Abstract
BACKGROUND AND PURPOSE:
In this prospective, multicenter, multireader study, we evaluated the impact on both image quality and quantitative image-analysis consistency of 60% accelerated volumetric MR imaging sequences processed with a commercially available, vendor-agnostic, DICOM-based, deep learning tool (SubtleMR) compared with that of standard of care.
MATERIALS AND METHODS:
Forty subjects underwent brain MR imaging examinations on 6 scanners from 5 institutions. Standard of care and accelerated datasets were acquired for each subject, and the accelerated scans were enhanced with deep learning processing. Standard of care, accelerated scans, and accelerated–deep learning were subjected to NeuroQuant quantitative analysis and classified by a neuroradiologist into clinical disease categories. Concordance of standard of care and accelerated–deep learning biomarker measurements were assessed. Randomized, side-by-side, multiplanar datasets (360 series) were presented blinded to 2 neuroradiologists and rated for apparent SNR, image sharpness, artifacts, anatomic/lesion conspicuity, image contrast, and gray-white differentiation to evaluate image quality.
RESULTS:
Accelerated–deep learning was statistically superior to standard of care for perceived quality across imaging features despite a 60% sequence scan-time reduction. Both accelerated–deep learning and standard of care were superior to accelerated scans for all features. There was no difference in quantitative volumetric biomarkers or clinical classification for standard of care and accelerated–deep learning datasets.
CONCLUSIONS:
Deep learning reconstruction allows 60% sequence scan-time reduction while maintaining high volumetric quantification accuracy, consistent clinical classification, and what radiologists perceive as superior image quality compared with standard of care. This trial supports the reliability, efficiency, and utility of deep learning–based enhancement for quantitative imaging. Shorter scan times may heighten the use of volumetric quantitative MR imaging in routine clinical settings.
Deep learning (DL) is a subset of machine learning that uses convolutional neural networks to process large volumes of data.1-6 While traditional reconstruction techniques can be limited by long scan times, SNR constraints, and motion artifacts, the recent application of DL to image reconstruction can enable faster image acquisitions with equal or enhanced image quality.2,3 While DL can boost SNR among other advantages over conventional methods,2-6 concerns exist over whether postprocessing can mask or alter pathology and whether the quantitative values derived are consistent with those obtained from routine standard of care (SOC) scans over the gamut of scanner vendors and across field strengths.
MR imaging depicts brain anatomy with high spatial and contrast resolution, qualities crucial when applying anatomic segmentation and quantitative volumetric analysis. Quantitative volumetric analysis requires 3D radiofrequency spoiled gradient-echo T1-weighted scans, and k-space acceleration opportunities have limits. Reduced excitations and undersampling techniques like compressed sensing accelerate scans at a cost of increased image noise (reduced SNR). Acceleration via decreases in the imaging matrix can improve both SNR and contrast resolution but reduce image sharpness. For this study, both undersampling and reduced imaging matrices were used for the accelerated scans (FAST), which were then processed with a commercially available DL tool (SubtleMR; Subtle Medical) that provides both denoising and sharpness enhancement (FAST-DL). Our goal was to match or exceed SOC image SNR and spatial resolution while maintaining clinical and quantitative integrity.7
In this prospective, multireader, multicenter study, we explored the impact of DL-based enhancement of 60% accelerated 3D T1-weighted brain MR image acquisitions. We found that the DL-processed images demonstrated high volumetric quantification accuracy and matched clinical disease status predictability and provided what readers perceived as superior image quality compared with the longer SOC examinations, suggesting good generalizability, accuracy, and potential utility of DL enhancement in routine clinical settings.
MATERIALS AND METHODS
Participants
With Western Institutional Review Board approval and patient consent, 40 consecutive subjects (mean age, 69 [SD, 17] years; 21 men, 19 women) undergoing clinically indicated brain MR imaging examinations for subjective memory loss were prospectively recruited during an 8-month period.
Image Acquisition
Imaging was obtained on 6 scanners (3T Skyra, Siemens; 3T Discovery 750 and 3T Discovery 750w, 3T Signa Premier, and 1.5T HDxt, GE Healthcare; 3T Verio, Siemens) at 3T (n = 32) and 1.5T (n = 8) from 5 different institutions in New York (n = 8) and California (n = 32).
The image acquisitions consisted of paired 3D T1-weighted sagittally acquired datasets: 1 SOC image set (mean scan time, 6 minutes, 56 seconds for 3T and 4 minutes for 1.5T), and 1 FAST image set (mean scan time, 2 minutes, 44 seconds for 3T and 2 minutes, 40 seconds for 1.5T). Native sagittal images were acquired and quantitatively postprocessed at 1-mm section thickness (for 3T) and 1.2-mm section thickness (for 1.5T). The spatial-resolution matrix was 256 × 256 for SOC and 128 × 256 for FAST. Acquisition parameters included MPRAGE (flip angle = 9°, TI = 1100 ms, section thickness = 1.0–1.2 mm); fast-spoiled gradient recalled (flip angle = 8°, TI = 900 ms, section thickness = 1.0–1.2 mm); and BRAVO (GE Healthcare; flip angle = 12°, TI = 450 ms, section thickness = 1.0–1.2 mm). The average pooled (3T + 1.5T) 3D T1WI scan times for all 40 patients (n = 32/3T and n = 8/1.5T) were 6 minutes, 1 second for SOC (range, 4–7 minutes) and 2 minutes, 43 seconds for the FAST dataset (range, 2–2 minutes, 50 seconds), representing a 60% sequence scan-time reduction for the FAST acquisition.
Image Processing
FAST-DL was performed off-line using an FDA-cleared, vendor-agnostic, DICOM-based, convolutional neural networks–dependent deep learning artificial intelligence image-enhancement software product, SubtleMR (Version 1.2). The training set included hundreds of thousands of MR imaging datasets from a variety of vendors (GE Healthcare, Philips Healthcare, Siemens, Hitachi, and so forth), scanner models, field strengths, and clinical sites, as well as a variety of disease states/clinical indications, thus experiencing a range of tissue contrasts, acquisition parameters, patient anatomies, and variable image quality.
The DL network was trained on paired low-/high-resolution images to impart structure-preserving noise reduction and sharpness enhancement to newly acquired images.7 Processing does not use proprietary raw k-space input (DICOM-based) and is, thus, vendor-agnostic. For the study, DL processing required <1 minute per series on a scanner-connected GPU server and finished before the next sequence acquisition was completed, thus not impacting overall examination time. Images were gathered from different sites and presented to the reviewers on a commercial DICOM viewer.
The SOC, FAST, and FAST-DL image sets were processed with a machine learning–based FDA-cleared quantitative volumetric software product, NeuroQuant (Cortechs.ai). The hippocampal occupancy score (HOC), a biomarker to predict the progression of neurodegenerative diseases, as well as the volumes of the hippocampi (HV), superior lateral ventricles (SLV), and inferior lateral ventricles (ILV) were analyzed for this study.
Radiologic Assessment
For the image-quality assessment, 2 experienced board-certified neuroradiologists (>17 years’ experience each) were presented with 40 paired side-by-side multiplanar 3D T1-weighted series datasets (360 series). The blinded datasets (SOC versus FAST, SOC versus FAST-DL, FAST-DL versus FAST) were randomized in disease classification, image plane, and left-right display order. The readers evaluated 2 images side-by-side and provided a single Likert scale ranking between 1 and 5 that described whether the left or right image was superior (3 = both images were preferred equally; 2 or 4 = right/left mildly preferred; 1 or 5 = right/left strongly preferred) for the following: 1) perceived SNR; 2) perceived spatial resolution (sharpness); 3) imaging artifacts; 4) anatomic/lesion conspicuity; 5) image contrast; and 6) gray-white matter differentiation. Anatomic conspicuity of brain structures such as the deep gray nuclei was used in cases in which a lesion was not conspicuous on the 3D T1-weighted images. Sample lesions in our datasets included infarcts and prominent white matter ischemic disease.
To assess clinical classification performance, we categorized the quantitative biomarkers obtained from 80 datasets (40 SOC and 40 FAST-DL) in a blinded, randomized fashion. Each dataset was rated using a binary predictive classification system (healthy/mild cognitive impairment [MCI] versus dementia) with ground truth established according to the statistical significance of 3 biomarkers falling >2 SDs from the mean: HOC (<5%), HV (<5%), and ILV (>95%) based on an (n > 4000) age- and sex-matched normative data base, with 0/3 and 1/3 statistically significant biomarkers categorized as healthy/MCI; and 2/3 and 3/3 categorized as likely dementia.
Following qualitative feature ranking and quantitative analysis, both readers were presented the SOC and FAST-DL datasets in a side-by-side fashion, randomized in right-left orientation to qualitatively assess the overall diagnostic quality of the 3D T1-weighted images before postprocessing with NeuroQuant and again after postprocessing with NeuroQuant, with the goal of visually assessing the quality of matched color-coded segmentation of the latter. The FAST scans were excluded from this analysis because they would not be typically used as input to quantitative segmentation software given their lower spatial resolution.
Statistical Analysis
Wilcoxon rank sum tests were performed to assess the equivalence or superiority of the image quality for each feature (Table 1). Statistically significant superiority for a feature was determined by a P value < .05.
Table 1:
Wilcoxon rank sum test results—both readers combineda
| Feature | SOC vs FAST |
FAST-DL vs SOC |
FAST-DL vs FAST |
|||
|---|---|---|---|---|---|---|
| Mean | P Value | Mean | P Value | Mean | P Value | |
| Perceived SNR | 4.1 (SD, 0.8) | <.001 | 3.5 (SD, 1.3) | <.001 | 4.3 (SD, 1.0) | <.001 |
| Sharpness | 4.5 (SD, 0.7) | <.001 | 3.5 (SD, 1.5) | .005 | 4.7 (SD, 0.9) | <.001 |
| Artifacts | 3.9 (SD, 0.8) | <.001 | 3.5 (SD, 1.1) | <.001 | 4.1 (SD, 0.9) | <.001 |
| Anatomic/lesion conspicuity | 4.2 (SD, 0.7) | <.001 | 3.3 (SD, 1.1) | .006 | 4.3 (SD, 0.7) | <.001 |
| Image contrast | 4.0 (SD, 0.7) | <.001 | 3.4 (SD, 1.1) | .004 | 4.1 (SD, 0.8) | <.001 |
| GM/WM differentiation | 4.3 (SD, 0.7) | <.001 | 3.4 (SD, 1.2) | .009 | 4.5 (SD, 0.8) | <.001 |
SOC is superior to FAST for all criteria (P values <.001). Numbers higher than 3 represent preference for the first of the 2 sequences listed in the upper row. FAST-DL is superior to SOC for all criteria (P values < .008), except for GM/WM differentiation. While this metric trended to be superior for FAST-DL versus SOC, it did not reach statistical significance after Bonferroni correction (P = .009). FAST-DL is superior to FAST for all criteria (P values <.001).
Adjustment for significance tests for multiple comparisons was made using the Bonferroni correction, which adjusts the significance level to P < .05/.06 (0.00833).
Paired t test analysis was performed to test the equivalence of quantitative data on both the SOC versus FAST-DL images (Table 2) and SOC versus FAST images (Table 3). Linear regression graphs (Figs 1 and 2) and Bland-Altman analysis (Figs 3 and 4) were performed to assess quantitative volumetric biomarker equivalence of the datasets. The Spearman rank correlation test was applied to assess interreader agreement between the 2 neuroradiologists on image-quality ratings. Additionally, interrater reliability analysis was performed using an equal-spacing weighted Cohen κ statistic to measure the consistency of the 2 readers’ evaluation of image quality.
Table 2:
Paired t test for SOC versus FAST-DLa
| SOC | FAST-DL | Paired t Test | |
|---|---|---|---|
| HOC (mean) | 0.68 (SD, 0.16) | 0.68 (SD, 0.16) | 0.58 |
| HV (mean) (cm3) | 6.45 (SD, 1.70) | 6.47 (SD, 1.69) | 0.77 |
| SLV volume (mean) (cm3) | 44.30 (SD, 20.60) | 43.63 (SD, 20.35) | <0.05 |
| ILV volume (mean) (cm3) | 3.07 (SD, 1.78) | 3.04 (SD, 1.69) | 0.27 |
There is excellent agreement between SOC and FAST-DL for quantitative assessment of HOC, HV, SLV volume, and ILV volume.
Table 3:
Paired t test for SOC versus FASTa
| SOC | FAST | Paired t Test | |
|---|---|---|---|
| HOC (mean) | 0.68 (SD, 0.16) | 0.68 (SD, 0.17) | 0.63 |
| HV (mean) (cm3) | 6.45 (SD, 1.70) | 6.56 (SD, 1.88) | 0.60 |
| SLV volume (mean) (cm3) | 44.30 (SD, 20.60) | 43.44 (SD, 20.01) | <0.05 |
| ILV volume (mean) (cm3) | 3.07 (SD, 1.78) | 3.17 (SD, 1.85) | 0.93 |
There is less optimal agreement between SOC versus FAST (compared with SOC versus FAST-DL) for quantitative assessment of HOC, HV, SLV volume, and ILV volume.
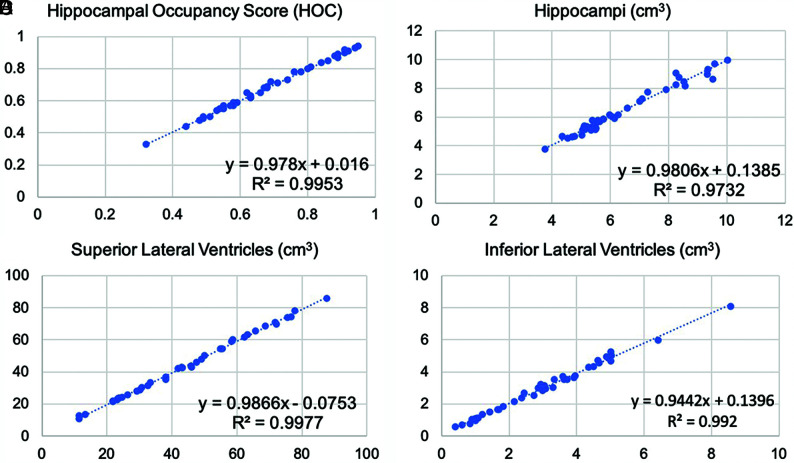
FIG 1.
Linear regression results for SOC versus FAST-DL. The plot graphs demonstrate linear distribution without scatter, indicating consistent concordance between SOC (x-axis) and FAST-DL (y-axis) in quantitative assessment of HOC (A), HV (B), SLV volume (C), and ILV volume (A).
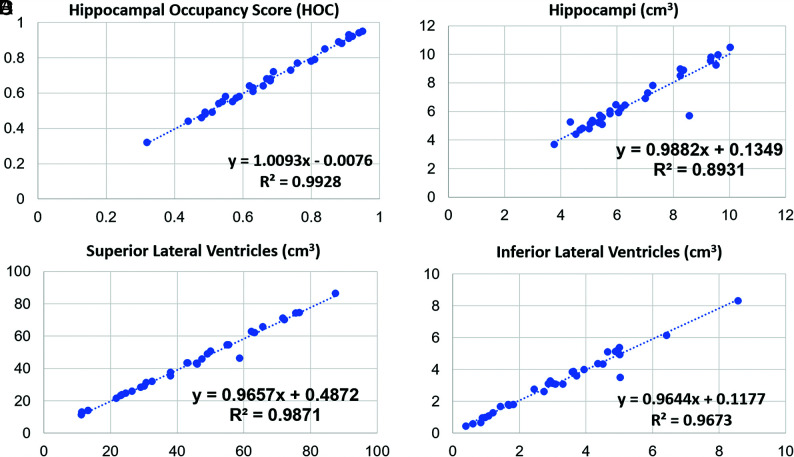
FIG 2.
Linear regression results for SOC versus FAST. The plot graphs demonstrate a modestly linear distribution though some scatter is present, indicating less optimal concordance of the cross-correlation factor between SOC (x-axis) and FAST (y-axis) (compared with SOC versus FAST-DL) in a quantitative assessment of HOC (A), HV (B), SLV volume (C), and ILV volume (A).
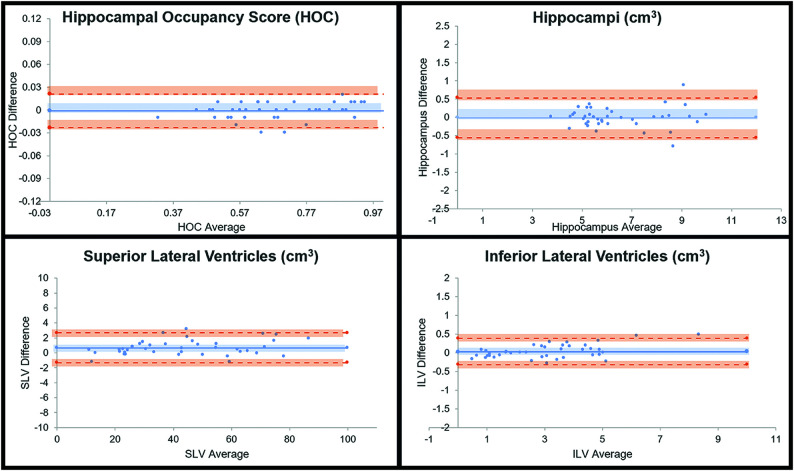
FIG 3.
Bland-Altman results for SOC versus FAST-DL. The plot graphs demonstrate a linear distribution without significant scatter, indicating consistent concordance between SOC and FAST-DL in the quantitative assessment of HOC, HV, SLV volume, and ILV volume.
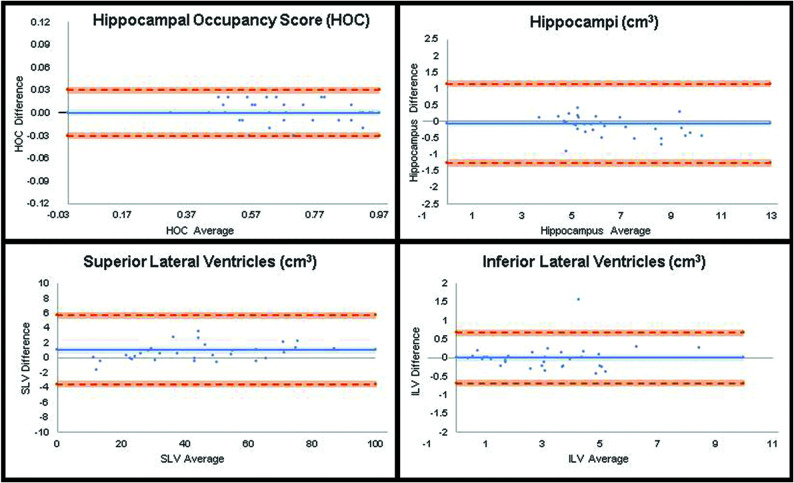
FIG 4.
Bland-Altman results for SOC versus FAST. The plot graphs demonstrate a modestly linear distribution though some scatter is present, indicating less optimal concordance of the cross-correlation factor between SOC versus FAST (compared with SOC versus FAST-DL) in the quantitative assessment of HOC, HV, SLV volume, and ILV volume.
RESULTS
Qualitative and Quantitative Performance
FAST-DL was statistically superior to SOC in subjective image quality for perceived SNR, sharpness, artifact reduction, anatomic/lesion conspicuity, and image contrast (all P values < .008), despite a 60% reduction in sequence scan time. Both FAST-DL and SOC were statistically superior to FAST for all analyzed features (all P values < .001). Wilcoxon rank sum statistical results are collectively summarized in Table 1.
Paired t test analysis demonstrated excellent agreement of quantitative data on both the SOC and FAST-DL images (Table 2). As expected, there was less agreement between the SOC and FAST datasets (Table 3) due to the lower spatial resolution of the FAST images. There was no statistically significant difference between mean HOC values in the SOC (0.68 [SD, 0.16]) and the mean FAST-DL (0.68 [SD, 0.16]) datasets. The difference of the HV, SLV volumes, and ILV volumes was also negligible (<2%) for the SOC and FAST-DL datasets. The linear regression graphs (Fig 1) and Bland-Altman plot graph analysis (Fig 3) further demonstrated strong agreement between quantitative values in each dataset across the range of conditions (normal, MCI, Alzheimer disease) with the HOC ranging from 0.32 to 0.95 mL. There was 100% agreement in clinical disease classification of both the SOC and FAST-DL datasets (n = 29 healthy/MCI and n = 11 dementia). The cross-correlation factor and degree of scatter was consistently worse for the SOC and FAST images compared with the SOC and FAST-DL images as demonstrated on the linear regression graphs (Fig 2) and Bland Altman plot graph analysis (Fig 4).
There was excellent interreader agreement between the 2 neuroradiologists on the Spearman rank correlation test applied to the Likert image quality ratings, with a Spearman ρ value of 0.725 (P < .01). The κ value of 0.62 (P < .001) also confirms substantial interrater agreement on the Likert scale rankings.
Overall Diagnostic Quality
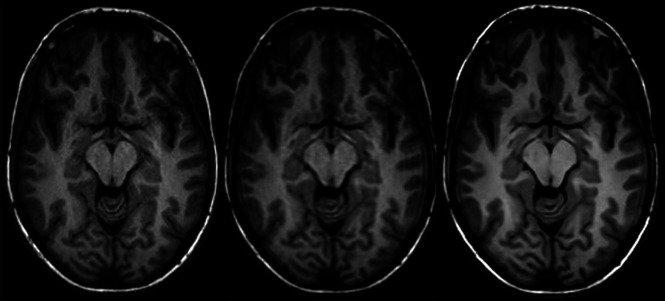
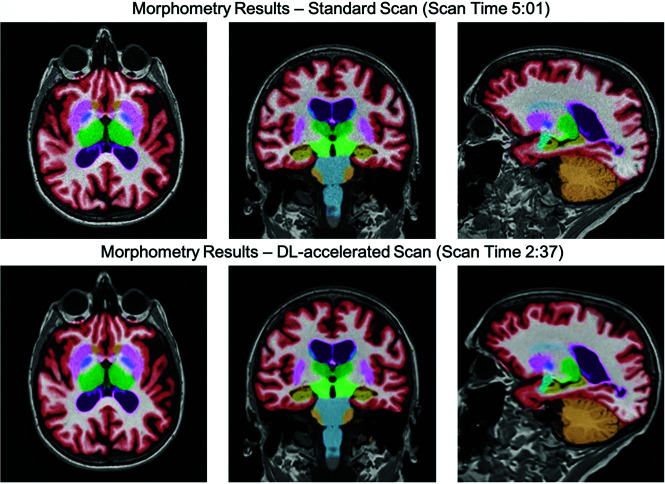
All SOC and FAST-DL datasets were rated of diagnostic quality by both interpreting neuroradiologists. Both readers determined that there was similar quality of segmentation for both the SOC and FAST-DL datasets. Representative imaging examples for image-quality analysis for SOC, FAST, and FAST-DL are shown in Fig 5. Sample volumetric segmentation of the SOC and FAST-DL images are shown in Fig 6.
FIG 5.
Representative axial 3D T1-weighted images on a 3T scanner. Left to right, SOC (scan time, 4 minutes, 55 seconds), FAST (scan time, 2 minutes, 10 seconds), FAST-DL (scan time, 2 minutes, 10 seconds).
FIG 6.
Representative 3D T1-weighted multiplanar images with volumetric segmentation on a 3T scanner. Left to right, Axial, coronal, sagittal T1-weighted images with SOC (scan time, 5 minutes, 01 second) on the upper row (A) and FAST-DL (scan time, 2 minutes, 37 seconds) on lower row (B).
DISCUSSION
DL enhancement of MR images is known to provide multiple benefits, including increased SNR,2,3 but questions remain about reliability in general clinical use.8 The approach used in our multicenter, multivendor study explored the impact on the image quality and consistency of quantitative volumetric analysis results obtained with FAST-DL compared with that obtained with SOC scans.
Quantitative volumetric MR imaging analytical tools are in widespread clinical use for the evaluation of patients with dementia, seizures, multiple sclerosis, traumatic brain injury, and pediatric brain disorders. The software segments, labels, and calculates the volumes of substructures (including lesions) in the brain. The derived quantitative values are compared with a large normative age and sex-matched data base, aiding in the diagnosis and longitudinal follow-up of clinical conditions such as Alzheimer’s disease. Quantitative assessment reduces reader subjectivity.9-12
MR imaging provides excellent anatomic detail and superb contrast resolution but involves trade-offs in SNR, spatial resolution, and scan duration.7 While DL-based augmentation is a recognized solution for accelerated MR imaging, it is important to validate the reliability and generalizability of its enhancement capabilities with quantitative biomarker accuracy.7
The MR imaging experience is uncomfortable and associated with frank anxiety reactions in up to 30% of patients.13,14 Faster MR image acquisition can thus increase patient satisfaction and may reduce motion artifacts. Motion is a significant challenge in MR imaging, occurring in 29% of inpatient/emergency department examinations and 7% of outpatient studies and can lead to repeat portions of or even complete examinations.15 Andre et al16 found that 19.8% of all MR imaging sequences need to be repeated due to motion artifacts, a $592 revenue loss per hour and $115,000 loss annually per scanner due to motion artifacts. DL-based reconstruction solutions promise to enable shorter examinations with decreased patient motion and improved patient comfort.14
In our study, we achieved a scan-time reduction of 60% while exceeding perceived routine 3D T1-weighted image quality. If DL-enhanced fast protocols were used on all pulse sequences across every study, one could anticipate a proportional increase in examination-based workflow efficiency for an imaging facility. One recent trial explored DL enhancement across all pulse sequences in clinical spine MR imaging, with preservation of quantitative features using a structural similarity index measure as well as gains in perceived SNR and artifact reduction, despite a 40% scan-time reduction.2 Future research could explore whether scan-time reduction of this scale results in a true-positive impact on workflow, eg, the ability to scan more patients per day.
In this trial, the SOC images serve as the standard for image preference. Our randomized blinded assessment of the imaging features is meant to reflect human subjective perception of comparative image quality. A radiologist’s qualitative assessment of noninferiority is critical before a DL-enhanced alternative would be considered acceptable for clinical use. On the other hand, processed images should satisfy both qualitative and quantitative measures to ensure that diagnostically relevant features are not altered and the integrity of the processed image information is maintained.
Concerns exist about DL postprocessing introducing instabilities in an image, in which tiny perturbations in the sampling domain have been shown to be capable of translating into noticeable artifacts on the reconstructed image.8 This issue has been shown for highly-contrived noise additions to k-space data, but it is unclear whether such effects occur under normal operating conditions. The current method starts from image-based DICOM data rather than within the k-space and is likely less susceptible to this effect.
To our knowledge, this is the first prospective, randomized, multicenter study of DL reconstruction capabilities assessing the impact on the integrity of quantitative volumetric analysis of clinical brain MR imaging examinations. The DL tool applied in this study shifts the usual MR imaging trade-off equation by imprinting a boost in spatial resolution on the target FAST series, which, due to inherently larger native voxel sizes, can have a higher SNR and contrast-to-noise ratio than even the basis SOC series.7 Along with sharpness enhancement, DL offers structure-preserving denoising, contributing to statistically significant gains in perceived SNR compared with SOC.
Blinded subjective assessment by the neuroradiologists found that the 60% accelerated, DL-enhanced 3D T1-weighted brain MR images delivered consistent clinical classification and were superior to standard of care MR imaging across essentially all analyzed quality features (perceived SNR, perceived spatial resolution, artifact reduction, anatomic/lesion conspicuity, and image contrast). These findings offer confidence that DL processing can add value and efficiency to clinical MR imaging brain examinations.
The quantitative biomarkers of HV, HOC, SLV volume, and ILV volume were statistically equivalent for the FAST-DL sequences and the SOC, supporting the absence of corruption by DL processing and demonstrating the robustness of the DL tool in maintaining quantitative integrity and enhancing image quality despite significant scan-time acceleration. Not unexpectedly, the cross-correlation factor was inferior for the SOC versus the lower resolution FAST dataset.
The strengths of this study include the use of a prospective, randomized, multicenter, multireader study design with images obtained on magnets from multiple vendors and of variable ages and field strengths, with preserved accuracy of quantitative volumetric measures and clinical predictive categories. The results of this trial support the use of DL enhancement to shorten clinical MR imaging brain examinations, even when additional quantitative tools such as volumetric analysis are applied.
Weaknesses include the small number of imaging subjects and the use of only a single DL and quantitative brain-analysis tool. The DL-enhancement tool used in this study is a vendor-agnostic, DICOM-based, commercially available solution. It is possible that alternative vendor-specific, k-space-based, commercially available DL enhancement tools may perform differently, though preliminary data suggest observed gains in acceleration and perceived quality with other DL-enhancement tools as well.17,18 However, this is the first study that the authors are aware of that specifically confirms the quantitative volumetric accuracy and consistent clinical disease categorization of the DL-enhanced dataset.
Future investigations might explore different methods and tools. Another area of future research might include a similar methodology applied to different clinical scenarios that demand accurate segmentation but where scan time acceleration would be desirable, such as in patients with multiple sclerosis, intracranial metastases, epilepsy, and traumatic brain injury. Follow-up studies could also assess whether the difference between the FAST and FAST-DL datasets was significant enough to impact the correct clinical diagnosis or alter the reader’s ability to detect a lesion.
CONCLUSIONS
DL can enable 60% faster brain MR image acquisitions with matched clinical disease status predictability and statistically superior perceived image quality, while maintaining high quantitative accuracy compared with the longer SOC examinations. This trial supports the reliability, efficiency, and utility of DL-based enhancement for quantitative imaging. Shorter scan times may boost the use of volumetric quantitative MR imaging in routine clinical settings.
ABBREVIATIONS:
- DL
deep learning
- FAST
accelerated scan
- HOC
hippocampal occupancy score
- HV
hippocampal volumes
- ILV
inferior lateral ventricles
- MCI
mild cognitive impairment
- SLV
superior lateral ventricles
- SOC
standard of care
Footnotes
Disclosures: Suzie Bash—UNRELATED: Consultancy: Cortechs.ai, icometrix, Subtle Medical, Guerbet, GE Pharmaceuticals, Comments: 1) honoraria in the past for consultancy work for each entity listed, and 2) Medical Advisory Board member for Cortechs.ai (stipend and stock options) and icometrix (stipend only); Payment for Lectures Including Service on Speakers Bureaus: icometrix, Subtle Medical, Guerbet, Comments: Honoraria for webinars/talks for the entities listed; Stock/Stock Options: Cortechs.ai, Comments: stock options as member of MAB; Travel/Accommodations/Meeting Expenses Unrelated To Activities Listed: Cortechs.ai, GE Pharmaceuticals, Comments: travel/accommodations for consultancy work. Greg Zaharchuk—UNRELATED: Board Membership: Subtle Medical*; Grants/Grants Pending: various National Institutes of Health projects, GE Healthcare, Bayer Healthcare*; Patents (Planned, Pending or Issued): assorted patents on deep learning; Royalties: Cambridge University Press; Stock/Stock Options: equity in Subtle Medical. Ajit Shankaranarayanan—UNRELATED: Employment: Subtle Medical; Stock/Stock Options: Subtle Medical, Comments: equity in the company. Lawrence Tanenbaum—UNRELATED: Consultancy: Subtle Medical, GE Healthcare, Philips Healthcare, Hitachi, Siemens, Fuji Healthcare, Bracco Diagnostics; Payment for Lectures Including Service on Speakers Bureaus: GE Healthcare, Fuji, Subtle Medical, Guerbet, Bracco, Philips Healthcare, Money paid to the individual; Stock Options: Subtle Medical; Other: Aidoc, Enlitic, Agamon, iMedis, Comments: consultant. Long Wang—UNRELATED: Employment: Subtle Medical. Enhao Gong—UNRELATED: Board Membership: Subtle Medical; Employment: Subtle Medical; Stock/Stock Options: Subtle Medical. *Money paid to the institution.
Paper previously presented at: Annual Meetings and Exhibitions of the International Society for Magnetic Resonance in Medicine and the Society for Magnetic Resonance Technologists, August 8–14, 2020; Virtual; and May 14–20, 2021, Virtual; Annual Meetings of the American Society of Neuroradiology, May 30 to June 4, 2020; Virtual; and May 15–20, 2021; Virtual; and Annual Meeting of the European Congress of Radiology, May 3–7, 2021, Porto Alegre, Brazil.
References
- 1.Radlak K, Malinski L, Smolka B. Deep learning-based switching filter for impulsive noise removal in color images. Sensors 2020;20:2782 10.3390/s20102782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanenbaum LN, Bash S, Johnson B, et al. Deep learning reconstructed, 40% faster spine MR examinations match or exceed the quality of standard of care exams. In: Proceedings of the Annual Meeting of the Radiological Society of North America; November 29 to December 5, 2020; Virtual
- 3.Tanenbaum LN, Bash S, Gibbs W, et al. CNN based deep learning enhances brain 3D FLAIR perceived quality, SNR and resolution at ∼30% less scan time. In: Proceedings of the Annual Meeting of the American Society of Neuroradiology, May 30 to June 4, 2020; Virtual [Google Scholar]
- 4.Tian C, Xu Y, Li Z, et al. Attention-guided CNN for image denoising. Neural Netw 2020;124:117–29 10.1016/j.neunet.2019.12.024 [DOI] [PubMed] [Google Scholar]
- 5.Zhang K, Zuo W, Chen Y, et al. Beyond a Gaussian denoiser: residual learning of deep CNN for image denoising. IEEE Trans Image Process 2017;26:3142–55 10.1109/TIP.2017.2662206 [DOI] [PubMed] [Google Scholar]
- 6.Lunderwold AS, Lunderwold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019;29:102–27 10.1016/j.zemedi.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 7.Chaudhari A, Zhongnan F, Lee J, et al. Deep learning super-resolution enables rapid simultaneous morphological and quantitative magnetic resonance imaging. In: Proceeding of Machine Learning for Medical Image Reconstruction Workshop at MICCA, Granada, Spain; September 16, 2018 [Google Scholar]
- 8.Antun V, Renna F, Poon C, et al. On instabilities of deep learning in image reconstruction and the potential costs of AI. Proc Natl Acad Sci U S A 2020;117:30088–95 10.1073/pnas.1907377117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bash S. Eye on AI: enhancing neuroimaging with artificial intelligence. Appl Radiol 2020;49:20–21 [Google Scholar]
- 10.Bryant M. Eye on AI: the potential and reality of AI in clinical application. Appl Radiol 2020;49:10–11 [Google Scholar]
- 11.Tanenbaum LN, Bash S, Davis M. Appl Radiol (AR Connect Expert Discussions); AI: clinical applications. In: Proceedings of the Annual Meeting of the Radiological Society of North America, Chicago, Illinois; December 1–6, 2019 [Google Scholar]
- 12.Bryant M. Bringing AI and genetics together to support clinical decisions. Appl Radiol July 1, 2020. https://appliedradiology.com/communities/Artificial-Intelligence/bringing-ai-and-genetics-together-to-support-clinical-decisions. Accessed July 1, 2020
- 13.Melendez JC, McCrank E. Anxiety-related reactions associated with magnetic resonance imaging examinations. JAMA 1993;270:745–47 10.1001/jama.1993.03510060091039 [DOI] [PubMed] [Google Scholar]
- 14.Tanenbaum L. Quality, efficiency and survival with patient centric imaging. In: Proceedings of the 7th Snowmass 2019: Hot Topics in Radiology: Advanced Applications and Artificial Intelligence, Snowmass, Colorado; February 10–15, 2019 [Google Scholar]
- 15.Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: a complex problem with many partial solutions. J Magn Reson Imaging 2015;42:887–901 10.1002/jmri.24850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andre J, Bresnahan B, Mossa-Basha M, et al. Toward quantifying the prevalence, severity, and cost associated with patient motion during clinical MR examinations. J Am Coll Radiol 2015;12:689–95 10.1016/j.jacr.2015.03.007 [DOI] [PubMed] [Google Scholar]
- 17.Bash S, Thomas M, Fung M, et al. K-space based deep learning reconstruction empowers 60-70% acceleration of MR imaging of the spine. In: Proceedings of the Annual Meeting of the Radiological Society of North America, Chicago, Illinois; December 1–6, 2019 [Google Scholar]
- 18.Tanenbaum LN, Bash S, Thomas M, et al. K-space based deep learning reconstruction empowers 50% acceleration of MR spine imaging: a prospective, multicenter, multireader trial. In: Proceedings of the European Congress of Radiology, February 26 to March 1, 2020; Virtual [Google Scholar]