRNA loci according to siRNA abundance in each cell type (Fig. 2A). In endosperm and ovaries (pre- and postfertilization), ∼0.1% (n = 73, 213 and 102, respectively) of the siRNA loci accounted for 60% of the total siRNA accumulation in all siRNA loci for each tissue type (Fig. 2A). Similarly, in egg cell and zygote, ∼1% (n = 1881 and 1429, respectively) of the siRNA loci accounted for 60% of the total siRNA accumulation in all siRNA loci for each cell type (Fig. 2A). We call these highly expressing loci siren loci, independently of siRNAs in endosperm. In fact, the siren loci in rice ovaries, egg, and zygote showed little correlation with the siren loci reported in rice endosperm, at least for the specific endosperm stage described; that is, 7–8 d after fertilization (Rodrigues et al. 2013; Li et al. 2020). Egg siren siRNAs did not show a significant difference in relative abundance between egg and zygote, without a significant decrease after fertilization (Fig. 2B), a factor that was taken into account in subsequent analysis (see below).
Figure 2.
Zygote siren loci are similar to siren loci detected in ovary and egg cell, and stably expressed between egg and zygote, but dissimilar to siren loci detected in endosperm. (A) x-axis is the rank order of siRNA loci. siRNA loci with highest siRNA abundances are ranked first. y-axis is cumulative relative abundance of siRNA in all siRNA loci. Axis values are scaled between 0% and 100%. 0.1% of siRNA loci accounted for 60% of siRNA reads in all siRNA loci in endosperm and ovary; 1% of siRNA loci accounted for 60% of siRNA reads in all siRNA loci in egg and zygote. (B) Bar plot showing relative abundances of 24-nt siRNAs at egg siren loci. Each data point is an siRNA transcriptome. Bar heights are averages. x-axis values are relative to total 24-nt siRNAs. (C) Stacked bar plots showing mean fraction of locus length overlapped by TEs or genes. (TIR) Terminal inverted repeat transposons, CACTA superfamily excluded; (Gypsy) Gypsy retrotransposons. (D) Box plots showing 24-nt siRNA relative abundances across siren classes across cell types. Middle lines are medians. Whiskers span 2.5th and 97.5th percentiles. Boxes span interquartile range. y-axis values are relative to per million total 24-nt siRNAs in each siRNA transcriptome. Letter grouping (α = 0.05) and P-values are based on Tukey tests. Embryo and endosperm data from Rodrigues et al. (2013). Seedling, gametes, and pre-fertilization ovary data from Li et al. (2020).
Next, we compared the similarity among different siren loci categories based on their genomic distributions (Supplemental Fig. S2). On a principal component (PC) plot, endosperm siren loci were well separated from all the others along PC1, which accounts for 62% of the variance in their genomic distributions. The rest of the siren loci categories were separated along PC2, which accounts for 27% of the variance, much less than what was explained by PC1. Egg and zygote siren loci also had nearly the same genomic distribution, clustering closely together (Supplemental Fig. S2). All siren categories have distinct genomic distributions from distributions of genes (Supplemental Fig. S2). Consistent with its unique genomic distribution, endosperm siren loci were more likely to overlap a gene (Fig. 2C). On average, ∼30% of the locus length of an endosperm siren locus was covered by a gene, whereas ∼15% of the locus length for all the other siren categories was covered by a gene (∼13%, P < 1.4 × 10−4, Tukey tests). Lastly, we compared the relative abundances of 24-nt siRNAs at different siren categories across different cell types. At endosperm siren loci, endosperm had the highest 24-nt siRNA expression, ∼10-fold higher than the level in embryo and more than 100-fold higher than the levels in all other cell types we examined (Fig. 2D). In contrast, the other siren classes shared an siRNA accumulation pattern across cell types (Fig. 2D). Ovaries (pre- and postfertilization), egg cell, and zygote all had high abundances of 24-nt siRNAs at ovary/egg/zygote siren loci, consistent with the stable expression of egg siren siRNAs in zygote (Fig. 2B), whereas seedling, sperm, embryo, and endosperm all had lower abundances of 24-nt siRNAs at these siren loci. Taken together, these distinct siRNA accumulation patterns reveal that zygote siRNAs were concentrated at discrete sites similar to egg and ovary and that the persistence of egg siren siRNAs explains the overall similarity between zygote and egg (Fig. 1A).
The genome distribution pattern of zygote 24-nt siRNA loci is distinct from egg
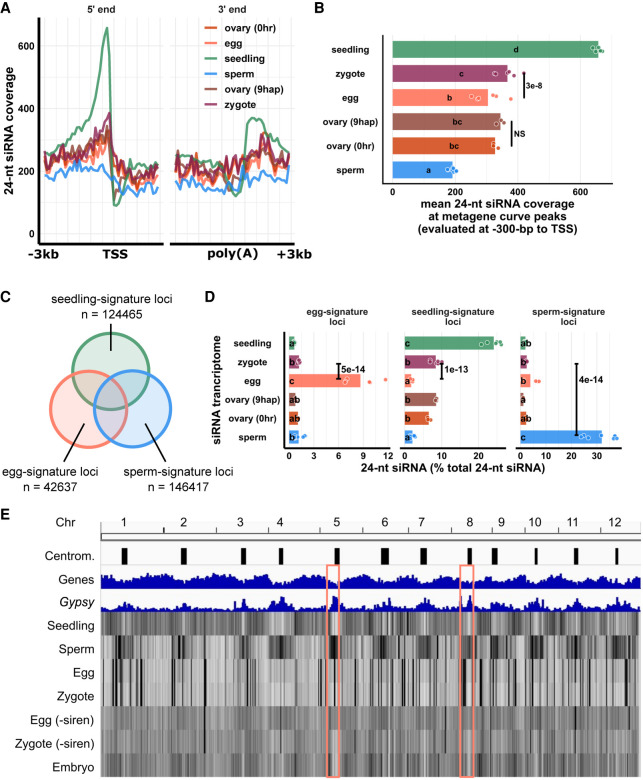
Although the siren siRNAs result in similarity of the zygote siRNA profile in the zygote to that of the egg cell in terms of overall patterns of abundance, a deeper analysis revealed significant differences from the egg cell. We produced metagene siRNA coverage plots for seedling, gametes, and zygote, as well as pre- and postfertilization ovaries (Fig. 3A). Seedling had a strong peak upstream of the transcription start site (TSS), corresponding to where short transposons are enriched in the genome, such as non-CACTA TIR transposons (Han et al. 2013), and such a peak was absent in gametes and ovaries. Zygote had a significant increase in 24-nt siRNA coverage at the peak of the metagene curve relative to egg cells (P = 3 × 10−8, Tukey tests) (Fig. 3A,B). In contrast, there were no significant changes between pre- and postfertilization ovaries (P = 0.98, Tukey tests) (Fig. 3A,B). Thus, the differences between zygote and egg could not be because of trafficking of the newly transcribed siRNAs from ovary. To analyze the abundance of siRNAs at the level of individual genomic loci, we classified seedling-signature loci as seedling siRNA loci that did not overlap any egg siRNA loci or sperm siRNA loci (seedling loci ∉ egg loci ∉ sperm loci) (Fig. 3C). Overlapping siRNA loci were defined as at least 1-bp overlap in genomic coordinates (see also Methods). Likewise, we classified sperm-signature loci as sperm siRNA loci that did not overlap any egg or seedling siRNA loci (sperm loci ∉ egg loci ∉ seedling loci) (Fig. 3C), and lastly, egg-signature loci as egg siRNA loci that did not overlap any seedling or sperm siRNA loci (egg loci ∉ seedling loci ∉ sperm loci) (Fig. 3C). At egg-signature loci, zygote experienced a 10-fold reduction of 24-nt siRNAs (P = 5 × 10−14, Tukey tests) (Fig. 3D). At seedling-signature loci, zygote had 4.7-fold more 24-nt siRNAs than egg cell (P = 1 × 10−13, Tukey tests) (Fig. 3D). Gaining siRNAs at gene-proximal regions and seedling-signature loci is consistent with an increase of TIR siRNAs in zygote (Fig. 1C). Because these seedling-signature loci did not overlap any egg siRNA loci or sperm siRNA loci, the increase of 24-nt siRNAs at seedling-signature loci in zygote was unlikely caused by carryover from either gamete. At sperm-signature loci, zygote had very few 24-nt siRNAs (zygote vs. sperm P = 4 × 10−14, Tukey tests) (Fig. 3D), much like the results for miRNAs (Fig. 1E), confirming small RNA contribution from sperm cell is very limited relative to egg. There was little difference in the ovaries before and after fertilization for any of these locus categories (P = 0.76, P = 0.84, and P = 0.84 at egg-, seedling-, and sperm-signature loci, respectively) (Fig. 3D). It is important to note that the zygote siRNA transcriptome was not used to define these locus categories. Lastly, we bioinformatically removed siren siRNAs from egg and zygote libraries (Fig. 3E) and re-analyzed their genome-wide 24-nt siRNA distributions. This analysis revealed that, outside of the siren loci (which as defined previously constitute ∼1% of all 24-nt siRNA loci), zygote and egg were indeed distinct from each other in genome-wide 24-nt siRNA distribution (P < 2.2 × 10−16, Tukey tests) (Fig. 3E; Supplemental Fig. S3F). Egg cell has a slight enrichment of 24-nt siRNAs at centromeric regions, whereas zygote showed a relative depletion of siRNAs at centromeric regions (egg vs. zygote P < 2.2 × 10−16, Tukey tests) (Supplemental Fig. S3G), much like embryo (zygote vs. embryo P = 0.5, Tukey tests) (Fig. 3E, pink boxes indicate two examples; see also Supplemental Fig. S3G). Taken together, these results indicate that the zygote has an siRNA transcriptome that is distinct from that of the egg cell and further, that the changes from egg cell to zygote were independent of postfertilization changes in the ovary.
Figure 3.
Changes in the zygote siRNA transcriptome after fertilization are independent from the ovary. (A) Metagene coverage plot for 24-nt siRNAs. Coverage is measured over 100-bp intervals and normalized per 1000 24-nt siRNAs. Vertical grid lines are 500-bp intervals. TSS transcription start site, poly(A) polyadenylation site. (B) Quantification of A at the interval from 300 to 200 bp upstream of TSS, corresponding to the peaks of metagene curves. Each data point is an siRNA transcriptome and bar heights are averages. x-axis values are normalized per 1000 24-nt siRNAs. (C) Venn diagram illustrating egg-signature loci (egg siRNA loci that do not overlap any seedling or sperm siRNA loci), seedling-signature loci (seedling siRNA loci that do not overlap any egg or sperm siRNA loci), and sperm-signature loci (sperm siRNA loci that do not overlap any egg or seedling siRNA loci). Sizes of overlap in Venn diagrams are not to scale. (D) Bar plot showing relative abundances of 24-nt siRNA across siRNA loci categories defined in C. The zygote siRNA transcriptome was not used to define these locus categories. Each data point is an siRNA transcriptome. Bar heights are averages. x-axis values are normalized to total 24-nt siRNAs. (E) Heat map showing abundance of 24-nt siRNA across the genome at 50-kb resolution. The first three tracks are centromeres (as defined by Mizuno et al. 2018), genes, and Gypsy retrotransposons. “−siren” refers to siren siRNAs removed. Pink boxes highlight examples of centromeric regions. Letter grouping (α = 0.05) and P-values are based on Tukey tests. Zygote and 9-hap ovary data are from this study; embryo (7–8 DAF) from Rodrigues et al. (2013); all other data from Li et al. (2020).
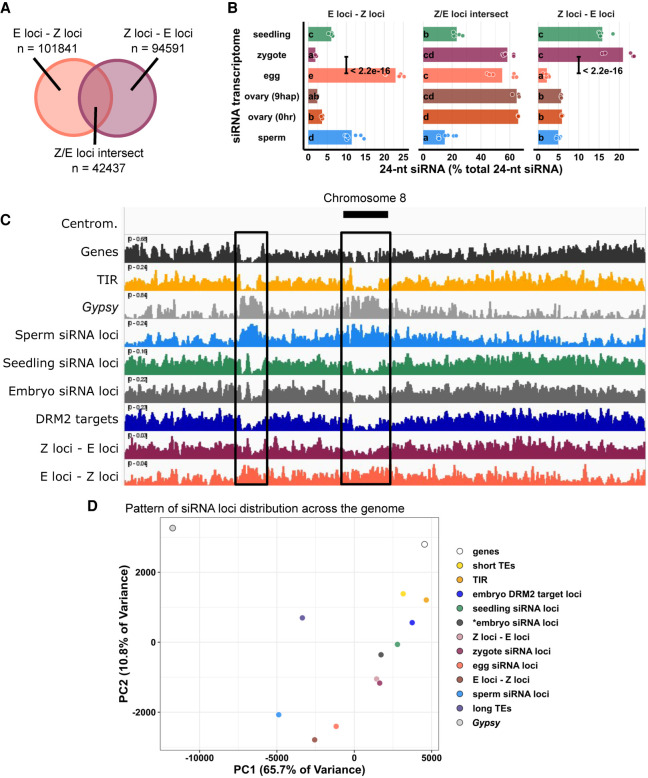
To further characterize the differences between the zygote siRNA transcriptome and that of the egg cell, we next defined zygote siRNA loci using Shortstack with zygote siRNAs. We then classified Z-E loci as zygote siRNA loci that did not overlap any egg cell siRNA loci (Z loci ∉ E loci in set operation), E-Z loci as egg siRNA loci that did not overlap any zygote siRNA loci (E loci ∉ Z loci), and Z/E loci intersect as zygote siRNA loci that overlapped egg siRNA loci (Z loci ∩ E loci) (Fig. 4A). Despite the similarities between egg and zygote at the high abundance siRNA loci (Figs. 1A, 2B), widespread distinct siRNA loci were detected in one cell type but not the other. There were 101,841 E-Z loci (newly diminished siRNA loci in zygote), 94,591 Z-E loci (newly detected siRNA loci in zygote), but only 42,437 Z/E loci intersect.
Figure 4.
Widespread distribution of newly detected zygote siRNA loci across the rice genome. (A) Venn diagram illustrating E-Z loci (egg siRNA loci that do not overlap any zygote siRNA loci, E loci ∉ Z loci), Z-E loci (zygote siRNA loci that do not overlap any egg siRNA loci, Z loci ∉ E loci), and Z/E loci intersect (zygote siRNA loci that overlap egg siRNA loci, Z loci ∩ E loci). Sizes of overlap in Venn diagrams are not to scale. (B) Quantification of 24-nt siRNA relative abundances for A. Each data point is an siRNA transcriptome. Bar heights are averages. x-axis values are relative to total 24-nt siRNA reads. Letter grouping (α = 0.05) and P-values are based on Tukey tests. (C) Distribution of siRNA loci along a chromosome. Chromosome 8 is chosen because it is one of the chromosomes with a completed sequenced centromeric region (Mizuno et al. 2018). (Centrom) Centromeric regions, black boxes highlight regions with abundant Gypsy elements and relative depletion of genes, (TIR) seedling siRNA loci, embryo siRNA loci, DRM2 targets, and Z-E loci. (D) Principal component plot showing siRNA loci distribution across the genome. Distributions are evaluated at 50-kb resolution across the genome. Each data point is the distribution of a loci category. Short TEs are TE of lengths between 50 bp and 250 bp. Long TEs are TEs of length >250 bp. Zygote and 9-hap ovary data are from this study; all other data from Li et al. (2020).
When 24-nt siRNA reads at individual loci were tallied and normalized to total 24-nt siRNAs, as expected, we found that at E-Z loci, egg had ∼10-fold more 24-nt siRNAs than zygote (P < 2.2 × 10−16, Tukey tests) (Fig. 4B); at Z-E loci, zygote had ∼10-fold more 24-nt siRNAs than egg (P < 2.2 × 10−16, Tukey tests) (Fig. 4B); and no difference at Z/E loci intersect. There were also siRNAs not captured by siRNA loci. These siRNAs resided at genomic regions with insufficient siRNAs and did not meet the 0.5 RPM threshold for assignment as loci by Shortstack (see also Methods), which explains the small number of egg siRNAs at Z-E loci and the small number of zygote siRNAs at E-Z loci. There were no differences between ovaries before and after fertilization in any of the three locus categories (Fig. 4B), again suggesting changes in the zygote siRNA transcriptome were not coupled with the ovary (Fig. 3). In addition, the abundance of seedling siRNAs in Z-E loci and scarcity in E-Z loci revealed the emergence of a seedling-like siRNA pattern in zygote (Fig. 4B). Because the seedling siRNA transcriptome was not used to classify Z-E loci, this observation suggests that the zygote had initiated a return to the canonical siRNA profile, consistent with the increase in 24-nt siRNAs from TIR transposons (Fig. 1C) as well as at gene-proximal regions (Fig. 3A,B) and seedling-signature loci in zygote (Fig. 3D).
During ZGA of mRNA transcriptomes, genes expressed in zygote but not in egg cell all had initially low expression relative to a background of abundant maternal transcript carryover (Anderson et al. 2017; Chen et al. 2017; Zhao et al. 2019). Thus, if the siRNA transcriptome transitions similarly in the zygote, one would expect to see an initial widespread detection of low abundance 24-nt siRNAs at new loci, relative to a background of more abundant maternal carryover siRNAs corresponding to egg siren loci. Indeed, in contrast to the high abundance siRNAs of intersect loci, Z-E loci and E-Z loci overall had lower siRNA abundances than zygote/egg intersect loci (Fig. 4B). Nevertheless, on average, one in every five zygote 24-nt siRNAs (∼20%) resided at Z-E loci in the zygote. Together with the numerical abundance of Z-E loci (70% of all zygote loci), these results suggest that newly detected siRNA loci in zygote are widespread and explain a substantial fraction of 24-nt siRNAs in zygote.
The highly expressed siren loci in egg and zygote raise the concern of whether the apparent up-regulation of Z-E loci could be explained by down-regulation of egg siren loci. Consistent with the stable expression of egg siren loci in zygote, including or excluding siren siRNAs did not change the results of analyses (Figs. 3A–D, 4A,B; Supplemental Fig. S3), supporting the distinct distributions of nonsiren 24-nt siRNAs in egg and zygote (Fig. 3E). Taken together, changes in the zygote siRNA transcriptome are not explained by down-regulation of egg siren siRNAs but are because of up- and down-regulation of other siRNAs that are widely distributed across the genome.
Newly detected siRNA loci in zygote diverge from gamete siRNA loci and resemble canonical siRNA loci in genomic location and DNA methylation
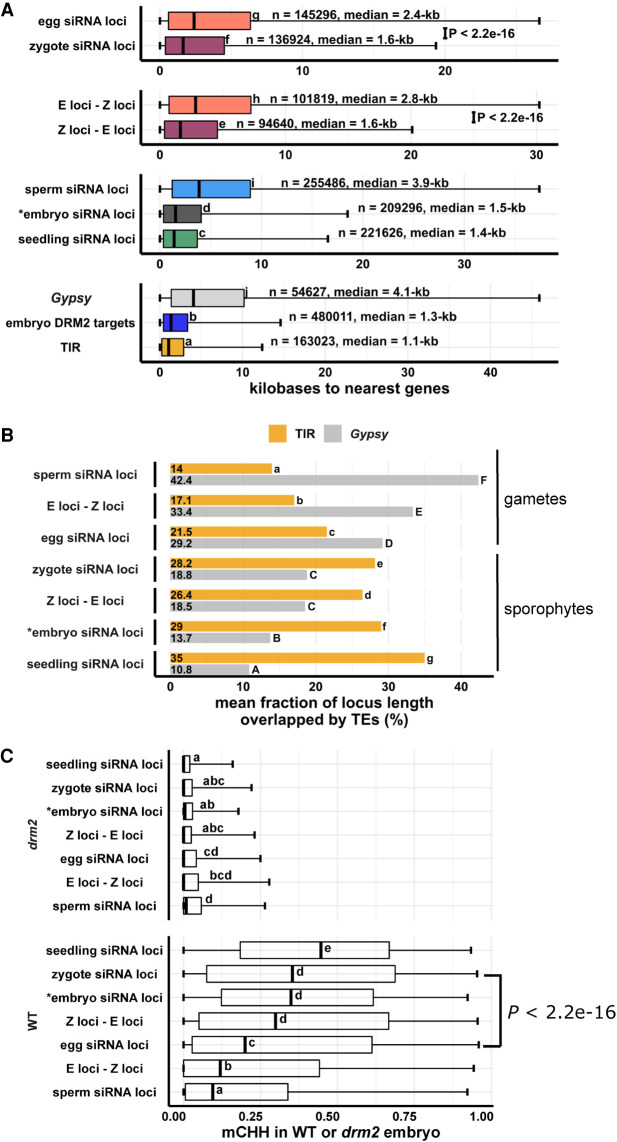
To investigate the patterns and characteristics of zygote siRNA loci, we compared the genomic distribution of zygote siRNA loci and Z-E loci against a set of different siRNA loci categories, including E-Z loci, egg siRNA loci, embryo siRNA loci (Rodrigues et al. 2013), seedling siRNA loci, and sperm siRNA loci. Our efforts to generate robust DNA methylome profiles for zygotes were not successful, possibly because zygotes are fragile compared with egg cells, and the methylation level readout from random-priming based, post-bisulfite adapter tagging (PBAT) sequencing methods are highly sensitive to library preparation conditions (Li et al. 2019b). We have previously shown that the sperm and egg methylomes are similar and that gamete-specific 24-nt siRNA loci are associated with sites of DDM1-dependent methylation and not DRM2-dependent methylation (Li et al. 2020). To determine whether the zygote siRNAs were correlated with DRM2-dependent methylation during embryogenesis, we compared mCHH between mature wild-type and drm2 embryos in a set of DRM2-dependent methylation sites previously identified in the embryo (Li et al. 2020). As reference points in the genome-wide analysis, we also included genes, TIR and Gypsy TEs (Kawahara et al. 2013). We observed that the genome-wide distribution of Z-E loci follows the distribution of short TIRs, as well as embryo DRM2 targets, and resembles the distribution of seedling and embryo siRNA loci (Fig. 4C). In contrast, the E-Z loci representing the newly diminished loci in zygote had a distinct pattern, more similar to sperm siRNA loci, which are predominantly heterochromatic (Fig. 4C). There was a relative depletion of siRNA loci from centromeric regions for zygote siRNA loci, much like canonical siRNA loci, and unlike egg siRNA loci (P = 5 × 10−13, Tukey tests) (Supplemental Fig. S4A). Consistent with a more similar distribution to canonical siRNA loci, zygote siRNA loci and Z-E loci had higher degrees of overlap with seedling siRNA loci and embryo DRM2 targets, whereas egg siRNA and E-Z loci had low degrees of overlap, as did sperm siRNA loci (Supplemental Fig. S4B). Zygote siRNA loci and Z-E loci overlapped larger numbers of DRM2 targets per Mb genome space, much like seedling siRNA loci, and unlike egg siRNA loci, E-Z loci, or sperm siRNA loci (Supplemental Fig. S4C).
To gain more information on the factors underlying the variation in siRNA loci distributions, we used principal component analysis (PCA) to cluster the genomic distributions of the above locus categories based on their abundance in 50-kb windows genome-wide (Fig. 4D). The separation of locus categories along PC1 had a near-perfect rank order correlation with median distance to nearest genes (rho = −0.97, P < 2.2 × 10−16) (Supplemental Fig. S4D), and PC2 was correlated with median length of locus categories (rho = 0.91, P < 2.2 × 10−16) (Supplemental Fig. S4E). PC1, which explained 65.7% of variance in genomic distributions across loci categories, was strongly correlated with various aspects of rice genome organization. PC1 was strongly correlated with TIR overlap as well as mCHH level in wild-type embryo (Supplemental Fig. S6A,C). PC1 was also strongly anticorrelated with Gypsy overlap and mCG and, to a lesser extent, mCHG in wild-type embryo (Supplemental Fig. S6B,D,E). These genomic features are mutually correlated (Supplemental Fig. S6F), consistent with the prior understanding of cereal genome organization (Gent et al. 2013; Han et al. 2013). In plants, RdDM occurs more frequently at short and fragmented TEs than in long or intact ones (for review, see Erdmann and Picard 2020). As expected, given the abundance of TIR elements that exist as truncated MITEs (Han et al. 2013), we found that the genomic distribution of short TEs (50–250 bp) closely mirrors the distribution of non-CACTA TIR elements and that of long TEs (>250 bp) closely mirrors the distribution of Gypsy elements (Fig. 4D; Supplemental Fig. S5A). CACTA TIR elements, which we excluded from other TIR analysis because of their more heterochromatic distribution than the other TIR elements, have an intermediate length distribution (Supplemental Fig. S5B). Short TEs and long TEs clustered as expected with TIRs and Gypsy elements in the PCA comparison with siRNA loci (Fig. 4D).
Strong correlations between PC1 (gene proximity), TE overlap, and DNA methylation led us to statistically assess the differences of these attributes among siRNA loci categories. TIR and short TEs were gene-proximal, consistent with the gene-proximal distribution of embryo DRM2 targets (Fig. 5A; Supplemental Fig. S5C). Canonical siRNA loci, such as seedling siRNA loci and embryo siRNA loci, were closer to genes than noncanonical siRNA loci, such as sperm siRNA loci (Fig. 5A). E-Z loci, the newly diminished siRNA loci in zygote, were on average much farther away from genes than Z-E loci were (2.8 kb vs. 1.6 kb, P < 2.2 × 10−16, Tukey tests) (Fig. 5A), consistent with their heterochromatic genomic distributions (Fig. 4C,D; Supplemental Fig. S4A). Total zygote siRNA loci were closer to genes than total egg siRNA loci (P < 2.2 × 10−16, Tukey tests). From egg cell to zygote, there was a 30% decrease in median distance (2.4 kb vs. 1.6 kb). In contrast, from zygote to embryo (7 d after fertilization) (data from Rodrigues et al. 2013), there was a 6% decrease (1.6 kb vs. 1.5 kb). In gametes, siRNA loci were more likely to overlap a Gypsy retrotransposon than a TIR transposon (Fig. 5B). However, in sporophytes, including zygote itself, siRNA loci are more likely to overlap a TIR transposon than a Gypsy retrotransposon (Fig. 5B). Consistent with this, in gametes, siRNA loci were on average far more overlapped by long TEs than by short TEs (Supplemental Fig. S5D). However, as the zygote is formed from gametes and sequentially develops into embryo and seedling, long TE overlaps steadily decreased whereas short TE overlaps steadily increased (Supplemental Fig. S5D). These observations are consistent with the results where zygote had more TIR siRNAs and less Gypsy siRNAs (Fig. 1C) and more gene-proximal 24-nt siRNAs than egg cell (Fig. 3A,B), zygote had increased siRNAs in seedling-signature loci (Fig. 3C), and seedling had comparable siRNA level with zygote in Z-E loci (Fig. 4B).
Figure 5.
Newly detected siRNA loci in zygote reset to the canonical siRNA profile and predict CHH methylation in embryo in an RdDM-dependent manner. (A) Box plots showing distance of siRNA loci to nearest genes. Middle lines are medians. Boxes span interquartile range. Whiskers span 2.5th and 97.5th percentiles. (B) Bar plots showing mean locus length overlapped by TIR transposon or Gypsy retrotransposons across siRNA loci categories. Statistical comparisons are made across siRNA loci categories within a TE length category. (C) Box plots showing CHH methylation level in mature wild-type and drm2 mutant embryos. Middle lines are medians. Boxes span interquartile range. Whiskers span 2.5th and 97.5th percentiles. E-Z loci: n = 101,841, Z-E loci: n = 94,591 (69% of all zygote siRNA loci). Letter groupings (α = 0.05) and P-values are based on Tukey tests. (*) Embryo siRNA data from Rodrigues et al. (2013), which was based on a single replicate. Seedling, gametes, and prefertilization ovary data from Li et al. (2020).
Resetting to a canonical siRNA pattern might suggest that the newly detected siRNA loci in the zygote are targeted for CHH methylation during embryogenesis in an RdDM-dependent manner. We compared DNA methylation levels across different siRNA loci categories in mature wild-type and drm2 embryos (Fig. 5C; see also Supplemental Fig. S7). Although all siRNA loci categories were associated with RdDM during embryogenesis, as median mCHH levels were all higher in wild-type embryo than drm2 embryo, zygote siRNA loci had a much higher level of mCHH than egg siRNA loci did in wild-type embryo (median mCHH level 0.35 vs. 0.2, P < 2.2 × 10−16, Tukey tests) (Fig. 5C), much like embryo and seedling siRNA loci. Newly detected siRNA loci (as represented by Z-E loci) had high mCHH levels in wild-type embryo (median mCHH level 0.3, P < 2.2 × 10−16, Tukey tests), whereas newly diminished siRNA loci (E-Z loci) had low mCHH levels in wild-type embryo (median mCHH level 0.11). In addition, whereas there were smaller differences in mCHG and especially in mCG across locus categories in wild-type embryo when compared with mCHH, the full extent of methylation across siRNA loci categories also depended on DRM2 for all three contexts (Supplemental Fig. S6). It is important to note that zygote siRNA loci had high degrees of overlap with canonical RdDM loci (seedling siRNA loci and embryo DRM2 targets) (Supplemental Fig. S4B) and that zygote siRNA loci overlapped a large number of DRM2 targets per Mb genome space (Supplemental Fig. S4C). Together with an elevated mCHH level in embryo, these results suggest that newly detected zygote siRNA loci mark canonical siRNA loci that will undergo hypermethylation during embryogenesis, rather than defining hypermethylated regions that are distinct from canonical siRNA loci.
DNA methylomes of rice zygote based on a random-priming PBAT sequencing method were recently published (Zhou et al. 2021). We examined the methylation levels across siRNA locus categories in zygote and found that the patterns were consistent with mature embryo (Supplemental Fig. S8). Specifically, in the zygote methylome, zygote siRNA loci had higher mCHH than egg siRNA loci (median mCHH level 0.037 vs. 0.02, P < 2.2 × 10−16, Tukey tests). Z-E loci also had higher mCHH than E-Z loci (median mCHH level 0.014 vs. 0, P < 2.2 × 10−16, Tukey tests) (Supplemental Fig. S8A). The PBAT methylomes from egg in the Zhou et al. study yielded lower methylation values compared with other studies (Park et al. 2016; Li et al. 2020), raising the possibility of systemic underestimation of methylation levels because of the extreme low input or other factors, which could also affect the zygote results (Supplemental Fig. S9). The fact that we are comparing methylation levels between loci rather than between methylomes, however, provides an internal control. Although possibly not quantitative, the differences in zygote mCHH between zygote siRNA loci and egg siRNA loci do suggest that the newly detected 24-nt siRNAs in zygote can direct DNA methylation in the zygote.
Lastly, the fact that siren loci were defined by abundant 24-nt siRNAs (Fig. 2A) led us to speculate that they would have high mCHH. In fact, we found that was not the case (Supplemental Figs. S11, S12). In ovary, siren loci had an mCHH level comparable with that of canonical siRNA loci (seedling siRNA loci) and siRNA loci that were not siren loci (Supplemental Fig. S11). Similar results were found for egg cell, zygote, and embryo as well (Supplemental Figs. S11B, S12A,B). At siren loci as well as siRNA loci that were not siren loci, DRM2 was required for mCHH in embryo (Supplemental Fig. S12B). Lastly, both wild-type and drm2 endosperm had overall low mCHH methylation including in endosperm siren loci (Supplemental Fig. S12C). Unlike mCHH, mCG and mCHG did not produce any notable pattern in siren loci across cell types (Supplemental Figs. S11, S12). Taken together, these results suggest that, although the highly abundant siRNAs produced by siren loci may also target DNA methylation in cis in a DRM2-dependent manner, they do so inefficiently compared with siRNAs from canonical RdDM loci.
Discussion
The parental gametes have unequal contributions to the zygote siRNA transcriptome in rice
The zygote is an indispensable stage of the plant life cycle, in which the genomes of the differentiated gametes have to be reprogrammed to specify a totipotent cell that will regenerate a new plant (McClintock 1984). However, the epigenetic changes that accompany this critical transition are poorly understood, because of the technical challenges of working with a cell type that presents major difficulties arising from the highly limiting and fragile nature of the material. We present here the successful characterization of the small RNA transcriptome of plant zygotes. The results have notable implications for the current models of postfertilization silencing through the male germline. It has been proposed that sperm-transmitted siRNAs regulate TEs and balance parental contribution in the endosperm, as RdDM-mutants affect endosperms from 2n maternal × 4n paternal crosses (Erdmann et al. 2017; Borges et al. 2018; Martinez et al. 2018; Satyaki and Gehring 2019) in Arabidopsis. A recent publication found that siRNAs are trafficked from tapetum into meiocytes to repress TEs and genes via DNA methylation (Long et al. 2021). Our findings that the zygote is depleted for sperm-signature siRNAs (Fig. 3D) as well as sperm-enriched miRNAs (Fig. 1E; Supplemental Fig. S1C) indicate that there is negligible paternal contribution to the zygote small RNA transcriptome. Thus, at least in rice embryogenesis, any proposed effects of sperm-transmitted siRNAs on embryos are likely to be indirect. Assuming sperm-derived siRNAs are also diluted by the larger central cell, we speculate that the effect of the sperm-transmitted siRNAs may act through sperm chromatin modifications and not siRNAs themselves. In the newly formed endosperm, sperm-derived histone variants are passively diluted through nuclear divisions (Ingouff et al. 2007, 2010). In contrast, in the zygote, histone variants are actively replaced in a replication-independent manner before the first embryonic division (Ingouff et al. 2007, 2010).
A special class of highly abundant maternal siRNAs persists in the zygote
A small number of loci accounted for most of the siRNAs in egg cells and zygotes (Fig. 2A). Egg siren siRNAs remained similarly highly expressed in the zygote (Fig. 2B). Thus, up-regulation of siRNAs at newly detected zygote siRNA loci cannot be explained by large down-regulation of egg siren loci (Supplemental Fig. S3). In Arabidopsis and Brassica rapa (Grover et al. 2020), siren loci detected in ovules are also highly expressed in the endosperm; however, siren loci in rice ovary are distinct from those detected in rice endosperm (Fig. 2C,D; Supplemental Fig. S2). Siren loci in the zygote were distinct from endosperm siren loci in endosperm collected 7–8 d after fertilization, instead coinciding with siren loci detected in ovary and egg cell (Fig. 2C,D). However, it remains possible that the central cell and earlier stages of endosperm have an siRNA transcriptome more like that of the zygote. It has been proposed that the Arabidopsis embryo receives siRNAs from the endosperm (Hsieh et al. 2009; Martinez and Köhler 2017). This does not appear to be the case in 7- to 8-d rice seeds, because rice embryos had low siRNA abundance at endosperm siren loci at this stage (Rodrigues et al. 2013). It also appears that the Brassica napa embryo directs its own methylation independently of endosperm siRNAs (Chakraborty et al. 2021). A recent publication demonstrated that trans-acting siRNAs from ARFs (tasiR-ARF) traffic across ovule cell layers to regulate megaspore mother cell (MMC) identity in Arabidopsis (Su et al. 2020). Arabidopsis also has siren RNAs in female reproduction, at least in the ovule (Zhou et al. 2020). It has also been proposed that siRNAs may traffic from the seed coat into the embryo during seed development (Grover et al. 2018, 2020). Likewise, it is possible that siren siRNAs in the egg cell and zygotes are produced in the ovary tissue instead. Although siRNAs at siren loci may direct some CHH methylation in cis in ovary or during embryogenesis, they appeared to have an mCHH level comparable with if not lower than that of their nonsiren siRNA loci counterparts (Supplemental Figs. S11, 12). A recent preprint reported that, in B. rapa ovules, a fraction of siren siRNAs can silence genes with sequence homology in trans via DNA methylation (Burgess et al. 2021). We cannot exclude that the same process might also occur in rice egg cells and zygotes, regardless of the cell type of origin of siren siRNAs.
The 24-nt siRNA landscape of rice zygotes reveals resetting toward a canonical siRNA pattern
We detected widespread new zygote siRNA loci relative to the egg cell, representing ∼69% of all zygote siRNA loci (Fig. 4A). In relative abundance, most of the siRNA reads were accounted for by egg siRNA carryover and stably expressed egg siren siRNAs (Fig. 2B), and thus siRNA abundance was lower at Z-E loci than at Z/E intersect loci that contain the siren siRNAs (Fig. 4B). However, the relative abundance of the zygote siRNAs at Z-E loci (∼20%) is similar to that of egg cell siRNAs at E-Z loci (∼22.5%) (Fig. 4B), but they differ significantly in their genome-wide distribution, as discussed below.
Several lines of evidence indicate the zygote has initiated a resetting toward the canonical siRNA pattern and that such resetting is independent from the ovary. First, the zygote had increased 24-nt siRNAs from TIR and short transposons and decreased siRNAs from Gypsy and long transposons, as compared with the egg cell (Fig. 1C). Second, zygote had increased 24-nt siRNAs at seedling-signature loci (Fig. 3D), as compared with the egg cell. In contrast, there were no significant changes to the 24-nt siRNAs at seedling-signature loci in ovaries pre- and postfertilization (Fig. 3D). Moreover, seedling had 24-nt siRNAs comparable with those of zygote at Z-E loci (Fig. 4B). As the zygote siRNA transcriptome was not used to define seedling-signature loci, and the seedling siRNA transcriptome was not used to define Z-E loci, these results serve as an objective indication that the zygote shifted toward a more seedling-like siRNA transcriptome. Third, zygote had increases relative to the egg cell in 24-nt siRNAs at the TSS region upstream of genes, while there was lack of a corresponding change in the ovary after fertilization (Fig. 3A,B). High 24-nt siRNA coverage upstream of genes around the TSS is a feature of a canonical siRNA transcriptome, as exemplified by seedling (Fig. 3A). Fourth, the genomic distribution of Z-E loci is more similar to TIR and short TEs, embryo siRNA loci, embryo DRM2 targets, and seedling siRNA loci, whereas that of E-Z loci is not (Fig. 4C,D). Fifth, similarities in genomic distribution were confirmed by distances from siRNA loci to the nearest genes and by TE overlaps (Fig. 5A,B). There was a 30% decrease in median distance from siRNA loci to genes from egg to zygote, which took place over the course of less than one cell cycle. Only a 6% decrease occurred during the transition from zygote to embryo (7 d after fertilization) over the course of numerous cell cycles.
Newly detected zygote siRNAs mark future CHH hypermethylation sites in mature embryos
Hypermethylation of embryo has been reported in a number of angiosperm species, including Arabidopsis, soybean, chickpea, Brassica rapa, and rice (Bouyer et al. 2017; Kawakatsu et al. 2017; Lin et al. 2017; Chakraborty et al. 2021; Li et al. 2020; Rajkumar et al. 2020; Zhou et al. 2021). We found that newly detected siRNA loci have abundant CHH methylation in embryos that is dependent on the RdDM methyltransferase DRM2 (Fig. 5C). These results indicate that newly detected siRNA loci in zygote not only reset to the canonical siRNA pattern but also that the corresponding 24-nt siRNAs are capable of targeting high CHH methylation during embryogenesis. This methylation pattern also appears to be consistent within the zygote itself (Supplemental Fig. S8A).
Zygote siRNAs are associated with high CHH methylation at regions similar to seedling siRNA loci and embryo DRM2 targets. Thus, resetting of the gametic 24-nt siRNA loci to a distribution that results in embryo hypermethylation appears to be initiated in the zygote before the first embryonic division. Reminiscent of the increased heterochromatic siRNAs in rice gametes, a recent paper in Arabidopsis revealed that heterochromatin becomes decondensed during embryogenesis and promotes a transient production of siRNAs from heterochromatic TEs at the preglobular stage (Papareddy et al. 2020). The Arabidopsis embryo siRNAs from euchromatic TE and canonical siRNA loci peaked toward the end of embryogenesis. Because neither egg nor zygote siRNAs have been sequenced yet in Arabidopsis, the relationship of the heterochromatic siRNAs in multicellular Arabidopsis embryos to the reprogramming of siRNAs in egg cells and zygotes, as described here for rice, remains to be determined.
Plant gametes are highly dimorphic in terms of size, chromatin (Ingouff et al. 2010; Borg and Berger 2015; Wang and Köhler 2017), and gene expression (Anderson et al. 2013), consistent with a differential reprogramming of gamete epigenomes prior to fertilization inferred from their siRNA profiles (Li et al. 2020). In mammals, studies have found a progressive change in epigenomes after the two-cell embryo stage and concluded by the blastocyst stage (Xu and Xie 2018). As detailed above, this study indicates that in plants, the zygote inherits primarily maternal 24-nt siRNAs and initiates a resetting toward a canonical siRNA pattern that sets the stage for the CHH methylation pattern in the embryo. Lastly, as siRNA expression is influenced by histone modifications, and siRNAs can either reinforce or initiate DNA methylation and histone modifications, the siRNA transcriptome is an indicator and output of the epigenome. Thus, it appears likely that resetting of the other features of the epigenome, such as histone modifications and chromatin conformation, may also be initiated in plant zygotes as a component of their transition to totipotency. More generally, such epigenetic resetting might be associated with other examples of acquisition of pluripotency in plants, especially from germline cells; for example, the regeneration of haploid plants from anther cells after heat stress that is widely used in plant breeding (Ibáñez et al. 2020).
Methods
Plant growth condition and zygote collection
Rice (Oryza sativa) variety Kitaake was grown in soil in a greenhouse under natural light conditions. Zygote isolation was performed as described (Anderson et al. 2017; Li et al. 2019a). Briefly, rice flowers were hand-pollinated. At 8–9 h postpollination, ovaries were dissected. A transverse cut was made at the middle region of the ovary in a droplet of 0.3 M mannitol. The lower part of the cut ovary was gently pushed using an acupuncture needle to separate selected cells under a phase contrast inverted microscope. Once the zygote was separated and floated out of the ovary incision, it was captured by a fine glass capillary and immediately frozen in liquid nitrogen. We routinely culled any unfertilized egg cells that did not conform to zygotic cell morphology during our collections (Anderson et al. 2017). Fifty zygotes were collected for each replicate, and six replicates were collected. Intact ovaries at 8–9 h after pollination were collected separately for the ovary small RNA analysis. Ten ovaries were collected for each replicate, and three replicates were collected (Supplemental Table S1).
RNA extraction and small RNA library construction
RNA extractions were performed using Ambion RNAqueous Total RNA kit (AM1931), including an on-column DNase I treatment using Qiagen DNase I (#79254). Total RNA was analyzed using a Bioanalyzer (Agilent) to check for RNA integrity, with the eukaryotic total RNA-pico program. RNA input for library construction was ∼30 ng. Small RNA libraries were made using the NEXTflex small RNA-seq kit v3 (PerkinElmer NOVA-5132-05), with the following modifications. One-quarter dilution of adapters was used. The 3′ adapter ligation step was done overnight at 20°C. Zygote libraries were amplified at 24 cycles. Postfertilization ovary libraries were amplified at 20 cycles, as prefertilization ovaries (Li et al. 2020). The library product was size-selected using PippinHT (Sage Science) 3% agarose gel cassettes.
Small RNA sequencing analysis
Analyses were based on the Os-Nipponbare-Reference-IRGSP-1.0 reference genome (Kawahara et al. 2013). Genome annotations for transposable elements, genes, miRNAs, 5S rRNA, tRNA, NOR, CentO repeats, and phasiRNA loci were performed as described (Li et al. 2020). Quality filtering, adapter trimming, PCR duplicate removal, and alignment were performed as described (Li et al. 2020). Small RNA-seq reads were quality-filtered and trimmed of adapters using cutadapt (Martin 2011), parameters “-q 20 -a TGGAATTCTCGGGTGCCAAGG -e .05 -O 5 ‐‐discard-untrimmed -m 28 -M 33.” PCR duplicates were then removed using PRINSEQ, parameters “prinseq-lite.pl -fastq out_format 3 -out_ good -derep 1” (Schmieder and Edwards 2011). The four random nucleotides at each end were then removed using cutadapt “-u4” followed by cutadapt “-u -4.” Reads were aligned to the genome with BWA-backtrack (version 0.7.15) (Li and Durbin 2009), parameters “aln -t 8 -l 10.” Except where indicated otherwise, multimapping reads were included in all analyses. The uniquely mapping subset of siRNAs was defined by having MAPQ values of at least 20 using SAMtools (Li et al. 2009). Except where indicated otherwise, siRNAs used for analyses were small RNA reads (20–25 nt) not overlapping 90% or more of their lengths with miRNA, 5S rRNA, tRNA, NOR, and phasiRNA loci as determined by the BEDTools coverage tool (Quinlan and Hall 2010). For analysis of overlaps of siRNAs at CentO centromeric tandem repeats, transposable elements, and 24-nt siRNA loci, only siRNAs that overlapped by at least 50% of their lengths were counted. Distances to closest genes were obtained using the BEDTools closest tool. Whole-genome small RNA heat maps were made on 50-kb intervals using IGVtools (Thorvaldsdottir et al. 2013). For better visualization of midrange values, heat map intensity was maxed out at 1.25× coverage per 10 million 24-nt siRNAs. Pairwise correlation coefficients between zygote and ovary replicates were calculated using the cor() function in R (R Core Team 2021; Supplemental Fig. S13).
miRNA analysis
To measure miRNA accumulation, the BEDTools coverage tool was used to count the number of 20- to 25-nt reads that overlapped at least 90% of their length with annotated miRNA positions (Supplemental Data Set 1). R package edgeR was used to analyze miRNA accumulation (McCarthy et al. 2012). Individual miRNA counts were normalized by total mapped small RNAs and filtered for >1 count per million reads (CPM) in at least three libraries. Differential expression analyses were performed under |log2FC| > 1 and FDR < 0.05 cutoffs. Differential expressing miRNA genes were visualized under counts per million miRNAs.
Definition of siRNA loci
Small RNA loci were identified from the initial 20- to 25-nt total small RNA alignment BAM files using Shortstack (Axtell 2013) after merging replicates using default parameters. Each cell type was downsampled to 3.5 million small RNAs first. For each tissue type (pre- and postfertilization ovary, egg cell, sperm cell, zygote, seedling, embryo, and endosperm), siRNA loci were defined as RPM > 0.5, 24-nt-dominant and not detected as an miRNA locus (“DicerCall = 24; MIRNA = N”). Endosperm siren loci were defined as the highest expressing loci that accounted for 60% of the cumulative RPM in the endosperm. Similarly, pre- and postfertilization ovary siren loci as well as egg and zygote siren loci were defined as the highest expressing loci that accounted for 60% of the cumulative RPM in the ovary. The 60% cutoff was selected based on the turning point of cumulative expression versus percentage rank plot of ovary (Fig. 2A). Seedling-signature loci were identified as seedling siRNA loci that did not overlap any sperm siRNA loci or egg siRNA loci (seedling loci ∉ egg loci ∉ sperm loci) (Fig. 3C) using the BEDTools intersect tool (Quinlan and Hall 2010). Overlaps were defined as at least 1-bp overlapping genomic coordinates. Similarly, sperm-signature loci were identified as sperm siRNA loci that did not overlap any egg siRNA loci or seedling siRNA loci (sperm loci ∉ egg loci ∉ seedling loci) (Fig. 3C). Egg-signature loci were identified as egg siRNA loci that did not overlap any seedling siRNA loci or sperm siRNA loci (egg loci ∉ seedling siRNA loci ∉ sperm siRNA loci) (Fig. 3C). Z-E loci were zygote siRNA loci that did not overlap egg siRNA loci (Z loci ∉ E loci). E-Z loci were egg siRNA loci that did not overlap zygote siRNA loci (E loci ∉ Z loci). Z/E loci intersect were zygote siRNA loci that overlapped egg siRNA loci (Z loci ⋂ E loci) (Fig. 4A).
PCR and Sanger sequencing validation of abundant siRNAs
We designed PCR primers for 15 individual siRNAs (Supplemental Table S3) from zygote or egg siRNA loci, including Z-E loci. All PCRs were performed with the following conditions: initial 1 min at 95°C, and cycling condition of 30 sec at 95°C, 15 sec at 58°C, and 15 sec a 72°C for 30 cycles, and a final elongation of 1 min at 72°C, without optimization for each target siRNA. The siRNAs were classified into two groups: high expression group with expression values ranging from 26 to ∼100 RPM in zygote (n = 7 loci) and low expression group with expression values ranging from 0 to 2 RPM in the zygote (n = 8 loci), including two that were detected as E-Z loci, which were used as controls. PCR amplification was performed with the Illumina universal P5 sequence as the forward primer, an siRNA-specific reverse primer, and a ready-to-sequence small RNA library as template (Supplemental Fig. S14A). When designing siRNA-specific primers, 2–4 nt at the start of the 24-nt siRNA sequence were omitted from the 3′ end of the primer, such that the identity of the amplicon can be confirmed by Sanger sequencing (Supplemental Fig. S14A). Four siRNAs in the high expression group produced correct size products, which were confirmed by Sanger sequencing traces (Supplemental Fig. S14B–E), and none in the low expression group produced products of the correct size or sequence. A highly expressed miRNA miRNA159b was used as a positive control (Supplemental Fig. S14F). The higher success rate in the high expression siRNA group also provides indirect confirmation of the relative expression levels in the small RNA transcriptome inferred from RNA-seq reads.
DNA methylation analyses
Methylation values were calculated for each locus using the mtr function of CGmapTools v0.1.2 (Guo et al. 2018) using the CGmap files generated in our previous study (Li et al. 2020) or ones we created using the same methods from the recently published zygote and gamete methylomes (Zhou et al. 2021). Only loci with more than three (mC + C) calls were included in the analyses.
Statistical analyses
Tukey tests were performed using the R package emmeans (Searle et al. 1980) with multiple comparison correction using Tukey's method. Letter groupings were done at α = 0.05, where differences between means sharing the same letter were not statistically significant. For multifactorial analyses, multiple comparisons were applied to families of tests at each interacting factor level: at the level of each TE/locus category for Figures 1C, 2D, 3D, 4B, and 5B and Supplemental Figures S1A; S3, D and F; and S4B, and at the level of genotype and context for Figure 5C and Supplemental Figures S7–S10), or at the level of context × cell type combination (Supplemental Figs. S11, S12). For analyses of siRNA relative abundances or siRNA coverage across siRNA locus category across siRNA transcriptomes, a linear model was fitted using logit transformation to correct for heteroscedasticity (Figs. 1C, 2B, 3A,B,D, 4B; Supplemental Figs. S1A, S3B,D, S3F). For analyses of siRNA counts or locus counts, a linear model was fitted using log(RPM + 1) transformation to correct for heteroscedasticity (Fig. 2D; Supplemental Figs. S3G, S4A). For analyses of distances to nearest genes, a generalized linear model was fitted using log link function to correct for heteroscedasticity (Fig. 5A). For analyses of fraction of locus length covered by genes, a generalized linear model of quasibinomial family with logit link function was fitted to accommodate the mean–error relationship of fractional data (Figs. 2C, 5B; Supplemental Fig. S4B). For analyses of DNA methylation levels across different locus categories, a generalized linear model of quasibinomial family with logit link function was fitted to accommodate the mean-error relationship of proportion data (Fig. 5C; Supplemental Figs. S7–S12). For analysis of correlations between PC1 (Fig. 4D), distance to nearest genes, TE overlaps, and DNA methylation, Spearman's rank order correlation was used (Supplemental Figs. S4D,E, S6). P-values < 2.2 × 10−16, which is the smallest positive floating point number R can store or display, were reported as “P < 2.2 × 10−16,” instead of the exact values.
Data access
All small RNA data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA533115. All R codes regarding data visualization and statistical analyses are available at GitHub (https://github.com/cxli233/zygote_smRNA) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Zachary Liechty and Christian Santos for assistance in R programming; and Alina Yalda, Jake Anichowski, and Michelle Binyu Cui for greenhouse maintenance and technical assistance. The UC Davis Genome Center provided Illumina sequencing, library quality control, and size selection services. C.L. also acknowledges partial support by an Elsie Taylor Stocking Memorial Fellowship from the Department of Plant Biology at University of California, Davis. This study was supported in part by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia's Office of the Vice President for Research and Office of the Vice President for Information Technology. This research was funded by the National Science Foundation (IOS-1547760) and the U.S. Department of Agriculture (USDA) Agricultural Experiment Station (CA-D-XXX-6973-H).
Author contributions: C.L., J.I.G., S.D.R., and V.S. designed the study. H.X. and H.F. collected zygotes. S.D.R. supervised zygote collections. C.L. produced small RNA sequencing libraries. C.L. and J.I.G. analyzed data. V.S. supervised data collection and analyses. C.L. wrote the manuscript with input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275981.121.
Competing interest statement
The authors declare no competing interests.
References
- Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V. 2013. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76: 729–741. 10.1111/tpj.12336 [DOI] [PubMed] [Google Scholar]
- Anderson SN, Johnson CS, Chesnut J, Jones DS, Khanday I, Woodhouse M, Li C, Conrad LJ, Russell SD, Sundaresan V. 2017. The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev Cell 43: 349–358.e4. 10.1016/j.devcel.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Armenta-Medina A, Gillmor CS. 2019. Genetic, molecular and parent-of-origin regulation of early embryogenesis in flowering plants. Curr Top Dev Biol 131: 497–543. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. 2011. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. 10.1016/j.cell.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Berger F. 2015. Chromatin remodelling during male gametophyte development. Plant J 83: 177–188. 10.1111/tpj.12856 [DOI] [PubMed] [Google Scholar]
- Borg M, Jacob Y, Susaki D, LeBlanc C, Buendía D, Axelsson E, Kawashima T, Voigt P, Boavida L, Becker J, et al. 2020. Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat Cell Biol 22: 621–629. 10.1038/s41556-020-0515-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Parent J, Van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA. 2018. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat Genet 50: 186–192. 10.1038/s41588-017-0032-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V. 2017. DNA methylation dynamics during early plant life. Genome Biol 18: 179. 10.1186/s13059-017-1313-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D, Chow HT, Grover JW, Freeling M, Mosher RA. 2021. Ovule siRNAs methylate and silence protein-coding genes in trans. bioRxiv 10.1101/2021.06.10.447945 [DOI] [PMC free article] [PubMed]
- Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo A, Becker D, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. 10.1016/j.cell.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Kendall T, Grover JW, Mosher RA. 2021. Embryo CHH hypermethylation is mediated by RdDM and is autonomously directed in Brassica rapa. Genome Biol 22: 140. 10.1186/s13059-021-02358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Strieder N, Krohn NG, Cyprys P, Sprunck S, Engelmann JC, Dresselhaus T. 2017. Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29: 2106–2125. 10.1105/tpc.17.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D, Slotkin RK. 2016. Non-canonical RNA-directed DNA methylation. Nat Plants 2: 16163. 10.1038/NPLANTS.2016.163 [DOI] [PubMed] [Google Scholar]
- Ding J, Shen J, Li W, Yang H. 2009. Cytological observation of double fertilization and its duration in Oryza sativa. Chin Bull Bot 44: 473–483. 10.3969/j.issn.1674-3466.2009.04.009 [DOI] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL. 2000. Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12: 2101–2118. 10.1105/tpc.12.11.2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann RM, Picard CL. 2020. RNA-directed DNA methylation. PLoS Genet 16: e1009034. 10.1371/journal.pgen.1009034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann RM, Satyaki PRV, Klosinska M, Gehring M, Erdmann RM, Satyaki PRV, Klosinska M, Gehring M. 2017. A small RNA pathway mediates allelic dosage in endosperm. Cell Rep 21: 3364–3372. 10.1016/j.celrep.2017.11.078 [DOI] [PubMed] [Google Scholar]
- Gehring M. 2019. Epigenetic dynamics during flowering plant reproduction: evidence for reprogramming? New Phytol 224: 91–96. 10.1111/nph.15856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK. 2013. CHH islands : de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 628–637. 10.1101/gr.146985.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover JW, Kendall T, Baten A, Burgess D, Freeling M, King GJ, Mosher RA. 2018. Maternal components of RNA -directed DNA methylation are required for seed development in Brassica rapa. Plant J 94: 575–582. 10.1111/tpj.13910 [DOI] [PubMed] [Google Scholar]
- Grover JW, Burgess D, Kendall T, Baten A, Pokhrel S, King GJ, Meyers BC, Freeling M, Mosher RA. 2020. Abundant expression of maternal siRNAs is a conserved feature of seed development. Proc Natl Acad Sci 117: 15305–15315. 10.1073/pnas.2001332117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhu P, Pellegrini M, Zhang MQ, Wang X, Ni Z. 2018. CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics 34: 381–387. 10.1093/bioinformatics/btx595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Qin S, Wessler SR. 2013. Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics 14: 71. 10.1186/1471-2164-14-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houri-Zeevi L, Rechavi O. 2017. A matter of time: small RNAs regulate the duration of epigenetic inheritance. Trends Genet 33: 46–57. 10.1016/j.tig.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Hsieh T-F, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. 10.1126/science.1172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, He S, Buttress T, Gao H, Couchman M, Fischer RL, Zilberman D, Feng X. 2016. Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc Natl Acad Sci 113: 15132–15137. 10.1073/pnas.1619074114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez S, Carneros E, Testillano PS, Pérez-Pérez JM. 2020. Advances in plant regeneration: shake, rattle and roll. Plants 9: 897. 10.3390/plants9070897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh T-F, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. 10.1126/science.1224839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. 2007. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037. 10.1016/j.cub.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. 2010. zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20: 2137–2143. 10.1016/j.cub.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Kao P, Nodine MD. 2019. Transcriptional activation of Arabidopsis zygotes is required for initial cell divisions. Sci Rep 9: 17159. 10.1038/s41598-019-53704-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6: 4. 10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR. 2017. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18: 171. 10.1186/s13059-017-1251-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Ono A, Scholten S, Kinoshita T, Zilberman D, Okamoto T, Fischer RL. 2019. DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc Natl Acad Sci 116: 9652–9657. 10.1073/pnas.1821435116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. 10.1038/nature03368 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47: D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz E, Bautor J, Lörz H. 1991. In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sex Plant Reprod 4: 12–16. 10.1007/BF00194565 [DOI] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci 112: 14728–14733. 10.1073/pnas.1514680112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu H, Russell SD, Sundaresan V. 2019a. Step-by-step protocols for rice gamete isolation. Plant Reprod 32: 5–13. 10.1007/s00497-019-00363-y [DOI] [PubMed] [Google Scholar]
- Li X, Chen L, Zhang Q, Sun Y, Li Q, Yan J. 2019b. BRIF-seq: bisulfite-converted randomly integrated fragments sequencing at the single-cell level. Mol Plant 12: 438–446. 10.1016/j.molp.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Li C, Xu H, Fu F-F, Russell SD, Sundaresan V, Gent JI. 2020. Genome-wide redistribution of 24-nt siRNAs in rice gametes. Genome Res 30: 173–184. 10.1101/gr.253674.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-Y, Le BH, Chen M, Henry KF, Hur J, Hsieh T-F, Chen P-Y, Pelletier JM, Pellegrini M, Fischer RL, et al. 2017. Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc Natl Acad Sci 114: E9730–E9739. 10.1073/pnas.1716758114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Walker J, She W, Aldridge B, Gao H, Deans S, Vickers M, Feng X. 2021. Nurse cell–derived small RNAs define paternal epigenetic inheritance in Arabidopsis. Science 373: eabh0556. 10.1126/science.abh0556 [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD. 2002. The mechanisms of pollination and fertilization in plants. Annu Rev Cell Dev Biol 18: 81–105. 10.1146/annurev.cellbio.18.012502.083438 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 3. [Google Scholar]
- Martinez G, Köhler C. 2017. Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr Opin Plant Biol 36: 22–28. 10.1016/j.pbi.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK. 2016. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2: 16030. 10.1038/nplants.2016.30 [DOI] [PubMed] [Google Scholar]
- Martinez G, Wolff P, Wang Z, Moreno-Romero J, Santos-González J, Conze LL, DeFraia C, Slotkin RK, Köhler C. 2018. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat Genet 50: 193–198. 10.1038/s41588-017-0033-4 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 224: 792–801. 10.1126/science.15739260 [DOI] [PubMed] [Google Scholar]
- Mérai Z, Chumak N, García-Aguilar M, Hsieh T-F, Nishimura T, Schoft VK, Bindics J, Ślusarz L, Arnoux S, Opravil S, et al. 2014. The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc Natl Acad Sci 111: 16166–16171. 10.1073/pnas.1418564111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. 2014. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28: 812–828. 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Matsumoto T, Wu,J. 2018. Composition and structure of rice centromeres and telomeres. In Rice genomics, genetics and breeding (ed. Sasaki T, Ashikari M.), pp. 37–52. Springer Singapore, Singapore. 10.1007/978-981-10-7461-5_3 [DOI] [Google Scholar]
- Moritoh S, Eun C-H, Ono A, Asao H, Okano Y, Yamaguchi K, Shimatani Z, Koizumi A, Terada R. 2012. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation: osdrm2 affects DNA methylation and development. Plant J 71: 85–98. 10.1111/j.1365-313X.2012.04974.x [DOI] [PubMed] [Google Scholar]
- Okada T, Endo M, Singh MB, Bhalla PL. 2005. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3: histone H3 gene family in Arabidopsis. Plant J 44: 557–568. 10.1111/j.1365-313X.2005.02554.x [DOI] [PubMed] [Google Scholar]
- Papareddy RK, Páldi K, Paulraj S, Kao P, Lutzmayer S, Nodine MD. 2020. Chromatin regulates expression of small RNAs to help maintain transposon methylome homeostasis in Arabidopsis. Genome Biol 21: 251. 10.1186/s13059-020-02163-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J-S, Cahn J, Herridge RP, Grimanelli D, Martienssen RA. 2021. Small RNAs guide histone methylation in Arabidopsis embryos. Genes Dev 35: 841–846. 10.1101/gad.343871.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim MY, Vickers M, Park J-S, Hyun Y, Okamoto T, Zilberman D, Fischer RL, Feng X, Choi Y, et al. 2016. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci 113: 15138–15143. 10.1073/pnas.1619047114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M, Baroux C, Arteaga Vazquez M, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D. 2010. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22: 307–320. 10.1105/tpc.109.071647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar MS, Gupta K, Khemka NK, Garg R, Jain M. 2020. DNA methylation reprogramming during seed development and its functional relevance in seed size/weight determination in chickpea. Commun Biol 3: 340. 10.1038/s42003-020-1059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D. 2013. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci 110: 7934–7939. 10.1073/pnas.1306164110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. 2012. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139: 15–31. 10.1242/dev.050849 [DOI] [PubMed] [Google Scholar]
- Satyaki PRV, Gehring M. 2019. Paternally acting canonical RNA-directed DNA methylation pathway genes sensitize Arabidopsis endosperm to paternal genome dosage. Plant Cell 31: 1563–1578. 10.1105/tpc.19.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. 2009. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. Sci Rep 10: 1015–1021. 10.1038/embor.2009.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten S, Lörz H, Kranz E. 2002. Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 32: 221–231. 10.1046/j.1365-313X.2002.01418.x [DOI] [PubMed] [Google Scholar]
- Searle SR, Speed FM, Milliken GA. 1980. Population marginal means in the linear model: an alternative to least squares means. Am Stat 34: 216–221. 10.1080/00031305.1980.10483031 [DOI] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 461–472. 10.1016/j.cell.2008.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wang N, Hou Z, Li B, Li D, Liu Y, Cai H, Qin Y, Chen X. 2020. Regulation of female germline specification via small RNA mobility in Arabidopsis. Plant Cell 32: 2842–2854. 10.1105/tpc.20.00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tan F, Zhou C, Zhou Q, Zhou S, Yang W, Zhao Y, Li G, Zhou D-X. 2016. Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol 171: 2041–2054. 10.1104/pp.16.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Lu Y, Jiang W, Wu T, Zhang R, Zhao Y, Zhou D-X. 2018. DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol 177: 1187–1197. 10.1104/pp.18.00352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. 2016. Specification and epigenetic programming of the human germ line. Nat Rev Genet 17: 585–600. 10.1038/nrg.2016.88 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative enomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Köhler C. 2017. Epigenetic processes in flowering plant reproduction. J Exp Bot 68: 797–807. 10.1093/jxb/erw486 [DOI] [PubMed] [Google Scholar]
- Wang Z, Butel N, Santos-González J, Borges F, Yi J, Martienssen RA, Martinez G, Köhler C. 2020. Polymerase IV plays a crucial role in pollen development in Capsella. Plant Cell 32: 950–966. 10.1105/tpc.19.00938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Xie W. 2018. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol 28: 237–253. 10.1016/j.tcb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. 2012. Cytoplasmic assembly and selective nuclear import of Arabidopsis ARGONAUTE4/siRNA complexes. Mol Cell 46: 859–870. 10.1016/j.molcel.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. 2013. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. 10.1016/j.cell.2013.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhou X, Shen K, Liu Z, Cheng T, Liu D, Cheng Y, Peng X, Sun M. 2019. Two-step maternal-to-zygotic transition with two-phase parental genome contributions. Dev Cell 49: 882–893.e5. 10.1016/j.devcel.2019.04.016 [DOI] [PubMed] [Google Scholar]
- Zhou M, Coruh C, Xu G, Bourbousse C, Lambolez A, Law JA. 2020. The CLASSY family controls tissue-specific DNA methylation patterns in Arabidopsis. bioRxiv 10.1101/2021.01.23.427869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Li X, Liu Q, Zhao Y, Jiang W, Wu A, Zhou D-X. 2021. DNA demethylases remodel DNA methylation in rice gametes and zygote and are required for reproduction. Mol Plant 14: 1569–1583. 10.1016/j.molp.2021.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.